This study investigates the spatial processing and activation of the Nodal ligand during epiblast patterning in early post-implantation mouse development. The endoproteases Furin and Pace4, which are secreted by the extraembryonic environment, are found to be enzymatically active in the adjacent epiblast, where they activate Nodal to maintain pluripotency.
Keywords: progenitor cells, microenvironment, proprotein convertases, Furin, Nodal, gastrulation
Abstract
The fate of pluripotent cells in early mouse embryos is controlled by graded Nodal signals that are activated by the endoproteases Furin and Pace4. Soluble forms of Furin and Pace4 cleave proNodal in vitro and after secretion in transfected cells, but direct evidence for paracrine activity in vivo is elusive. Here, we show that Furin and Pace4 are released by the extraembryonic microenvironment, and that they cleave a membrane-bound reporter substrate in adjacent epiblast cells and activate Nodal to maintain pluripotency. Secreted Pace4 and Furin also stimulated mesoderm formation, whereas endoderm was only induced by Pace4, correlating with a difference in the spatiotemporal distribution of these proteolytic activities. Our analysis of paracrine Furin and Pace4 activities and their in vivo functions significantly advances our understanding of how the epiblast is patterned by its microenvironment. Adding cell–cell communication to the pleiotropic portfolio of these proteases provides a new framework to study proprotein processing also in other relevant contexts.
The early implanted mouse embryo consists of pluripotent epiblast cells nested between the extraembryonic ectoderm (ExE) and the embryonic visceral endoderm (EmVE). Upon interaction with this microenvironment, epiblast cells proliferate and adopt a columnar epithelial shape, followed by differentiation into ectoderm, mesoderm, or definitive endoderm (DE) during gastrulation. Pluripotency and differentiation of epiblast cells and their cross-talk with the microenvironment are regulated by Nodal, a secreted protein of the TGFβ family. Nodal signals through complexes of the coreceptors Cripto or Cryptic with Acvr1b and Acvr2 receptors to activate Smad2 and Smad3 transcription factors, thereby inducing endodermal and mesodermal target genes in a dose-dependent manner (Arnold and Robertson 2009). Graded Nodal signals also specify endoderm and mesoderm cell fates in other vertebrates (Schier 2009). In zebrafish, the Nodal homolog Squint forms a morphogen gradient to induce distinct cell fates, depending on the cumulative dose of its local concentration and signal duration (Chen and Schier 2001; Hagos and Dougan 2007). In contrast, in the mouse, Smad-mediated autoinduction triggers uniform Nodal expression throughout the epiblast and EmVE (Arnold and Robertson 2009). Nodal signaling at this stage maintains pluripotency and suppresses precocious neural differentiation in the epiblast, and it specifies and patterns EmVE (Brennan et al. 2001; Camus et al. 2006; Mesnard et al. 2006). Anterior EmVE cells in turn secrete the feedback inhibitors Lefty1 and Cer1, which extinguish Nodal expression in the adjacent epiblast, thereby limiting induction of mesoderm and endoderm to the opposite pole (Perea-Gomez et al. 2002). Thus, an overt Nodal activity gradient in the epiblast is only evident at the level of signal duration, which is regulated by negative feedback from the microenvironment.
To activate autoregulation, the Nodal precursor protein must be cleaved after the sequence RQRR by proteases of the proprotein convertase (PC) family (Constam and Robertson 1999; Ben Haim et al. 2006). Pace4 and Furin are the first PCs expressed in implanted embryos (Beck et al. 2002; Mesnard and Constam 2010). They are already active at the blastocyst stage in epiblast progenitors (Mesnard and Constam 2010; Granier et al. 2011) and are necessary to activate Nodal signaling (Beck et al. 2002). However, when and where a direct interaction with Nodal might occur is unclear, since loss of zygotic Furin and Pace4 can be partially compensated by an unknown redundant activity in the uterus (Mesnard and Constam 2010). Furthermore, upon implantation, Furin and Pace4 transcripts are confined to the ExE (Beck et al. 2002) and, in the case of Furin, also to the VE (Mesnard et al. 2006). Thus, a direct effect on pluripotent epiblast cells after implantation would depend on secreted forms of Furin or Pace4 acting in a paracrine manner. However, whether PCs perform long-range activity in these or other physiological target cells or whether they act strictly cell-autonomously remains to be determined.
Both Pace4 and Furin mature from zymogenic precursors by autocatalytic cleavage in the endoplasmic reticulum (Seidah et al. 2008), but distinct trafficking in more distal compartments potentially limits functional redundancy. Pace4 is soluble by default and enriched in extracellular matrix by heparan sulfate proteoglycans (HSPGs) (Tsuji et al. 2003; Mayer et al. 2008). In contrast, Furin has a transmembrane domain and binds cytosolic adapters that mediate cycling between the trans-Golgi network, the cell surface, and endosomes (Thomas 2002). The acidic pH of the trans-Golgi network/endosomal system favors a second autocatalytic cleavage displacing an inhibitory propeptide (Anderson et al. 2002), and cleavage of the membrane anchor by an unknown “sheddase” in acidic compartments can give rise to a soluble form of activated Furin (Wise et al. 1990; Vidricaire et al. 1993; Denault et al. 2002). Shed Furin has been observed in the milk of transgenic mice (Paleyanda et al. 1997) and in epididymal fluid (Thimon et al. 2006). Soluble Furin (or Pace4) can also activate Nodal in a complex with its coreceptor, Cripto, in cultured cells (Blanchet et al. 2008). However, a role for secreted forms of Furin and Pace4 or of any other PC has not been demonstrated in vivo.
To assess the functions of secreted Pace4 and Furin, GFP fusions of these proteases were specifically expressed in the ExE of wild-type and double knockout (DKO) embryos alone or together with a transgenic reporter substrate. We show that active forms of both proteases reach the epiblast and are sufficient to sustain determinants of pluripotency and to promote mesoderm formation. However, only Pace4GFP also stimulated endoderm formation. Correlating with this difference in activity, FurinGFP and Pace4GFP also showed strikingly different distributions outside the ExE, suggesting that distinct trafficking itineraries compartmentalize interactions with substrates.
Results
Complementary distribution of ExE-derived Furin and Pace4 in the epiblast
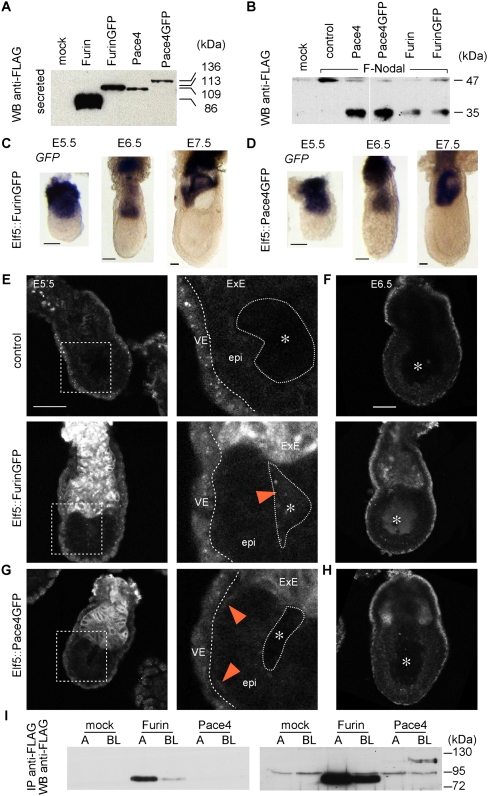
To image Furin and Pace4, we generated GFP-tagged forms that are secreted and active similarly to wild-type forms (Fig. 1A,B). To direct expression to the ExE, FurinGFP or Pace4GFP cDNAs were cloned into a BAC transgene of the Elf5 locus (Donnison et al. 2005), which was then introduced into wild-type and Furin;Pace4 DKO embryos (Supplemental Fig. S1A,B). Anti-GFP Western blot analysis of embryonic day 6.5 (E6.5) embryos showed similar expression levels of the two fusion proteins (Supplemental Fig. S1C). Whole-mount in situ hybridization and GFP imaging confirmed expression of FurinGFP or Pace4GFP starting at E4.5 until E7.5 specifically in the ExE (Fig. 1C–H; data not shown). In addition, FurinGFP was detected outside the ExE in the proamniotic cavity faintly at E5.5 (Fig. 1E) and more strongly at E6.5 (Fig. 1F). In contrast, Pace4GFP did not accumulate in the proamniotic cavity (Fig. 1G,H), but at the interface between the epiblast and the EmVE at E5.5 (Fig. 1G). The Pace4GFP signal outside the ExE was lost by E6.5 (Fig. 1H). These results show that GFP-tagged Furin and Pace4 are secreted by the ExE and are able to reach the epiblast.
Figure 1.
Pace4GFP and FurinGFP are secreted by the ExE and reach the epiblast by distinct routes. (A) Western blot analysis of GFP-tagged Furin and Pace4 in conditioned medium of transiently transfected COS-1 cells. (B) Western blot analysis of Nodal processing in conditioned medium of COS-1 cells cotransfected with Furin, Pace4, or empty vector (control). Note that both native and GFP-tagged Furin and Pace4 cleave off a Flag-tagged prodomain (35 kDa) from the Nodal precursor (47 kDa). (C,D) Expression of FurinGFP (C) and Pace4GFP (D) mRNA in BAC transgenic embryos between E5.5 and E7.5. (E–H) Confocal imaging of GFP fluorescence in Elf5::FurinGFP (E,F) and Elf5::Pace4GFP (G,H) transgenic embryos at E5.5 (E,G) and E6.5 (F,H). The right panels in E and G show magnifications of the dashed box in the left panels. GFP fluorescence above background outside the ExE is indicated (red arrowheads). Dashed outlines indicate the boundaries between the VE, epiblast (epi), and proamniotic cavity. Asterisks indicate the center of the proamniotic cavity. The top panels show fluorescent background of nontransgenic embryos. Bar, 50 μm. (I) Western blot analysis of Flag-tagged Furin and Pace4 in apical (A) and basolateral (BL) conditioned medium of stably infected MDCK epithelial cells. The left and right panels show short and long exposures, respectively.
The complementary spatiotemporal distribution of ExE-derived Furin and Pace4 suggests distinct mechanisms of secretion. To test this, we monitored polarized secretion in Madin-Darby canine kidney (MDCK) cells that were infected with Furin or Pace4 lentiviruses. Consistent with our in vivo results, Furin and Pace4 were enriched on the apical or basolateral side, respectively (Fig. 1I). This result shows that Pace4 and shed Furin are differentially secreted in epithelial cells.
Secreted FurinGFP and Pace4GFP from the ExE are active in the epiblast
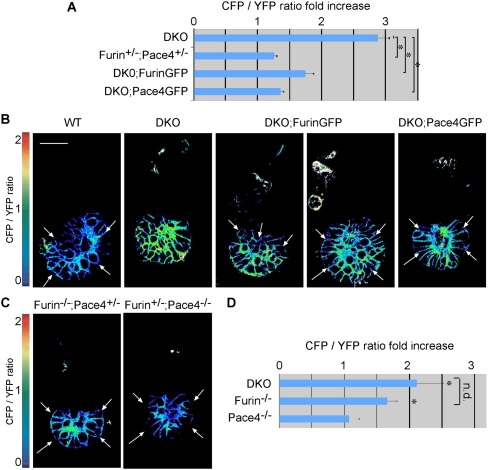
To determine whether ExE-derived FurinGFP and Pace4GFP are active in the epiblast, we introduced a transgene encoding the cell surface-linked indicator of proteolysis CLIP. This biosensor consists of secreted CFP linked by a PC recognition motif to glycosylphosphatidylinositol-modified citrine (henceforth called YFP), so that cleavage by PCs alters the ratio of CFP versus YFP fluorescence at the cell surface (Mesnard and Constam 2010). Previous analysis established that Furin and Pace4 are responsible for cleaving CLIP in the inner cell mass of the blastocyst, but direct evidence for cell-nonautonomous activity in the epiblast is lacking. To address this, we compared CLIP fluorescence in DKO and wild-type embryos that were isolated at E5.25 and cultured for several hours, thereby eliminating cleavage by an uncharacterized compensatory PC-like activity in the uterus (Mesnard and Constam 2010). After applying a mask to select for CLIP levels above an arbitrary threshold (Supplemental Fig. S2), CFP/YFP ratios were quantified and plotted as heat maps (Fig. 2). In this assay, DKO embryos increased the CFP/YFP ratio of CLIP in the epiblast by 2.9-fold (n = 9) compared with wild type (Fig. 2A,B). In contrast, this increase was reduced to 1.7-fold (n = 4) or 1.3-fold (n = 3) in DKO embryos carrying a FurinGFP or Pace4GFP transgene, respectively, and to 1.2-fold in Furin+/−; Pace4+/− double heterozygous embryos (Fig. 2A). Thus, in good agreement with our Nodal cleavage assays in transfected cells (Fig. 1B), the activities of FurinGFP and Pace4GFP resemble in magnitude those of the endogenous proteases. Furthermore, CFP/YFP heat maps of CLIP revealed that processing by FurinGFP at least transiently was more efficient in proximal than in distal cells, whereas Pace4GFP was active throughout the epiblast (Fig. 2B). FurinGFP eventually became active also in more distal regions (Fig. 2B, fourth panel). CLIP was also widely processed in single mutants carrying at least one copy of endogenous Furin or Pace4 (Fig. 2C; Supplemental Fig. S3), suggesting that endogenous Furin activity, like Pace4 and Pace4GFP, is more broadly distributed than FurinGFP. Overall, these results corroborate our conclusion that Furin and Pace4 are released by the ExE, albeit with distinct spatiotemporal profiles. They also demonstrate, in a physiologically relevant context, that Pace4 and shed Furin cleave a membrane-bound substrate in distant target cells.
Figure 2.
Imaging of the membrane-anchored reporter substrate CLIP reveals paracrine activities of Furin and Pace4 in the epiblast. (A–C) Normalized CFP/YFP ratios (A) and heat maps (B,C) of the transgenic PC biosensor CLIP in epiblasts of wild-type or Furin- and Pace4-deficient embryos with or without the ExE-specific FurinGFP or Pace4GFP transgenes. To reduce cleavage of CLIP by a PC activity in the uterus, the embryos were isolated at E5.25 and cultured for 8 h prior to analysis. Heat maps represent CFP/YFP ratio at a pixel level, with green and blue colors corresponding to a value close to 1 and 0, respectively. As the ratio is inversely correlated to PC cleavage, green/red and blue colors represent low and high cleavage activity, respectively. Due to detection limitations, reliable analysis of cleavage could only be performed in the epiblast. Areas where CLIP has been processed are indicated (arrows). Note that the fluorescence of GFP is not sufficiently intense to interfere with CLIP imaging. (D) Relative CFP/YFP ratios in the epiblast of E6.5 embryos expressing CLIP. Fold increase was measured relative to CLIP cleavage in wild-type littermates. Asterisks indicate significant difference as determined by t-test (P < 0.05). “n.d.” indicates no significant difference as determined by t-test (P < 0.05). Error bars represent standard error of the mean. Bar, 50 μm.
We hypothesized that endogenous Furin is more broadly active at E5.5 than FurinGFP because it is transiently expressed until that stage by the EmVE and able to sustain Nodal activity independently of the ExE (Mesnard et al. 2006). Therefore, we also compared Furin and Pace4 activities at E6.5, when the expression of endogenous Furin outside the ExE has subsided. At E6.5, CLIP processing was decreased in Furin−/− but not in Pace4−/− single knockout (KO) (Fig. 2D), concurring with the observation that the ExE continues to release FurinGFP, but not Pace4GFP, at this later stage (Fig. 1F,H). Overall, these data suggest that Pace4 activity in the epiblast entirely depends on its expression in the ExE and ceases after E5.5. In contrast, Furin is initially produced by both the ExE and the VE and continues to be secreted by the ExE at E6.5.
Expression of Pace4 or Furin in the ExE is sufficient to activate Nodal in the epiblast
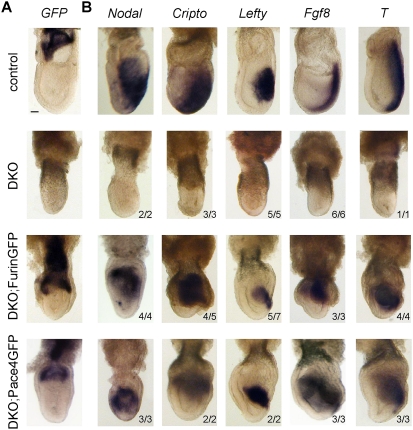
To evaluate the biological function of ExE-derived Furin and Pace4 activities, we assessed whether the Elf5::FurinGFP and Elf5::Pace4GFP BAC transgenes rescue the expression of Nodal target genes in DKO embryos. Importantly, DKO embryos maintained expression of Elf5 until E7.5, demonstrating that Elf5 promoter activity in the ExE is independent of endogenous Furin and Pace4 until E7.5 (Supplemental Fig. S1B). DKO embryos also maintained expression of Elf5::FurinGFP and Elf5::Pace4GFP (Fig. 3A). Each of these transgenes was sufficient to restore induction of Nodal and Cripto, and of the Nodal target genes Lefty2, Fgf8, and Brachyury (Fig. 3B). Although the shape of the embryos and patterning of the VE were not fully rescued (see below), these results strongly suggest that both ExE-derived Furin and Pace4 sufficiently activate Nodal to rescue autoinduction and mesoderm formation.
Figure 3.
Secretion of Pace4GFP or FurinGFP by the microenvironment sufficiently activates Nodal in the epiblast to induce mesoderm. (A) Whole-mount in situ hybridization of a GFP antisense RNA probe reveals expression of the Elf5::FurinGFP or Elf5::Pace4GFP BAC transgenes in DKO embryos and in control litter mates at E7.5. (B) Markers of Nodal signaling and mesoderm formation in E7.5 DKO and control embryos carrying the Elf5::FurinGFP or Elf5::Pace4GFP transgene. Bar, 50 μm.
Expression of Pace4 or Furin in the ExE suppresses neural differentiation
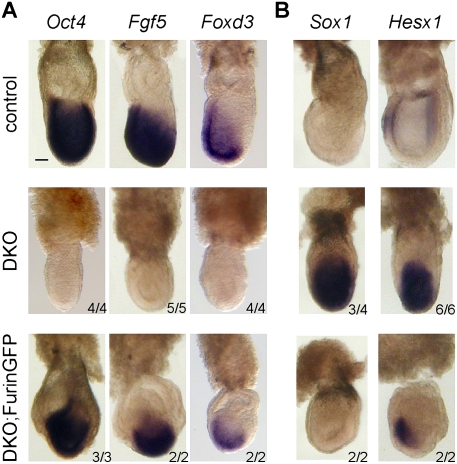
In mutant embryos lacking Nodal, epiblast cells prematurely differentiate into neurectoderm (Camus et al. 2006; Mesnard et al. 2006). Therefore, we expected a similar phenotype in DKO embryos. Indeed, the neurectoderm markers Sox1 and Hesx1 were ectopically up-regulated throughout the epiblast of E7.5 DKO embryos at the expense of the pluripotency markers Oct4, Foxd3, and Fgf5 (Fig. 4). In contrast, DKO embryos carrying the FurinGFP BAC transgene maintained high expression levels of Oct4, Fgf5, and Foxd3, and the distribution of Sox1 and Hesx1 mRNAs resembled that of the wild type (Fig. 4). Analogous results were obtained for DKO;Pace4GFP embryos (Supplemental Fig. S4). These data establish that expression of Furin or Pace4 in the ExE is sufficient to maintain pluripotency and prevent precocious neural differentiation until E7.5, consistent with a non-cell-autonomous role in Nodal processing after implantation.
Figure 4.
Expression of FurinGFP in the ExE suppresses precocious differentiation of pluripotent epiblast cells toward neurectoderm. Analysis of the indicated markers of pluripotency (A) and neurectoderm differentiation (B) in E7.25–E7.5 DKO mutants in the absence or presence of the Elf5::FurinGFP transgene. Bar, 50 μm.
Pace4, but not Furin, provided by the ExE induces EmVE and endoderm formation
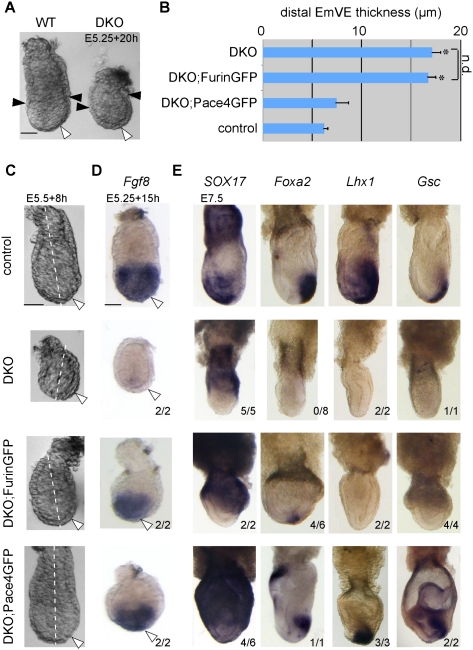
Besides its role in pluripotent cells, processed Nodal also acts on the microenvironment (Brennan et al. 2001; Norris et al. 2002; Ben-Haim et al. 2006), especially to induce EmVE and its differentiation from a columnar to a squamous epithelium (Mesnard et al. 2006). This early Nodal function is independent of zygotic Furin and Pace4, because of a compensatory PC activity in the uterus (Mesnard and Constam 2010). In contrast, when isolated from the uterus at E5.25 and cultured in vitro, DKO embryos were recognized within 8 h by a 2.8-fold increase in the thickness of EmVE, from 6 μm ± 1 μm (n = 8) to 17 μm ± 2 μm (n = 6), and by a shortened proximo–distal axis characteristic of impaired Nodal signaling (Fig. 5A–C). In addition, they failed to up-regulate Fgf8 in the epiblast and in the EmVE (Fig. 5D). Introducing the Elf5::Pace4GFP transgene was sufficient to rescue the flattening of EmVE (7 μm ± 2 μm, n = 3) and expression of Fgf8. In contrast, if FurinGFP was present instead, the EmVE remained abnormally thick (16 μm ± 2 μm, n = 3), and Fgf8 expression was only rescued in the epiblast (Fig. 5B–D). These data show that ExE-derived Pace4GFP, but not FurinGFP, can pattern the VE.
Figure 5.
Pace4, but not Furin, provided by the ExE induces EmVE and DE. (A) Morphology of DKO embryos cultured at E5.25 for 20 h. The EmVE (white arrows) and its proximal boundary (black arrows) are indicated. Asterisks indicate significant difference as determined by t-test (P < 0.05). “n.d.” indicates no significant difference as determined by t-test (P < 0.05). Error bars represent standard error of the mean. (B) Quantification of the thickness of EmVE in DKO cultured in the presence or absence of the Elf5::Pace4GFP or Elf5::FurinGFP transgenes. (C) Sites of measurement were at the intersection of the EmVE and the proximo–distal axis (white dotted line). (C,D) Morphology (C) and Fgf8 mRNA whole-mount in situ hybridization analysis (D) of E5.5 wild-type (WT) embryos, DKO embryos, and DKO embryos carrying an Elf5::Pace4GFP or Elf5::FurinGFP transgene after in vitro culture. (E) In situ hybridization for markers of DE in DKO, DKO;FurinGFP, and DKO;Pace4GFP embryos at E7.5. Bar, 50 μm.
During gastrulation, the EmVE intercalates with nascent DE cells (Kwon et al. 2008) recruited from the epiblast by Nodal signaling (Conlon et al. 1994; Brennan et al. 2001). DE marked by Sox17β at E7.5 was absent in DKO and DKO;FurinGFP embryos, but significantly rescued by Pace4GFP. Pace4GFP also restored expression of the high-threshold Nodal target genes Foxa2, Gsc, and Lhx1 in axial mesoderm, whereas FurinGFP had little or no effect on these markers (Fig. 5E). Overall, these data demonstrate that ExE-derived Pace4 can support EmVE, DE, and mesoderm formation, whereas ExE-derived Furin only stimulates the induction of mesoderm.
Discussion
Insight into the function of secreted proteases generally is limited by a lack of reliable information as to when and where they are active and colocalizing with substrates. Here, we showed that functional GFP fusions of Furin and Pace4 secreted by the ExE can cleave the membrane-anchored fluorescent reporter substrate CLIP far away from their source and rescue various aspects of Nodal signaling in the epiblast. Our findings also reveal unexpected differences in the dispersal of these two proteases that likely account for distinct roles in regulating the fate of progenitor cells.
The spatiotemporal distribution of FurinGFP in the epiblast differs from that of Pace4GFP
Expression of FurinGFP or Pace4GFP specifically in the ExE revealed that both proteases are secreted into the embryonic region, although with distinct spatiotemporal profiles. FurinGFP accumulated in the proamniotic cavity, where it reached maximal levels at E6.5 during gastrulation. In contrast, outside the ExE, Pace4GFP was only observed between the epiblast and EmVE around E5.5. This striking difference indicates that ExE cells dispatch Furin and Pace4 by distinct routes. Polarized epithelial cells sort Furin to the basolateral membrane when its cytosolic tail binds the adaptor protein AP-4 (Simmen et al. 1999, 2002), although it can also be found apically (Mayer et al. 2004). In particular, a truncated form of Furin lacking the transmembrane domain and cytosolic tail is secreted from the apical side in MDCK cells (Fasciotto et al. 2008). Here, expression of the full-length protein in polarized MDCK cells revealed a similar apical enrichment of shed Furin, whereas Pace4 was preferentially secreted from the basolateral side. It is likely, therefore, that distinct polarized secretion also accounts for the differential distribution of extracellular Furin and Pace4 in the embryo. Unlike Furin, Pace4 contains a cysteine-rich repeat region that mediates high-affinity binding to HSPGs (Tsuji et al. 2003; Nour et al. 2005; Mayer et al. 2008). HSPGs in general are sorted basolaterally (Mertens et al. 1996; Tveit et al. 2005). It is tempting to speculate, therefore, that HSPGs sort Pace4 to a basolateral secretory route. Similarly, the HSPGs dally and dally-like in Drosophila are thought to deliver associated morphogens such as Dpp, Hh, and Wnt to the basal side (Constam 2009; Yan and Lin 2009). HSPGs may also recruit Pace4 to target cells, although alternative candidate receptors in receiving cells include EGF-CFC coreceptors of Nodal (Blanchet et al. 2008).
CLIP imaging reveals non-cell-autonomous activities of ExE-derived Furin and Pace4
Our conclusion that ExE-derived FurinGFP and Pace4GFP directly act on cells in the epiblast was independently confirmed by live imaging of the transgenic reporter substrate CLIP. Imaging of CLIP revealed strikingly different spatiotemporal activity profiles of these proteases. While Pace4GFP cleaved CLIP uniformly throughout the epiblast already at E5.5, FurinGFP activity initially was graded and limited to the proximal region. During the following day, processing of CLIP by Furin was still detected, whereas the activity of Pace4 subsided below detectable levels. Thus, the non-cell-autonomous activities of these proteases correlated well with their relative abundance outside the ExE as observed by GFP fluorescence.
ExE-derived Furin is less potent than Pace4 in activating Nodal signaling
Consistent with their predicted non-cell-autonomous function, expression of FurinGFP or Pace4GFP in the ExE also rescued Nodal signaling in Furin−/−;Pace4−/− double mutants. Interestingly, however, while both proteases stimulated mesoderm formation, only Pace4 also induced DE and EmVE. A comparison of GFP fluorescence intensities or protein levels indicated that FurinGFP and Pace4GFP were expressed at comparable levels. Thus, a likely explanation is that Furin provided by the ExE alone is less potent than Pace4 in generating the elevated Nodal signaling threshold that distinguishes endoderm from mesoderm (Vincent et al. 2003; Schier 2009) because of lower enzymatic activity, differences in trafficking, or both. In keeping with this interpretation, Nodal is more potently activated by Pace4 than by Furin in cell-free assays and in transfected cells (Beck et al. 2002; Blanchet et al. 2008), and knockdown of endogenous Pace4 but not Furin diminished processing of a Nodal-related precursor in Xenopus (Birsoy et al. 2005). Possibly, Furin is less efficient because it does not bind HSPGs, a property that may be required to generate an early high-threshold Nodal signal.
In the mouse, a prolonged Nodal signal after rapid autoinduction is sustained by delayed feedback mediated by Bmp4 through induction of Wnt3, which in turn maintains Nodal expression during gastrulation independently of Nodal precursor processing (Ben-Haim et al. 2006). Since Pace4GFP and FurinGFP equally rescued Wnt3 expression in DKO mutants (Supplemental Fig. S5), we speculate that their potencies differ during an earlier phase of Nodal autoinduction. Consistent with this interpretation, the fluorescence of secreted FurinGFP peaked at E6.5, whereas Pace4GFP was only detected in the epiblast compartment at E5.5, but not beyond.
Unlike FurinGFP expression in the ExE, endogenous Furin is sufficient to induce endoderm in the absence of Pace4 (Constam and Robertson 2000). A likely explanation of this striking difference is that Furin endogenously relies on its complementary transient expression in the VE to support endoderm formation (Fig. 6). In keeping with this interpretation, Furin can activate Nodal independently of the ExE in epiblast explants if they are cultured at E5.5 with intact VE (Mesnard et al. 2006). Expression of Furin in the EmVE may also be important to efficiently convert proximal–distal patterning in the epiblast into antero–posterior asymmetry, since this process was never completely rescued in DKO embryos by either Pace4GFP or FurinGFP. However, incubation with conditioned medium of SNH fibroblasts containing shed Furin or Pace4 alone was not sufficient to maintain lacZ expression in isolated NodallacZ/+ epiblast explants (D. Mesnard., unpubl.), indicating that both VE and ExE support Nodal signaling in part by additional factors or indirectly by maintaining the epiblast architecture.
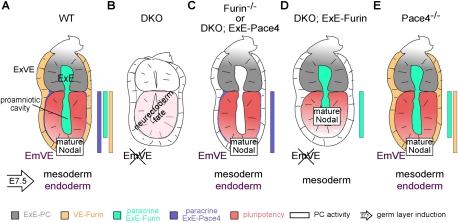
Figure 6.
Diagram summarizing the distribution and function of paracrine Furin and Pace4 activities in implanted mouse embryos. (A) Furin is released by the ExE (gray) into the proamniotic cavity (green) at E5.5–E6.5. At E5.5, Furin is also produced in the EmVE (yellow). In contrast, ExE-derived Pace4 (blue) is enriched at the interface between the epiblast and EmVE. Colored rectangles depict the contributions of these proteases to CLIP processing in the epiblast. (B) In the absence of secreted PC activity, EmVE fails to be induced and epiblast cells lose pluripotency and differentiate into neurectoderm, reminiscent of a Nodal-null phenotype. (C,D) ExE-derived Pace4 (C) and Furin (D) each sufficiently process Nodal to maintain pluripotency (red) and to induce mesoderm. In contrast, only ExE-derived Pace4 supports induction of EmVE and DE (cf. C and D). (E) Endogenous Furin, however, is also sufficient to induce EmVE, indicating that Furin from the EmVE and ExE complement each other to activate Nodal.
Differences between Pace4 and Furin point to a role for EmVE in DE formation
Our data provide the first direct evidence that paracrine Furin and Pace4 directly act on the epiblast. Based on their distinct activities and spatiotemporal distribution, we propose that they complement each other to regulate the strength and the duration of Nodal signaling (Fig. 6). In this model, the timely onset of PC action is particularly important to reach the high signaling threshold required for DE formation. In addition, the timing of PC action may be critical because of the role of Nodal in the EmVE. Until recently, EmVE was thought to be displaced by a contiguous layer of DE emanating from the primitive streak. However, genetic lineage tracing revealed that DE cells instead rapidly intercalate with the EmVE, suggesting a more intricate relationship (Kwon et al. 2008). Indeed, while both DE and EmVE are specified by Nodal, deletion of Smad2 in extraembryonic tissues is sufficient to abrogate either of these fates (Waldrip et al. 1998). In contrast, embryos lacking Smad2 specifically in the epiblast only lack the most anterior DE (Vincent et al. 2003), indicating that Nodal/Smad2 signaling stimulates DE formation at least in part through its effect on EmVE. Possibly, FurinGFP failed to induce DE simply because it is less potent than Pace4GFP in stimulating EmVE formation. It will be important to investigate how terminal differentiation of DE progenitors might be linked to specific interactions with the EmVE.
Neural differentiation of pluripotent epiblast cells is suppressed by Furin and Pace4 from the microenvironment
Our study also revealed a role for ExE-derived Furin and Pace4 in regulating neural differentiation. Neural differentiation of pluripotent progenitors in the epiblast is suppressed by Nodal (Brennan et al. 2001; Camus et al. 2006; Mesnard et al. 2006). In keeping with this notion, pharmacological inhibition of Nodal/Activin/TGFβ receptors blocks self-renewal and triggers neural differentiation in cultured mouse epiblast stem cells (Brons et al. 2007). Here, we found that embryos lacking Pace4 and Furin display a similar phenotype, since neurectodermal genes were broadly up-regulated at the expense of pluripotency markers. Importantly, this defect was fully suppressed by ExE-derived FurinGFP or Pace4GFP, concurring with a rescue of Nodal signaling. This finding further highlights the importance of Furin and Pace4 in the microenvironment for coordinating cell fate decisions in the epiblast.
Precocious neural differentiation also occurs in Bmpr1a mutant epiblasts (Di-Gregorio et al. 2007). This has been attributed in part to premature down-regulation of Nodal at E5.5, because suppression of BMP signaling alone is not sufficient to induce neural fates as long as Nodal remains active, at least in Xenopus (Chang and Harland 2007). Accordingly, neural differentiation in DKO embryos is not simply a consequence of impaired Bmp signaling. In keeping with this conclusion, the most important Bmpr1a ligand, Bmp4, can clearly mature independently of zygotic Furin and Pace4 (Beck et al. 2002; Nelsen and Christian 2009), and DKO embryos retain Nodal expression at E5.5, at least on the present outbred genetic background (Mesnard and Constam 2010).
Conclusion
Overall, our study proves that secreted Furin and Pace4 from the microenvironment directly reach the epiblast and modulate the fate of progenitor cells. Unexpectedly, we also found that Furin and Pace4 adopt distinct delivery routes, correlating with differences in their spatiotemporal activity profiles, monitored here with the fluorescent biosensor CLIP, and at the level of Nodal signaling. To our knowledge, imaging of paracrine activities of endoproteases and of their regulation in vivo has not been achieved previously. Our results provide the first direct evidence that secreted Furin and Pace4 can act over several cell diameters. As this considerably enlarges the spectrum of possible functions, it will be important to decipher in future studies the machinery regulating the distinct secretion routes of these versatile endoproteases and their interactions with specific substrates.
Materials and methods
Plasmids and expression vectors
FurinGFP and Pace4GFP expression vectors were generated by inserting eGFP after the P-domain or at the C terminus of Flag-tagged Furin or Pace4 cDNAs in pCR3.1 (Invitrogen) and pEGFP-N2 (Clontech) vectors, respectively. The Flag epitopes were introduced immediately after the zymogenic cleavage motifs by PCR (Constam and Robertson 1999). For Furin-GFP, the EGFP CDS was amplified by PCR and inserted in-frame between the residues G594 and C595 of Flag-tagged Furin. For Pace4-GFP, the full-length Flag-tagged Pace4 CDS was ligated in frame between the Sal1 and BamH1 restriction sites of pEGFP-N2 (Clontech). For direct comparison with FlagPace4 lacking a GFP tag, Pace4-GFP was subcloned into the pCS2+ plasmid.
The lentiviral Flag-Furin vector was generated by inserting Flag-Furin into Xho1–Mlu1 sites of pEF IRES-P (Hobbs et al. 1998). The resulting hEF1α-Furin cassette was excised using Age1 and Xba1 and inserted into Age1–Xba1 sites of pLenti hEF1α-MCS//SV40-PuroR (Fuerer and Nusse 2010). Flag-Pace4 (Constam and Robertson 1999) was directly inserted into Xho1–BamH1 sites of pLenti hEF1α-MCS//SV40-PuroR.
Lentivirus production and infection
Lentiviruses were produced as described (Fuerer and Nusse 2010) by cotransfecting HEK293T with lentiviral vectors and the helper plasmids pCMVΔR8.74 and pMD2.VSVG (Dull et al. 1998). MDCK cells were infected with less than one virus per cell and selected with 5 μg/mL puromycin.
Cell transfection and Western blot analysis
COS-1, HEK293T, and MDCK cells were cultured in Dulbecco's modified Eagle's medium containing 10% fetal calf serum, 100 μg/mL gentamycine, and 2 mM glutamine. For transient transfection, cells were plated on 24-well plates at a density of 5 × 104 cells per well. The following day, the cells were incubated with plasmids (0.1–1.5 μg per well) in Lipofectamine 2000 CD reagent (Invitrogen) during 4–6 h. Thereafter, the medium was replaced by OptiMEM (Invitrogen) containing 0.25% knockout serum replacement (Invitrogen). After 24 h, conditioned medium was harvested and cells were lysed in Laemmli buffer containing 5% β-mercaptoethanol. Expression and processing of wild-type and GFP-tagged Furin and Pace4 was monitored by chemiluminescent immunoblotting of the Flag tag using monoclonal anti-Flag M2 antibody (Sigma) and HRP-conjugated anti-mouse secondary antibody (Amersham).
For transwell assays, infected MDCK cells were seeded in a six-well transwell plate with 0.4-μm pores (Corning). Four days later, the upper and lower chambers were supplied with fresh medium (1.5 or 2.6 mL, respectively). Medium was conditioned for 4 d and cleared by centrifugation. Flag-tagged proteins were immunoprecipitated using M2 agarose beads (Sigma) and analyzed by SDS-PAGE. The tightness of the MDCK monolayer was confirmed by dye exclusion using phenol red measured at OD560 after addition of 1/10 NaOH 2.5N. Proteins were detected with the M2 antibody (Sigma).
BAC transgenes
Elf5::FurinGFP BAC and Elf5::Pace4GFP BAC were created by ET-based homologous recombination following published protocols (Muyrers et al. 1999; Liu et al. 2003). Recombination building vectors were assembled by inserting Furin-GFP or Pace4-GFP upstream of an SV40 PolyA-frt-Kan-frt region, and by flanking this cassette by 83- and 50-base-pair (bp) (for Furin) or 307- and 170-bp (for Pace4) Elf5 homology boxes. Recombination with mouse Elf5 BAC RP23-44A14 (C57Bl/6J strain; CHORI) resulted in the insertion of the flanked region 6 bp downstream from the splice acceptor junction of Elf5 Exon-2, while removing 62 bp comprising the Elf5 start codon. Constructs were verified by restriction mapping and sequencing of the modified regions. BAC DNAs were purified by CsCl density gradient centrifugation, precipitated twice in EtOH, resuspended, and dialyzed in 8 mM Tris.HCl (pH 7.5) containing 0.1 mM EDTA.
Mice
Transgenic mice were generated at the EPFL core facility by pronuclear injection of the ELF5::FurinGFP or ELF5::Pace4GFP BAC transgenes at a concentration of 2 ng/μL into zygotes of FVB and NRMI mice (Harlan and Janvier). Three and 10 founders, respectively, and their offspring were genotyped by 31 PCR amplification cycles at the annealing temperature of 63°C using the primer pairs 5′-ACCACATGACTACTCTGCTGATGGG-3′ and 5′-CTGCTTTCTGGAGGTGTAGAGAG C-3′ for FurinGFP, and 5′-GTGTCGAAGATGTGAGGAGAACTG CC-3′ and 5′-GCTGAACTTGTGGCCGTTTACGTCG-3′ for Pace4GFP. Fluorescence of the transgenes was monitored in embryos at the stages indicated. Furin+/−;Pace4+/− double heterozygotes on a C57Bl/6 inbred background (Beck et al. 2002) were serially backcrossed to an NMRI outbred background for more than eight generations.
Whole-embryo culture
Whole-embryo in vitro culture was performed as described (Beck et al. 2002) but in the absence of STO fibroblasts. In brief, embryos were recovered 5.25 d post-coitum. Reichert's membrane was mechanically removed using fine needles. Dissected embryos were transferred to Millipore filter inserts (pore size 12 μm) and incubated in OptiMEM I supplemented with 15% knockout serum replacement, 1% gentamycin, and glutamin sulfate in a water-saturated atmosphere containing 5% CO2. Protein expression was monitored at E6.5 by anti-GFP (Roche) Western blot analysis of pools of nine embryos of each genotype lysed in Laemmli buffer containing 1 mM DTT.
Whole-mount in situ hybridization
Whole-mount in situ hybridization was performed as described using DIG-labeled antisense probes (Varlet et al. 1997; Beck et al. 2002). The GFP antisense probe comprised the entire coding sequence of EGFP. Anti-DIG antibodies conjugated to alkaline phosphatase and the substrate BM purple were from Roche Diagnostics. Color reactions were developed until saturation at room temperature.
Imaging
Images were acquired on a Leica TCS SP II confocal microscope at the following settings: GFP channel: 458 nm excitation, 466–506 nm emission; CFP channel: 458 nm excitation, 462–510 nm emission; YFP channel: 514 nm excitation, 518/580 emission. Scanning was done at 400 Hz with a PlanApo 63× 1.3 NA glycerol objective. Heat maps of C/Y ratios were generated by Imaris 7.1.1 software using a Matlab script. In brief, a mask was created for YFP signals above a critical threshold to highlight cell surface staining. Each CFP/YFP ratio was then attributed a proportional value between 1 (CFP/YFP = 0; blue) and 255 (CFP/YFP = 2; red). Values above 2 were considered as artifactual (white). Areas not included in the mask were attributed a black color. To quantify CFP/YFP fluorescence ratios on embryos, using ImageJ 1.43g, a mask was extracted from the YFP channel by including pixels with a value between 2000 and 4000 on the 12-bit grayscale image. The mask was then applied on the CFP and YFP channels to integrate the total CFP and YFP signals, respectively, and determine the CFP/YFP ratio (Supplemental Fig. S2).
Acknowledgments
We thank Friedrich Beerman and the staff of the EPFL School of Life Sciences animal core facility for their support in deriving and maintaining transgenic mice, and the microscopy core facility (BiOP) for advice on image acquisition and analysis. We also thank Stephen Frankenberg for useful comments on the manuscript, and the Swiss National Science Foundation for their financial support (grants 3100A0-105583/1 and 3100A0-118080/1 to D.B.C.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.16738711.
References
- Anderson ED, Molloy SS, Jean F, Fei H, Shimamura S, Thomas G 2002. The ordered and compartment-specfific autoproteolytic removal of the furin intramolecular chaperone is required for enzyme activation. J Biol Chem 277: 12879–12890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold SJ, Robertson EJ 2009. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat Rev Mol Cell Biol 10: 91–103 [DOI] [PubMed] [Google Scholar]
- Beck S, Le Good JA, Guzman M, Ben Haim N, Roy K, Beermann F, Constam DB 2002. Extraembryonic proteases regulate Nodal signalling during gastrulation. Nat Cell Biol 4: 981–985 [DOI] [PubMed] [Google Scholar]
- Ben-Haim N, Lu C, Guzman-Ayala M, Pescatore L, Mesnard D, Bischofberger M, Naef F, Robertson EJ, Constam DB 2006. The nodal precursor acting via activin receptors induces mesoderm by maintaining a source of its convertases and BMP4. Dev Cell 11: 313–323 [DOI] [PubMed] [Google Scholar]
- Birsoy B, Berg L, Williams PH, Smith JC, Wylie CC, Christian JL, Heasman J 2005. XPACE4 is a localized pro-protein convertase required for mesoderm induction and the cleavage of specific TGFβ proteins in Xenopus development. Development 132: 591–602 [DOI] [PubMed] [Google Scholar]
- Blanchet MH, Le Good JA, Mesnard D, Oorschot V, Baflast S, Minchiotti G, Klumperman J, Constam DB 2008. Cripto recruits Furin and PACE4 and controls Nodal trafficking during proteolytic maturation. EMBO J 27: 2580–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan J, Lu CC, Norris DP, Rodriguez TA, Beddington RS, Robertson EJ 2001. Nodal signalling in the epiblast patterns the early mouse embryo. Nature 411: 965–969 [DOI] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, et al. 2007. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448: 191–195 [DOI] [PubMed] [Google Scholar]
- Camus A, Perea-Gomez A, Moreau A, Collignon J 2006. Absence of Nodal signaling promotes precocious neural differentiation in the mouse embryo. Dev Biol 295: 743–755 [DOI] [PubMed] [Google Scholar]
- Chang C, Harland RM 2007. Neural induction requires continued suppression of both Smad1 and Smad2 signals during gastrulation. Development 134: 3861–3872 [DOI] [PubMed] [Google Scholar]
- Chen Y, Schier AF 2001. The zebrafish Nodal signal Squint functions as a morphogen. Nature 411: 607–610 [DOI] [PubMed] [Google Scholar]
- Conlon FL, Lyons KM, Takaesu N, Barth KS, Kispert A, Herrmann B, Robertson EJ 1994. A primary requirement for nodal in the formation and maintenance of the primitive streak in the mouse. Development 120: 1919–1928 [DOI] [PubMed] [Google Scholar]
- Constam DB 2009. Intracellular trafficking and signaling in development. F1000 Biol Rep 1: 59 doi: 10.3410/B1-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constam DB, Robertson EJ 1999. Regulation of bone morphogenetic protein activities by pro domains and proprotein convertases. J Cell Biol 144: 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constam DB, Robertson EJ 2000. SPC4/PACE4 regulates a TGFβ signaling network during axis formation. Genes Dev 14: 1146–1155 [PMC free article] [PubMed] [Google Scholar]
- Denault J, Bissonnette L, Longpre J, Charest G, Lavigne P, Leduc R 2002. Ectodomain shedding of furin: kinetics and role of the cysteine-rich region. FEBS Lett 527: 309–314 [DOI] [PubMed] [Google Scholar]
- Di-Gregorio A, Sancho M, Stuckey DW, Crompton LA, Godwin J, Mishina Y, Rodriguez TA 2007. BMP signalling inhibits premature neural differentiation in the mouse embryo. Development 134: 3359–3369 [DOI] [PubMed] [Google Scholar]
- Donnison M, Beaton A, Davey HW, Broadhurst R, L'Huillier P, Pfeffer PL 2005. Loss of the extraembryonic ectoderm in Elf5 mutants leads to defects in embryonic patterning. Development 132: 2299–2308 [DOI] [PubMed] [Google Scholar]
- Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, Naldini L 1998. A third-generation lentivirus vector with a conditional packaging system. J Virol 72: 8463–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasciotto BH, Kuhn U, Cohn DV, Gorr SU 2008. Secretory cargo composition affects polarized secretion in MDCK epithelial cells. Mol Cell Biochem 310: 67–75 [DOI] [PubMed] [Google Scholar]
- Fuerer C, Nusse R 2010. Lentiviral vectors to probe and manipulate the Wnt signaling pathway. PLoS ONE 5: e9370 doi: 10.1371/journal.pone.0009370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granier C, Gurchenkov V, Perea-Gomez A, Camus A, Ott S, Papanayotou C, Iranzo J, Moreau A, Reid J, Koentges G, et al. 2011. Nodal cis-regulatory elements reveal epiblast and primitive endoderm heterogeneity in the peri-implantation mouse embryo. Dev Biol 349: 350–362 [DOI] [PubMed] [Google Scholar]
- Hagos EG, Dougan ST 2007. Time-dependent patterning of the mesoderm and endoderm by Nodal signals in zebrafish. BMC Dev Biol 7: 22 doi: 10.1186/1471-213X-7-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs S, Jitrapakdee S, Wallace JC 1998. Development of a bicistronic vector driven by the human polypeptide chain elongation factor 1α promoter for creation of stable mammalian cell lines that express very high levels of recombinant proteins. Biochem Biophys Res Commun 252: 368–372 [DOI] [PubMed] [Google Scholar]
- Kwon GS, Viotti M, Hadjantonakis AK 2008. The endoderm of the mouse embryo arises by dynamic widespread intercalation of embryonic and extraembryonic lineages. Dev Cell 15: 509–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, Copeland NG 2003. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res 13: 476–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer G, Boileau G, Bendayan M 2004. Sorting of furin in polarized epithelial and endothelial cells: expression beyond the Golgi apparatus. J Histochem Cytochem 52: 567–579 [DOI] [PubMed] [Google Scholar]
- Mayer G, Hamelin J, Asselin MC, Pasquato A, Marcinkiewicz E, Tang M, Tabibzadeh S, Seidah NG 2008. The regulated cell surface zymogen activation of the proprotein convertase PC5A directs the processing of its secretory substrates. J Biol Chem 283: 2373–2384 [DOI] [PubMed] [Google Scholar]
- Mertens G, Van der Schueren B, van den Berghe H, David G 1996. Heparan sulfate expression in polarized epithelial cells: the apical sorting of glypican (GPI-anchored proteoglycan) is inversely related to its heparan sulfate content. J Cell Biol 132: 487–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesnard D, Constam DB 2010. Imaging proprotein convertase activities and their regulation in the implanting mouse blastocyst. J Cell Biol 191: 129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesnard D, Guzman-Ayala M, Constam DB 2006. Nodal specifies embryonic visceral endoderm and sustains pluripotent cells in the epiblast before overt axial patterning. Development 133: 2497–2505 [DOI] [PubMed] [Google Scholar]
- Muyrers JP, Zhang Y, Testa G, Stewart AF 1999. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res 27: 1555–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelsen SM, Christian JL 2009. Site-specific cleavage of BMP4 by furin, PC6 and PC7. J Biol Chem 284: 27157–27166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris DP, Brennan J, Bikoff EK, Robertson EJ 2002. The Foxh1-dependent autoregulatory enhancer controls the level of Nodal signals in the mouse embryo. Development 129: 3455–3468 [DOI] [PubMed] [Google Scholar]
- Nour N, Mayer G, Mort JS, Salvas A, Mbikay M, Morrison CJ, Overall CM, Seidah NG 2005. The cysteine-rich domain of the secreted proprotein convertases PC5A and PACE4 functions as a cell surface anchor and interacts with tissue inhibitors of metalloproteinases. Mol Biol Cell 16: 5215–5226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paleyanda RK, Drews R, Lee TK, Lubon H 1997. Secretion of human furin into mouse milk. J Biol Chem 272: 15270–15274 [DOI] [PubMed] [Google Scholar]
- Perea-Gomez A, Vella FD, Shawlot W, Oulad-Abdelghani M, Chazaud C, Meno C, Pfister V, Chen L, Robertson E, Hamada H, et al. 2002. Nodal antagonists in the anterior visceral endoderm prevent the formation of multiple primitive streaks. Dev Cell 3: 745–756 [DOI] [PubMed] [Google Scholar]
- Schier AF 2009. Nodal morphogens. Cold Spring Harb Perspect Biol 1: a003459 doi: 10.1101/cshperspect.a003459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidah NG, Mayer G, Zaid A, Rousselet E, Nassoury N, Poirier S, Essalmani R, Prat A 2008. The activation and physiological functions of the proprotein convertases. Int J Biochem Cell Biol 40: 1111–1125 [DOI] [PubMed] [Google Scholar]
- Simmen T, Nobile M, Bonifacino JS, Hunziker W 1999. Basolateral sorting of furin in MDCK cells requires a phenylalanine-isoleucine motif together with an acidic amino acid cluster. Mol Cell Biol 19: 3136–3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmen T, Honing S, Icking A, Tikkanen R, Hunziker W 2002. AP-4 binds basolateral signals and participates in basolateral sorting in epithelial MDCK cells. Nat Cell Biol 4: 154–159 [DOI] [PubMed] [Google Scholar]
- Thimon V, Belghazi M, Dacheux JL, Gatti JL 2006. Analysis of furin ectodomain shedding in epididymal fluid of mammals: demonstration that shedding of furin occurs in vivo. Reproduction 132: 899–908 [DOI] [PubMed] [Google Scholar]
- Thomas G 2002. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat Rev Mol Cell Biol 3: 753–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji A, Sakurai K, Kiyokage E, Yamazaki T, Koide S, Toida K, Ishimura K, Matsuda Y 2003. Secretory proprotein convertases PACE4 and PC6A are heparin-binding proteins which are localized in the extracellular matrix. Potential role of PACE4 in the activation of proproteins in the extracellular matrix. Biochim Biophys Acta 1645: 95–104 [DOI] [PubMed] [Google Scholar]
- Tveit H, Dick G, Skibeli V, Prydz K 2005. A proteoglycan undergoes different modifications en route to the apical and basolateral surfaces of Madin-Darby canine kidney cells. J Biol Chem 280: 29596–29603 [DOI] [PubMed] [Google Scholar]
- Varlet I, Collignon J, Robertson EJ 1997. nodal expression in the primitive endoderm is required for specification of the anterior axis during mouse gastrulation. Development 124: 1033–1044 [DOI] [PubMed] [Google Scholar]
- Vidricaire G, Denault JB, Leduc R 1993. Characterization of a secreted form of human furin endoprotease. Biochem Biophys Res Commun 195: 1011–1018 [DOI] [PubMed] [Google Scholar]
- Vincent SD, Dunn NR, Hayashi S, Norris DP, Robertson EJ 2003. Cell fate decisions within the mouse organizer are governed by graded Nodal signals. Genes Dev 17: 1646–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldrip WR, Bikoff EK, Hoodless PA, Wrana JL, Robertson EJ 1998. Smad2 signaling in extraembryonic tissues determines anterior–posterior polarity of the early mouse embryo. Cell 92: 797–808 [DOI] [PubMed] [Google Scholar]
- Wise RJ, Barr PJ, Wong PA, Kiefer MC, Brake AJ, Kaufman RJ 1990. Expression of a human proprotein processing enzyme: correct cleavage of the von Willebrand factor precursor at a paired basic amino acid site. Proc Natl Acad Sci 87: 9378–9382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Lin X 2009. Shaping morphogen gradients by proteoglycans. Cold Spring Harb Perspect Biol 1: a002493 doi: 10.1101/cshperspect.a002493 [DOI] [PMC free article] [PubMed] [Google Scholar]