Human telomere function is mediated by the shelterin complex. Smith and colleagues found that the shelterin component TIN2 associates with HP1γ via a conserved binding motif, which is required for S-phase sister chromatid cohesion and can impact telomere length maintenance. Mutations in this motif abrogate HP1γ binding and lead to defects in telomere maintenance, giving rise to dyskeratosis congenita.
Keywords: telomeres, TIN2, HP1, dyskeratosis congenita
Abstract
Human telomere function is mediated by shelterin, a six-subunit complex that is required for telomere replication, protection, and cohesion. TIN2, the central component of shelterin, has binding sites to three subunits: TRF1, TRF2, and TPP1. Here we identify a fourth partner, heterochromatin protein 1γ (HP1γ), that binds to a conserved canonical HP1-binding motif, PXVXL, in the C-terminal domain of TIN2. We show that HP1γ localizes to telomeres in S phase, where it is required to establish/maintain cohesion. We further demonstrate that the HP1-binding site in TIN2 is required for sister telomere cohesion and can impact telomere length maintenance by telomerase. Remarkably, the PTVML HP1-binding site is embedded in the recently identified cluster of mutations in TIN2 that gives rise to dyskeratosis congenita (DC), an inherited bone marrow failure syndrome caused by defects in telomere maintenance. We show that DC-associated mutations in TIN2 abrogate binding to HP1γ and that DC patient cells are defective in sister telomere cohesion. Our data indicate a novel requirement for HP1γ in the establishment/maintenance of cohesion at human telomeres and, furthermore, may provide insight into the mechanism of pathogenesis in TIN2-mediated DC.
Mammalian telomeres are heterochromatic structures comprised of duplex TTAGGG repeats that end as a 3′ single-strand overhang, and shelterin, a six-subunit complex that is required for the protection and replication of chromosome ends (Palm and de Lange 2008). TIN2 is at the heart of the shelterin complex, with binding sites to three subunits: TRF1 and TRF2 (the duplex DNA-binding proteins) (Houghtaling et al. 2004; Kim et al. 2004; Liu et al. 2004a; Ye et al. 2004a), and TPP1 (Houghtaling et al. 2004; Liu et al. 2004b; Ye et al. 2004b), which binds to POT1, the single-strand overhang-binding protein (Baumann and Cech 2001). TIN2 regulates telomere elongation (Kim et al. 1999) by telomerase, the reverse transcriptase that adds TTAGGG repeats to chromosome ends (Greider and Blackburn 1987). Telomerase is recruited to telomeres by TIN2-tethered TPP1 (Wang et al. 2007; Xin et al. 2007; Abreu et al. 2010). In humans, telomerase is expressed in the germline and during embryogenesis, but it is repressed in most somatic tissues (Wright et al. 1996). The resulting telomere shortening that occurs with aging contributes to human disease (Artandi and DePinho 2010).
In addition to its role in telomere length maintenance, TIN2 is required for cohesion of sister telomeres (Canudas and Smith 2009). Sister chromatids are held together from the time of their replication by cohesin complexes, ring structures formed by Smc1, Smc3, and Scc1, and a peripheral regulatory subunit, Scc3 (Anderson et al. 2002; Haering et al. 2002), that exists as two isoforms, SA1 and SA2, in vertebrate cells. SA1–cohesin is much less abundant than SA2–cohesin in somatic cells (Losada et al. 2000; Sumara et al. 2000). TIN2 and TRF1 were found to be associated with the SA1–cohesin complex (Canudas et al. 2007). Cells depleted of TIN2 or SA1 are unable to establish/maintain cohesion between sister telomeres in S phase and, as a result, are unable to repair sister chromatid breaks following DNA replication and suffer sister telomere loss (Canudas and Smith 2009).
TIN2 is mutated in a subset of cases of dyskeratosis congenita (DC) (Savage et al. 2008; Walne et al. 2008), an inherited bone marrow failure syndrome that is caused by defects in telomere maintenance due to mutations in telomerase and its associated factors (Bessler et al. 2010; Zhong et al. 2011). While mutations in the telomerase subunits are usually inherited, TIN2 mutations frequently occur de novo. Moreover, patients harboring TIN2 mutations have extremely short telomeres, correlating with early age of presentation and severe clinical presentation (Savage et al. 2008; Walne et al. 2008; Sasa et al. 2011). While the reason for the severity is not known, it suggests that the TIN2 DC mutations may uniquely influence aspects of telomere function that are distinct from telomerase. The DC mutation cluster in TIN2 is distinct from its TRF1-, TRF2-, and TPP1-binding sites, but the amino acids in the mutation cluster are highly conserved, raising the possibility of a yet to be identified binding partner.
In addition to shelterin, vertebrate telomeres also contain heterochromatic histone marks that are characteristic of repressive or silent chromatin (Schoeftner and Blasco 2009). Studies in mice indicate a role for these epigenetic marks in telomere length maintenance and telomere recombination (Garcia-Cao et al. 2004; Gonzalo et al. 2005). One such mark, histone H3 trimethylated Lys 9 (H3K9Me3), provides a high-affinity binding site for heterochromatin protein 1 (HP1) (Bannister et al. 2001; Lachner et al. 2001), a family of highly conserved nonhistone chromosomal proteins that regulate heterochromatin formation and maintenance (Lomberk et al. 2006). A role for HP1 in telomere function is firmly established in Drosophila (Fanti and Pimpinelli 2008) and is suggested by studies in mice (Garcia-Cao et al. 2004; Gonzalo et al. 2005). However, the function of HP1 at telomeres in human cells remains to be elucidated. Here we identify HP1γ as a new TIN2 partner that binds to the DC mutation cluster in TIN2 and is required to establish/maintain cohesion at telomeres.
Results
The HP1-binding site in TIN2 is required for sister telomere cohesion
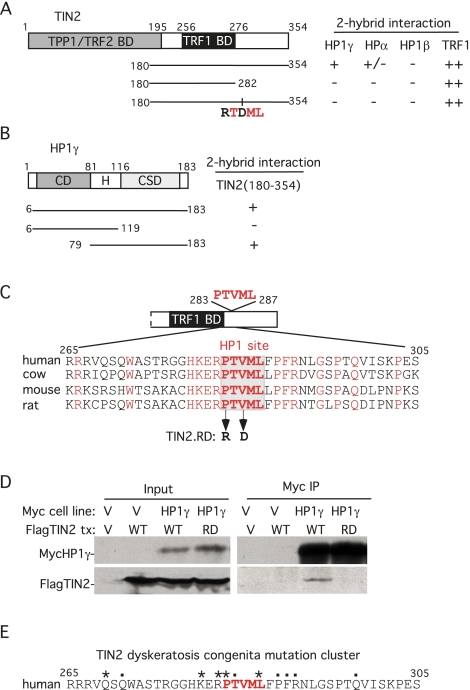
Human HP1α and HP1γ were identified in yeast two-hybrid screens with human TIN2 as bait. Three HP1 isoforms are found in human cells (HP1α, HP1β, and HP1γ), which, although highly conserved, show distinct subnuclear localization and function (Minc et al. 1999). Two-hybrid analysis showed that TIN2 binds strongly to HP1γ, weakly to HP1α, and not at all to HP1β (Fig. 1A). HP1 proteins contain an N-terminal chromodomain (CD) that binds H3K9Me3, an internal hinge region (H), and a C-terminal chromoshadow domain (CSD) to which TIN2 binds (Fig. 1B). The CSD exists as a homodimer that binds with high affinity to proteins containing the consensus pentapeptide PXVXL (Brasher et al. 2000; Smothers and Henikoff 2000). TIN2 contains a conserved PXVXL motif C-terminal to its TRF1-binding domain (Fig. 1C). C-terminal deletion or a double point mutation of PTVML to RTDML (designated as TIN2.RD) abrogated binding of TIN2 to HP1γ and HP1α, while interaction with TRF1 was retained (Fig. 1A). Immunoprecipitation analysis in human cells shows that TIN2.WT, but not TIN2.RD, coimmunoprecipitated with HP1γ (Fig. 1D).
Figure 1.
HP1 binds to the PTVML site in the DC-associated TIN2 mutation cluster. (A) Schematic representation of TIN2 and two-hybrid interaction with HP1. TPP1-, TRF2-, and TRF1-binding domains (BDs) and the double point mutation in TIN2 (RTDML) are indicated. (B) Schematic representation of HP1 and two-hybrid interaction with TIN2. The chromodomain (CD), hinge (H), and chromoshadow domain (CSD) in HP1 are indicated. (A,B) Two-hybrid interactions were scored according to the number of minutes required for the color change: 20 min (++), 45 min (+), and 150–180 min (+/−). (C) Alignment of the TIN2 domain containing the PTVML HP1-binding site. Identical amino acids are indicated in red. (D) HP1 binds to TIN2 in human cells. HT1080 stable cell lines expressing HP1γ or vector (V) were transfected with vector or FlagTIN2 (WT or RD). Cell lysates were immunoprecipitated with anti-myc beads and analyzed by immunoblotting with anti-myc or anti-TIN2 701 antibody. (E) The DC-associated mutation cluster in TIN2. DC mutations are indicated by dots and asterisks; asterisks indicate mutations shown to give rise to shortened telomeres.
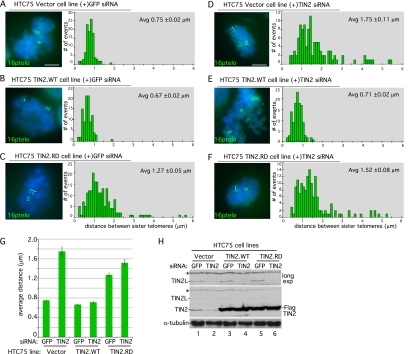
Strikingly, the PTVML site is embedded in the DC-associated TIN2 mutation cluster (Fig. 1E; Savage et al. 2008; Walne et al. 2008), raising the possibility that this site plays an essential role in TIN2 function in human cells. We thus asked whether an intact PTVML-binding site is required for sister telomere cohesion. Stable cell lines expressing Vector, TIN2.WT, or TIN2.RD were analyzed by measuring the distance between sister telomeres at mitosis by fluorescent in situ hybridization (FISH) with a subtelomere-specific probe, 16ptelo (Fig. 2A–G; Table 1). In Vector control cells, telomeres appeared as closely associated doublets (0.75-μm average distance) (Fig. 2A), indicating normal resolution of cohesion (Canudas and Smith 2009). Overexpression of TIN2.RD led to an increase in the distance between sister telomeres (1.27-μm average distance) (Fig. 2C), indicating a cohesion defect and suggesting a dominant-negative effect for the TIN2.RD mutant protein. The increase was not due to overexpression per se, since TIN2.WT was expressed to a similar level (Fig. 2H, cf. lanes 3 and 5), but did not lead to loss of cohesion (Fig. 2B).
Figure 2.
An intact HP1-binding site in TIN2 is required for sister telomere cohesion. (A–F) FISH analysis of HTC75 stable cell lines expressing Vector (A,D), TIN2.WT (B,E), or TIN2.RD (C,F) following transfection with GFP siRNA (A–C) or TIN2 siRNA (D–F) with a 16ptelo probe. DNA was stained with DAPI (blue). Bar, 5 μm. Histograms based on measurements from two independent experiments (Table 1) showing the distance between FISH signals are on the right, with the average (Avg) distance ± SEM indicated. (G) Graphical representation of the average distance between sister telomeres ± SEM. (H) Immunoblot analysis of extracts from stable HTC75 lines expressing Vector, TIN2.WT, or TIN2.RD transfected with GFP siRNA or TIN2 siRNA and probed with anti-TIN2 701 or anti-α-tubulin antibody. Asterisk (*) indicates a nonspecific band.
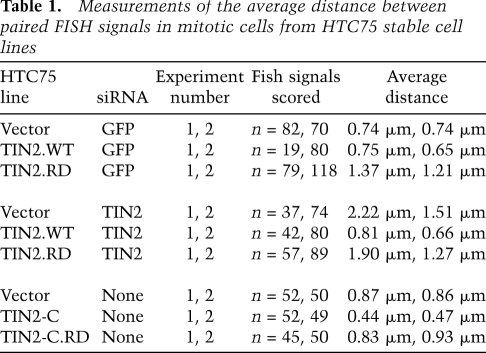
Table 1.
Measurements of the average distance between paired FISH signals in mitotic cells from HTC75 stable cell lines
To further query the requirement of the PTVML sequence in cohesion, we performed a rescue experiment. TIN2 was depleted from the stable cell lines with siRNA to the 3′ untranslated region that is not contained in the TIN2.WT or TIN2.RD transgenes. Immunoblot analysis indicated efficient depletion of TIN2 (64% knockdown) and TIN2L (75% knockdown; a higher-molecular-weight TIN2 isoform) (Fig. 2H; Kaminker et al. 2009). FISH analysis of TIN2-depleted vector control cells revealed a dramatic loss in sister telomere cohesion (Fig. 2D), consistent with our previous studies showing loss of cohesion using a different nonoverlapping siRNA directed against the TIN2 coding sequence (Canudas and Smith 2009). The loss in cohesion was rescued by expression of TIN2.WT (Fig. 2E), but not by TIN2.RD (Fig. 2F). These data demonstrate that an intact HP1-binding site in TIN2 is required to maintain sister telomere cohesion.
HP1 is required to establish telomere cohesion in S phase
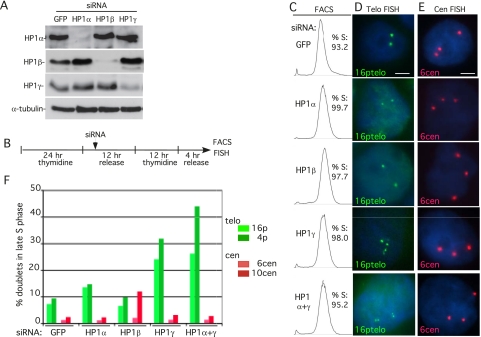
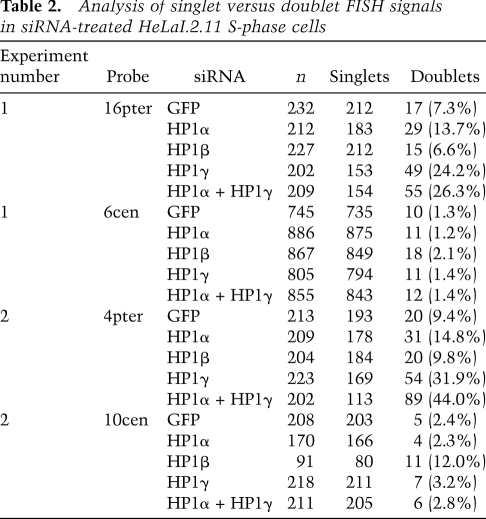
Sister telomere cohesion is normally established in S phase during DNA replication (Uhlmann and Nasmyth 1998; Sherwood et al. 2010). We showed previously that TIN2 was required to establish/maintain telomere cohesion in S phase (Canudas and Smith 2009). We thus asked whether HP1 is required to establish cohesion at telomeres in S phase. HeLaI.2.11 cells were transfected with siRNA directed against HP1α, HP1β, HP1γ, or GFP as control. Immunoblot analysis indicated efficient depletion of the proteins (Fig. 3A). To determine the status of cohesion after DNA replication, synchronized siRNA-treated cells were analyzed in mid-S phase. Cells were synchronized by double thymidine treatment; 4 h after release from the first thymidine treatment, cells were transfected with siRNA, and 4 h after the second thymidine treatment, cells were processed for FACS and FISH (Fig. 3B, experimental protocol). FACS analysis showed that cells were synchronized in S phase (Fig. 3C). Telomere FISH analysis of control (GFP) cells showed that, as expected, only a small fraction (7.3%) of telomeres lost cohesion (appeared as doublets) (Fig. 3D,F; Table 2). Depletion of HP1α led to a twofold increase in doublets (13.7%). HP1γ-depleted cells showed a more dramatic (3.3-fold) increase to 24.2%. The double depletion (HP1α and HP1γ) led to a slightly greater (26.3% doublets) increase. Centromere cohesion in HP1α- and HP1γ-depleted cells was unaffected (Fig. 3E,F; Table 2). Telomere cohesion was unaffected in HP1β-depleted cells, but centromere cohesion was slightly impaired (Fig. 3E,F;Table 2). Similar results were obtained with a different set of telomere and centromere probes (Fig. 3F; Table 2). Together, these data indicate that HP1γ (and, to a lesser extent, HP1α) is required for sister telomere cohesion in S phase.
Figure 3.
HP1 is required to establish/maintain sister telomere cohesion in S phase. (A) Immunoblot analysis of extracts from HeLaI.2.11 cells transfected for 48 h with GFP, HP1α, HP1β, or HP1γ siRNA. (B) Schematic representation of the protocol to analyze siRNA-treated synchronized cells. (C–E) FACS (C) and FISH (D,E) analysis 4 h after release from the second thymidine block. Cells were hybridized with a telomere 16ptelo (green) (D) or centromere 6cen (red) (E) probe. The cen locus is trisomic. DNA was stained with DAPI (blue). Bar, 5 μm. (F) Graphical representation of the frequency of doublets from two independent experiments: 16pter and 6cen probes (Experiment 1) and 4pter and 10cen probes (Experiment 2) (see Table 2).
Table 2.
Analysis of singlet versus doublet FISH signals in siRNA-treated HeLaI.2.11 S-phase cells
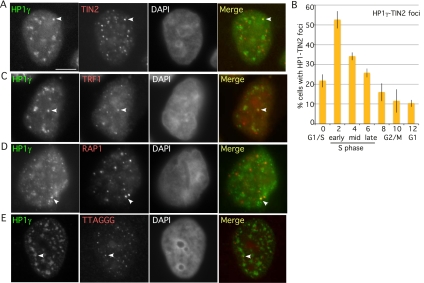
We next asked whether we could detect an association between HP1γ and TIN2 in cells by focusing on the window in S phase when TIN2 and HP1γ are required to establish cohesion. Cells were synchronized by a double thymidine block, collected 4 h after release, and analyzed by immunofluorescence. Cells were costained with antibodies to TIN2 and HP1γ. Individual foci where the HP1γ and TIN2 signal overlapped could be detected (Fig. 4A). We observed generally one focus per cell and only in a subset of cells. To determine whether the colocalization was regulated across the cell cycle, cells were synchronized by a double thymidine block, collected at 2-h time points after release (from 0 h to 12 h), and analyzed by immunofluorescence. As shown in Figure 4B, the HP1γ–TIN2 foci were highly enriched in early S phase, coincident with the time when cohesion is established. A similar colocalization of HP1γ could be detected with other shelterin subunits, TRF1 (Fig. 4C) and RAP1 (Fig. 4D), and with telomeres using FISH with a TTAGGG repeat probe (Fig. 4E), indicating that HP1γ association with TIN2 occurs at telomeres.
Figure 4.
HP1γ colocalizes with TIN2 at telomeres in S phase. (A,C–E) Immunofluorescence analysis of HeLaI.2.11 cells 4 h after release from a double thymidine block. Cells were extracted with Triton prior to fixation and dually stained with antibodies against HP1γ (green) (A,C–E), TIN2 (red) (A), TRF1 (red) (C), and RAP1 (red) (D), or processed for FISH with a PNA-TTAGGG repeat probe (red) (E). Merge is yellow. Bar, 5 μm. (B) Graphical representation of the frequency of cells containing colocalizing HP1γ–TIN2 foci across the cell cycle. Cells were synchronized by a double thymidine block and released for 0 h (G1/S), 2 h (early S), 4 h (mid S), 6 h (late S), 8 h (G2), 10 h (M), and 12 h (G1). Mean ± SD of three independent experiments (n, ∼100 cells each).
The HP1-binding site in TIN2 can impact telomere length maintenance by telomerase
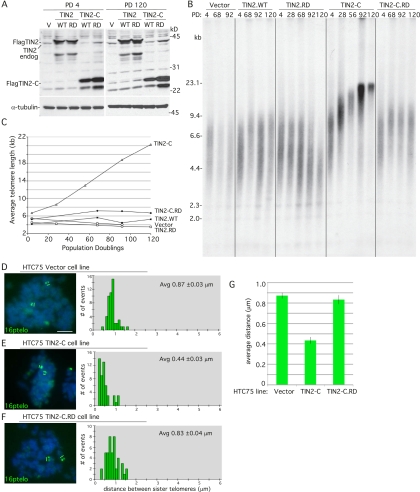
To determine whether HP1 binding to TIN2 influenced telomere length maintenance by telomerase, stable HTC75 cell lines expressing Vector, TIN2.WT, and TIN2.RD were passaged for 120 population doublings and subjected to immunoblot (Fig. 5A) and telomere length analysis (Fig. 5B,C). Vector and TIN2.WT cells maintained telomere length, whereas telomeres shortened in TIN2.RD cells, indicating that an intact HP1 site is required for telomere length maintenance. As these effects were subtle and only appeared after many of generations, we sought a more dramatic example. Previous studies showed that an N-terminally deleted allele of TIN2, TIN2-13 (amino acids 197–354), led to dramatic telomere elongation that was dependent on telomerase (Kim et al. 1999). We generated a similar N-terminally deleted allele, TIN2-C (amino acids 180–354), and a TIN2-C.RD allele and subjected them to telomere length analysis. As shown in Figure 5, B and C, the telomere lengthening observed in TIN2-C cells was abolished in TIN2-C.RD cells.
Figure 5.
TIN2 mutations in the HP1-binding site interfere with telomere length maintenance. (A) Immunoblot analysis of extracts from stable HTC75 cell lines expressing Vector (V), TIN2.WT, TIN2.RD, TIN2-C, or TIN2-C.RD at population doubling 4 (PD 4) or 120 (PD 120) probed with anti-TIN2 701 or anti-α-tubulin antibody. (B) Analysis of telomere restriction fragments isolated from the stable HTC75 cell lines at the indicated population doubling, fractionated on agarose gel, denatured, and probed with a 32P-labeled CCCTAA probe. (C) Graphical representation of telomere length changes in B. (D–F) FISH analysis of HTC75 stable cell lines expressing Vector (D), TIN2-C (E), or TIN2-C.RD (F) with a 16ptelo probe. DNA was stained with DAPI (blue). Bar, 5 μm. Histograms based on measurements from two independent experiments (Table 1) showing the distance between FISH signals are on the right, with the average (Avg) distance ± SEM indicated. (G) Graphical representation of the average distance between sister telomeres ± SEM.
The effect of mutating the HP1-binding site in TIN2 on telomere length maintenance was very striking in the context of the TIN2-C allele. Apart from its dependence on telomerase (Kim et al. 1999), the mechanism by which this allele induces telomere elongation is not known. We thus queried the cohesion status of sister telomeres in the TIN2-C cells lines (Fig. 5D–G). We observed a dramatic decrease in the distance between sister telomeres in TIN2-C cells (Fig. 5E)—0.44-μm average distance compared with 0.87-μm average distance in the vector control (Fig. 5D)—indicating an increased association between sister telomeres. This increased association was abolished in TIN2-C.RD cells (Fig. 5F).
DC mutations in TIN2 abrogate binding to HP1 and show a loss in telomere cohesion in human patient cells
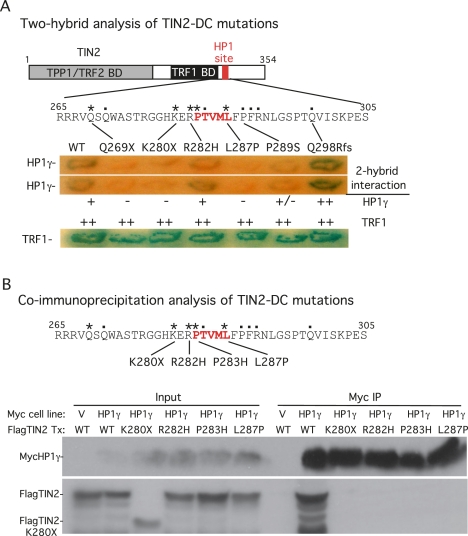
Our data indicate that the PTVML motif in TIN2 is required for HP1 binding. The observation that the PTVML site is embedded in the DC mutation cluster raises the possibility that TIN2-DC mutations are defective in HP1 binding. We used the two-hybrid assay in yeast and immunoprecipitation analysis in human cells to analyze interaction of the TIN2-DC mutations with HP1γ (Fig. 6). We focused mainly on those mutations associated with short telomeres, indicated by asterisks in Figure 6, A and B (Sasa et al. 2011; Vulliamy et al. 2011). The nonsense mutations Q269X and K280X result in truncated proteins that lack the HP1-binding site and thus, as expected, did not interact with HP1γ in the yeast two-hybrid assay (Fig. 6A). The missense mutations P283R (in the context of TIN2.RD) (see Fig. 1A), and L287P also did not bind HP1γ (Fig. 6A). The R282H mutation showed some binding in the two-hybrid assay (Fig. 6A). However, immunoprecipitation analysis in human cells showed that binding of TIN2.R282H as well as K280X, P283H, and L287P to HP1γ was severely diminished compared with TIN2.WT (Fig. 6B). Together, these data show that DC-associated TIN2 mutations are defective in HP1γ binding.
Figure 6.
DC-associated TIN2 mutations interfere with HP1 binding. (A) Two-hybrid analysis of the interaction between DC-associated TIN2 mutations and HP1γ and TRF1. Two-hybrid interactions were scored according to the number of minutes required for the color change: 20 min (++), 45 min (+), and 150–180 min (+/−). (B) Coimmunoprecipitation analysis of the interaction between DC-associated TIN2 mutations and HP1γ. HT1080 stable cell lines expressing HP1γ or vector (V) were transfected with the indicated FlagTIN2 plasmids. Cell lysates were immunoprecipitated with anti-myc beads and analyzed by immunoblotting with anti-myc or anti-TIN2 701 antibody.
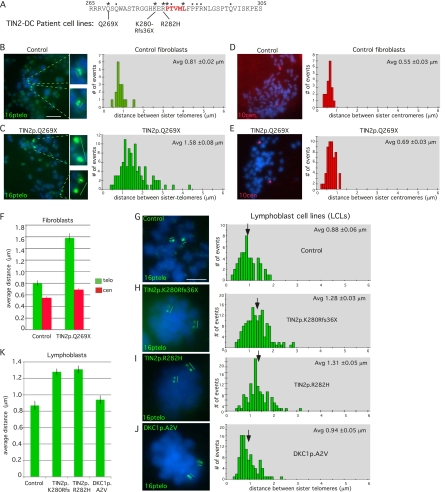
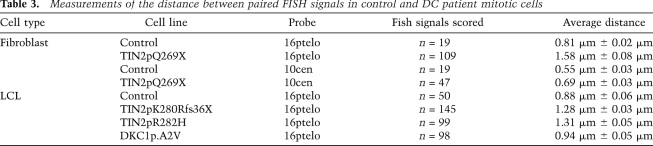
We next asked whether DC-associated TIN2 mutations were defective in sister telomere cohesion by analyzing patient cell lines using chromosome-specific FISH (Fig. 7; Table 3). Analysis of skin fibroblasts derived from a patient harboring the TIN2p.Q269X mutation (Sasa et al. 2011) revealed a dramatic loss in sister telomere cohesion compared with control fibroblasts (Fig. 7B,C), while centromere cohesion was unaffected (Fig. 7D,E). Additionally, analysis of lymphoblastoid cell lines (LCLs) from patients harboring the mutations TIN2p.K280Rfs36X (Sasa et al. 2011) and TIN2p.R282H (Savage et al. 2008) showed a loss in sister telomere cohesion compared with LCL control cells (Fig. 7G–I). In contrast, LCLs from a patient harboring a DC-associated mutation in dyskerin (DKC1p.A2V) showed normal telomere cohesion, similar to control LCLs (Fig. 7J), indicating that loss in telomere cohesion is not a general feature of DC or of short telomeres.
Figure 7.
DC patient cells harboring TIN2 mutations suffer loss in sister telomere cohesion. (A) DC mutations in patient cell lines are indicated. (B–E) Loss in sister telomere cohesion in DC patient fibroblasts harboring the TIN2p.Q269X mutation. FISH analysis with a 16ptelo (B,C) or 10cen (D,E) probe. DNA was stained with DAPI (blue). Bar, 5 μm. Histograms showing the distance between FISH signals (Table 3) are on the right, with the average (Avg) distance ± SEM indicated. (F) Graphical representation of the average distance ± SEM. (G–K) Loss in sister telomere cohesion in DC patient LCLs harboring the following mutations: TIN2p.K280Rfs36X, TIN2p.R282H, and DKC1p.A2V. (G–J) FISH analysis with a 16ptelo probe. DNA was stained with DAPI (blue). Bar, 5 μm. Histograms showing the distance between FISH signals (Table 3) are shown on the right, with the average (Avg) distance ± SEM indicated. (K) Graphical representation of the average distance ± SEM.
Table 3.
Measurements of the distance between paired FISH signals in control and DC patient mitotic cells
Discussion
We identified HP1γ as a novel binding partner for TIN2 and showed that it (like TIN2) is required to establish/maintain cohesion at telomeres in S phase. We show that HP1γ colocalizes with TIN2 at telomeres, peaking in early to mid-S phase (Fig. 4). Initially, we were surprised to detect only a single focus per cell and in only ∼50% of cells in early S phase. However, previous studies have shown that human telomeres replicate asynchronously during S phase (Wright et al. 1999) and that telomerase elongates telomeres immediately following DNA replication (Zhao et al. 2009), evidenced by localization of telomerase at one to two telomeres per cell in a fraction of S-phase cells (Jady et al. 2006; Tomlinson et al. 2006). Only 70% of HeLa cells express telomerase at any time (Bryan et al. 1998). Hence, the timing and frequency of HP1γ–TIN2 foci (one per cell in ∼50% of cells in S phase) (Fig. 4) suggest that they could correspond to replicating telomeres.
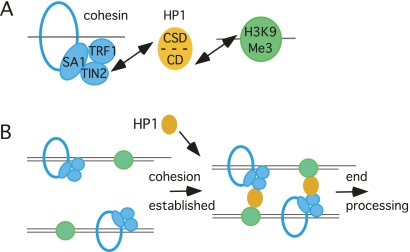
Precisely how cohesin links sister chromatids in vivo is not known. The embrace model posits a single cohesin complex entrapping the sisters (Haering et al. 2008). Alternatively, the handcuff model posits a cohesin complex per sister connected by the SA subunit (Zhang et al. 2008). We showed previously that depletion of SA1 leads to loss of sister telomere cohesion (Canudas and Smith 2009), consistent with the handcuff model. Studies indicate that heterochromatic domains may require distinct mechanisms for cohesion. For example, in budding yeast, cohesion between sister chromatids in silent chromatin relies on the SIR proteins (which might be considered functional orthologs of HP1) to bridge cohesin complexes that encircle each sister (Chang et al. 2005). It is interesting to speculate that HP1 could have a similar function in telomeric heterochromatin by binding to TIN2/SA1–cohesin via its CSD and to H3K9Me3 via its CD (Fig. 8A), thereby bridging cohesin rings on each sister to promote cohesion (Fig. 8B). In support of this model, overexpression of the TIN2-C allele (containing only a TRF1- and HP1-binding site) led to increased cohesion between sister telomeres (Fig. 5E), perhaps by increased recruitment of HP1γ to telomeres.
Figure 8.
Schematic diagram showing how HP1 might influence cohesion at telomeres. (A) HP1 could act as a bridge in telomeric heterochromatin by binding to the heterochromatin mark H3K9Me3 via its CD and to TIN2-SA1 cohesin via its CSD. (B) HP1 associates with telomeres in S phase to aid in establishment of cohesion. Once cohesion is established, chromosome ends from both sisters could undergo coordinated processing, including C-strand resection, telomerase-dependent elongation, and C-strand synthesis.
Overexpression of full-length TIN2 lacking an intact HP1-binding site (TIN2.RD) led to telomere shortening; however, the effect was subtle and took many population doublings to appear. We detected a more dramatic requirement for HP1 binding in telomere lengthening in the context of TIN2-C. Apart from the requirement for telomerase (Kim et al. 1999), the mechanism by which TIN2-C induces telomere elongation is not known. It is striking that telomeres in TIN2-C cells show an increased cohesion compared with control cells. Although TIN2-C is an artificial protein, these observations nonetheless raise the possibility that cohesion status may influence telomere lengthening by telomerase. Cohered sisters may be a preferred substrate for telomerase, which has been suggested to function as a dimer (Prescott and Blackburn 1997a,b; Wenz et al. 2001). Additionally, cohered sisters may serve to coordinate processing events (C-strand resection, telomerase-dependent elongation, and C-strand synthesis) that occur at sister chromatid ends following DNA replication (Fig. 8B; Zhao et al. 2009; Giraud-Panis et al. 2010). Recent work indicates that extension of human telomeres by telomerase can be influenced at multiple levels, including recruitment, processivity, and release of telomerase (Zhao et al. 2011). Sister telomere cohesion may impact any or all of these aspects.
How does the HP1-binding site in TIN2 impact our understanding of DC-associated TIN2 mutations? We showed that an intact HP1-binding site in TIN2 is required for sister telomere cohesion (Fig. 2). The HP1-binding site is embedded in the DC-associated TIN2 mutation cluster. We show that TIN2 proteins containing DC mutations are diminished in HP1γ binding (Fig. 6) and that DC-associated TIN2 patient cells suffer loss in sister telomere cohesion (Fig. 7). The HP1-binding site is distinct from TIN2's binding sites for TRF1, TRF2, and TPP1. We speculate that this distinction (in the context of human disease) allows TIN2 DC mutant proteins (despite their defect in HP1γ binding and telomere cohesion) to maintain the central organization of the shelterin complex, thereby avoiding catastrophic consequences like loss of end protection and cell death.
Finally, we consider how loss in sister telomere cohesion might impact on telomere function in DC patients. DC-associated TIN2 mutations are characterized by very short telomeres, shorter than most telomerase mutations (Savage et al. 2008; Walne et al. 2008; Bessler et al. 2010), suggesting that DC-associated TIN2 mutations could influence other aspects of telomere function. Indeed, recent studies have shown that while overexpression of missense DC-associated TIN2 mutations in tumor cells leads to telomere shortening (Yang et al. 2011), these length effects are subtle (similar to what we observed here with TIN2.RD) (Fig 5B,C) and thus may not fully account for the severe shortening observed in TIN2-DC patients. We thus consider the possibility that defective telomere cohesion might impact telomerase-independent aspects of telomere function. We showed previously that cells defective in sister telomere cohesion were unable to repair sister chromatid breaks following DNA replication and suffered sister telomere loss (Canudas and Smith 2009). Telomere cohesion might be important for telomere lengthening by homologous recombination between sister chromatids. Recent studies indicate that telomere lengthening in mouse embryos relies on a recombination-based mechanism that is independent of telomerase (Liu et al. 2007; Zalzman et al. 2010). Although highly speculative, one possibility is that defective sister telomere cohesion could hinder telomere lengthening by recombination during embryogenesis and thereby contribute to the extreme telomere shortening observed in the context of de novo DC-associated TIN2 mutations.
Materials and methods
Yeast two-hybrid screen and assays
Two-hybrid screens were performed with the yeast reporter strain L40 (Hollenberg et al. 1995) using a HeLa cell or human fetal liver cDNA library (Clontech) and the LexA-TIN2C bait plasmid (pBTTIN2-C;TIN2 amino acids 180–354 cloned into the pBTM116 vector) (Bartel et al. 1993) according to Clontech Matchmaker protocol. We identified four independent isolates of HP1γ from a human fetal liver cDNA library and one isolate of HP1α from a HeLa cell cDNA library. Two-hybrid interactions were scored according to the number of minutes required for the color change: 20 min (++), 45 min (+), and 150–180 min (+/−). Interactions were confirmed by two or more independent experiments.
Site-directed mutagenesis
TIN2.RD and TIN2-C.RD mutations were created by substituting the proline (P) and valine (V) residues at position 283 and 285 with arginine (R) and glutamic acid (D) residues by site-directed mutagenesis of pBTTIN2 and pBTTIN2C using the oligonucleotide 5′-GGCCATAAGGAGCGCCGCACAGACATGCTGTTTCCCTTT-3′. The following mutations were generated by site-directed mutagenesis of pBTTIN2C or FlagTIN2 using the indicated oligonucleotides: K280X, 5′-GGGGAGGCCATTAGGAGCGCCCC-3′; R282H, 5′-GGCCATAAGGAGCACCCCACAGTCATGC-3′; P283H: 5′-GGAGGCCATAAGGAGCGCCACACAGTCATGC-3′; L287P, 5′-CGCCCCACAGTCATGCCGTTTCCCTTTAGG-3′; P289S, 5′-CCCACAGTCATGCTGTTTTCCTTTAGGAATCTCGG-3′; and Q298Rfs, 5′-GGCTCACCAACCAGGTCATATCTAAGCC-3′. Mutagenesis was performed using the Stratagene QuikChange site-directed mutagenesis kit according to the manufacturer's instructions.
Immunofluorescence analysis
HeLaI.2.11 cells, a HeLa-derived clonal cell line (van Steensel et al. 1998), were permeabilized in Triton X-100 buffer (0.5% Triton X-100, 20 mM Hepes-KOH at pH 7.9, 50 mM NaCl, 3 mM MgCl2, 300 mM sucrose) for 5 min at room temperature and fixed in 3% paraformaldehyde (in phosphate-buffered saline [PBS], 2% sucrose) for 10 min at room temperature, followed by permeabilization in Triton X-100 buffer for 10 min at room temperature. Cells were blocked in 1% bovine serum albumin in PBS, followed by incubation with mouse anti-HP1γ (1.5 μg/mL) (Upstate Biotechnology) and rabbit anti-TRF1 415 (0.36 μg/mL) (Cook et al. 2002), rabbit anti-TIN2 701 (0.5 μg/mL) (Houghtaling et al. 2004), or rabbit anti-RAP1 (0.25 μg/mL) (Novus Biologicals). Primary antibodies were detected with fluorescein isothiocyanate-conjugated or tetramethyl rhodamine isothiocyanate-conjugated donkey anti-rabbit or anti-mouse antibodies (1:100) (Jackson Laboratories). DNA was stained with 4,6-diamino-2-phenylindole (DAPI) (0.2 μg/mL).
Immunofluorescence/PNA FISH analysis
HeLaI.2.11 cells were processed for immmunofluorescence exactly as described above. After the last step (incubation with the fluorescently labeled second antibody), coverslips were washed in PBS, fixed in 2% paraformaldehyde in PBS for 5 min; washed six times in PBS over 10 min; dehydrated in a 70%, 95%, 100% ethanol series for 5 min each; and air dried. Coverslips were incubated with 0.5 μg/mL of a Cy3-conjugated (CCCTAA)3 PNA probe in 10 mM NaHPO4, 10 mM NaCl, 20 mM Tris (pH 7.5), 70% formamide, 0.1 μg/mL tRNA, and 0.1 μg/mL herring sperm DNA for 5 min at 75°C, followed by a 2-h incubation at room temperature; they were then washed twice for 15 min each with 70% formamide in 10 mM Tris (pH 7.2), washed with PBS, and stained with DAPI.
Telomere length analysis
Genomic DNA was isolated as described (de Lange et al. 1990) and cleaved with HinfI and RsaI. Approximately 1 μg of DNA was fractionated on 0.7% agarose gels, and telomeric restriction fragments were detected by hybridization to a 32P-end-labeled (CCCTAA)4 oligonucleotide probe as described (Dynek and Smith 2004). The mean length of telomeric restriction fragments was determined by TELO, a macro for NIH Image written by the Research Computing Department at Fox Chase Cancer Center (http://www.fccc.edu), using scanned images of autoradiograms.
Retroviruses and cell lines
Amphotropic retroviruses were generated by transfecting pBABEpuro, pBABE-Flag-TIN2FL.WT pBABE-Flag-TIN2.RD, pBABE-Flag-TIN2C.WT, pBABE-Flag-TIN2C.RD, or pBABE-MycHP1γ into Phoenix amphotropic cells using calcium phosphate precipitation. HT1080 or HTC75 cells, an HT1080-derived clonal cell line (van Steensel and de Lange 1997), were infected essentially as described (Serrano et al. 1997). Infected cells were selected in 2 μg/mL puromycin, and population doublings were counted as described previously (Cook et al. 2002).
Transient transfection and immunoprecipitation
HT1080 stable cell lines expressing pBABE-puro (V) or pBABE-MycHP1γ were transfected with (Vector [pBABEpuro], pBABE-Flag-TIN2FL.WT, or pBABE-Flag-TIN2.RD) or (Vector [3XFlag-CMV-10; Sigma], 3XFlag-TIN2.WT, 3XFlag-K280X, 3XFlag-R282H, 3XFlag-P293H, or 3XFlag-L287P) using Lipofectamine 2000 reagent (Invitrogen) for 18 h and processed for immunoprecipitation as described previously (Houghtaling et al. 2004). Briefly, cells were lysed in 0.5 mL (per one 15-cm-diameter dish) of TNE buffer (10 mM Tris at pH 7.8, 1% Nonidet P-40, 0.15 M NaCl, 1 mM EDTA, protease inhibitor cocktail [Sigma]) for 1 h on ice, then pelleted at 8000g for 10 min. Supernatants were precleared with protein G-Sepharose, rotating for 30 min at 4°C. Nonspecific protein aggregates were removed by centrifugation, and the supernatant was used for immunoprecipitation analysis or fractionated directly on SDS-PAGE (indicated as input, ∼1% of the amount used in the immunoprecipitation). Supernatants were incubated with 35 μL of rabbit anti-Myc agarose bead conjugates (Sigma) for 3 h, washed three times with 1 mL of TNE buffer, fractionated on a 10% SDS-PAGE gel, and processed for immunoblotting as described below.
siRNA transfection
siRNA transfections were performed with Oligofectamine (Invitrogen) according to the manufacturer's protocol. The final concentration of siRNA was 100 nM. The following siRNAs (synthesized by Dharmacon Research, Inc.) were used: HP1α (5′-GCUUUGAGAGAGGACUGGAAC-3′), HP1β (5′-GACUCCAGUGGAGAGCUCAUG-3′), HP1γ (5′-GAGGCAGAGCCUGAAGAAU-3′) (HP1α, HP1β, and HP1γ were described previously) (du Chene et al. 2007), GFP Duplex I, and TIN2.L (5′-AGGAAUCUCUGGAAAACUA-3′). TIN2.L is directed against TIN2 long (TERF1 isoform 1), but also has 16 nucleotides (nt) of homology with TIN2 short (TERF1 isoform 2), and thus leads to depletion of both proteins (shown in Fig. 2H, lane 2). The amount of siRNA knockdown was quantified using ImageJ software.
Cell synchronization and siRNA transfection
For siRNA experiments, cells were synchronized essentially as described (Canudas and Smith 2009). Briefly, HeLaI.2.11 cells were grown in the presence of 2 mM thymidine for 24 h, washed three times with PBS, and released into fresh medium for 4 h. Cells were then transfected with siRNA as described above. After 4 h, the medium was replaced with fresh medium and cells were further incubated for 4 h. Two-millimolar thymidine was then added and the cells were incubated for 12 h, washed three times with PBS, and released into fresh medium. Cells were then harvested by trypsinization for 4 h for FACS analysis and for chromosome-specific FISH as described below.
For immunofluorescence analysis across the cell cycle, HeLaI.2.11 cells were treated with 2 mM thymidine for 24 h, washed three times with PBS, released into fresh medium for 11 h, treated again with 2 mM thymidine for 14 h, washed three times with PBS, and released into fresh medium for 0–12 h.
FACS analysis
siRNA transfected, trypsinized cells were washed twice with PBS containing 2 mM EDTA, fixed in cold 70% ethanol, stained with propidium iodide (50 μg/mL), and analyzed using a Becton-Dickenson FACSAN and FlowJo 8.8.6 software.
Immunoblot analysis
Whole-cell extracts were prepared and immunoblots were performed exactly as described (Canudas et al. 2007). Briefly, siRNA transfected HeLaI.2.11 cells were resuspended in 4 vol of buffer C (20 mM Hepes-KOH at pH 7.9, 420 mM KCl, 25% glycerol, 0.1 mm EDTA, 5 mM MgCl2, 0.2% NP40, 1 mM dithiothreitol, 2.5% protease inhibitor cocktail [Sigma]) and incubated for 1 h on ice. Suspensions were pelleted at 8000g for 10 min. Twenty-five micrograms (determined by Bio-Rad protein assay) of supernatant proteins was fractionated by SDS-PAGE and analyzed by immunoblotting. Immunoblots were incubated with the following primary antibodies: mouse anti-HP1α MAB3446 (1:1000) (Chemicon International Inc.), rabbit anti-HP1β (1:1000) (Cell Signaling Technology), mouse anti-HP1γ MAB3450 BL143G (1:1000) (Millipore), rabbit anti-TIN2 701 (0.5 μg/mL) (Houghtaling et al. 2004), rabbit anti-Myc (0.8 μg/mL) (Santa Cruz Biotechnologies), and mouse anti-α-tubulin ascites (1:50,000) (Sigma), followed by horseradish peroxidase-conjugated donkey anti-rabbit or anti-mouse IgG (Amersham) (1:2500). Bound antibody was detected with Super Signal West Pico (Thermo Scientific).
Skin fibroblasts
TIN2p.Q269X mutant (Sasa et al. 2011) human skin fibroblasts were cultured from a punch biopsy obtained from a research subject diagnosed with DC and enrolled on a research protocol approved by the Baylor College of Medicine institutional review board. Fibroblasts were cultured in α minimal essential medium with 10% fetal bovine serum. Control human skin fibroblasts were kindly provided by Olivia Pereira-Smith (University of Texas Health Science Center, San Antonio, TX).
LCLs
For the DKCp.A2V cells, a blood sample was obtained from a male with DC and a DKC1p.A2V mutation at Texas Children's Hospital, Houston, TX, following enrollment on a research protocol that was approved by the Baylor College of Medicine institutional review board. A DKC1p.A2V Epstein-Barr virus-transformed LCL was generated from the blood sample by the tissue culture core laboratory within the Department of Molecular and Human Genetics, Baylor College of Medicine. The DKCp.A2V mutation was reported previously (Knight et al. 1999; Safa et al. 2001). The development of TIN2p.R282fs36X and control unaffected sibling LCLs was described (Sasa et al. 2011). The R282H LCL was derived from a participant in the National Cancer Institute's institutional review board-approved protocol 02-C-0052 (NCT00056121, http://www.marrowfailure.cancer.gov) as described (Savage et al. 2008). LCLs were cultured in RPMI-1640 medium with 10% fetal bovine serum.
Chromosome-specific FISH
Cells were treated with 0.5 μg/mL colcemide (Invitrogen) for 1.5 h (siRNA transfected HTC75 cell lines) or 24 h (human skin fibroblasts and LCLs), collected by mitotic shake-off, fixed, and processed as described previously (Dynek and Smith 2004). Briefly, cells were fixed twice in methanol:acetic acid (3:1) for 15 min, cytospun (Shandon Cytospin) at 2000 rpm for 2 min onto slides, rehydrated in 2× SSC for 2 min at 37°C, and dehydrated in an ethanol series of 70%, 80%, and 95% for 2 min each. Cells were denatured for 2 min at 75°C and hybridized overnight at 37°C with a subtelomeric FITC-conjugated probe (16ptelo or 4ptelo), a chromosome 6-specific α-satellite TRITC-conjugated centromere probe (6cen), or a FITC-conjugated chromosome 10 centromere probe (10cen) (Cytocell). Cells were washed in 0.4× SSC for 2 min at 72°C, and in 2× SSC with 0.05% Tween 20 for 30 sec at room temperature. DNA was stained with 0.2 μg/mL DAPI. The distance between FISH signals was measured using Openlab software (Perkin Elmer).
Image acquisition
Images were acquired using a microscope (Axioplan 2, Carl Zeiss, Inc.) with a Plan Apochrome 63× NA 1.4 oil immersion lens (Carl Zeiss, Inc.) and a digital camera (C4742-95, Hamamatsu Photonics). Images were acquired and processed using Openlab software (Perkin Elmer).
Acknowledgments
We thank Tom Meier and members of the Smith laboratory for comments on the manuscript and helpful discussion. We are grateful to the patients for their participation in these studies. We thank Drs. Blanche Alter and Neelam Giri, National Cancer Institute, for collecting patient materials and data. Lisa Leathwood, RN, Westat, Inc. (NIH contracts N02-CP-11019, N02-CP-65504, and N02-CP-65501), provided outstanding study support. This work was supported by NIH grant R01 CA116352 to S.S. and by a NSF grant (0543553) to S.S. S.C. was the 2010 Helen and Martin Kimmel Senior Fellow in Stem Cell Biology. S.A.S. was supported by the intramural research program of the Division of Cancer Epidemiology, National Cancer Institute.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.17325211.
References
- Abreu E, Aritonovska E, Reichenbach P, Cristofari G, Culp B, Terns RM, Lingner J, Terns MP 2010. TIN2-tethered TPP1 recruits human telomerase to telomeres in vivo. Mol Cell Biol 30: 2971–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DE, Losada A, Erickson HP, Hirano T 2002. Condensin and cohesin display different arm conformations with characteristic hinge angles. J Cell Biol 156: 419–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artandi SE, DePinho RA 2010. Telomeres and telomerase in cancer. Carcinogenesis 31: 9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124 [DOI] [PubMed] [Google Scholar]
- Bartel PL, Chien C, Sternglanz R, Fields S 1993. Using the two-hybrid system to detect protein–protein interaction. In Cellular interactions in development: a practical approach (ed. Harley DA), pp. 153–179 IRL Press, Oxford [Google Scholar]
- Baumann P, Cech TR 2001. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292: 1171–1175 [DOI] [PubMed] [Google Scholar]
- Bessler M, Wilson DB, Mason PJ 2010. Dyskeratosis congenita. FEBS Lett 584: 3831–3838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasher SV, Smith BO, Fogh RH, Nietlispach D, Thiru A, Nielsen PR, Broadhurst RW, Ball LJ, Murzina NV, Laue ED 2000. The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J 19: 1587–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan TM, Englezou A, Dunham MA, Reddel RR 1998. Telomere length dynamics in telomerase-positive immortal human cell populations. Exp Cell Res 239: 370–378 [DOI] [PubMed] [Google Scholar]
- Canudas S, Smith S 2009. Differential regulation of telomere and centromere cohesion by the Scc3 homologues SA1 and SA2, respectively, in human cells. J Cell Biol 187: 165–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canudas S, Houghtaling BR, Kim JY, Dynek JN, Chang WG, Smith S 2007. Protein requirements for sister telomere association in human cells. EMBO J 26: 4867–4878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CR, Wu CS, Hom Y, Gartenberg MR 2005. Targeting of cohesin by transcriptionally silent chromatin. Genes Dev 19: 3031–3042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook BD, Dynek JN, Chang W, Shostak G, Smith S 2002. Role for the related poly(ADP-Ribose) polymerases tankyrase 1 and 2 at human telomeres. Mol Cell Biol 22: 332–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T, Shiue L, Myers RM, Cox DR, Naylor SL, Killery AM, Varmus HE 1990. Structure and variability of human chromosome ends. Mol Cell Biol 10: 518–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Chene I, Basyuk E, Lin YL, Triboulet R, Knezevich A, Chable-Bessia C, Mettling C, Baillat V, Reynes J, Corbeau P, et al. 2007. Suv39H1 and HP1γ are responsible for chromatin-mediated HIV-1 transcriptional silencing and post-integration latency. EMBO J 26: 424–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynek JN, Smith S 2004. Resolution of sister telomere association is required for progression through mitosis. Science 304: 97–100 [DOI] [PubMed] [Google Scholar]
- Fanti L, Pimpinelli S 2008. HP1: a functionally multifaceted protein. Curr Opin Genet Dev 18: 169–174 [DOI] [PubMed] [Google Scholar]
- Garcia-Cao M, O'Sullivan R, Peters AH, Jenuwein T, Blasco MA 2004. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat Genet 36: 94–99 [DOI] [PubMed] [Google Scholar]
- Giraud-Panis MJ, Teixeira MT, Geli V, Gilson E 2010. CST meets shelterin to keep telomeres in check. Mol Cell 39: 665–676 [DOI] [PubMed] [Google Scholar]
- Gonzalo S, Garcia-Cao M, Fraga MF, Schotta G, Peters AH, Cotter SE, Eguia R, Dean DC, Esteller M, Jenuwein T, et al. 2005. Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat Cell Biol 7: 420–428 [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH 1987. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell 51: 887–898 [DOI] [PubMed] [Google Scholar]
- Haering CH, Lowe J, Hochwagen A, Nasmyth K 2002. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell 9: 773–788 [DOI] [PubMed] [Google Scholar]
- Haering CH, Farcas AM, Arumugam P, Metson J, Nasmyth K 2008. The cohesin ring concatenates sister DNA molecules. Nature 454: 297–301 [DOI] [PubMed] [Google Scholar]
- Hollenberg SM, Sternglanz R, Cheng PF, Weintraub H 1995. Identification of a new family of tissue-specific basic helix–loop–helix proteins with a two-hybrid system. Mol Cell Biol 15: 3813–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houghtaling BR, Cuttonaro L, Chang W, Smith S 2004. A dynamic molecular link between the telomere length regulator TRF1 and the chromosome end protector TRF2. Curr Biol 14: 1621–1631 [DOI] [PubMed] [Google Scholar]
- Jady BE, Richard P, Bertrand E, Kiss T 2006. Cell cycle-dependent recruitment of telomerase RNA and Cajal bodies to human telomeres. Mol Biol Cell 17: 944–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminker PG, Kim SH, Desprez PY, Campisi J 2009. A novel form of the telomere-associated protein TIN2 localizes to the nuclear matrix. Cell Cycle 8: 931–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Kaminker P, Campisi J 1999. TIN2, a new regulator of telomere length in human cells. Nat Genet 23: 405–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Beausejour C, Davalos AR, Kaminker P, Heo SJ, Campisi J 2004. TIN2 mediates functions of TRF2 at human telomeres. J Biol Chem 279: 43799–43804 [DOI] [PubMed] [Google Scholar]
- Knight SW, Heiss NS, Vulliamy TJ, Greschner S, Stavrides G, Pai GS, Lestringant G, Varma N, Mason PJ, Dokal I, et al. 1999. X-linked dyskeratosis congenita is predominantly caused by missense mutations in the DKC1 gene. Am J Hum Genet 65: 50–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410: 116–120 [DOI] [PubMed] [Google Scholar]
- Liu D, O'Connor MS, Qin J, Songyang Z 2004a. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J Biol Chem 279: 51338–51342 [DOI] [PubMed] [Google Scholar]
- Liu D, Safari A, O'Connor MS, Chan DW, Laegeler A, Qin J, Songyang Z 2004b. PTOP interacts with POT1 and regulates its localization to telomeres. Nat Cell Biol 6: 673–680 [DOI] [PubMed] [Google Scholar]
- Liu L, Bailey SM, Okuka M, Munoz P, Li C, Zhou L, Wu C, Czerwiec E, Sandler L, Seyfang A, et al. 2007. Telomere lengthening early in development. Nat Cell Biol 9: 1436–1441 [DOI] [PubMed] [Google Scholar]
- Lomberk G, Wallrath L, Urrutia R 2006. The heterochromatin protein 1 family. Genome Biol 7: 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Yokochi T, Kobayashi R, Hirano T 2000. Identification and characterization of SA/Scc3p subunits in the Xenopus and human cohesin complexes. J Cell Biol 150: 405–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minc E, Allory Y, Worman HJ, Courvalin JC, Buendia B 1999. Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma 108: 220–234 [DOI] [PubMed] [Google Scholar]
- Palm W, de Lange T 2008. How shelterin protects mammalian telomeres. Annu Rev Genet 42: 301–334 [DOI] [PubMed] [Google Scholar]
- Prescott J, Blackburn EH 1997a. Functionally interacting telomerase RNAs in the yeast telomerase complex. Genes Dev 11: 2790–2800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott J, Blackburn EH 1997b. Telomerase RNA mutations in Saccharomyces cerevisiae alter telomerase action and reveal nonprocessivity in vivo and in vitro. Genes Dev 11: 528–540 [DOI] [PubMed] [Google Scholar]
- Safa WF, Lestringant GG, Frossard PM 2001. X-linked dyskeratosis congenita: restrictive pulmonary disease and a novel mutation. Thorax 56: 891–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasa G, Ribes-Zamora A, Nelson N, Bertuch A 2011. Three novel truncating TINF2 mutations causing severe dyskeratosis congenita in early childhood. Clin Genet. doi: 10.1111/j.1399-0004.2011.01658.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage SA, Giri N, Baerlocher GM, Orr N, Lansdorp PM, Alter BP 2008. TINF2, a component of the shelterin telomere protection complex, is mutated in dyskeratosis congenita. Am J Hum Genet 82: 501–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S, Blasco MA 2009. A ‘higher order' of telomere regulation: telomere heterochromatin and telomeric RNAs. EMBO J 28: 2323–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88: 593–602 [DOI] [PubMed] [Google Scholar]
- Sherwood R, Takahashi TS, Jallepalli PV 2010. Sister acts: coordinating DNA replication and cohesion establishment. Genes Dev 24: 2723–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smothers JF, Henikoff S 2000. The HP1 chromo shadow domain binds a consensus peptide pentamer. Curr Biol 10: 27–30 [DOI] [PubMed] [Google Scholar]
- Sumara I, Vorlaufer E, Gieffers C, Peters BH, Peters JM 2000. Characterization of vertebrate cohesin complexes and their regulation in prophase. J Cell Biol 151: 749–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson RL, Ziegler TD, Supakorndej T, Terns RM, Terns MP 2006. Cell cycle-regulated trafficking of human telomerase to telomeres. Mol Biol Cell 17: 955–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F, Nasmyth K 1998. Cohesion between sister chromatids must be established during DNA replication. Curr Biol 8: 1095–1101 [DOI] [PubMed] [Google Scholar]
- van Steensel B, de Lange T 1997. Control of telomere length by the human telomeric protein TRF1. Nature 385: 740–743 [DOI] [PubMed] [Google Scholar]
- van Steensel B, Smogorzewska A, de Lange T 1998. TRF2 protects human telomeres from end-to-end fusions. Cell 92: 401–413 [DOI] [PubMed] [Google Scholar]
- Vulliamy T, Beswick R, Kirwas MJ, Hossain U, Walne AJ, Dokal I 2011. Telomere length measurement can distinguish pathogenic from non-pathogenic variant in the shelterin component, TIN2. Clin Genet. doi: 10.1111/j.1399-0004.2010.01605.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walne AJ, Vulliamy T, Beswick R, Kirwan M, Dokal I 2008. TINF2 mutations result in very short telomeres: analysis of a large cohort of patients with dyskeratosis congenita and related bone marrow failure syndromes. Blood 112: 3594–3600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M 2007. The POT1–TPP1 telomere complex is a telomerase processivity factor. Nature 445: 506–510 [DOI] [PubMed] [Google Scholar]
- Wenz C, Enenkel B, Amacker M, Kelleher C, Damm K, Lingner J 2001. Human telomerase contains two cooperating telomerase RNA molecules. EMBO J 20: 3526–3534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright WE, Piatyszek MA, Rainey WR, Byrd W, Shay JW 1996. Telomerase activity in human germline and embryonic tissues and cells. Dev Genet 18: 173–179 [DOI] [PubMed] [Google Scholar]
- Wright WE, Tesmer VM, Liao ML, Shay JW 1999. Normal human telomeres are not late replicating. Exp Cell Res 251: 492–499 [DOI] [PubMed] [Google Scholar]
- Xin H, Liu D, Wan M, Safari A, Kim H, Sun W, O'Connor MS, Songyang Z 2007. TPP1 is a homologue of ciliate TEBP-β and interacts with POT1 to recruit telomerase. Nature 445: 559–562 [DOI] [PubMed] [Google Scholar]
- Yang D, He Q, Kim H, Ma W, Songyang Z 2011. TIN2 dyskeratosis congenital missense mutants are defective in association with telomerase. J Biol Chem doi: 10.1074/jbc.M111.225870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye JZ, Donigian JR, van Overbeek M, Loayza D, Luo Y, Krutchinsky AN, Chait BT, de Lange T 2004a. TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J Biol Chem 279: 47264–47271 [DOI] [PubMed] [Google Scholar]
- Ye JZ, Hockemeyer D, Krutchinsky AN, Loayza D, Hooper SM, Chait BT, de Lange T 2004b. POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev 18: 1649–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalzman M, Falco G, Sharova LV, Nishiyama A, Thomas M, Lee SL, Stagg CA, Hoang HG, Yang HT, Indig FE, et al. 2010. Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature 464: 858–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Kuznetsov SG, Sharan SK, Li K, Rao PH, Pati D 2008. A handcuff model for the cohesin complex. J Cell Biol 183: 1019–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Sfeir AJ, Zou Y, Buseman CM, Chow TT, Shay JW, Wright WE 2009. Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell 138: 463–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Abreu E, Kim J, Stadler G, Eskiocak U, Terns MP, Terns RM, Shay JW, Wright WE 2011. Processive and distributive extension of human telomeres by telomerase under homeostatic and nonequilibrium conditions. Mol Cell 42: 297–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong F, Savage SA, Shkreli M, Giri N, Jessop L, Myers T, Chen R, Alter BP, Artandi SE 2011. Disruption of telomerase trafficking by TCAB1 mutation causes dyskeratosis congenita. Genes Dev 25: 11–16 [DOI] [PMC free article] [PubMed] [Google Scholar]