Frolov and colleagues identify an intriguing example of an embedded Drosophila miRNA that directly regulates the transcriptional targets of its own host gene. Intronic miR-11 is coexpressed with its host gene, dE2F1, yet, in the context of DNA damage, miR-11 specifically limits the dE2F1 proapoptotic response by repressing dE2F1’s proapoptotic gene targets.
Keywords: cell death, apoptosis, E2F, microRNA, miR-11, DNA damage
Abstract
The E2F family of transcription factors regulates the expression of both genes associated with cell proliferation and genes that regulate cell death. The net outcome is dependent on cellular context and tissue environment. The mir-11 gene is located in the last intron of the Drosophila E2F1 homolog gene dE2f1, and its expression parallels that of dE2f1. Here, we investigated the role of miR-11 and found that miR-11 specifically modulated the proapoptotic function of its host gene, dE2f1. A mir-11 mutant was highly sensitive to dE2F1-dependent, DNA damage-induced apoptosis. Consistently, coexpression of miR-11 in transgenic animals suppressed dE2F1-induced apoptosis in multiple tissues, while exerting no effect on dE2F1-driven cell proliferation. Importantly, miR-11 repressed the expression of the proapoptotic genes reaper (rpr) and head involution defective (hid), which are directly regulated by dE2F1 upon DNA damage. In addition to rpr and hid, we identified a novel set of cell death genes that was also directly regulated by dE2F1 and miR-11. Thus, our data support a model in which the coexpression of miR-11 limits the proapoptotic function of its host gene, dE2f1, upon DNA damage by directly modulating a dE2F1-dependent apoptotic transcriptional program.
The E2F family of transcription factors regulates the expression of both genes associated with cell proliferation and genes that regulate cell death. While the conditions, requirements, and consequences of regulation of cell proliferation genes by E2F are well appreciated (for review, see Iaquinta and Lees 2007; van den Heuvel and Dyson 2008), less is understood about the circumstances under which E2F activates genes associated with cell death, either in normal cells or following DNA damage, and ultimately what dictates the fate of the cells.
The ability to control cell proliferation is a hallmark feature of each of eight mammalian E2F genes. In contrast, a proapoptotic function is associated primarily with E2F1. Early studies demonstrated that, in addition to driving the cell into the S phase, overexpression of E2F1 potently induces apoptosis in mammalian cells (Johnson et al. 1993; Kowalik et al. 1995). E2F1 promotes apoptosis by regulating genes that function prior to the commitment of the cell to undergo apoptosis and genes that are part of the core apoptotic machinery, such as caspases (Asano et al. 1996; Du et al. 1996; Muller et al. 2001; Nahle et al. 2002; Pediconi et al. 2003). E2F1 induces apoptosis in multiple settings. In response to DNA damage, the E2F1 protein is stabilized as a result of phosphorylation and/or acetylation, and these increased levels of E2F1 facilitate the induction of apoptosis (for review, see Iaquinta and Lees 2007). In other circumstances, deregulated E2F1 activity results in accumulation of p53 protein followed by apoptosis, although E2F1 can also induce apoptosis independently of p53 (Iaquinta and Lees 2007; Moon et al. 2008; van den Heuvel and Dyson 2008). The Retinoblastoma (Rb) gene mouse knockout models provided insights into the proapoptotic function of endogenous E2F. The pRB protein is a negative regulator of E2F, and therefore functional inactivation of pRB results in deregulated E2F activity. The loss of Rb, whether in cancer tissues, cell lines, or animal models, leads to inappropriate S-phase entry and sensitizes cells to apoptosis, and this is often dependent on E2F. However, while these findings shed light on the mechanism of E2F-induced apoptosis, there is still a lot to learn about what recruits E2Fs to the promoters of proapoptotic genes in different contexts and what additional factors are required to permit or prevent the expression of these genes.
Drosophila has been extensively used to study E2F-dependent apoptosis in a developmental context. Similar to a mammalian experimental system, the loss of the Drosophila pRB ortholog RBF resulted in increased apoptosis and ectopic S-phase entry (Du and Dyson 1999), while overexpression of dE2F1 led to unscheduled S phases and cell death (Asano et al. 1996; Du et al. 1996; Morris et al. 2006). Accordingly, dE2F1 has been shown to activate the expression of proapoptotic genes such as reaper (rpr) and head involution defective (hid). The ability of deregulated endogenous dE2f1 to induce apoptosis appears to be context-dependent. For example, EGFR signaling largely suppresses dE2F1-dependent apoptosis in rbf mutant cells of the eye imaginal disc. As a result, cell death is restricted to a specific region of the eye discs, called the morphogenetic furrow (MF), where the EGFR signaling is normally down-regulated during development (Moon et al. 2006). Additionally, dE2F1-dependent apoptosis occurs upon DNA damage, and the loss of rbf further enhances cell death (Zhou and Steller 2003; Moon et al. 2005). These studies illustrate that additional signals and/or factors exist to modulate the cellular response to dE2F1-dependent apoptosis in vivo.
While the dE2f1 gene was cloned in 1994, it was only recently discovered that the last intron of dE2f1 harbors the mir-11 gene (Lagos-Quintana et al. 2001; Sempere et al. 2003). MicroRNAs are transcribed as a ∼70-nucleotide (nt) stem–loop primary microRNA that then undergoes maturation and processing to yield a 20- to 23-nt ssRNA. The mature microRNA is incorporated into an RNA-induced silencing complex that binds to complementary sequences, usually in the 3′ untranslated region (UTR) of a target mRNA. This interaction regulates the expression of the mRNA and the protein for which it encodes, usually by repression (Carthew and Sontheimer 2009). miR-11 is a member of the largest family of microRNAs in Drosophila: the miR-2 family. Members of the miR-2 family share the first 8 nt, the seed sequence, which is a primary determinant of target specificity. Using overexpression systems and antisense oligonucleotides, these microRNAs were shown to down-regulate effectors of Notch signaling and proapoptotic genes rpr, hid, grim, and sickle (skl) to varying degrees (Brennecke et al. 2005; EC Lai et al. 2005; Leaman et al. 2005).
Despite extensive studies of dE2F1, little is known about the function of mir-11, which the dE2f1 gene harbors. Here, we investigated the role of miR-11 in the context of E2F regulation and showed that miR-11 specifically modulates the proapoptotic function of dE2F1. In genetic interaction tests, overexpression of miR-11 suppressed dE2F1-induced apoptosis in multiple tissues. Consistently, miR-11 mutant cells were sensitized to irradiation-induced, dE2F1-dependent apoptosis and expressed higher levels of proapoptotic genes hid and rpr. Surprisingly, we did not find any evidence that miR-11 affects the ability of dE2F1 to regulate cell proliferation, suggesting that these genes, mir-11 and dE2f1, interact specifically to modulate the apoptotic response. Furthermore, we identified a novel set of genes, in addition to rpr and hid, which can modulate cell death and was directly regulated by dE2F1 and miR-11. Thus, we uncovered a partial feed-forward loop between mir-11 and its host gene, dE2f1, which suggests a novel mechanism whereby miR-11 protects cells from dE2F1-dependent cell death upon DNA damage.
Results
Expression of miR-11 in dE2f1 mutant alleles
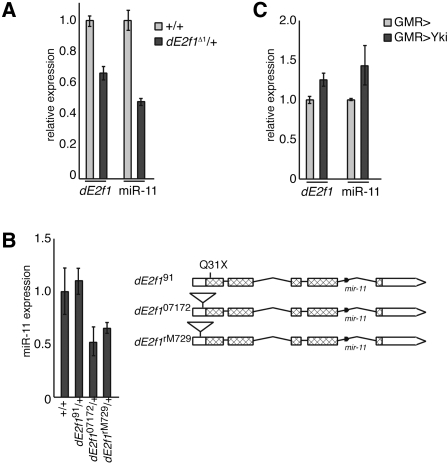
The Drosophila mir-11 gene is located within the last intron of the dE2f1 gene and is conserved in 12 Drosophila species (Ruby et al. 2007). Intronic microRNA genes are thought to be coexpressed with their protein-encoding hosts (Rodriguez et al. 2004; Baskerville and Bartel 2005; Kim and Kim 2007). Indeed, previous studies showed that dE2F1 and miR-11 have similar expression profiles throughout the different stages of development (Brook et al. 1996; Lagos-Quintana et al. 2001; Sempere et al. 2003). To investigate whether dE2f1 and miR-11 are cotranscribed, we used the TaqMan quantitative PCR (qPCR) assay to measure miR-11 levels in a series of dE2f1 mutant alleles. As expected, in flies heterozygous for a deficiency, dE2f1Δ1, which removes the entire dE2f1 genomic region, dE2f1 and miR-11 expression were approximately half that of wild type, indicating that a 50% reduction in miR-11 expression levels could be detected in this assay (Fig. 1A). The dE2f191 allele is a point mutant that results in a premature stop codon at amino acid 31 (Duronio et al. 1995). This allele does not generate dE2F1 protein; however, the dE2f1 transcript is produced. Similarly, miR-11 expression was not altered (Fig. 1B). The dE2f1rM729 and dE2f107172 alleles are loss-of-function alleles resulting from a P-element insertion within the 5′ end of the dE2f1 gene, which interferes with dE2f1 expression (Fig. 1B; Duronio et al. 1995). Importantly, while the mir-11 gene is intact in the dE2f1rM729 and dE2f107172 mutant alleles, the expression of mature miR-11 was reduced approximately twofold in heterozygous animals (Fig. 1B). These results suggest that dE2f1 and miR-11 are generated from the same transcript.
Figure 1.
Expression of miR-11 parallels expression of dE2f1. (A) cDNA was prepared from RNA extracted from Canton S (wild-type) or de2f1Δ1 adult flies. The expression of mir-11 and the de2f1 mRNA were measured using TaqMan or standard real-time PCR, respectively, and normalized to β-tubulin or RpP0 levels. Expression of normalized dE2f1 and miR-11 in wild type was designated as 1.0, and expression in de2f1Δ1 samples was compared. (B) RNA was extracted from third instar larvae. miR-11 expression in the indicated genotypes was determined and compared with wild type, following normalization to β-tubulin and RpP0 expression. A diagram of the dE2f1 exon/intron structure, mutant alleles, and mir-11 gene examined is shown. The dE2f1 ORF corresponds to hatched bars, while UTRs are indicated by white bars. Introns are represented by horizontal lines. The dE2f1rM729 P-element insertion is 48 nt upstream of the initiator methionine, the dE2f107172 P-element insertion is 33 nt upstream of the initiator methionine, and the dE2f191 allele is a C91T point mutation, giving a Q31X early translation termination codon (Mlodzik and Hiromi 1992; Duronio et al. 1995; Brook et al. 1996). (C) Flies carrying the GMR-Gal4 transgene were crossed to wild-type or UAS-ykiS168A flies. RNA was extracted from third instar larval eye discs, and miR-11, dE2f1, β-tubulin, and RpP0 expression was measured by qRT–PCR. Expression levels shown are relative to GMR-Gal4/+.
The dE2f1 gene has been shown to be a transcriptional target of the Hippo pathway (Goulev et al. 2008; Nicolay and Frolov 2008). Expression of a constitutively active transcriptional coactivator, YkiS196A, a downstream effector of the Hippo signaling pathway, in the posterior compartment of the larval eye disc resulted in the increased expression of the dE2f1 mRNA as revealed by qRT–PCR (Fig. 1C). Similarly, YkiS196A induced miR-11 expression, suggesting that the activation of transcription from the dE2f1 locus results in increased expression of both dE2f1 mRNA and miR-11. These results, as well as published miR-11 developmental expression patterns (Brook et al. 1996; Lagos-Quintana et al. 2001; Sempere et al. 2003), suggest that the expression of miR-11 parallels that of dE2f1: Animals with a lower level of dE2f1 have reduced expression of miR-11, while induction of dE2f1 expression also results in the increased expression of miR-11. These findings are consistent with the notion that dE2f1 and miR-11, which is embedded in the dE2f1 gene, are coexpressed.
In transgenic animals, expression of miR-11 suppressed dE2F1-induced apoptosis but did not affect dE2F1-induced proliferation
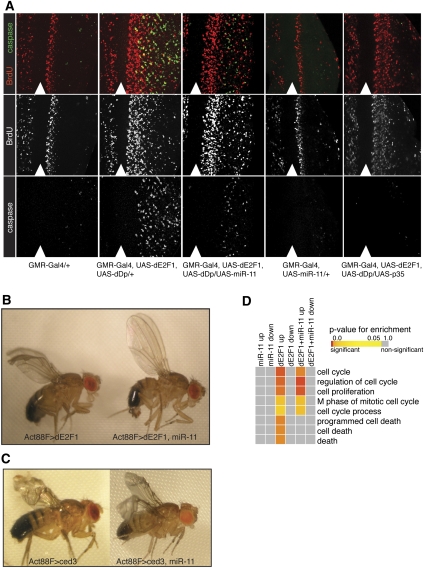
Given that the expression of miR-11 follows that of dE2f1, we examined genetic interactions between dE2f1 and mir-11 in transgenic flies. Overexpression or deregulation of dE2F1 caused unscheduled proliferation and/or apoptosis in several developmental contexts (Asano et al. 1996; Du et al. 1996; Royzman et al. 2002; Moon et al. 2005, 2006, 2008; Morris et al. 2006). We used the Gal4/UAS technique to induce the expression of dE2F1 and dDP, the heterodimeric DNA-binding partner of dE2F, in post-mitotic cells within the posterior compartment of the eye imaginal disc with the GMR-Gal4 driver. It had been previously shown that in these settings, dE2F1 potently induces both ectopic S phases and apoptosis (Du et al. 1996). In the larval eye imaginal disc, S phases could be readily visualized by BrdU labeling, while apoptotic cells could be distinguished by the presence of activated caspase, which was monitored with the C3 antibody (Fig. 2A). In wild-type eye discs, cells posterior to the MF synchronously enter the cell cycle for one last synchronous cell division, in what is called the second mitotic wave, before withdrawing from the cell cycle and committing to a differentiation program. Following induced dE2F1 expression by GMR-Gal4, cells posterior to the second mitotic wave, which are normally post-mitotic, were inappropriately incorporating BrdU, indicating that these cells were undergoing ectopic S phases. Coexpression of miR-11 with the GMR-Gal4 driver did not prevent the ectopic S phases induced by dE2F1 (Fig. 2A). Thus, in these settings, miR-11 did not modulate dE2F1-induced proliferation.
Figure 2.
Overexpression of miR-11 suppresses dE2f1-induced apoptosis in transgenic animals. (A) Third instar larval eye discs of various genotypes were incubated with BrdU for 90 min at room temperature, followed by fixing and staining with antibodies recognizing BrdU (red) and active caspase (C3) (green). Analysis was performed on a minimum of 10 larvae of each genotype. The position of the MF is marked with an arrowhead. (B,C) Flies carrying Act88F-Gal4 and UAS-dE2f1 (B) or UAS-ced3 (C) transgenes were crossed to either a wild-type chromosome (Canton S) or UAS-miR-11. (D) RNA was extracted from third instar larval eye discs from GMR-Gal4 crosses described in A, and transcriptional profiles were determined by Affymetrix gene expression microarrays (see the Materials and Methods). Differentially expressed genes were compared for enrichment of GOBP categories with a false discovery rate (FDR) standard cutoff of 0.05 for differentially expressed genes relative to GMR-Gal4/+. Heat map color values represent statistical significance of enrichment (see the scale bar).
In addition to, and independently of, ectopic S phases, overexpression of dE2F1 also induced apoptosis (Asano et al. 1996; Brook et al. 1996), as was evident from the presence of cells that express activated caspases (Fig. 2A). As expected, the number of cells with C3 staining was strongly reduced following coexpression of the p35 baculoviral caspase inhibitor (Fig. 2A). Strikingly, coexpression of miR-11 potently suppressed dE2F1-dependent apoptosis, as the number of cells with active caspases was dramatically reduced in the GMR>dE2F1/dDP/miR-11 eye discs in comparison with the GMR>dE2F1/dDP eye discs (Fig. 2A). This effect is not due to reduced expression of dE2F1, as dE2F1 proteins levels were not affected by coexpression of mir-11 in the GMR>dE2F1/dDP/mir-11 eye discs (data not shown). Therefore, we concluded that coexpression of miR-11 specifically suppressed dE2F1-induced apoptosis, but did not influence dE2F1-dependent cell proliferation in transgenic animals.
The inhibition of E2F-induced apoptosis by miR-11 expression could be specific to the eye, or it could reflect a more general role of miR-11 in suppressing dE2F1-induced apoptosis. Act88F-Gal4 directs expression in wing and thoracic muscles, and when dE2F1 expression is under the control of this driver, the wings look gnarled, downward-curved, and/or blistered. Importantly, this phenotype was shown to arise exclusively from dE2F1-dependent apoptosis and not from increased proliferation (Fig. 2B; Morris et al. 2006; Moon et al. 2008). While overexpression of miR-11 using the Act88F-Gal4 driver had no effect on its own (data not shown), miR-11 strongly suppressed the apoptotic phenotype in the wing induced by dE2F1 (Fig. 2B). Therefore, coexpression of miR-11 suppressed dE2F1-induced apoptosis in multiple tissues. Interestingly, miR-11 did not block cell death induced by ced3, the Caenorhabditis elegans caspase homolog (Fig. 2C). This suggested that miR-11 blocks dE2F1-induced cell death at a point prior to a cell's commitment to undergo apoptosis.
miR-11 specifically alters the proapoptotic gene expression signature induced by dE2F1
To gain further insight into the suppression of dE2F1-induced apoptosis by miR-11 at the molecular level, we performed gene expression array analysis of third instar larval eye discs from GMR-Gal4-driven expression of dE2F1/dDP/mir-11, dE2F1/dDP, or mir-11 alone, and compared expression profiles with that of GMR-Gal4 heterozygotes. Gene Ontology (GO) of biological processes (GOBP) analysis revealed overrepresentation of genes associated with proliferation and apoptosis when dE2F1 and dDP were expressed (Fig. 2D). Coexpression of miR-11 with dE2F1 and dDP also showed overrepresentation of proliferation-related genes, but genes associated with apoptosis were no longer enriched (see Supplemental Tables S1 [for statistical details], S2 [for the enriched gene list]). Measurement of gene expression by microarray provided an unbiased analysis of differential gene expression, and the trends observed from this analysis were in agreement with and complemented genetic interaction tests (Fig. 2A–C). Therefore, in different tissues, and using different analysis methods, we found that coexpression of miR-11 suppressed dE2F1-induced apoptosis, but did not affect dE2F1-induced cell proliferation. However, these conclusions were drawn using transgenic animals, and it was not clear whether these interactions accurately reflected the endogenous function of miR-11. Therefore, we generated a mir-11 mutant and examined the consequence of the loss of miR-11 on dE2F1-dependent apoptosis.
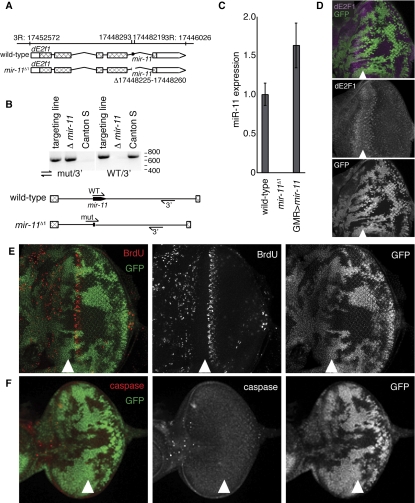
Generation and characterization of a mir-11 mutant
The mir-11 gene is located within a 1.1-kb intron in the dE2f1 gene, and there were no pre-existing mutant alleles of mir-11 that did not also affect the dE2f1 gene. Therefore, we used ends-in homologous recombination (Rong and Golic 2000) to produce a small deletion, mir-11Δ1, which specifically removed the mir-11 gene in the context of an otherwise intact dE2f1 gene (Fig. 3A,B; for details, see the Materials and Methods). Using the TaqMan assay, we confirmed that miR-11 was not expressed in mir-11Δ1 (Fig. 3C), and therefore mir-11Δ1 is a null allele. Importantly, the following data strongly argue that the function and expression of the dE2f1 host gene was not affected in the mir-11Δ1 mutants. In wild-type larval eye imaginal discs, dE2F1 is expressed in a highly distinct pattern (Fig. 3D; Frolov et al. 2005; Nicolay and Frolov 2008). The cells within the MF are paused in the G1 phase of the cell cycle and express high levels of dE2F1. Immediately posterior to the MF, cells re-enter in S phase, at which time dE2F1 expression decreases to nearly undetectable levels. We used the FLP/FRT technique to induce clones of miR-11 homozygous mutant cells in the eye imaginal disc. In this approach, the wild-type cells are distinguished by the presence of GFP, while the mutant tissue lacks GFP expression. As shown in Figure 3D, endogenous dE2F1 expression was not affected in the mir-11Δ1 mutant tissue. Importantly, genetic complementation tests revealed that the mir-11Δ1 mutant fully complemented two null alleles of dE2f1 [Df(3R)dE2f1Δ1 and dE2f191] and two strong loss-of-function dE2f1 alleles (dE2f1rM729 and dE2f107172). Thus, we concluded that the mir-11Δ1 deletion specifically removes miR-11 in the context of a functional dE2f1 host gene.
Figure 3.
Generation and initial characterization of mir-11 mutant. (A) Genomic organization of the mir-11 region. (B) Genomic DNA was extracted from wild type (Canton S); the targeting line used for homologous recombination, which contains wild-type and mir-11 mutant intron sequences (targeting line); or the homozygous mir-11 mutant (Δ mir-11) flies. PCR was performed using primers binding to the regions indicated to follow the presence and loss of the mir-11 gene. (C) RNA was extracted from wild-type (Canton S), mir-11 mutant, or GMR>mir-11 larvae. The dme-miR-11 TaqMan assay was used to measure mature miR-11 expression. (D–F) Clones of mir-11 mutant tissue were generated using ey-FLP. Wild-type tissue is marked by GFP (green). (D) Third instar larval eye discs were dissected, and dE2F1 expression (magenta) was detected by immunofluorescence. (E) To mark proliferating cells, eye discs were incubated with BrdU for 60 min prior to fixation and antibody staining. (F) Apoptotic cells were labeled with the C3 antibody, which recognizes the active forms of caspases.
Homozygous mir-11Δ1 mutant animals were viable and showed no obvious developmental defects when raised under normal conditions. Consistently, the patterns of cell proliferation were not affected in mir-11 mutant tissue, as revealed by BrdU labeling (Fig. 3E) and by staining with a mitotic marker, phosphorylated histone H3 (data not shown). No spontaneous apoptosis was observed in clones of mir-11 mutant cells in larval eye or wing imaginal discs (Fig. 3F; data not shown). Thus, the mir-11Δ1 mutant allele provides us with the opportunity to study the mir-11 function in normal developmental settings.
Loss of miR-11 sensitizes cells to dE2F1-dependent apoptosis induced by DNA damage
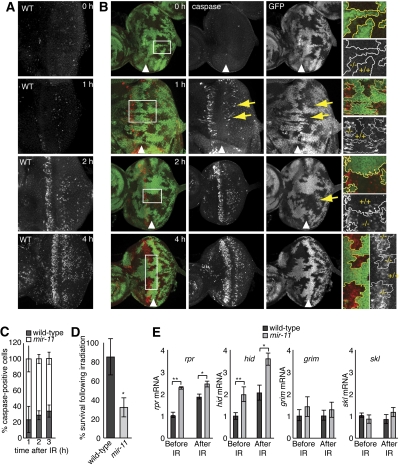
Normally, there is little, if any, apoptosis in developing third instar larval eye imaginal discs. However, exposure of third instar larvae to high levels of γ-irradiation (40 Gy) causes DNA damage and subsequent apoptosis, which occurs in a dE2F1-dependent manner (Moon et al. 2008; Wichmann et al. 2010). The apoptotic cells are commonly revealed with an antibody specifically recognizing cleaved, active caspases (C3). In wild-type discs, apoptotic cells first appeared 2 h following irradiation, primarily in a narrow stripe immediately anterior to the MF (Fig. 4A). The number of apoptotic cells in the stripe increased at 4-h post irradiation, and additional C3-positive cells were detected posterior to the MF among post-mitotic cells (Fig. 4A; Moon et al. 2008). Importantly, cell death following irradiation reflects a proapoptotic function of dE2F1, and apoptosis is fully blocked in the eye imaginal discs lacking dE2F1 (Moon et al. 2008; Wichmann et al. 2010). Therefore, DNA damage-induced apoptosis represents an ideal system to investigate the requirement of the endogenous mir-11 in dE2F1-dependent apoptosis in physiological settings.
Figure 4.
mir-11 mutant cells are highly sensitive to dE2F1-dependent irradiation-induced apoptosis and have a higher level of expression of hid and rpr. (A,B) Wild-type (A) or mir-11 mosaic mutant (B) third instar larvae were exposed to 40 Gy of irradiation, and eye discs were harvested at the indicated times following exposure. Apoptosis was detected using the C3 antibody (red), which recognizes active caspases. Clones of mir-11 mutant tissue were generated using ey-FLP. Wild-type tissue is marked by GFP (green). Yellow arrows indicate patches of apoptotic cells that appear in the mir-11 mutant cells at an earlier time point than in the adjacent wild-type tissue. At 4 h after irradiation, the stripe of C3-positive cells anterior to the MF is expanded in the mir-11 mutant tissue. The position of the MF is marked with the arrowhead. (C) C3-positive cells in wild-type or mutant tissue were counted at the indicated times after irradiation. An average of a minimum of six eye discs was counted. Standard deviations are indicated. (D) Canton S or mir-11Δ1 third instar larvae were exposed to 40 Gy of irradiation and then allowed to develop. The number of adults to emerge was counted, and the percentage of survival was calculated. The average and standard deviation of three separate experiments are shown. (*) P = 0.032 in a paired t-test. (E) RNA was extracted from wild-type or mir11 mosaic mutant larval eye discs 1 h following irradiation (40 Gy), and rpr, hid, grim, and skl expression was measured by qRT–PCR. β-tubulin and RpP0 were used as controls in normalization. Expression of rpr and hid is elevated in mir-11 mutant mosaic eye discs before and after irradiation. (**) P < 0.001; (*) P < 0.05 in paired t-tests.
We used the FLP/FRT technique to generate clones of mir-11 mutant cells in the eye imaginal disc. Following irradiation, mir-11 mutant cells showed increased sensitivity to apoptosis, as C3 staining appeared more quickly in the mutant cells. As early as 1 h after irradiation, apoptosis was already detected in mir-11 mutant tissue, while little or no cell death was observed in neighboring wild-type tissue (Fig. 4B). Two hours after irradiation, abnormally high levels of cell death were readily observed in mutant tissue posterior to the MF with less apoptosis in the adjacent wild-type tissue (Fig. 4B). Four hours after exposure to irradiation, elevated levels of C3 staining persisted in the mir-11 mutant tissue. In particular, there was a broadening of the stripe of apoptotic cells anterior to the MF within clones of mir-11 mutant cells, a hallmark of elevated dE2F1-dependent apoptosis (Fig. 4B; Moon et al. 2008). Quantification confirms that the number of C3-positive cells is increased in mir-11 mutant tissue in comparison with the adjacent wild-type tissue upon irradiation (Fig. 4C). This is not a tissue-specific effect, as an increased sensitivity of mir-11 mutant cells to irradiation was observed in the wing imaginal disc (Supplemental Fig. S1). Importantly, mir-11 homozygous mutant animals were severely compromised in their ability to recover following irradiation. As shown in Figure 4D, the survival of irradiated mir-11 mutant larvae was significantly lower than that of wild-type animals. Therefore, the loss of mir-11 sensitizes cells to dE2F1-dependent, DNA damage-induced apoptosis in multiple tissues and compromises the survival of the animal.
Next, we examined the expression of four proapoptotic genes, rpr, hid, grim, and skl, in mir-11 mutants following irradiation. RNA was extracted from mosaic eye imaginal discs containing mir-11 mutant tissue, before and after irradiation, and the mRNA levels of these genes were measured using real-time qRT–PCR. Strikingly, even prior to irradiation, the mir-11 mutant mosaic eye discs had elevated levels of rpr, hid, and, to a lesser extent, grim in comparison with their levels in control, wild-type discs (2.3-fold, 2.0-fold, and 1.4-fold, respectively) (Fig. 4E). One hour after irradiation, rpr and hid were up-regulated in wild-type and in mosaic mir-11 mutant eye discs. However, the extent of induction was significantly higher in mir-11 mutants compared with wild-type unirradiated controls (2.5-fold vs. 1.9-fold for rpr, and 3.6-fold vs. 2.1-fold for hid) (Fig. 4E). In contrast, grim and skl were not induced in mir-11 mosaic mutant discs after irradiation. This is significant, since previous studies suggested that rpr and hid are important mediators of DNA damage-induced apoptosis, while the roles of grim and skl are less clear (Brodsky et al. 2000, 2004; Jassim et al. 2003; Moon et al. 2005, 2008). Consistently, overexpression of miR-11 by transient transfection in Drosophila S2 cells resulted in decreased levels of endogenous hid and rpr mRNAs (Supplemental Fig. S2). Thus, in agreement with the increased sensitivity of mir-11 mutants to DNA damage-induced apoptosis, the expression of rpr and hid was induced to a higher level in the mutant mosaic eye discs than in wild-type eye discs following irradiation.
rpr and hid are direct targets of dE2F1 and miR-11
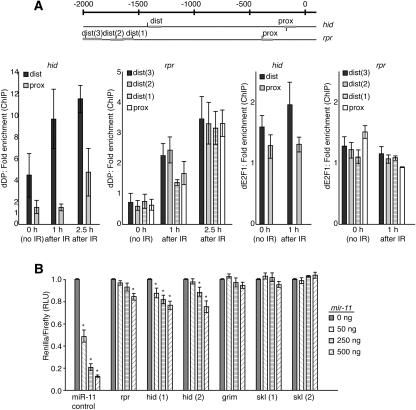
dE2F1 had been previously shown to activate the expression of hid and rpr and to bind to the hid promoter in asynchronously dividing S2 cells (Asano et al. 1996; Moon et al. 2008; Nicholson et al. 2009). To investigate the role of dE2F1 in the regulation of these genes during DNA damage-induced apoptosis, we examined the occupancy of dE2F1 on the rpr and hid promoters following irradiation. Genome-wide location analysis revealed that most mammalian E2F-binding sites occur close to the transcription start site (Xu et al. 2007), so we first identified putative binding sites between −2 kb and +100 base pairs (bp) relative to the transcription start site of these genes using Regulatory Sequence Analysis Tools (RSAT) (van Helden 2003) and the degenerate E2F-binding site (NWTSSCSS) (Fig. 5A). dE2F1 binds to DNA only as a heterodimer with dDP. It has been previously shown that dDP occupancy reflects that of dE2F1 (Dimova et al. 2003; Frolov et al. 2005). Therefore, we performed chromatin immunoprecipitation (ChIP) using a dDP antibody to determine the in vivo occupancy of dE2F1 at the rpr and hid promoters. Consistent with previous observations, in asynchronously dividing S2 cells under normal growth conditions, dE2F1/dDP occupied two putative binding sites in the hid promoter, with greater enrichment on the more distal binding site (Fig. 5A; Moon et al. 2005). Strikingly, following irradiation, we observed increased recruitment of dE2F1/dDP to the hid gene promoter, suggesting that dE2F1 directly activates the expression of hid in response to DNA damage (Fig. 5A). This conclusion was confirmed when dE2F1 antibodies were used in ChIP experiments. We note, however, that in our hands dE2F1 antibodies were far less efficient than dDP antibody in ChIP assays. There are four putative E2F-binding sites in the proximal promoter of the rpr gene, as shown by dDP ChIP. We did not observe significant enrichment for dE2F on the rpr promoter under normal growth conditions. However, as early as 1 h following irradiation, dE2F1/dDP was recruited to the rpr promoter, and even greater enrichment was observed 2 h following irradiation. These results suggested that dE2F1 is recruited to induce the expression of rpr specifically in response to irradiation.
Figure 5.
miR-11 and dE2F directly regulate the expression of hid and rpr. (A) Schematics of promoter regions (−2 kb to +100 bp relative to transcription start site) of hid and rpr. RSAT binding site analysis was performed using the NWTSSCSS consensus E2F-binding site (van Helden 2003). Predicted binding sites are shown as vertical lines above and below the promoter region, representing forward and reverse binding site orientation, respectively. Thick gray horizontal bars represent PCR products from primers tested in ChIP. S2R+ cells were exposed to 40 Gy of irradiation, and cells were fixed 1 or 2.5 h later. The 0-h sample was not exposed to irradiation. Sonicated chromatin was immunoprecipitated with an antibody that recognizes dDP, dE2F1, or an IgG control. The level of enrichment from the DP and dE2F1 antibodies was compared with that from the IgG control, and enrichment values were normalized to the RpP0 gene control. (B) 3′ UTR sensor assays were performed in HeLa cells using sequences predicted to be regulated by miR-11 (Leaman et al. 2005). Fifty nanograms of the indicated 3′ UTR sensor plasmid was transfected with the indicated amounts of pcDNA3/empty or pcDNA3/mir-11 plasmids. Cells were harvested 40–48 h post-transfection, and luciferase assay was measured. Error bars represent the standard deviation. (*) P < 0.05 in paired t-tests. A minimum of three independent transfections was performed for each sensor construct.
As described above, rpr, hid, and, to a lesser extent, grim were up-regulated in mir-11 mutants. In order to determine whether these effects were due to direct regulation by miR-11, we constructed Renilla luciferase sensors with putative seed sequence-targeted regions identified by Leaman et al. (2005) in the 3′ UTR. In each construct, Firefly luciferase was constitutively expressed from the same plasmid and served as an internal control for transfection efficiency. Since miR-11 and other miR-2 family members are endogenously expressed in Drosophila-derived cell lines such as S2, we used human HeLa cells, which do not express miR-11. Increasing amounts of pcDNA3/mir-11 were cotransfected with each of the luciferase sensors. Expression of miR-11 was confirmed by the TaqMan assay (data not shown). In a control experiment, miR-11 repressed, in a dose-dependent manner, a control sensor containing two copies of the exact miR-11 complement, confirming that miR-11 was active and could function to repress its targets in these cells (Fig. 5B). As shown in Figure 5B, miR-11 repressed the rpr and hid 3′ UTRs in a dose-dependent manner, but had no effect on the grim or skl 3′ UTRs (Fig. 5B). As miR-11 belongs to the miR-2 family, this result is consistent with previous findings that miR-2 family members regulated, to varying degrees, the rpr and hid 3′ UTRs (Brennecke et al. 2005; Leaman et al. 2005). Thus, we concluded that the 3′ UTRs of rpr and hid can be directly repressed by miR-11, which might explain why the loss of mir-11 resulted in their elevated expression. Since rpr and hid are also direct targets of dE2F1 (Moon et al. 2005; see above), this suggests an intriguing scenario in which miR-11 limits dE2F1-dependent DNA damage-induced apoptosis by directly regulating dE2F1 targets such as rpr and hid.
Cell death genes are overrepresented among predicted targets common to miR-11 and dE2F1
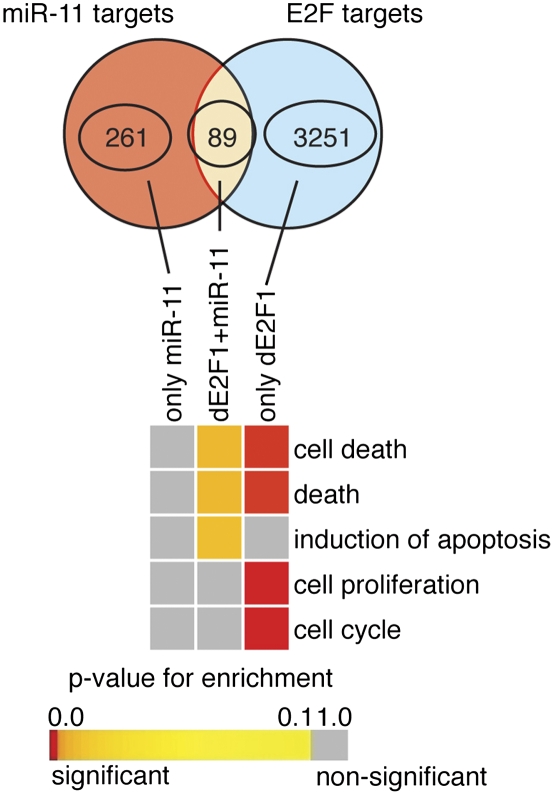
We considered the possibility that rpr and hid are not the only targets directly regulated by both miR-11 and dE2F1, and asked whether a functional relationship between miR-11 and dE2F1 extends beyond the regulation of these two genes and involves additional targets. To address this question, we used an orthology mapping approach and identified a set of 3340 Drosophila genes whose mammalian orthologs were directly bound by E2F transcription factors (Xu et al. 2007). Consistent with the known role of E2F, GOBP analysis identified E2F targets with functions in cell proliferation and cell death (Fig. 6). Next, we compared this set of putative 3340 Drosophila E2F target genes with 350 putative miR-11 targets identified by the TargetScan prediction algorithm (Ruby et al. 2007). We found that one-quarter of the predicted miR-11 targets were also predicted E2F targets (89 out of 350) (Fig. 6). GOBP analysis of these 89 genes revealed a statistically significant overrepresentation for genes having a role associated with cell death (Fig. 6; see Supplemental Tables S3 [for statistical details], S4 [for enriched gene list]). In agreement with our genetic and microarray analysis described above, there was no enrichment for genes associated with cell proliferation among targets common to E2F and miR-11. Thus, the bioinformatics analysis suggests that miR-11 and dE2F1 may regulate a set of putative common genes that function in apoptosis and cell death. We next asked which, if any, of these newly identified predicted common targets were directly regulated by dE2F1 and miR-11.
Figure 6.
miR-11 and E2F share common target genes with roles in cell death. Comparison of predicted miR-11 targets using TargetScan database targets (Ruby et al. 2007) with Drosophila homologs of mammalian E2F targets (Xu et al. 2007). Venn diagrams of predicted miR-11 targets and orthology mapping prediction of dE2F targets. Heat map of GOBP enrichment analysis for predicted miR-11 and dE2F targets (FDR ≤ 0.1).
Validation of putative common targets of miR-11 and dE2F
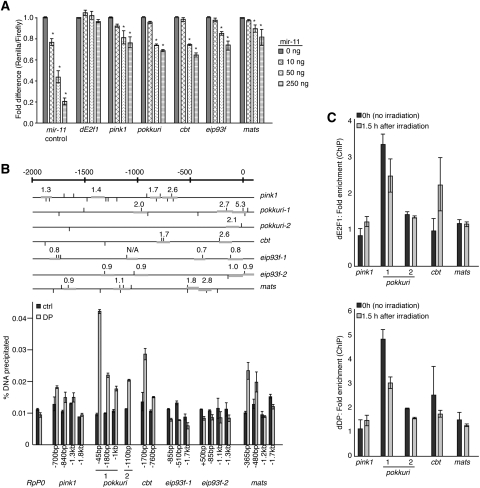
In order to determine whether predicted miR-11 targets with roles in modulating cell death were directly regulated by miR-11, we constructed Renilla luciferase sensors with the putative target sequence, flanked on each side by 25–30 bases (pink1, pokkuri, cbt, eip93f, mats, and scylla). As expected, a sensor containing the 3′ UTR of the dE2f1 gene, which is not a predicted miR-11 target, was not repressed by miR-11 (Fig. 7A). In contrast, we observed significant, dose-dependent repression of predicted miR-11 target sequences from the pink1, pokkuri, cbt, eip93f, and mats (Fig. 7A), but not those from scylla (data not shown). Consistently, pokkuri, pink1, mats, and cbt were elevated in mir-11 mutant larvae (Supplemental Fig. S3). Therefore, five out of six predicted targets were regulated by miR-11 in sensor assays, suggesting that miR-11 did indeed directly repress genes associated with cell death.
Figure 7.
miR-11 and dE2F1 directly regulate a common set of genes involved in the regulation of cell death. (A) 3′ UTR sensor assays were performed in HeLa cells using control or dE2F1/miR-11 shared targets identified by bioinformatic prediction algorithms. Fifty nanograms of the indicated 3′ UTR sensor plasmid was transfected with the indicated amounts of pcDNA3/empty or pcDNA3/mir-11 plasmids. Cells were harvested 40–48 h post-transfection, and luciferase assay was measured. Error bars represent the standard deviation. A minimum of three independent transfections was performed for each sensor construct. (*) P < 0.05 in a paired t-test compared with 0 ng pcDNA3/mir-11. (B) Schematics of promoter regions (−2 kb to +100 bp relative to transcription start site) of predicted targets of both miR-11 and dE2F1. RSAT binding site analysis was performed using the NWTSSCSS consensus E2F-binding site (van Helden 2003). Predicted binding sites are shown as vertical lines above and below promoter region, representing forward and reverse binding site orientation, respectively. Thick gray horizontal bars represent PCR products from primers tested in ChIP. Numbers above PCR products indicate fold enrichment from ChIP with DP antibody versus ChIP with nonspecific antibody. ChIP from S2R+ cells using a dDP or nonspecific control antibody (ctrl). Real-time qPCR analysis was done on the recovered chromatin samples, and the total amount of DNA precipitated with each antibody was quantified. RpP0 served as a negative control. Error bars represent the standard deviation from the mean. (C) S2R+ cells were exposed to 40 Gy of irradiation, and cells were fixed 1.5 h later. The 0-h sample was not exposed to irradiation. Sonicated chromatin was immunoprecipitated with an antibody that recognizes dDP, dE2F1, or an IgG control. The level of enrichment from the DP and dE2F1 antibodies was compared with that from the IgG control, and enrichment values were normalized to the RpP0 gene control.
Next we used ChIP assays to ask whether dE2F is present in vivo on the promoters of the miR-11 targets identified above. As shown in Figure 7B, dE2F/dDP was enriched on the pokkuri, pink1, cbt, and mats gene promoters. Importantly, enrichment was observed within particular regions of the promoter, not on all binding sites, indicating that the binding was likely to be specific. Furthermore, a nonspecific antibody did not immunoprecipitate significant amounts of target regions. We did not detect dE2F/dDP presence on the promoter of eip93f. It is possible that dE2F binds sites outside of the promoter region analyzed, or that dE2F does not occupy and regulate this gene in asynchronously dividing S2R cells, but we cannot exclude the possibility that dE2F does not directly regulate eip93f. Next, we asked whether binding of dE2F1/dDP to these promoters responds to irradiation similarly to what we observed for the hid and rpr promoters (Fig. 5). ChIP assays using dDP and dE2F1 antibodies showed a modest increase in the occupancy of dE2F1/dDP on the pink1 and cbt promoters upon irradiation (Fig. 7C). In contrast, binding of dE2F1/dDP to the mats and pokkuri promoters remained unaltered in irradiated cells. One possibility is that a different type of stress signal is needed to induce dE2F1 binding to these genes. In conclusion, combining 3′ UTR reporter assays (Fig. 7A) with ChIP experiments (Fig. 7B,C), we identified cell death genes that were previously unknown to be directly regulated by both miR-11 and dE2F1.
Discussion
Coupled expression of a microRNA and its host transcript creates a unique situation where the microRNA can modulate the function of its host (Barik 2008; Callis et al. 2009; Najafi-Shoushtari et al. 2010). Here, we found a novel relationship in which an embedded microRNA directly repressed targets that were directly regulated by the host gene transcription factor itself. We show that miR-11 specifically down-regulated the expression of rpr and hid, which were also directly targeted by dE2F1. However, dE2F1 also activates the expression of genes associated with cell proliferation, and this function was not modulated by miR-11. Therefore, what we identified is a selective, or partial, negative feed-forward loop in which one of the functions of dE2F1 is modulated by miR-11.
Target regulation by microRNAs has been shown to follow an incoherent feed-forward loop in which microRNAs buffer against the stochastic fluctuation of expression of their targets, and this facilitates the robustness of changes of expression in response to different cues (Stark et al. 2005; Cohen et al. 2006; Hornstein and Shomron 2006; Tsang et al. 2007; Li et al. 2009). For example, miR-7 modulates the expression of its targets in response to environmental fluctuation, while miR-9a modulates proneural signaling to ensure proper specification of sensory organ precursors (Li et al. 2006, 2009). However, miR-11 and dE2F1 are part of a somewhat unusual incoherent feed-forward loop in which the expression of both miR-11 and dE2F1 can be induced by the same signal, after which miR-11 negatively regulates only a subset of dE2F1 targets. In this sense, miR-11 imparts robustness to one of the functions of dE2F1, the regulation of expression of proapoptotic targets, while not influencing the function of dE2F1 in modulating the cell cycle. In doing so, miR-11 prevents an apoptotic response due to dE2F1 activity unless it is warranted, such as in the case of response to irradiation-induced DNA damage. Under these circumstances, dE2F1 would be recruited to proapoptotic gene promoters, and miR-11-mediated repression would be released, which could accelerate the apoptotic response. Therefore, our results are consistent with the view that microRNAs function in the canalization of development and response to environmental cues, and demonstrate that the proapoptotic function of the dE2F1 transcription factor is intrinsically modulated by the miR-11 gene, which is embedded in the dE2f1 locus.
The critical role of dE2F1 in irradiation-induced apoptosis is well established and is conserved between flies and mammals (Pediconi et al. 2003; Moon et al. 2008; Wichmann et al. 2010). Elimination of dE2F1 activity by a dDP mutation fully blocked DNA damage-induced cell death. Conversely, rbf mutant discs were sensitized to irradiation-induced apoptosis, and this sensitivity was due to elevated activity of dE2F1 (Moon et al. 2008). Consistent with the idea that miR-11 limits the proapoptotic function of dE2F1, the response of mir-11 mutant cells to irradiation closely parallels the response of rbf-deficient cells. Like rbf mutants (Moon et al. 2008), mir-11 mutant cells undergo apoptosis more quickly following irradiation than wild-type cells, and the stripe of cells with caspase activity anterior to the MF is expanded in both rbf and mir-11 mutants. Such a strikingly similar phenotype is likely due to abnormally high levels of two key mediators of DNA damage-induced apoptosis, rpr and hid, in both mutants. We note, however, that mir-11 and rbf mutations do not fully phenocopy each other. Unirradiated rbf mutant cells are prone to apoptosis within the MF during larval eye disc development, which was attributed to a requirement of EGFR signaling for survival of rbf mutant cells (Moon et al. 2006). In contrast, unirradiated mir-11 mutant cells did not undergo apoptosis in the MF, suggesting that miR-11 is not required to limit the proapoptotic function of dE2F1 in this developmental context.
It is well established that dE2F1 induces proliferation and apoptosis, and that the balance between these two opposing activities must be tightly regulated in a context-dependent manner. However, the precise mechanism that determines the net outcome has remained elusive. E2Fs cooperate with different transcription factors in the binding and regulation of different gene promoters, and this was suggested to impart specificity to E2F-mediated transcription (Schlisio et al. 2002; Giangrande et al. 2003; Hallstrom and Nevins 2003; Truscott et al. 2008). However, E2F has been shown to activate cell death genes in proliferating cells (Nahle et al. 2002); therefore, post-transcriptional regulation must be in place to limit the function of these cell death genes. Our data suggest that such regulation could be also mediated by microRNAs, and in Drosophila, mir-11 would play this role. Although there are other microRNAs that inhibit apoptosis (Brennecke et al. 2003; Xu et al. 2003; Leaman et al. 2005; Jaklevic et al. 2008), the coexpression of mir-11 with dE2f1 puts miR-11 in a special position, since it would be expressed precisely where and when dE2F1 is expressed. To our knowledge, this is the first example of an imbedded intronic microRNA regulating direct gene targets of its transcription factor host.
Intriguingly, the ability of miR-11 to regulate dE2F1-specific proapoptotic targets extends beyond rpr and hid. In this study, we identified a novel set of genes involved in apoptosis that could be directly regulated by both dE2F1 and miR-11. While the described functions of some of these genes are not linked to determining whether or not a cell should die (Tei et al. 1992; Lee et al. 2000; Gorski et al. 2003; ZC Lai et al. 2005; Park et al. 2006; Kim et al. 2010), their deregulation has been associated with cell death (Gene Ontology Consortium 2006). Therefore, fluctuations in the expression levels of genes in this set of common targets could stress the cells, and we propose that the presence of miR-11 buffers against such fluctuations, thereby contributing to the maintenance of the overall stability of the cell, while still permitting normal cellular activities.
Our data suggest that, in normal cells, while the cell death targets of dE2F1 may be transcribed and poised for translation, miR-11 would prevent the accumulation of an excess of such transcripts. The presence of such transcripts is not sufficient to trigger cell death, as we do not find an elevated level of apoptosis in mir-11 mutants during normal development. However, it appears that the loss of mir-11 sensitizes cells to apoptotic signals such as irradiation. Given that miR-11 does not seem to regulate cell death at the level of the core executioners, the caspases, and that at least one target shared by miR-11 and dE2F1, pink1, carries anti-apoptotic functions, cells that express miR-11 would not be unable to die. Indeed, overexpression of miR-11 was not as effective as p35 at blocking dE2F1-induced apoptosis, indicating that some level of apoptosis can be induced even in the presence of a high level of miR-11. One could extend this idea and speculate that, in the case of replicative stress resulting from deregulated dE2F1 activity, for example, miR-11 itself would be subject to negative regulation, thus permitting high expression of the proapoptotic dE2F1 targets, and if necessary, cell death would follow.
While there is no mammalian homolog of miR-11, E2F-induced cell death can be inhibited by microRNAs in mammalian cells. However, the precise mechanism appears to be different. In humans, E2F induces the expression of let-7a-d, let-7i, mir-15b-16-2, and mir-106b-25 at the G1-to-S-phase transition (Bueno et al. 2010). These microRNAs down-regulate critical cell cycle regulators, which are also targets of E2Fs and also directly down-regulate E2F1 and E2F2. In addition, E2Fs induce the expression of the mir-17∼92 cluster of microRNAs (O'Donnell et al. 2005; Woods et al. 2007). mir-17-5-p and mir-20a, in turn, target E2F factors, thereby limiting E2F activity at the level of E2F expression levels (Sylvestre et al. 2007; Pickering et al. 2009). By inducing these microRNAs, E2F initiates a negative feedback mechanism that limits the activation of E2F cell cycle gene targets directly and indirectly, thereby preventing replicative stress-induced cell death (Bueno et al. 2010). Therefore, excessive E2F activity could be limited by coexpressed microRNAs in flies and mammals.
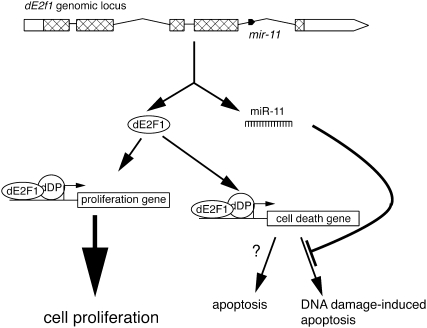
In summary, our data suggest a model whereby coexpression of miR-11 with dE2F1 from the same locus permits transcriptional activation of cell cycle genes by dE2F1 in the absence of apoptosis (Fig. 8). In this scenario, miR-11 initiates a partial negative feed-forward loop in which miR-11 specifically limits the dE2F1 proapoptotic transcriptional program. Intriguingly, although this regulation occurs in normal proliferating cells and leads to up-regulation of dE2F1 proapoptotic targets rpr and hid, the loss of mir-11 was not sufficient to trigger spontaneous apoptosis. Instead, mir-11 becomes important in specific settings, such as protecting cells from dE2F1-dependent DNA damage-induced apoptosis. Therefore, miR-11 buffers against apoptosis, in part by directly modulating the proapoptotic function of its host gene, dE2f1.
Figure 8.
miR-11 specifically represses a proapoptotic function of its host gene, dE2f1, upon DNA damage. Our results suggest that miR-11 is coexpressed with dE2F1 and specifically inhibits dE2F1-induced cell death targets upon DNA damage, thereby protecting cells from unwanted cellular consequences of dE2F1 activation in this context.
Materials and methods
Fly stocks
All fly crosses were done at 25°C. The following stocks were obtained from the Bloomington Drosophila Stock Center at Indiana University: GMR-Gal4, UAS-dE2F1, and UAS-dDP/TM6B. The following additional stocks were previously published: UAS-miR-11 (Brennecke et al. 2005); UAS-p35, GMR-rpr, and GMR-hid (generous gifts from Kristin White); Act88F-Gal4 and Act88F-Gal4,UAS-ced3 (generous gifts from Erick Morris and Teiichi Tanimura); and dE2f1rM729/TM6B (Duronio et al. 1995). dE2F1Δ1 is a deletion that lacks the genomic region between P-element insertion P[XP]E2fd01508 and piggyBac insertion PBac[RB]InRe01952 and was generated according to Parks et al. (2004). The following double-balanced recombinant stocks were created for these studies: UAS-miR-11; UAS-dE2F1, UAS-dDP/TM6B and UAS-p35; UAS-dE2F1, UAS-dDP/TM6B.
Construction of miR-11 mutant donor transgene
A donor construct was built in pBluescript and then subcloned into pTargetB (Radford et al. 2005). A 5.2-kb left arm fragment was generated by PCR with the primers ATCGGGTACCGCTTCTCTTTTGCGTTTGTTGC, and ATCGGGATCCCACGCGGGTCACAGAGAAAG, and was cloned into KpnI and BamHI of pBluescript. A 2.7-kb right arm fragment was generated with the primers ATCGGGATCCCTGAGTGCGAAATCCTCG and ATCGGCGGCCGCCTGCTTTTTGGGGTTTG, and was cloned into BamHI and NotI sites. Underlined sequences correspond to dE2f1 genomic sequence. miR-11 was replaced by a BamHI restriction site. The sequence removed was 17448225–17448260. To introduce the I-SceI site, annealed oligos carrying the I-SceI site were ligated into the unique XbaI site in dE2f1 intron 3. This cassette was then transferred to pTargetB using KpnI and NotI restriction sites. Sequencing confirmed that no mutations had been introduced. This construct was used to generate transgenic animals by P-element-mediated germline transformation (The Best Gene).
Generation of mir-11 mutant by homologous recombination
Ends-in homologous recombination was performed according to the procedure described by Rong et al. (2002). Briefly, a second-chromosome insertion of pTargetB(dE2f1 ΔmiR-11) was used as a donor. y1 w*; P(70FLP)11 P(70I-SceI)2B nocSco/CyO, S2 virgins were crossed to TargetB(dE2f1 ΔmiR-11) males, and their progeny were heat-shocked on the third day after egg laying for 1 h at 37°C. Potential targeting events were crossed with w1118; P(70FLP)10 to screen for the presence of a w+ element that does not mobilize in the presence of constitutive FLP expression, which indicates that homologous recombination occurred. PCR was used to identify and verify the targeted mutant using primers binding in mini-white (GGTGTTTGCTGCCTCCGCGA) and dE2f1, exon 4 (TCCGCCTTCACGTAAATCTCGC) (4.2-kb PCR product), and a second PCR reaction using primers specific for ΔmiR-11(ctgtgacccgcgtgGGATCCct) and FRT (TCTCTAGAAAGTATAGGAACTTCggatcca) (2.7-kb PCR product). I-CreI-mediated reduction to a single allele was carried out using the following crosses: targeted TargetB(dE2f1 ΔmiR-11) virgins were crossed to w1118; P(hs-I-CreI.R)1A Sb1/TM6 males, and TargetB(dE2f1 ΔmiR-11)/70I-CreI, Sb males were crossed to w; TM2/TM6B virgin females. After 3 d, the vials were heat-shocked for 1 h at 37°C. Ubx, non-Sb, or Hu, non-Sb progeny were analyzed by PCR for reduction events leaving either wild-type miR-11 or ΔmiR-11. The dE2f1 coding region and 5′ end of the adjacent gene InR were sequenced, and no mutations were detected. It was also confirmed that the targeted chromosome complemented the InR mutant and dE2f1 mutants.
3′ UTR sensor plasmid construction
Sequences were cloned downstream from the Renilla luciferase coding sequence in the psiCheck2 (Promega) plasmid using standard cloning techniques. Primer sequences will be provided on request.
Cell culture, transfection, and luciferase assay
Transfections were performed with the FuGene HD transfection reagent (Roche) according to manufacturers' protocol. Cells were harvested 24–48 h post-transfection. The psiCheck2 vector contains the Firefly luciferase gene under control of HSV-TK promoter, and the Renilla luciferase is under control of SV40 promoter. Neither of these promoters function efficiently in S2 cells, and as a result the basal activities of Firefly and Renilla luciferase were very low. Therefore, we used human HeLa cells in reporter assays. To overexpress miR-11, the region containing the miR-11 microRNA from the dE2f1 gene was cloned into pcDNA3 vector using the following primers: GTGGCTGACTGCGATGCCAAC and GGAGTTGAAGTGCCTATAATATCAC. Correct sequence was confirmed by sequencing. HeLa cells were cultured in DMEM + 10% FBS. Firefly and Renilla luciferase activity were measured using the Dual Luciferase Assay protocol (Promega). S2 cells were maintained in Schneider's medium + 10% FBS. pIE4 and pIEx-7/mir-11 were used in the transfection of S2 cells. pIEx-7 contains the same mir-11 sequence used in pcDNA3 (see above). Correct sequence was confirmed by sequencing.
Immunohistochemistry
Antibodies used were as follows: rabbit anti-C3 (Cleaved Caspase3), lot 26, 1:100 (Cell Signaling), mouse anti-BrdU 1:50 (Beckton Dickinson), and Cy3- and Cy5-conjugated anti-mouse and anti-rabbit secondary antibodies (Jackson Immunoresearch Laboratories). Larval tissues were fixed in 4% formaldehyde in phosphate-buffered saline (PBS) for 30 min on ice, permeabilized in 0.3% Triton X-100 in PBS twice for 10 min each, blocked in PBS with 0.1% Triton X-100 for 30 min at 4°C, and then incubated with antibodies overnight at 4°C in 10% normal goat serum and 0.3% Triton X-100 in PBS. After washing three times for 10 min each at room temperature in 0.1% Triton X-100 (in PBS), samples were incubated with appropriate conjugated secondary antibodies for 45 min at room temperature in 10% normal goat serum and 0.3% Triton X-100 (in PBS). After washing with 0.1% Triton X-100 (in PBS), tissues were stored in glycerol + anti-fade reagents and then mounted on glass slides.
To detect S phases, dissected larval eye discs were labeled with BrdU for 2 h at room temperature and then fixed overnight in 1.5% formaldehyde and 0.2% Tween 20 in PBS at 4°C. Samples were then digested with DNase (Promega) for 30 min at 37°C. Samples were then probed with primary and secondary antibodies as described above. All immunofluorescence was done on a Zeiss Confocal microscope, and images were prepared using Adobe Photoshop CS4. All images are confocal single-plane images unless otherwise stated as projection images. A minimum of 10 larvae was used for each analysis.
miR-11 target prediction
We used miR-2a-1/6/11/13/308 family target predictions from http://www.targetscan.org/fly (Ruby et al. 2007).
Microarray data analysis and enrichment analysis
Extracted RNA samples were subjected to Affymetrix GeneChip microarray analysis. After normalization by Bioconductor package Affy, differentially expressed genes (RankProd) were analyzed for GO enrichment using GiTools (http://www.gitools.org) (Perez-Llamas and Lopez-Bigas 2011). For details, see the Supplemental Material. The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (GEO) (Edgar et al. 2002) and are accessible through GEO Series accession number GSE25267 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE25267).
qRT–PCR
Total RNA was isolated from 10 adult heads, 10 larvae, or 30–50 eye discs with TRIzol (Invitrogen). Reverse transcription to measure standard mRNAs was performed using the iScript kit (Bio-Rad) according to the manufacturer's specifications. qPCR was performed with the iQ SYBR Green kit (Bio-Rad) on a MyiQ iCycler (Bio-Rad). miR-11 was measured using a TaqMan assay for dme-miR-11 (Applied Biosystems).
ChIP
S2R+ cells (2 × 107) were cross-linked for 10 min at room temperature in 1.8% formaldehyde. Cells were lysed (15 mM HEPES at pH 7.61, 40 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% sodium deoxycholate, 1% Triton-X-100, 0.5 mM DTT, 0.1% SDS, 0.5% lauroylsarcosine, with protease inhibitor cocktail [Complete, Roche]), and sheared using a Branson 450 digital sonifier. Chromatin was immunoprecipitated with rabbit anti-dDP, anti-dE2F1 (Seum et al. 1996; Dimova et al. 2003), anti-Myc (9E10), or anti-rabbit IgG as a control. Complexes were pulled down with Protein G Dynabeads (Invitrogen), washed with lysis buffer four times, washed twice with TE (pH 8), eluted, and decross-linked for 5 h at 65°C, and protein was digested with proteinase K for 2 h at 50°C. DNA was purified by phenol-chloroform extraction, followed by overnight ethanol precipitation. DNA was analyzed by real-time qPCR, and enrichment was calculated for each antibody relative to input DNA.
Acknowledgments
We are grateful to S.M. Cohen, E.J. Morris, T. Tanimura, K. White, and the Bloomington Stock Center for fly stocks. We are thankful to C. Reinke and R.W. Carthew for discussion, suggestions, and technical efforts. We thank N. Dyson, A. Katzen, G. Ramsay, B. Nicolay, A. Ambrus, and B. Bayarmagnai for helpful discussions, and S.M. Cohen for sharing unpublished data and exchanging mir-11 mutant lines. We are grateful to M.G. Frost, V.I. Rasheva, and R. Suckling for technical assistance. This research was supported by the National Institutes of Health (M.V.F.), by a Scholar Award from the Leukemia and Lymphoma Society (to M.V.F.), and by a Post-doctoral Fellowship (to M.T.) from the Fonds de Recherches en Santé Québec. N.L.B. acknowledges funding from the Spanish Ministerio de Educación y Ciencia grant number SAF2009-06954. A.I. is supported by a fellowship from AGAUR of the Catalonian Government.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.16947411.
References
- Asano M, Nevins JR, Wharton RP 1996. Ectopic E2F expression induces S phase and apoptosis in Drosophila imaginal discs. Genes Dev 10: 1422–1432 [DOI] [PubMed] [Google Scholar]
- Barik S 2008. An intronic microRNA silences genes that are functionally antagonistic to its host gene. Nucleic Acids Res 36: 5232–5241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskerville S, Bartel DP 2005. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 11: 241–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM 2003. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113: 25–36 [DOI] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM 2005. Principles of microRNA-target recognition. PLoS Biol 3: e85 doi: 10.1371/journal.pbio.0030085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky MH, Nordstrom W, Tsang G, Kwan E, Rubin GM, Abrams JM 2000. Drosophila p53 binds a damage response element at the reaper locus. Cell 101: 103–113 [DOI] [PubMed] [Google Scholar]
- Brodsky MH, Weinert BT, Tsang G, Rong YS, McGinnis NM, Golic KG, Rio DC, Rubin GM 2004. Drosophila melanogaster MNK/Chk2 and p53 regulate multiple DNA repair and apoptotic pathways following DNA damage. Mol Cell Biol 24: 1219–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook A, Xie JE, Du W, Dyson N 1996. Requirements for dE2F function in proliferating cells and in post-mitotic differentiating cells. EMBO J 15: 3676–3683 [PMC free article] [PubMed] [Google Scholar]
- Bueno MJ, Gomez de Cedron M, Laresgoiti U, Fernandez-Piqueras J, Zubiaga AM, Malumbres M 2010. Multiple E2F-induced microRNAs prevent replicative stress in response to mitogenic signaling. Mol Cell Biol 30: 2983–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callis TE, Pandya K, Seok HY, Tang RH, Tatsuguchi M, Huang ZP, Chen JF, Deng Z, Gunn B, Shumate J, et al. 2009. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest 119: 2772–2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ 2009. Origins and mechanisms of miRNAs and siRNAs. Cell 136: 642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SM, Brennecke J, Stark A 2006. Denoising feedback loops by thresholding–a new role for microRNAs. Genes Dev 20: 2769–2772 [DOI] [PubMed] [Google Scholar]
- Dimova DK, Stevaux O, Frolov MV, Dyson NJ 2003. Cell cycle-dependent and cell cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes Dev 17: 2308–2320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W, Dyson N 1999. The role of RBF in the introduction of G1 regulation during Drosophila embryogenesis. EMBO J 18: 916–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W, Xie JE, Dyson N 1996. Ectopic expression of dE2F and dDP induces cell proliferation and death in the Drosophila eye. EMBO J 15: 3684–3692 [PMC free article] [PubMed] [Google Scholar]
- Duronio RJ, O'Farrell PH, Xie JE, Brook A, Dyson N 1995. The transcription factor E2F is required for S phase during Drosophila embryogenesis. Genes Dev 9: 1445–1455 [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov MV, Moon NS, Dyson NJ 2005. dDP is needed for normal cell proliferation. Mol Cell Biol 25: 3027–3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology Consortium 2006. The Gene Ontology (GO) project in 2006. Nucleic Acids Res 34: D322–D326 doi: 10.1093/nar/gkj021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giangrande PH, Hallstrom TC, Tunyaplin C, Calame K, Nevins JR 2003. Identification of E-box factor TFE3 as a functional partner for the E2F3 transcription factor. Mol Cell Biol 23: 3707–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski SM, Chittaranjan S, Pleasance ED, Freeman JD, Anderson CL, Varhol RJ, Coughlin SM, Zuyderduyn SD, Jones SJ, Marra MA 2003. A SAGE approach to discovery of genes involved in autophagic cell death. Curr Biol 13: 358–363 [DOI] [PubMed] [Google Scholar]
- Goulev Y, Fauny JD, Gonzalez-Marti B, Flagiello D, Silber J, Zider A 2008. SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Curr Biol 18: 435–441 [DOI] [PubMed] [Google Scholar]
- Hallstrom TC, Nevins JR 2003. Specificity in the activation and control of transcription factor E2F-dependent apoptosis. Proc Natl Acad Sci 100: 10848–10853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornstein E, Shomron N 2006. Canalization of development by microRNAs. Nat Genet 38: S20–S24 doi: 10.1038/ng1803 [DOI] [PubMed] [Google Scholar]
- Iaquinta PJ, Lees JA 2007. Life and death decisions by the E2F transcription factors. Curr Opin Cell Biol 19: 649–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaklevic B, Uyetake L, Wichmann A, Bilak A, English CN, Su TT 2008. Modulation of ionizing radiation-induced apoptosis by bantam microRNA in Drosophila. Dev Biol 320: 122–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jassim OW, Fink JL, Cagan RL 2003. Dmp53 protects the Drosophila retina during a developmentally regulated DNA damage response. EMBO J 22: 5622–5632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DG, Schwarz JK, Cress WD, Nevins JR 1993. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature 365: 349–352 [DOI] [PubMed] [Google Scholar]
- Kim YK, Kim VN 2007. Processing of intronic microRNAs. EMBO J 26: 775–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YI, Ryu T, Lee J, Heo YS, Ahnn J, Lee SJ, Yoo O 2010. A genetic screen for modifiers of Drosophila caspase Dcp-1 reveals caspase involvement in autophagy and novel caspase-related genes. BMC Cell Biol 11: 9 doi: 10.1186/1471-2121-11-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalik TF, DeGregori J, Schwarz JK, Nevins JR 1995. E2F1 overexpression in quiescent fibroblasts leads to induction of cellular DNA synthesis and apoptosis. J Virol 69: 2491–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T 2001. Identification of novel genes coding for small expressed RNAs. Science 294: 853–858 [DOI] [PubMed] [Google Scholar]
- Lai EC, Tam B, Rubin GM 2005. Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes Dev 19: 1067–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai ZC, Wei X, Shimizu T, Ramos E, Rohrbaugh M, Nikolaidis N, Ho LL, Li Y 2005. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell 120: 675–685 [DOI] [PubMed] [Google Scholar]
- Leaman D, Chen PY, Fak J, Yalcin A, Pearce M, Unnerstall U, Marks DS, Sander C, Tuschl T, Gaul U 2005. Antisense-mediated depletion reveals essential and specific functions of microRNAs in Drosophila development. Cell 121: 1097–1108 [DOI] [PubMed] [Google Scholar]
- Lee CY, Wendel DP, Reid P, Lam G, Thummel CS, Baehrecke EH 2000. E93 directs steroid-triggered programmed cell death in Drosophila. Mol Cell 6: 433–443 [DOI] [PubMed] [Google Scholar]
- Li Y, Wang F, Lee JA, Gao FB 2006. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev 20: 2793–2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW 2009. A microRNA imparts robustness against environmental fluctuation during development. Cell 137: 273–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlodzik M, Hiromi Y 1992. Enhancer trap method in Drosophila: its application to neurobiology. Methods Neurosci 9: 397–414 [Google Scholar]
- Moon NS, Frolov MV, Kwon EJ, Di Stefano L, Dimova DK, Morris EJ, Taylor-Harding B, White K, Dyson NJ 2005. Drosophila E2F1 has context-specific pro- and antiapoptotic properties during development. Dev Cell 9: 463–475 [DOI] [PubMed] [Google Scholar]
- Moon NS, Di Stefano L, Dyson N 2006. A gradient of epidermal growth factor receptor signaling determines the sensitivity of rbf1 mutant cells to E2F-dependent apoptosis. Mol Cell Biol 26: 7601–7615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon NS, Di Stefano L, Morris EJ, Patel R, White K, Dyson NJ 2008. E2F and p53 induce apoptosis independently during Drosophila development but intersect in the context of DNA damage. PLoS Genet 4: e1000153 doi: 10.1371/journal.pgen.1000153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris EJ, Michaud WA, Ji JY, Moon NS, Rocco JW, Dyson NJ 2006. Functional identification of Api5 as a suppressor of E2F-dependent apoptosis in vivo. PLoS Genet 2: e196 doi: 10.1371/journal.pgen.0020196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H, Bracken AP, Vernell R, Moroni MC, Christians F, Grassilli E, Prosperini E, Vigo E, Oliner JD, Helin K 2001. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev 15: 267–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahle Z, Polakoff J, Davuluri RV, McCurrach ME, Jacobson MD, Narita M, Zhang MQ, Lazebnik Y, Bar-Sagi D, Lowe SW 2002. Direct coupling of the cell cycle and cell death machinery by E2F. Nat Cell Biol 4: 859–864 [DOI] [PubMed] [Google Scholar]
- Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Naar AM 2010. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science 328: 1566–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson SC, Gilbert MM, Nicolay BN, Frolov MV, Moberg KH 2009. The archipelago tumor suppressor gene limits rb/e2f-regulated apoptosis in developing Drosophila tissues. Curr Biol 19: 1503–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolay BN, Frolov MV 2008. Context-dependent requirement for dE2F during oncogenic proliferation. PLoS Genet 4: e1000205 doi: 10.1371/journal.pgen.1000205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT 2005. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435: 839–843 [DOI] [PubMed] [Google Scholar]
- Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, Bae E, Kim J, Shong M, Kim JM, et al. 2006. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature 441: 1157–1161 [DOI] [PubMed] [Google Scholar]
- Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, et al. 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet 36: 288–292 [DOI] [PubMed] [Google Scholar]
- Pediconi N, Ianari A, Costanzo A, Belloni L, Gallo R, Cimino L, Porcellini A, Screpanti I, Balsano C, Alesse E, et al. 2003. Differential regulation of E2F1 apoptotic target genes in response to DNA damage. Nat Cell Biol 5: 552–558 [DOI] [PubMed] [Google Scholar]
- Perez-Llamas C, Lopez-Bigas N 2011. Gitools: analysis and visualisation of genomic data using interactive heat-maps. PLoS ONE 6: e19541 doi: 10.1371/journal.pone.0019541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering MT, Stadler BM, Kowalik TF 2009. miR-17 and miR-20a temper an E2F1-induced G1 checkpoint to regulate cell cycle progression. Oncogene 28: 140–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford SJ, Goley E, Baxter K, McMahan S, Sekelsky J 2005. Drosophila ERCC1 is required for a subset of MEI-9-dependent meiotic crossovers. Genetics 170: 1737–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A 2004. Identification of mammalian microRNA host genes and transcription units. Genome Res 14: 1902–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong YS, Golic KG 2000. Gene targeting by homologous recombination in Drosophila. Science 288: 2013–2018 [DOI] [PubMed] [Google Scholar]
- Rong YS, Titen SW, Xie HB, Golic MM, Bastiani M, Bandyopadhyay P, Olivera BM, Brodsky M, Rubin GM, Golic KG 2002. Targeted mutagenesis by homologous recombination in D. melanogaster. Genes Dev 16: 1568–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royzman I, Hayashi-Hagihara A, Dej KJ, Bosco G, Lee JY, Orr-Weaver TL 2002. The E2F cell cycle regulator is required for Drosophila nurse cell DNA replication and apoptosis. Mech Dev 119: 225–237 [DOI] [PubMed] [Google Scholar]
- Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC 2007. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res 17: 1850–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlisio S, Halperin T, Vidal M, Nevins JR 2002. Interaction of YY1 with E2Fs, mediated by RYBP, provides a mechanism for specificity of E2F function. EMBO J 21: 5775–5786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempere LF, Sokol NS, Dubrovsky EB, Berger EM, Ambros V 2003. Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and broad-Complex gene activity. Dev Biol 259: 9–18 [DOI] [PubMed] [Google Scholar]
- Seum C, Spierer A, Pauli D, Szidonya J, Reuter G, Spierer P 1996. Position-effect variegation in Drosophila depends on dose of the gene encoding the E2F transcriptional activator and cell cycle regulator. Development 122: 1949–1956 [DOI] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM 2005. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell 123: 1133–1146 [DOI] [PubMed] [Google Scholar]
- Sylvestre Y, De Guire V, Querido E, Mukhopadhyay UK, Bourdeau V, Major F, Ferbeyre G, Chartrand P 2007. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem 282: 2135–2143 [DOI] [PubMed] [Google Scholar]
- Tei H, Nihonmatsu I, Yokokura T, Ueda R, Sano Y, Okuda T, Sato K, Hirata K, Fujita SC, Yamamoto D 1992. pokkuri, a Drosophila gene encoding an E-26-specific (Ets) domain protein, prevents overproduction of the R7 photoreceptor. Proc Natl Acad Sci 89: 6856–6860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truscott M, Harada R, Vadnais C, Robert F, Nepveu A 2008. p110 CUX1 cooperates with E2F transcription factors in the transcriptional activation of cell cycle-regulated genes. Mol Cell Biol 28: 3127–3138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang J, Zhu J, van Oudenaarden A 2007. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell 26: 753–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel S, Dyson NJ 2008. Conserved functions of the pRB and E2F families. Nat Rev Mol Cell Biol 9: 713–724 [DOI] [PubMed] [Google Scholar]
- van Helden J 2003. Regulatory sequence analysis tools. Nucleic Acids Res 31: 3593–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichmann A, Uyetake L, Su TT 2010. E2F1 and E2F2 have opposite effects on radiation-induced p53-independent apoptosis in Drosophila. Dev Biol 346: 80–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods K, Thomson JM, Hammond SM 2007. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem 282: 2130–2134 [DOI] [PubMed] [Google Scholar]
- Xu P, Vernooy SY, Guo M, Hay BA 2003. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol 13: 790–795 [DOI] [PubMed] [Google Scholar]
- Xu X, Bieda M, Jin VX, Rabinovich A, Oberley MJ, Green R, Farnham PJ 2007. A comprehensive ChIP-chip analysis of E2F1, E2F4, and E2F6 in normal and tumor cells reveals interchangeable roles of E2F family members. Genome Res 17: 1550–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Steller H 2003. Distinct pathways mediate UV-induced apoptosis in Drosophila embryos. Dev Cell 4: 599–605 [DOI] [PubMed] [Google Scholar]