Protein deubiquitination is a critical regulatory mechanism employed in the multistep response to DNA damage. The deubiquitinating complex USP1/UAF1 coordinately regulates the homologous recombination pathway (via FANCD2/FANC1) and the translesion synthesis pathway (via hELG1 and PCNA). USP1/UAF1 is targeted to its substrates by a SUMO-like domain on UAF1, which binds to SIM motifs present on both FANC1 and the PCNA-binding protein hELG1.
Keywords: Fanconi anemia, deubiquitinating enzymes, PCNA, SUMO-like domains
Abstract
The USP1/UAF1 complex deubiquitinates the Fanconi anemia protein FANCD2, thereby promoting homologous recombination and DNA cross-link repair. How USP1/UAF1 is targeted to the FANCD2/FANCI heterodimer has remained unknown. Here we show that UAF1 contains a tandem repeat of SUMO-like domains in its C terminus (SLD1 and SLD2). SLD2 binds directly to a SUMO-like domain-interacting motif (SIM) on FANCI. Deletion of the SLD2 sequence of UAF1 or mutation of the SIM on FANCI disrupts UAF1/FANCI binding and inhibits FANCD2 deubiquitination and DNA repair. The USP1/UAF1 complex also deubiquitinates PCNA-Ub, and deubiquitination requires the PCNA-binding protein hELG1. The SLD2 sequence of UAF1 binds to a SIM on hELG1, thus targeting the USP1/UAF1 complex to its PCNA-Ub substrate. We propose that the regulated targeting of USP1/UAF1 to its DNA repair substrates, FANCD2-Ub and PCNA-Ub, by SLD–SIM interactions coordinates homologous recombination and translesion DNA synthesis.
Protein ubiquitination plays a key role in the regulation of a variety of DNA repair mechanisms (Huang and D'Andrea 2006; Bergink and Jentsch 2009). Protein ubiquitination is controlled by the opposing activity of ubiquitin ligases and deubiquitinating enzymes (DUBs) (Nijman et al. 2005b; Komander et al. 2009; Reyes-Turcu et al. 2009). Failure to regulate the proper ubiquitination state of a DNA repair protein can result in a DNA repair defect and genomic instability, the hallmark of many human cancers.
The Fanconi anemia (FA) pathway and the translesion DNA synthesis (TLS) pathway are two DNA repair pathways controlled by protein ubiquitination. FA is a recessive genetic disease characterized by developmental abnormalities, cancer susceptibility, and cellular hypersensitivity to DNA interstrand cross-linking (ICL) agents. There are 15 known FA proteins, and the monoubiquitination of one of these proteins, FANCD2, by the FA core complex is a critical step in the pathway (Al-Hakim et al. 2010; Kee and D'Andrea 2010). Monoubiquitination of FANCD2 is required for the recruitment of additional downstream proteins, such as the FAN1 nuclease and SLX4, to the site of ICL repair (Cybulski and Howlett 2011; Yamamoto et al. 2011). In contrast, a critical step in the translesion synthesis pathway is the monoubiquitination of the processivity factor PCNA by the ubiquitin E3 ligase RAD18. Monoubiquitination of PCNA is required for the recruitment of a translesion polymerase to the site of DNA repair (Bienko et al. 2010).
The DUB USP1 regulates DNA repair through its association with the binding partner UAF1 (Cohn et al. 2007) and through its deubiquitination of two critical DNA repair substrates: FANCD2-Ub (Nijman et al. 2005a) and PCNA-Ub (Huang et al. 2006). The cellular level of FANCD2-Ub controls homologous recombination (HR) repair (Nakanishi et al. 2005), and the level of PCNA-Ub controls TLS repair (Kannouche et al. 2004; Bienko et al. 2010). Knockdown of USP1 or UAF1 results in elevated levels of FANCD2-Ub and PCNA-Ub (Cohn et al. 2007; Oestergaard et al. 2007), leading to defective HR and TLS (Kim et al. 2009). FANCD2-Ub and PCNA-Ub are coordinately deubiquitinated following DNA damage and during the normal cell cycle (Huang et al. 2006). How the USP1/UAF1 complex recruits these two specific substrates and coordinates their deubiquitination under specific cellular conditions is unknown.
UAF1 has an N-terminal WD40 domain, with eight WD40 propeller sequences and a C-terminal coiled-coil domain. The N-terminal WD40 region of UAF1 binds and stimulates the ubiquitin protease activity of USP1 (Cohn et al. 2007). This region of UAF1 also binds and stimulates at least two other DUB enzymes, USP12 and USP46 (Cohn et al. 2009; Sowa et al. 2009), although the substrates of the USP12/UAF1 and USP46/UAF1 complexes remain unknown. The function of the C-terminal region of UAF1 is also previously unknown.
In the current study, we determined that the C-terminal region of UAF1 is required for targeting the USP1/UAF1 complex to its substrates. Specifically, we showed that this region of UAF1 contains a tandem repeat of SUMO-like domains (SLD1 and SLD2). The SLD2 domain binds to a SIM (SUMO-like domain-interacting motif) on protein complexes containing FANCD2-Ub and/or PCNA-Ub and thereby regulates the deubiquitination of these substrates. Disruption of these SLD–SIM interactions blocks the deubiquitination of FANCD2-Ub and/or PCNA-Ub, rendering cells DNA repair-deficient. SLD–SIM interactions are therefore critical for the timed delivery of the USP1/UAF1 deubiquitinating complex to its substrates and for proper coordination of DNA ICL repair.
Results
The C-terminal region of UAF1 is required for HR repair
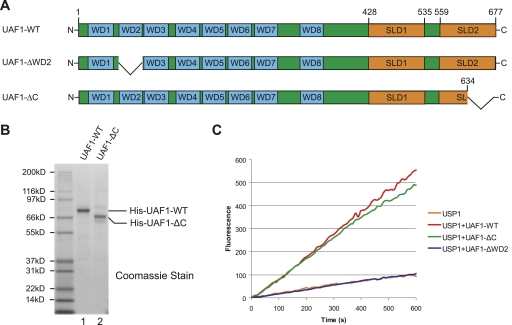
We initially tested whether the C terminus of UAF1 is required for stimulating USP1 activity in vitro (Fig. 1). We purified wild-type UAF1 or a C-terminal-truncated UAF1 protein (UAF1-ΔC) (Fig. 1A,B) from insect cells, mixed the proteins with purified USP1 in vitro, and measured deubiquitinating activity (Fig. 1C). The UAF1-ΔC protein was able to bind and stimulate USP1, with activity comparable with full-length UAF1 protein (Supplemental Fig. S1). Another UAF1 mutant protein, UAF1-ΔWD2, with an internal deletion of the WD40 domain, failed to stimulate USP1, consistent with our previous report (Cohn et al. 2009).
Figure 1.
The C-terminal region of UAF1 is dispensable for the activation of USP1 activity in vitro. (A) Schematic representation of the domain structure of UAF1-WT and two deletion mutants: ΔWD2 and ΔC. (B) Coomassie stain of purified wild-type and C-terminal truncated human UAF1 proteins from Sf9 insect cells. (C) The indicated wild-type or mutant human UAF1 proteins were mixed with purified USP1, and deubiquitination enzyme activity was monitored using Ub-AMC as the substrate.
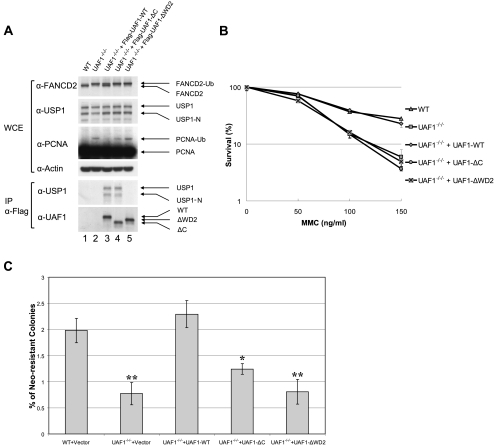
Previous studies have demonstrated that the USP1/UAF1 complex deubiquitinates FANCD2-Ub, FANCI-Ub, and PCNA-Ub in mammalian cells (Cohn et al. 2007). Accordingly, siRNA knockdown of USP1 or UAF1 in HeLa cells results in elevated FANCD2-Ub and FANCI-Ub levels. In order to test the cellular function of the C terminus of UAF1, we next generated null mutants of UAF1 and USP1 in the chicken lymphoblast line DT40 (Supplemental Fig. S2A; Oestergaard et al. 2007; Murai et al. 2011). As predicted, the UAF1-deficient DT40 cells (UAF1−/−/− cells) had elevated FANCD2-Ub and PCNA-Ub levels (Fig. 2A, lane 2), consistent with the known cellular role of USP1/UAF1 in deubiquitinating these substrates (Cohn et al. 2007), and the cells were hypersensitive to MMC (Fig. 2B). Thus, the phenotype of UAF1-deficient DT40 cells is similar to that of USP1-deficient DT40 cells previously described (Oestergaard et al. 2007). Transfection of the UAF1-deficient cells with the cDNA encoding wild-type human UAF1 reduced the level of FANCD2-Ub and PCNA-Ub (Fig. 2A, lane 3; Supplemental S2B, lane 3) and complemented their MMC hypersensitivity (Fig. 2B). Interestingly, the mutant form of UAF1 protein, lacking the C terminus, failed to restore deubiquitination of these substrates and failed to complement MMC hypersensitivity, although it still bound to intracellular USP1 (Fig. 2A, lane 4) and stimulated its activity (data not shown).
Figure 2.
The C-terminal region of UAF1 is required for DNA repair in vivo. (A) UAF1-ΔC failed to correct the increased FancD2-Ub and PCNA-Ub levels in UAF1−/−/− chicken DT40 cells. Plasmids expressing human UAF1-WT, UAF1-ΔC, or UAF1-ΔWD2 were stably transfected into UAF1−/−/− DT40 cells. Whole-cell lysates were prepared and immunoprecipitated using anti-Flag agarose beads. The protein level of UAF1, USP1, FANCD2, PCNA, and Actin were measured by Western blot. (B) UAF1-ΔC failed to correct the MMC sensitivity of UAF1−/−/− chicken DT40 cells. MMC clonogenic assays were performed on the stable correction clones from A. (C) To investigate the involvement of UAF1 in HR-mediated repair, we integrated the SCneo substrate into the Ovalbumin locus of the UAF1-deficient cells and measured the efficiency of I-SceI-induced gene conversion. Wild-type human UAF1 rescued the HR defect in the transfected cells, but UAF1-ΔC and UAF1-ΔWD2 mutants failed to rescue. The difference between the single-asterisk (*) cells and the wild-type DT40 cells (WT + Vector) was statistically significant (P < 0.01). The difference between the double-asterisk (**) cells and the wild-type DT40 cells (WT + Vector) was also statistically significant (P < 0.001).
The UAF1-deficient DT40 cells also exhibited a defect in HR repair (Fig. 2C). To investigate the involvement of UAF1 in HR-mediated repair directly, we measured gene conversion induced by the rare-cutting endonuclease I-SceI, using the SCneo substrate (Hochegger et al. 2006). Specifically, we integrated the SCneo substrate into the Ovalbumin locus of the DT40 cells (Fukushima et al. 2001) and measured the efficiency of I-SceI-induced gene conversion. While 1.98% of the wild-type DT40 cells successfully underwent gene conversion and reconstituted neomycin resistance, the same reaction occurred in only 1.24% of the UAF1−/−/− cells expressing the human UAF1-ΔC protein. UAF1−/−/− cells expressing human wild-type UAF1 protein were fully complemented (2.29% of the cell population). Taken together, these results suggested that the C terminus of UAF1 is not required for USP1 stimulation but is required for targeting the USP1/UAF1 complex to its substrates and for HR repair.
The USP1/UAF1 complex binds to FANCI via a SLD–SIM interaction
We next analyzed the C terminus of UAF1 using various bioinformatic tools. Interestingly, we found that the C terminus of human UAF1 (amino acids 425–677) contains a tandem repeat of two SLDs (shown schematically in Fig. 1A; Supplemental Fig. S3A). Each SUMO-like sequence, referred to as SLD1 and SLD2, is ∼100 amino acids in length and has homology with SUMO2 and SUMO3 (see alignment in Supplemental Fig. S3B). Specifically, the SLD1 sequence has 59% similarity to SUMO2, and the SLD2 sequence has 55% similarity to SUMO2. SLD1 and SLD2 each have a conserved C-terminal diglycine of SUMO2, although there is no evidence that UAF1 undergoes endoproteolytic cleavage after these GG sequences. Previous studies have identified other proteins with tandem SLDs (Novatchkova et al. 2005; Prudden et al. 2009), although for these proteins the SLDs are more similar to SUMO1 than to SUMO2 or SUMO3.
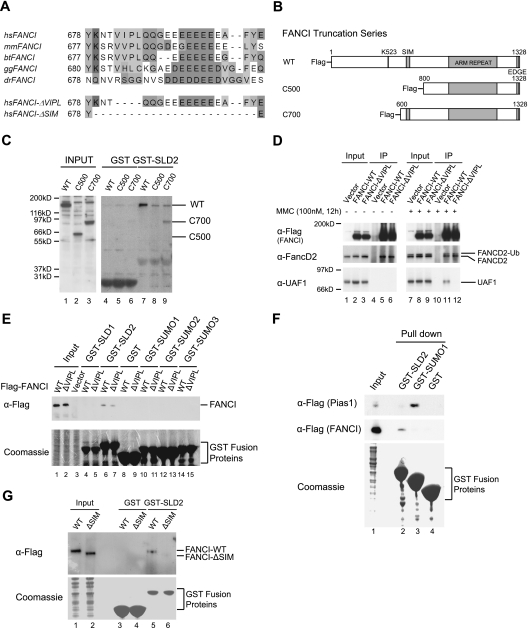
Recent studies indicate that the SUMO moiety of sumoylated proteins can mediate enzyme/substrate interactions (Prudden et al. 2007; Meulmeester et al. 2008; Denuc et al. 2009). Accordingly, we reasoned that the SLD1 and SLD2 of UAF1 might target the USP1/UAF1 complex to its substrates, FANCD2-Ub and PCNA-Ub, either directly or through a binding partner (Fig. 3). SUMO binds to a peptide sequence on target proteins, referred to as a SIM. This sequence conforms to the consensus VLXXEEEEE (Song et al. 2004; Hecker et al. 2006). Recent studies suggest that other Ubiquitin-like and SUMO-like sequences may bind to a similar consensus sequence (Sanchez-Pulido et al. 2008; Noda et al. 2010). Sequence analysis of FANCD2 and FANCI revealed a highly conserved candidate SIM sequence on FANCI (amino acids 681–697) (Fig. 3A). To test whether this sequence was required for binding to UAF1, we generated GST fusion proteins containing either the SLD1 or SLD2 domain of UAF1. Interestingly, GST-SLD2 specifically pulled down FANCI proteins (WT [wild type] and C700) containing the putative SIM sequence (Fig. 3B,C, lanes 7,9), but did not pull down a FANCI mutant protein in which the SIM sequence was deleted (C500) (Fig. 3C, lane 8).
Figure 3.
The C-terminal SLD2 of UAF1 binds to a conserved SIM sequence on FANCI. (A) Identification of a conserved SIM in the primary amino acid sequence of human FANCI, aligned with the homologous regions of FANCI from other species: Homo sapiens (hs), Mus musculus (mm), Bos taurus (bt), Gallus gallus (gg), and Danio rerio (dr). (B) Schematic representation of FANCI-WT and two deletion mutants. FANCI-WT and C700 both contain the SIM region, while the SIM was absent in the C500 mutant. The N-terminal Flag epitope was indicated. (C) GST-SLD2 pulled down FANCI-WT and C700, but not C500. Plasmids expressing the indicated deletion mutants of human FANCI were transiently transfected into HEK293T cells. Cell lysates were used for the pull-down experiment with the indicated GST or GST-SLD2 fusion protein. Western blot was then performed using anti-Flag antibody. (D) FANCI interacted with full-length UAF1 in vivo. Flag-tagged wild-type FANCI and a mutant form of full-length FANCI (ΔVIPL) were transfected into HEK293T cells and immunoprecipitated using anti-Flag agarose beads. Wild-type FANCI specifically interacted with UAF1 after MMC treatment, and this interaction was impaired in the SIM mutant (ΔVIPL). (E) GST fusion proteins containing the indicated SUMO or SLD protein sequences were prepared and used to pull down wild-type and the ΔVIPL mutant form of full-length FANCI overexpressed in HEK293T cells. Only SLD2, but not SLD1 or SUMO1/2/3, pulled down wild-type FANCI, and this interaction was dependent on the SIM. By densitometry, the relative band intensities of lanes 6 and 7 were 1.00 and 0.57, respectively. (F) GST fusion proteins containing either SLD2 or SUMO1 were prepared and used to pull down wild-type Flag-FANCI or wild-type Flag-Pias1 protein, which were overexpressed in HEK293T cells. GST-SLD2 specifically pulled down FANCI and GST-SUMO1 specifically pulled down Pias1. (G) GST-SLD2 pull-down of wild-type FANCI but not of FANCI-ΔSIM (another FANCI mutant with a deletion of the entire SIM sequence).
To further examine the role of the SIM sequence of FANCI in this interaction, we next generated Flag-tagged wild-type protein or mutant FANCI protein with a deletion of the SIM sequence (Fig. 3D) and expressed these proteins in HEK293T cells. Specifically, for this study, we used a FANCI mutant in which four residues (VIPL) had been removed from the SIM (so-called FANCI-ΔVIPL) (Figure 3A; Supplemental Fig. S5A). Flag-FANCI-WT but not Flag-FANCI-ΔVIPL coimmunoprecipitated with endogenous UAF1 (Fig. 3D, cf. lanes 11 and 12). Coimmunoprecipitation of FANCI-WT and endogenous UAF1 was enhanced by pretreatment of the cells with MMC (Fig. 3D, lane 11). DNA damage therefore appears to increase the SLD2–SIM interaction, perhaps by activating an ATM- or ATR-dependent phosphorylation of FANCI. Phosphorylation of residues adjacent to SIM sequences have previously been shown to enhance SUMO/SIM interactions. Consistent with these results, inhibition of ATM and/or ATR activity with caffeine appears to decrease the FANCI/UAF1 interaction (Supplemental Fig. S4).
We also determined whether GST fusion proteins containing SUMO1, SUMO2, or SUMO3 can pull down the wild-type FANCI protein versus a FANCI-ΔVIPL mutant (Fig. 3E). GST-SLD2 specifically bound to FANCI (wild-type) protein and had diminished binding to FANCI-ΔVIPL. Interestingly, the other GST fusion proteins, including GST-SUMO1, GST-SUMO2, GST-SUMO3, and GST-SLD1, did not pull down wild-type or mutant FANCI. Therefore, we propose using the term “SLIM” for SUMO-like domain-interacting motif for the binding site on the FANCI protein (see the Discussion section below).
To further evaluate the specificity of these binding interactions, we compared the ability of GST-SLD2 versus GST-SUMO1 to pull down full-length proteins containing either a SLIM or a SIM (Fig. 3F). Interestingly, GST-SUMO1 precipitated the Pias1 protein (Hecker et al. 2006), but GST-SLD2 did not interact with Pias1. Also, GST-SLD2 pulled down the FANCI protein containing the SLIM motif, but did not precipitate Pias1. Taken together, these results demonstrate that the SLD2 sequence has a specific binding interaction with the SLIM sequence of FANCI, and that this interaction is distinct from the known SUMO1/SIM interaction. To further confirm these results, we generated another mutant form of the FANCI protein with a deletion of the entire SIM sequence (FANCI-ΔSIM) (Fig. 3A). As predicted, the GST-SLD2 fusion protein failed to pull down this FANCI mutant protein (Fig. 3G).
UAF1 binding to FANCI is required for function of the FA pathway
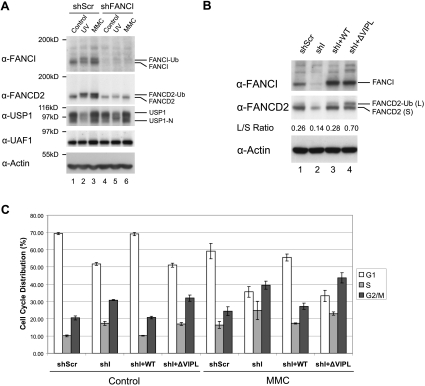
In order to functionally test the SIM sequence of FANCI, we then generated a HeLa cell line with a stable shRNA knockdown of FANCI. These FANCI-deficient human cells displayed the characteristic phenotype of FA-I cells previously described (Smogorzewska et al. 2007). Specifically, the FANCI-deficient HeLa cells had reduced levels of FANCD2 and FANCI and a reduced level of FANCD2-Ub (Fig. 4A, lanes 4–6), and the cells exhibited an increased level of G2/M accumulation (Supplemental Fig. S5B), consistent with their underlying defect in DNA ICL repair (Pulsipher et al. 1998). Transfection of these cells with a shRNA-resistant cDNA encoding wild-type human FANCI restored FANCI expression (Fig. 4B, lane 3), stabilized FANCD2 levels, and promoted the MMC-inducible accumulation of FANCI-Ub and FANCD2-Ub in the chromatin fraction (Supplemental Fig. S5C). Transfection with the cDNA encoding a nonubiquitinated form of FANCI (FANCI-K523R mutant) stabilized the unubiquitinated FANCD2 protein in the transfected cells but failed to promote FANCD2 monoubiquitination and failed to rescue the G2/M accumulation defect (data not shown).
Figure 4.
Deletion of the SIM domain of FANCI blocks deubiquitination by USP1/UAF1 and disrupts the FA pathway. (A) FANCD2 ubiquitination was impaired in shFANCI cells. HeLa cells were stably transfected with an shRNA specific for the FANCI gene product. Cell lysates were immunoblotted with the indicated antibodies. (B) Both WT and ΔVIPL mutant FANCI rescued FANCD2 monoubiquitination. Cells with stable FANCI knockdown were transfected with an shRNA-resistant cDNA encoding the indicated Flag-tagged wild-type or mutant FANCI proteins. Lysates from stably transfected cells were immunoblotted with the indicated antibodies. The ratio of FANCD2-Ub to FANCD2 (L/S ratio) was indicated. (C) The indicated HeLa cell transfectants were analyzed by the G2/M accumulation assay. The percentage of cells in different phases of the cell cycle was determined by FACS analysis.
We next transfected these FANCI-deficient cells with the cDNAs encoding either the wild-type FANCI protein or the FANCI-ΔVIPL mutant. The FANCI-ΔVIPL mutant protein increased the monoubiquitination of FANCD2 (Fig. 4B, lane 4). Importantly, the cells expressing the FANCI mutant protein remained DNA repair-deficient, as indicated by their persistent high level of G2/M accumulation (Fig. 4C). The elevated G2/M accumulation was further increased by exposure of the transfected cells to MMC. Taken together, these results suggest that the FANCI-ΔVIPL mutant protein can bind and enhance the monoubiquitination of its FANCD2-binding partner but fails to bind to the UAF1/USP1 complex and therefore fails to promote FANCD2-Ub deubiquitination. These results are consistent with previous studies indicating that the absence of USP1/UAF1-mediated deubiquitination of FANCD2 causes hypersensitivity to DNA damage (Oestergaard et al. 2007; Kim et al. 2009).
The SLD2 domain of UAF1 binds to the hELG1/PCNA complex
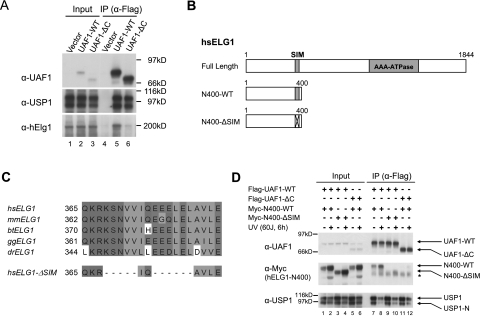
Previous studies have indicated that USP1/UAF1 also regulates the level of PCNA-Ub (Huang et al. 2006; Lee et al. 2010; Terai et al. 2010). Moreover, PCNA-Ub interacts with FANCD2-Ub at stalled replication forks (Howlett et al. 2005). We therefore hypothesized that the USP1/UAF1 complex may bind to a SIM sequence on PCNA-Ub or a PCNA-binding partner. Recent studies indicate that PCNA binds to hELG1 (Lee et al. 2010; Parnas et al. 2010), an ATPase in the RFC family of proteins. We also conducted a serial deletion of hELG1 and identified a region near its N terminus required for coimmunoprecipitation with the USP1/UAF1 complex (Lee et al. 2010). Interestingly, this binding site on hELG1 (KSNVVIQEEELELAVLE) also conforms to the consensus sequence of a SIM, and the sequence is highly conserved among hELG1 proteins from multiple species (Fig. 5C).
Figure 5.
The SLD2 region of UAF1 is required for binding to a SIM on hELG1. (A) SLD2 is required for UAF1/hELG1 interaction. HEK293T cells were transfected with the cDNA encoding either empty vector (lanes 1,4), Flag-UAF1-WT (lanes 2,5), or Flag-UAF1-ΔC (lanes 3,6). As indicated, immunoprecipitation with anti-Flag agarose beads was performed, and proteins were immunoblotted with the indicated antibodies. (B) Schematic representation of hELG1 mutants. The N400 deletion contains only the first 400 residues of hELG1 protein. The ΔSIM mutation was described previously (Lee et al. 2010). (C) Sequence alignment of the SIM region of hELG1 from multiple species (Homo sapiens [hs], Mus musculus [mm], Bos taurus [bt], Gallus gallus [gg], and Danio rerio [dr]), showing a high level of sequence conservation. The sequence of ΔSIM mutant was also shown. (D) SIM is required for UAF1/hELG1 interaction. HEK293T cells were cotransfected with the cDNAs encoding Flag-tagged wild-type (or ΔC mutant) UAF1 and/or Myc-tagged wild-type hELG1 or a ΔSIM mutant (sequence shown in C), which contains a deletion of 11 residues in the SIM region (Lee et al. 2010). For the experiment, we examined the 1- to 400-amino-acid fragment of hELG1. An anti-Flag immunoprecipitation was performed (lanes 7–12), and proteins were immunoblotted with the indicated antibodies. The asterisk (*) denotes a nonspecific band.
We hypothesized that the SLD2 domain of UAF1 may also bind to this SIM sequence on hELG1 (Fig. 5). Cells were transfected with the cDNA encoding a Flag-tagged UAF1-WT or UAF1-ΔC lacking the SLD2 region (Fig. 5A). As predicted, both wild-type UAF1 and the ΔC mutant coimmunoprecipitated with endogenous USP1. This result further indicates that the WD40 region of UAF1 binds USP1 (Fig. 5A, lanes 5,6). Interestingly, the UAF1-ΔC protein failed to pull down hELG1 (Fig. 5A, lane 6). Moreover, the UAF1/hELG1 interaction was also dependent on the SIM of hELG1 (Fig. 5B,D). For this study, we generated the cDNA encoding a truncated form of hELG1, containing residues 1–400 (Fig. 5B). This truncated protein, containing the SIM, coimmunoprecipitated with UAF1 (Fig. 5D, lanes 7,8). Deletion of the SIM region of hELG1 (see hELG1-ΔSIM in Fig. 5C) resulted in loss of UAF1 binding (Fig. 5D, lanes 9,10). These results demonstrate that UAF1 binds to the DUB USP1 and that the SLD2 sequence delivers the complex to multiple monoubiquitinated protein complexes, each containing a SIM sequence.
A point mutation of SLD2 blocks the interaction between UAF1 and its substrates
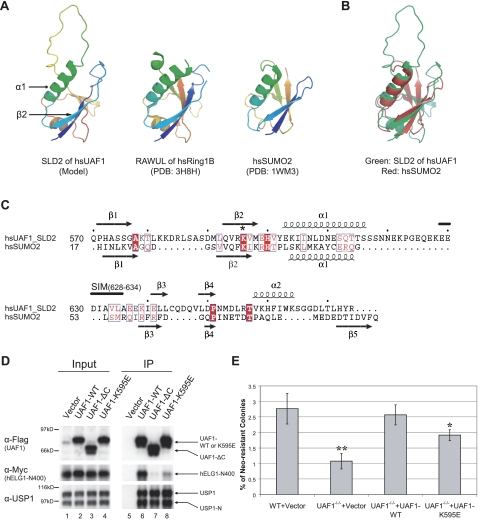
To further assess the interaction between the SLD2 sequence of UAF1 and the SIM sequences of hELG1 or FANCI, we identified a nonbinding point mutant of the SLD2 (Fig. 6). To accomplish this, we initially generated a structural model of the SLD2 domain (Fig. 6A). The crystal structure of the highly related RAWUL domain of hsRing1B (Sanchez-Pulido et al. 2008; Bezsonova et al. 2009) was previously determined. Based on this structure, we modeled the three-dimensional (3D) structure of SLD2 (Fig. 6A). Interestingly, the SLD2 domain contains a nonstructured loop of 23 residues (the loop between α1 and β3) (Fig. 6C). Moreover, this loop contains an inverted SIM sequence (Fig. 6C, residues 628–634) that may function as an autoinhibitory domain or an oligomerization domain of the USP1/UAF1 complex.
Figure 6.
Structural similarity between the SLD2 of UAF1 and SUMO2. (A) Predicted model structure of SLD2 domain of hsUAF1. The structures of the RAWUL domain of hsRing1B (PDB: 3H8H) and hsSUMO2 (PDB: 1WM3) were also shown for comparison. (B) The superposed view of the model structure of the SLD2 domain from hsUAF1 (green) and the structure of hsSUMO2 (red). (C) Structure-based sequence alignment of the SLD2 domain from hsUAF1 and hsSUMO2. The major secondary structures were well conserved, especially the β2/α1 region. The 23-residue sequence of SLD2 (amino acids 616–638), which is not conserved in SUMO2, contains an inverted SIM sequence. This sequence is shown as the long loop in A and B. The asterisk indicates the conserved lysine (K) residue targeted by mutagenesis. (D) The Flag-UAF1-K595E mutant protein binds weakly to the SIM sequence on hELG1. HEK293T cells were cotransfected with the cDNAs encoding Flag-tagged wild-type, ΔC mutant, or K595E mutant UAF1 and Myc-tagged wild-type hELG1 (the 1- to 400-amino-acid fragment of hELG1). Cells were treated with UV (60J/m2, 6 h) before being harvested (Lanes 5–8). An anti-Flag immunoprecipitation was performed, and proteins were immunoblotted with the indicated antibodies. (E) The gene conversion assay was performed as described in Figure 2C. The difference between the single-asterisk (*) cells and the wild-type DT40 cells (WT + Vector) was statistically significant (P < 0.05). The difference between the double-asterisk (**) cells and the wild-type cells (WT + Vector) was also statistically significant (P < 0.001).
Then we superimposed the model structure of SLD2 onto the known structure of SUMO2 (Fig. 6B; Huang et al. 2004) and generated the structure-based sequence alignment between the SLD2 sequence and hsSUMO2 (Fig. 6C). This alignment revealed conserved residues, found in overlapping secondary structures, suitable as potential target sites for mutagenesis.
We next generated a point mutation of a conserved lysine residue (K595E) in SLD2 (indicated with an asterisk in Fig. 6C), as the corresponding lysine on SUMO2 (K33) was critical in SUMO–SIM interaction (Sekiyama et al. 2008). Compared with the wild-type Flag-UAF1 protein, the K595E mutant exhibited a much weaker interaction with the SIM sequence of hELG1, although the same mutant protein bound normally to USP1 (Fig. 6D). The UAF1-K595E mutant protein also exhibited impaired activity in the functional gene conversion assay in UAF1−/−/− DT40 cells (Fig. 6E). Taken together, these results further confirm that the SLD2 sequence of UAF1 has conserved binding characteristics with a SIM sequence, similar to the known SUMO–SIM interactions previously described (Hecker et al. 2006).
SLD2 delivers the USP1/UAF1 complex independently to multiple monoubiquitinated substrates
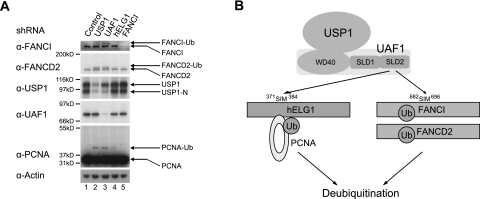
We determined whether the USP1/UAF1 deubiquitinating complex interacts independently with its various substrates or whether the interaction with one substrate is influenced by another. shRNA knockdown of USP1 or UAF1 resulted in an increase in the monoubiquitination levels of FANCD2 and PCNA-Ub (Fig. 7A, lanes 2,3, respectively). In contrast, knockdown of hELG1 resulted in an elevation only in PCNA-Ub levels and did not affect the level of FANCD2-Ub in the cell (Fig. 7A, lane 4). Knockdown of FANCI has no effect on PCNA-Ub (Fig. 7A, lane 5). These results further confirmed a model in which SLD2 delivers the USP1/UAF1 complex independently to multiple monoubiquitinated substrates (Fig. 7B).
Figure 7.
The USP1/UAF1 complex regulates the ubiquitination level of FANCD2/FANCI and PCNA. (A) The USP1/UAF1 complex regulates the ubiquitination level of both FANCD2/FANCI and PCNA, while hELG1 specifically regulates the ubiquitination level of PCNA. Retroviral shRNAs targeting the genes as indicated were used to stably infect HeLa cells, and the effects on FANCD2/FANCI and PCNA ubiquitination were examined by Western blot. By densitometry, the relative band intensities for the PCNA-Ub band for lanes 1–4 were 1.00, 4.92, 4.63, and 2.53, respectively. (B) Schematic interaction between the SLD2 region of UAF1 and the SIM sequences on hELG1 and FANCI. Most intracellular USP1 is constitutively bound to UAF1. UAF1 is a more abundant protein than USP1, and it has multiple binding partners. The WD40 domain of UAF1 binds and stimulates USP1. The SLD2 region of UAF1 binds the SIM sequences of hELG1 and of FANCI.
Our results suggested that the USP1/UAF1 complex is targeted to multiple DNA repair substrates and that this targeting is essential for the successful completion of DNA repair. A recent proteomic survey indicated that the UAF1 subunit (also referred to as WDR48) forms a complex with other DNA repair proteins (Sowa et al. 2009). For instance, UAF1 binds to RAD51AP1, another DNA repair protein required for efficient RAD51 D-loop formation and HR (Wiese et al. 2007). Interestingly, the SLD2 sequence of UAF1 binds directly to a SIM sequence found at the N terminus of RAD51AP1 (Supplemental Fig. S6A–C), although the functional importance of this targeting event remains untested.
Discussion
At least 95 DUBs are encoded by the human genome (Nijman et al. 2005b; Komander et al. 2009; Sowa et al. 2009). Only a few of these ubiquitin proteases have genetically confirmed substrates, and little is known regarding the molecular mechanism of substrate recognition and recruitment. DUB targeting to substrates may occur, at least in part, through SUMO modifications (Meulmeester et al. 2008), similar to the mechanism of some E3 ligase substrate targeting (Prudden et al. 2007, 2009). Extensive genetic evidence indicates that the USP1/UAF1 complex specifically deubiquitinates the substrates FANCD2-Ub and PCNA-Ub and regulates HR repair and TLS repair, respectively. The mechanism by which the USP1/UAF1 complex is targeted to its specific substrates has remained elusive. Here we show that the USP1/UAF1 deubiquitinating complex is targeted directly to these monoubiquitinated substrates by unique SLD–SIM interactions.
Our study indicates that, while the N-terminal WD40 repeat region of UAF1 binds constitutively to USP1, the C-terminal region contains a tandem repeat of SLDs required for substrate targeting. Specifically, the SLD2 sequence of UAF1 binds to a SIM-like sequence found on the FANCI/FANCD2 complex and on the hELG1/PCNA-Ub complex. Importantly, while GST-SLD2 binds to the SIM-like sequence on FANCI and hELG1, the GST-SUMO1, GST-SUMO2, and GST-SUMO3 fusion proteins do not bind to this peptide sequence. Conversely, GST-SUMO1 binds to a SIM sequence on the protein Pias1 (Hecker et al. 2006), but GST-SLD2 does not interact with this protein (Fig. 3F). We therefore propose using the term SLIM to describe these peptide sequences on FANCI and hELG1. Based on our results and published proteomic studies (Sowa et al. 2009), we predict that the UAF1 C-terminal SLD2 will bind to other proteins containing SLIM sequences. Future crystallographic and NMR studies will be required to determine whether the SLD2–SLIM interaction has structural features conserved with the known SUMO–SIM interaction. For the SUMO–SIM binding, the hydrophobic core of the SIM interacts with second β strand and the first α helix on SUMO. The acidic residues of the SIM form additional electrostatic interactions with positively charged residues on the binding surface of SUMO. Since the SLIM sequence also has a hydrophobic core and a stretch of glutamic acid or aspartic acid residues, its interaction with SLD2 may turn out to have similar structural features.
We propose that these SLD–SLIM interactions contribute to, but are not solely responsible for, substrate recruitment by the USP1/UAF1 complex. Only a low level of interaction between the endogenous USP1/UAF1 complex and its substrates is evident at any given time (K Yang and AD D'Andrea, unpubl.). USP1/UAF1 is primarily a soluble nuclear protein complex, while its monoubiquitinated substrates are chromatin-bound. DNA damage and cell cycle transitions may further up-regulate these interactions through phosphorylation of the USP1/UAF1 complex or its substrates. Indeed, recent studies have shown that regulated phosphorylation of amino acid residues within or adjacent to SIM sequences can enhance SUMO/SIM-binding interactions (Stehmeier and Muller 2009). Accordingly, USP1, FANCD2, FANCI, and hELG1 are phosphorylated by the kinase ATM (Matsuoka et al. 2007), and phosphorylation of sites adjacent to the SLIM sequence on FANCI and hELG1 may further increase these SLD–SLIM interactions in vivo.
Consistent with this idea, we have shown that DNA damage by MMC or UV light can increase the interaction of the USP1/UAF1 complex with its substrates. For instance, MMC enhances the interaction of the complex with FANCI (Fig. 3D). It will be interesting to determine whether this enhanced binding of USP1/UAF1 with its substrates is dependent on ATM or ATR. Interestingly, our data indicate that caffeine, a known inhibitor of ATM and ATR, can inhibit the FANCI/UAF1 interaction.
Recent studies also indicate that DNA ICL repair requires a FANCD2-Ub-mediated recruitment of a nuclease and a TLS polymerase (Knipscheer et al. 2009). PCNA-Ub is recruited to FANCD2-Ub complexes (Song et al. 2010) and may recruit a TLS polymerase to the ICL site. Therefore, we speculate that the USP1/UAF1 complex may play a central role in coordinating ICL repair through regulated SLD–SLIM interactions and a ubiquitination/deubiquitination cycle of its FANCD2-Ub and PCNA-Ub substrates (model in Supplemental Fig. S7).
Finally, several features of the SLD–SLIM targeting mechanism for the USP1/UAF1 deubiquitinating complex remain unresolved, and these features are topics for future experimentation. First, the function of the SLD1 domain of UAF1 is unknown. The presence of a tandem repeat of SLD domains may further increase the affinity of the UAF1 C terminus for binding partner complexes with multiple SLIMs. Although we were unable to detect binding of the isolated SLD1 to a SLIM in vitro, SLD1 may contribute to substrate binding when it is positioned in a tandem repeat with SLD2. Second, UAF1 is an abundant protein, and it has multiple DUB-binding partners. For instance, UAF1 binds, via its N-terminal WD40 domain, to USP1, USP12, and USP46 (Cohn et al. 2009). However, USP12/UAF1 and USP46/UAF1 complexes do not deubiquitinate FANCD2/FANCI or hELG1/PCNA complexes in vivo. Since USP12/UAF1 and USP46/UAF1 complexes contain additional subunits, such as WDR20 (Kee et al. 2010), these subunits may specifically direct the DUB complexes to other ubiquitinated substrates. Last, UAF1 not only delivers DUBs to their substrates, but also may have a more general role in sorting intracellular proteins. Consistent with this notion, UAF1 has previously been shown to bind and recruit the Lck tyrosine kinase to lysosomes for degradation (Park et al. 2002). Hence, SLD–SLIM interactions may have a broader role in cellular transport mechanisms beyond their role in DNA repair.
Materials and methods
Western blot analysis and antibodies
Cells were harvested, and total protein extracts were prepared using radioimmune precipitation assay buffer (50 mM Tris-HCl at pH 7.3, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1% sodium deoxycholate, 0.05% SDS). Protein extracts were normalized using Bradford assay, loaded onto polyacrylamide gels, and transferred onto nitrocellulose membranes. Immunoblotting was performed using antibodies as indicated: human USP1 (Cohn et al. 2007), chicken USP1 (A301-699A, Bethyl Laboratories), human and chicken UAF1 (cross-react with proteins from both species) (Cohn et al. 2007), human FANCI (Vinciguerra et al. 2010), human FANCD2 (FI-17, Santa Cruz Biotechnology), chicken FANCD2 (Ishiai et al. 2008), human PALB2 (Xia et al. 2006), human ELG1 (Lee et al. 2010), human and chicken PCNA (PC-10, Santa Cruz Biotechnology; cross-react with both species), Actin (AC-40, Sigma), Flag (M2, Sigma), c-Myc (sc-40, Santa Cruz Biotechnology), and pChk1 (Ser317, #2344, Cell Signaling Technology).
Immunoprecipitation
Cells were harvested, and total protein extracts were prepared using a lysis buffer containing 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 1% Triton X-100. The total protein concentration of the lysates was normalized using Bradford assay. Anti-Flag M2 agarose beads (A2220, Sigma) were incubated with indicated cell lysates for 3 h at 4°C. Beads were then washed with lysis buffer three times and boiled in 1× SDS loading buffer.
Protein purification and in vitro deubiquitination assay
Preparation of USP1 and UAF1 recombinant proteins in Sf9 cells and Ub-AMC deubiquitination assay were previously described (Cohn et al. 2007). Briefly, the cDNAs encoding UAF1 wild type and mutants were cloned into pFastBac-HTa vector (Invitrogen). Baculoviruses were made and used to infect fresh Sf9 insect cells. Overexpressed proteins were purified using Ni-NTA beads. The in vitro deubiquitination assays using Ub-AMC (U-550, Boston Biochem) as the substrate were performed in 100 μL of reaction buffer (20 mM HEPES-KOH at pH 7.8, 20 mM NaCl, 0.1 mg/mL ovalbumin [A7641, Sigma], 0.5 mM EDTA, 10 mM DTT) at 37°C. The fluorescence signal was monitored in a FluoStar Galaxy fluorometer (BMG Labtech).
Complementation of UAF1−/−/− DT40 cells
DT40 cells were cultured and transfected as described previously (Sonoda et al. 1998). The cDNAs encoding human UAF1 wild type and deletion mutants were cloned into pcDNA3.1 vector, linearized, and stably transfected into UAF1−/−/− cells. Individual clones were picked, and the expression of exogenous proteins was confirmed by Western blot.
Acknowledgments
We thank Minoru Takata for chicken anti-FANCD2 antibody, David Livingston for anti-PALB2 antibody, Kyungjae Myung for anti-hELG1 antibody, David Schild for anti-RAD51AP1 antibody, and Ivan Dikic for cDNAs of Pias1, GST-SUMO1, GST-SUMO2, and GST-SUMO3. We thank Kay Hofmann, Fernando Bazan, Jeff Perry, John Tainer, David Pellman, Daniel Finley, Lee Zou, Johannes Walter, Geoffrey Shapiro, and members of the D'Andrea laboratory for helpful discussions. K.Y. is a Harvard University Presidential Scholar. G.L.M. is supported by a post-doctoral fellowship from the International Human Frontier Science Program Organization. P.V. was supported by a fellowship from the Swiss Foundation for grants in biology and medicine, and by Swiss National Science Foundation grant PASMA3-119584. J.M. is funded by long-term research grants from the TOYOBO Biotechnology Foundation. This work was supported by NIH grants R01DK43889, R01HL52725, P01CA092584 to A.D.D.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.17020911.
References
- Al-Hakim A, Escribano-Diaz C, Landry MC, O'Donnell L, Panier S, Szilard RK, Durocher D 2010. The ubiquitous role of ubiquitin in the DNA damage response. DNA Repair (Amst) 9: 1229–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergink S, Jentsch S 2009. Principles of ubiquitin and SUMO modifications in DNA repair. Nature 458: 461–467 [DOI] [PubMed] [Google Scholar]
- Bezsonova I, Walker JR, Bacik JP, Duan S, Dhe-Paganon S, Arrowsmith CH 2009. Ring1B contains a ubiquitin-like docking module for interaction with Cbx proteins. Biochemistry 48: 10542–10548 [DOI] [PubMed] [Google Scholar]
- Bienko M, Green CM, Sabbioneda S, Crosetto N, Matic I, Hibbert RG, Begovic T, Niimi A, Mann M, Lehmann AR, et al. 2010. Regulation of translesion synthesis DNA polymerase eta by monoubiquitination. Mol Cell 37: 396–407 [DOI] [PubMed] [Google Scholar]
- Cohn MA, Kowal P, Yang K, Haas W, Huang TT, Gygi SP, D'Andrea AD 2007. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol Cell 28: 786–797 [DOI] [PubMed] [Google Scholar]
- Cohn MA, Kee Y, Haas W, Gygi SP, D'Andrea AD 2009. UAF1 is a subunit of multiple deubiquitinating enzyme complexes. J Biol Chem 284: 5343–5351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cybulski KE, Howlett NG 2011. FANCP/SLX4: a Swiss army knife of DNA interstrand crosslink repair. Cell Cycle 10: 1757–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denuc A, Bosch-Comas A, Gonzalez-Duarte R, Marfany G 2009. The UBA–UIM domains of the USP25 regulate the enzyme ubiquitination state and modulate substrate recognition. PLoS ONE 4: e5571 doi: 10.1371/journal.pone.0005571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima T, Takata M, Morrison C, Araki R, Fujimori A, Abe M, Tatsumi K, Jasin M, Dhar PK, Sonoda E, et al. 2001. Genetic analysis of the DNA-dependent protein kinase reveals an inhibitory role of Ku in late S–G2 phase DNA double-strand break repair. J Biol Chem 276: 44413–44418 [DOI] [PubMed] [Google Scholar]
- Hecker CM, Rabiller M, Haglund K, Bayer P, Dikic I 2006. Specification of SUMO1- and SUMO2-interacting motifs. J Biol Chem 281: 16117–16127 [DOI] [PubMed] [Google Scholar]
- Hochegger H, Dejsuphong D, Fukushima T, Morrison C, Sonoda E, Schreiber V, Zhao GY, Saberi A, Masutani M, Adachi N, et al. 2006. Parp-1 protects homologous recombination from interference by Ku and Ligase IV in vertebrate cells. EMBO J 25: 1305–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett NG, Taniguchi T, Durkin SG, D'Andrea AD, Glover TW 2005. The Fanconi anemia pathway is required for the DNA replication stress response and for the regulation of common fragile site stability. Hum Mol Genet 14: 693–701 [DOI] [PubMed] [Google Scholar]
- Huang TT, D'Andrea AD 2006. Regulation of DNA repair by ubiquitylation. Nat Rev Mol Cell Biol 7: 323–334 [DOI] [PubMed] [Google Scholar]
- Huang WC, Ko TP, Li SS, Wang AH 2004. Crystal structures of the human SUMO-2 protein at 1.6 A and 1.2 Å resolution: implication on the functional differences of SUMO proteins. Eur J Biochem 271: 4114–4122 [DOI] [PubMed] [Google Scholar]
- Huang TT, Nijman SM, Mirchandani KD, Galardy PJ, Cohn MA, Haas W, Gygi SP, Ploegh HL, Bernards R, D'Andrea AD 2006. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol 8: 339–347 [DOI] [PubMed] [Google Scholar]
- Ishiai M, Kitao H, Smogorzewska A, Tomida J, Kinomura A, Uchida E, Saberi A, Kinoshita E, Kinoshita-Kikuta E, Koike T, et al. 2008. FANCI phosphorylation functions as a molecular switch to turn on the Fanconi anemia pathway. Nat Struct Mol Biol 15: 1138–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannouche PL, Wing J, Lehmann AR 2004. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol Cell 14: 491–500 [DOI] [PubMed] [Google Scholar]
- Kee Y, D'Andrea AD 2010. Expanded roles of the Fanconi anemia pathway in preserving genomic stability. Genes Dev 24: 1680–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y, Yang K, Cohn MA, Haas W, Gygi SP, D'Andrea AD 2010. WDR20 regulates activity of the USP12·UAF1 deubiquitinating enzyme complex. J Biol Chem 285: 11252–11257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Parmar K, Huang M, Weinstock DM, Ruit CA, Kutok JL, D'Andrea AD 2009. Inactivation of murine Usp1 results in genomic instability and a Fanconi anemia phenotype. Dev Cell 16: 314–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, Scharer OD, Elledge SJ, Walter JC 2009. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science 326: 1698–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Clague MJ, Urbe S 2009. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol 10: 550–563 [DOI] [PubMed] [Google Scholar]
- Lee KY, Yang K, Cohn MA, Sikdar N, D'Andrea AD, Myung K 2010. Human ELG1 regulates the level of ubiquitinated proliferating cell nuclear antigen (PCNA) through Its interactions with PCNA and USP1. J Biol Chem 285: 10362–10369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. 2007. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316: 1160–1166 [DOI] [PubMed] [Google Scholar]
- Meulmeester E, Kunze M, Hsiao HH, Urlaub H, Melchior F 2008. Mechanism and consequences for paralog-specific sumoylation of ubiquitin-specific protease 25. Mol Cell 30: 610–619 [DOI] [PubMed] [Google Scholar]
- Murai J, Yang K, Dejsuphong D, Hirota K, Takeda S, D'Andrea AD 2011. The USP1/UAF1 complex promotes double-strand break repair through homologous recombination. Mol Cell Biol 31: 2462–2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi K, Yang YG, Pierce AJ, Taniguchi T, Digweed M, D'Andrea AD, Wang ZQ, Jasin M 2005. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc Natl Acad Sci 102: 1110–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijman SM, Huang TT, Dirac AM, Brummelkamp TR, Kerkhoven RM, D'Andrea AD, Bernards R 2005a. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell 17: 331–339 [DOI] [PubMed] [Google Scholar]
- Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R 2005b. A genomic and functional inventory of deubiquitinating enzymes. Cell 123: 773–786 [DOI] [PubMed] [Google Scholar]
- Noda NN, Ohsumi Y, Inagaki F 2010. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett 584: 1379–1385 [DOI] [PubMed] [Google Scholar]
- Novatchkova M, Bachmair A, Eisenhaber B, Eisenhaber F 2005. Proteins with two SUMO-like domains in chromatin-associated complexes: the RENi (Rad60–Esc2–NIP45) family. BMC Bioinformatics 6: 22 doi: 10.1186/1471-2105-6-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestergaard VH, Langevin F, Kuiken HJ, Pace P, Niedzwiedz W, Simpson LJ, Ohzeki M, Takata M, Sale JE, Patel KJ 2007. Deubiquitination of FANCD2 is required for DNA crosslink repair. Mol Cell 28: 798–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Lee BS, Choi JK, Means RE, Choe J, Jung JU 2002. Herpesviral protein targets a cellular WD repeat endosomal protein to downregulate T lymphocyte receptor expression. Immunity 17: 221–233 [DOI] [PubMed] [Google Scholar]
- Parnas O, Zipin-Roitman A, Pfander B, Liefshitz B, Mazor Y, Ben-Aroya S, Jentsch S, Kupiec M 2010. Elg1, an alternative subunit of the RFC clamp loader, preferentially interacts with SUMOylated PCNA. EMBO J 29: 2611–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudden J, Pebernard S, Raffa G, Slavin DA, Perry JJ, Tainer JA, McGowan CH, Boddy MN 2007. SUMO-targeted ubiquitin ligases in genome stability. EMBO J 26: 4089–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudden J, Perry JJ, Arvai AS, Tainer JA, Boddy MN 2009. Molecular mimicry of SUMO promotes DNA repair. Nat Struct Mol Biol 16: 509–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulsipher M, Kupfer GM, Naf D, Suliman A, Lee JS, Jakobs P, Grompe M, Joenje H, Sieff C, Guinan E, et al. 1998. Subtyping analysis of Fanconi anemia by immunoblotting and retroviral gene transfer. Mol Med 4: 468–479 [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Ventii KH, Wilkinson KD 2009. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem 78: 363–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Devos D, Sung ZR, Calonje M 2008. RAWUL: a new ubiquitin-like domain in PRC1 ring finger proteins that unveils putative plant and worm PRC1 orthologs. BMC Genomics 9: 308 doi: 10.1186/1471-2164-9-308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiyama N, Ikegami T, Yamane T, Ikeguchi M, Uchimura Y, Baba D, Ariyoshi M, Tochio H, Saitoh H, Shirakawa M 2008. Structure of the small ubiquitin-like modifier (SUMO)-interacting motif of MBD1-containing chromatin-associated factor 1 bound to SUMO-3. J Biol Chem 283: 35966–35975 [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER III, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D'Andrea AD, et al. 2007. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell 129: 289–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y 2004. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci 101: 14373–14378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song IY, Palle K, Gurkar A, Tateishi S, Kupfer GM, Vaziri C 2010. Rad18-mediated translesion synthesis of bulky DNA adducts is coupled to activation of the Fanconi anemia DNA repair pathway. J Biol Chem 285: 31525–31536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E, Sasaki MS, Buerstedde JM, Bezzubova O, Shinohara A, Ogawa H, Takata M, Yamaguchi-Iwai Y, Takeda S 1998. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J 17: 598–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW 2009. Defining the human deubiquitinating enzyme interaction landscape. Cell 138: 389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehmeier P, Muller S 2009. Phospho-regulated SUMO interaction modules connect the SUMO system to CK2 signaling. Mol Cell 33: 400–409 [DOI] [PubMed] [Google Scholar]
- Terai K, Abbas T, Jazaeri AA, Dutta A 2010. CRL4(Cdt2) E3 ubiquitin ligase monoubiquitinates PCNA to promote translesion DNA synthesis. Mol Cell 37: 143–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinciguerra P, Godinho SA, Parmar K, Pellman D, D'Andrea AD 2010. Cytokinesis failure occurs in Fanconi anemia pathway-deficient murine and human bone marrow hematopoietic cells. J Clin Invest 120: 3834–3842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C, Dray E, Groesser T, San Filippo J, Shi I, Collins DW, Tsai MS, Williams GJ, Rydberg B, Sung P, et al. 2007. Promotion of homologous recombination and genomic stability by RAD51AP1 via RAD51 recombinase enhancement. Mol Cell 28: 482–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, Liu X, Jasin M, Couch FJ, Livingston DM 2006. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell 22: 719–729 [DOI] [PubMed] [Google Scholar]
- Yamamoto KN, Kobayashi S, Tsuda M, Kurumizaka H, Takata M, Kono K, Jiricny J, Takeda S, Hirota K 2011. Involvement of SLX4 in interstrand cross-link repair is regulated by the Fanconi anemia pathway. Proc Natl Acad Sci 108: 6492–6496 [DOI] [PMC free article] [PubMed] [Google Scholar]