Abstract
We present here a new method to enhance the detection of secreted cytokines and chemokines from single human mononuclear cells. The technique uses a hybridization chain reaction (HCR) to amplify signals resulting from sandwich immunoassays. This immuno-HCR employs oligonucleotide-based initiators covalently linked to antibodies to propagate a chain reaction of hybridization events involving a pair of complementary hairpin oligomers bearing fluorescent labels. Integrating this strategy for signal amplification with microengraving—a soft lithographic method for printing arrays of secreted proteins from thousands of single cells—improves both the limits of detection and sensitivity for cytokines and chemokines captured from individual cells by an average of 200-fold relative to methods for direct detection by fluoresence. This approach should enhance the utility of microengraving for defining the immunological signatures of diseases and responses to interventional therapies based on multiplexed single-cell analysis.
Keywords: Hybridization chain reaction, microengraving, single-cell analysis, peripheral blood mononuclear cells
Assessing the quality and evolution of an immune response to a pathological condition often relies on scoring the numbers of cells responding to a defined stimulus. In particular, understanding the types of proteins secreted by single cells is important for evaluating the breadth and nature of an immune response.1 Any individual cell, however, only secretes small quantities of molecules, such as antibodies, cytokines and chemokines. Sensitive assays are required, therefore, to detect them accurately. Here, we describe a new method that adapts an isothermal, enzyme-free, hybridization chain reaction (HCR) of DNA to enhance the sensitivity and accuracy of signals detected from sandwich immunoassays for analytes captured from single viable cells.
The most common method to detect secreted proteins from single cells is the enzyme-linked immunosorbent spot (ELISPOT) assay.2 This assay relies on the capture of secreted proteins from cells that rest on top of analyte-specific antibodies. The analytes are then detected by a second antibody in combination with either an enzyme-based amplification of the signal or a direct measure by fluorescence. This method typically requires long incubation times (12–24h) to capture sufficient analytes for detection, however. Adaptations of this approach that use microsystems to confine individual cells spatially have also used antibodies conjugated directly with fluorescent labels to detect analytes in a microarray-based format for both convenience and multiplexed analysis.3–5
One strategy to amplify signals associated with specific proteins captured on microarrays is rolling circle amplification (RCA). This method uses an isothermal polymerase and a circular primer to extend an oligomer that is attached to the detection antibody used for a particular antigen.6, 7 The extended DNA can then hybridize with multiple oligonucleotides bearing fluorescence probes. The degree of labeling exceeds that for an antibody directly conjugated with fluorescent labels, and therefore, improves the limit of detection. One disadvantage of RCA, however, is that it depends on inefficient enzymes, and the steps required to prepare circular oligomer templates are time consuming and costly.
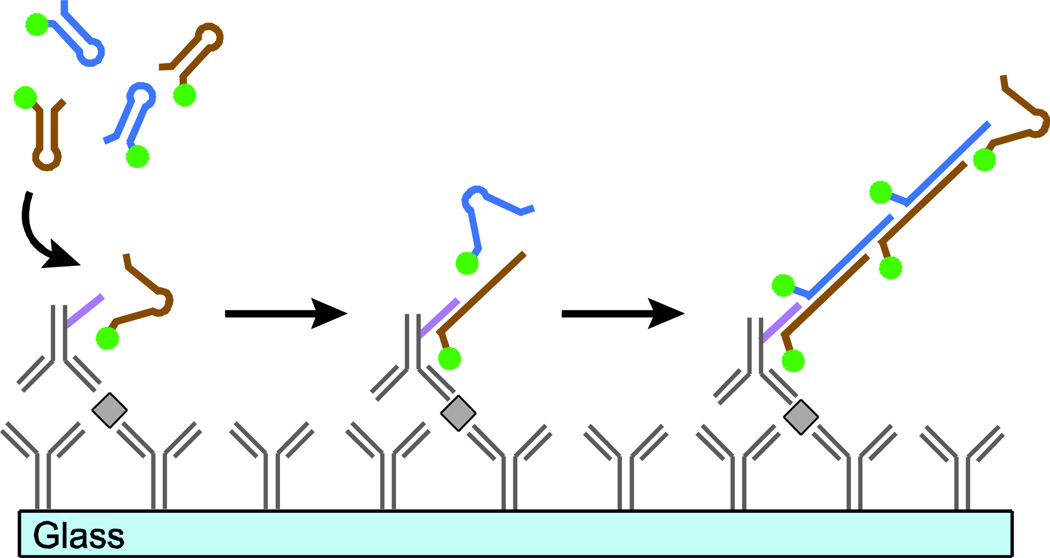
An approach for amplifying short sequences of oligonucleotides called hybridization chain reaction (HCR) was recently reported that allows for the selective and specific extension at room temperature without enzymes.8, 9 HCR uses a pair of complementary, kinetically-trapped hairpin oligomers to propagate a chain reaction of hybridization events. Here, we report a new modification of this method called immuno-HCR that amplifies specific DNA sequences conjugated to antibodies to improve the detection of cytokines secreted from immune cells and captured on a pre-functionalized glass surface (Figure 1). We show that this method enhances the sensitivity of detecting multiple cytokines secreted from human peripheral mononuclear cells (PBMC) and improves the lower limit of detection for each analyte relative to detection by antibodies directly labeled with fluorescent dyes.
Figure 1.
Schematic illustration of immuno-HCR. An antibody-coated glass surface captures analytes of interest. Secondary antibodies labeled with an oligonucleotide initiator are introduced and bind to the target analyte. Fluorescently-labeled DNA hairpins then hybridize with their complementary oligonucleotide initiator. The fluorescence signal for each bound antibody increases with additional incorporation of hairpins bearing fluorophores during the HCR.
EXPERIMENTAL METHODS
Design of Immuno-HCR initiator and hairpins
Hairpins and initiators for HCR were designed according to previously reported methods8–10 and were obtained from IDT. Hairpins used for fluorescence were obtained with one of four non-overlapping fluorophores (6-FAM, TYE563, TYE615 or TYE665) conjugated at their 5’ ends. Each hairpin within a pair (H1/H2, H3/H4, etc.) was modified with the same fluorophore.
Solution-based extension of oligonucleotide chains by HCR
1.2 µM of oligonucleotide initiators in 1× SPSC (0.1 M sodium phosphate, 1 M sodium chloride) buffer was heated at 95 °C and cooled to room temperature. 12 µM hairpin DNAs were heated separately to 95°C for 2 min, then immediately placed on ice for 1 min and kept at room temperature. Next, initiator and hairpins were mixed and incubated at room temperature for 6 h with constant mixing. The conjugated HCR products were loaded onto a precast 2% agarose gel in 1× TBE buffer (90 mM Tris-borate, 2 mM EDTA, pH = 8.3) and gel electrophoresis was run for 3h at 40 V. After the gel electrophoresis, the gel was viewed using a UV lightsource at 350 nm. The lengths of HCR-extended chains were estimated by comparing with a DNA molecular weight marker (Roche Applied Science).
Four-color HCR with designed initiator and hairpin sets
2 µM each of four different oligonucleotide initiators were resuspended separately in 1× TBE buffer and heated to 95°C for 5 min, then cooled to room temperature. 20 µM solutions of each hairpin were heated separately to 95°C for 2 min, then immediately placed on ice for 1 min and kept in room temperature. Drops of each initiator (~1 µL) were spotted on a poly-L-lysine glass slide and vacuum-dried. A mixture of the four sets of snap-cooled hairpins was introduced on top of the glass slide and the entire surface of the slide was covered with a lifter slip (Electron Microscopy Sciences) for 2 h at room temperature. Next, the slide was washed sequentially with PBS/Tween (0.05%) and PBS. Slides were scanned using a Genepix 4200AL microarray scanner (Molecular Devices) to detect the four unique fluorescent markers from the HCR-extended oligomer chains. Commercial software (Genepix Pro 6.1) was used to extract the relative fluorescence units for each spot.
Conjugation of oligonucleotide initiators to capture antibodies
Oligonucleotide initiators containing 3’ thiol groups were dissolved in 1× SSC buffer (pH at 7.3) and introduced to microcentrifuge tubes containing 100 µL of a tris(2-carboxyethyl)phosphine (TCEP) reducing gel slurry (Pierce) washed with 1× SSC. Tubes were kept at room temperature for 3 h and then centrifuged to recover reduced initiators. 100 µL of each detection antibody (1 mg/mL) was added to each tube along with 1 µl of 5 mg/mL sulfo-SMCC crosslinker (Pierce). Tubes were incubated for 60 min at room temperature before removing excess crosslinkers using a desalting column (Pierce) equilibrated with PBS. Desalted antibodies with crosslinkers were treated with reduced initiators at a 1:1 molar ratio for 12h at 4°C with constant mixing. Initiator conjugated antibodies were isolated and concentrated by filtering them through membrane spin columns (100kD MWCO, Vivaspin 500, GE Healthcare). Conjugation reactions were monitored by gel electrophoresis. 20 µL of conjugation reaction mixture was loaded onto an 8% tris/glycine blue native gel and run for 3h at 150 V. Gels were imaged by silver stain. Control samples were neat antibodies without conjugated initiators.
Estimation of the efficiency of conjugation reaction between initiators and antibodies
Dilution series of oligonucleotide initiators (A1) bound to anti human IgG antibodies were reacted with a pair of 6-FAM-modified hairpins (H1/H2) at different molar ratios from 1:20 to 1:200 (A1-hIgG : H1/H2) for 6 h and loaded into 2% agarose gel containing 0.5 µg/mL ethidium bromide. Free initiators (A1) with a known concentration (1 µM) were mixed with a pair of hairpins (1 µM, 10 µL; H1/H2) at different volume ratio (1:1 – 1:5, A1 : H1/H2) for 6 h and loaded into the same gel to estimate the average number of initiators conjugated to the antibodies. The gel electrophoresis was run for 1 h at 100 V and imaged under UV. The concentration of initiator in the initiator-antibody conjugates was estimated based on the profile of free initiator and hairpin reaction. The estimated concentration of initiator was compared to the starting concentration of initiator and the efficiency of conjugation was calculated based on the difference between them.
Direct conjugation of fluorescent dyes to capture antibodies
Monoclonal antibodies were used for capture and detection of IL-13, IL-2 (R&D Systems), IFN-γ, (Mabtech), IL-10, IL-9 (BD), TNF-α, IL-4 (BioLegend), IL-17, IL-21 (eBioscience). A biotinylated DUOset ELISA kit was used for capture and detection of MIP-1β (R&D Systems). For detection, antibodies for each of the above cytokines and chemokines were labeled by dye conjugation using NHS-ester activated fluorescent dyes (Invitrogen; Alexa Fluor 488, Alexa Fluor 532, Alexa Fluor 594, and Alexa Fluor 647) and purified by a desalting column (Pierce). The average degree of dye labeling was determined to be 3–4 dyes per antibody. For detection of MIP-1β, biotinylated antibodies and fluorescent streptavidin (Alexa Fluor 647) were used.
Cell culture
PBMCs were obtained by Percoll (Sigma-Aldrich) isolation from heparinized whole blood (Research Blood Components, LLC). Frozen PBMCs were thawed and cultured in a HL-1 medium (Mediatech), supplemented with 10% FBS (PAA Laboratories), 100 U/mL penicillin, 100 mg/mL streptomycin, 10 mM HEPES, 50 mM 2-mercaptoethanol, 1 mM sodium pyruvate and 0.1mM non-essential amino acids. To stimulate PBMCs, a mixture of lipopolysaccharide (LPS, 0.5 µg/mL), cytosine-phosphate-guanosine (CpG, 5 µg/mL) and tetanus toxoid (TT, 1.5 µg/mL) was added to cells in round bottom 96-well microtiter plates and then incubated at 37 °C with 5% CO2 for 24h.
Preparation of the microwell arrays
Arrays of microwells were prepared as previously described.11 Briefly, an array of microwells was manufactured by injecting a silicone elastomer (polydimethylsiloxane, PDMS; 10:1 = base : catalyst; Dow Corning) into a custom-built mold and cured at 80 °C for 2 h. The arrays were adhered directly to a standard glass slide. Each array consisted of 72 × 24 blocks that contained a 7 × 7 array of microwells, each measuring 50 µm in all dimensions. The arrays of microwells were sterilized in an oxygen plasma chamber (Harrick) for 30 s prior to use.
Cell staining and image cytometry
PBMCs were washed twice with PBS after stimulation and counted to maintain 105 cells per mL. Viable PBMCs were labeled by adding 1 mM of Calcein violet AM (Invitrogen) for 30 min. The cells were then washed with PBS prior to loading onto the array. A suspension of cells was then placed onto the array of microwells, and the cell were allowed to settle by gravity into the wells at a density of about one cell per well, as described previously.11 Image-based cytometry of the cells in the microwells was acquired on an automated inverted epifluorescence microscope (Zeiss) equipped for live-cell imaging (temperature and CO2 control). Transmitted light and epifluorescence micrographs were collected block-by-block (7 × 7 microwells per block). The resulting collection of images was analyzed using a custom software program to determine the number of cells present in each well.
Microengraving cytokines from PBMCs
Poly-L-lysine-functionalized glass slides were coated with a mixture of up to four capture antibodies (10 µg/mL for each) and 10 µg/mL goat anti-human IgG (Invitrogen). The functionalized slide was then used for microengraving with PBMC-loaded arrays of microwells as reported for 1 h.11 Following microengraving, functionalized slides were blocked with 3% milk in PBS and washed with PBS/T (PBS with 0.05% Tween) and PBS.
Analysis of printed protein microarrays
For detection using antibody-dye conjugates, the printed slides were incubated with fluorescent dye-conjugated detection antibodies (1 µg/mL) for 1 h at room temperature. For immuno-HCR, slides were incubated with initiator-conjugated detection antibodies for 1 h at room temperature. Following either incubation, slides were briefly washed with PBS/T and PBS and then dried. For immuno-HCR, the slide was incubated with a mixture of fluorescently labeled hairpins for 2h. Slides were extensively washed multiple times with 2× SSC buffer, PBS/Tween and PBS and then dried. Slides were scanned using a Genepix 4200AL microarray scanner (Molecular Devices). Prior to the scanning, the gain and power for each laser channel were adjusted to maximize a signal without saturation. The accompanying commercial software (Genepix Pro 6.1) was used to extract the relative fluorescence of each feature. These data were filtered to remove features that contained saturated pixels (% Sat. > 1) or that exhibited a high degree of covariance (CV > 60).
Statistical analysis
The limit of detection (LOD) for each cytokine assayed by either immuno-HCR or direct antibody detection was calculated from the individual calibration curves as 3*σ/s, where σ is the standard deviation of the lowest concentration of analyte detected and s is the slope of the calibration curve in the linear range. The Fisher’s test and Student’s t-test each were performed using GraphPad Prism (Version 5.0b for Mac OS X, GraphPad Software, San Diego California USA).
RESULTS AND DISCUSSION
Design and validation of hairpins
In order to adapt HCR to detect cytokines secreted from cells in a multiplexed manner, we designed four different oligomers (initiators A1–A4; Table S-1) based on previously reported examples for HCR8, 9 along with four pairs of complementary hairpin oligomers (H1/H2 – H7/H8) to hybridize selectively with their respective initiators. Each pair of hairpins was synthesized with non-overlapping fluorescent dyes to enable multiplexed analysis (6-FAM, TYE563, TYE615 or TYE665). We used fluorescent dyes that could be incorporated during the synthesis, and sought to minimize the costs of the reagents, in order to improve the practicality of the technique.
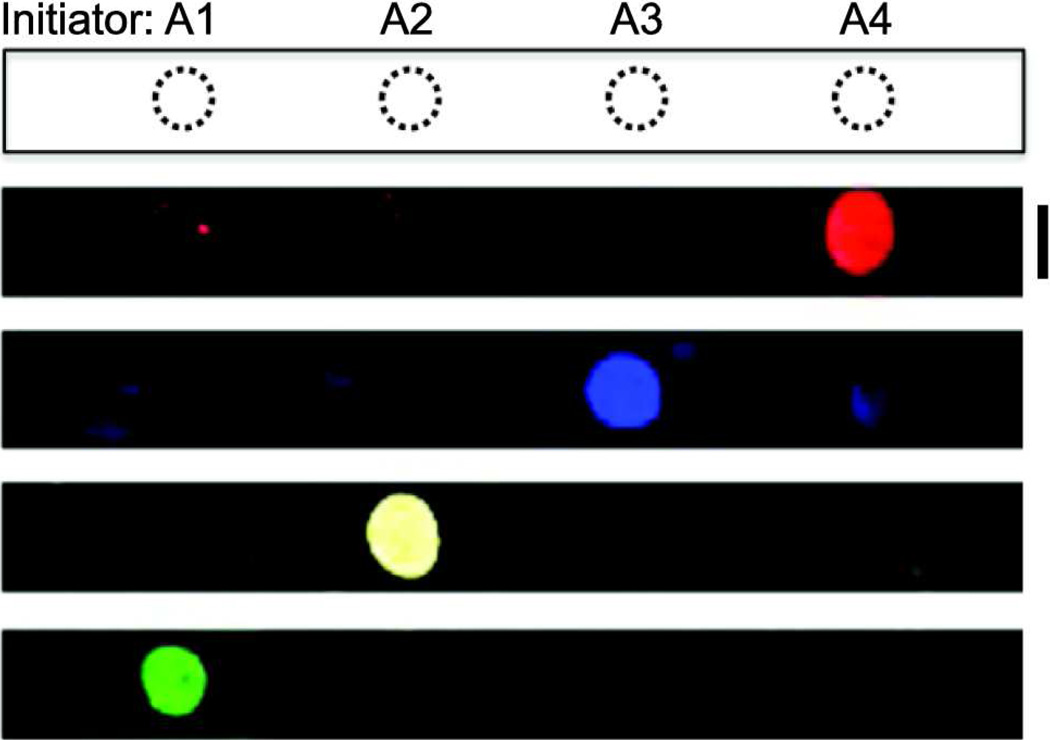
We tested each set of initiators and hairpins to ensure that they underwent specific propagation reactions. In solution, the initiators induced chain extension with the proper pairs of hairpins (Figure S-1). To confirm the specificity of the designed initiators and hairpins on a solid substrate, we then spotted each initiator on a poly-L-lysine-coated glass slide and incubated them with the mixture of all four sets of hairpins for 2 h. Hairpins only hybridized with their complementary initiators, and fluorescent signals appeared only in the expected fluorescence channels with no detectable crossover among initiators and pairs of hairpin (Figure 2, Figure S-1). These results suggested that unique initiators adsorbed to a solid substrate in a microarray can initiate specific extensions of hairpins within a mixture of multiple pairs.
Figure 2.
Immuno-HCR amplifies hairpin-specific signals from captured analytes after binding of an oligonucleotide-conjugated antibody. Custom-designed initiators (A1–A4) were spotted separately on a glass slide and a mixture of initiator-specific hairpins (H1/H2 – H7/H8) was introduced on the surface. (Each pair was labeled with an orthogonal fluorophore.) Following incubation for 2 h, the glass slide was scanned using four different excitation wavelengths (488, 555, 594, 647 nm) to detect the resulting fluorescence. Scale bar is 5 mm.
Evaluation of immuno-HCR to enhance signals for cytokine microarrays
Microarrays used to detect cytokines typically employ a sandwich immunoassay to capture and detect one or more analytes.12 The antibodies used for detecting captured analytes are often conjugated directly with fluorophores because the approach is simple and allows multiplexed analysis within the same array.13, 14 Here, for immuno-HCR, we replaced the fluorophores with unique initiators, and compared the resulting signals detected following HCR to those from directly-labeled antibodies as a reference. To test this principle, the validated initiators were covalently linked to monoclonal antibodies specific for three human cytokines (IL-2, IL-9, or IL-10) and one chemokine (MIP-1β). The efficiency of the conjugation reaction between initiators and antibodies was analyzed by gel electrophoresis to determine the yield and average number of initiators per modified antibody. The yield for conjugation was 33–50% (Figure S-2), and the average number of initiators conjugated per antibody was approximately 1–2 (Figure S-3).
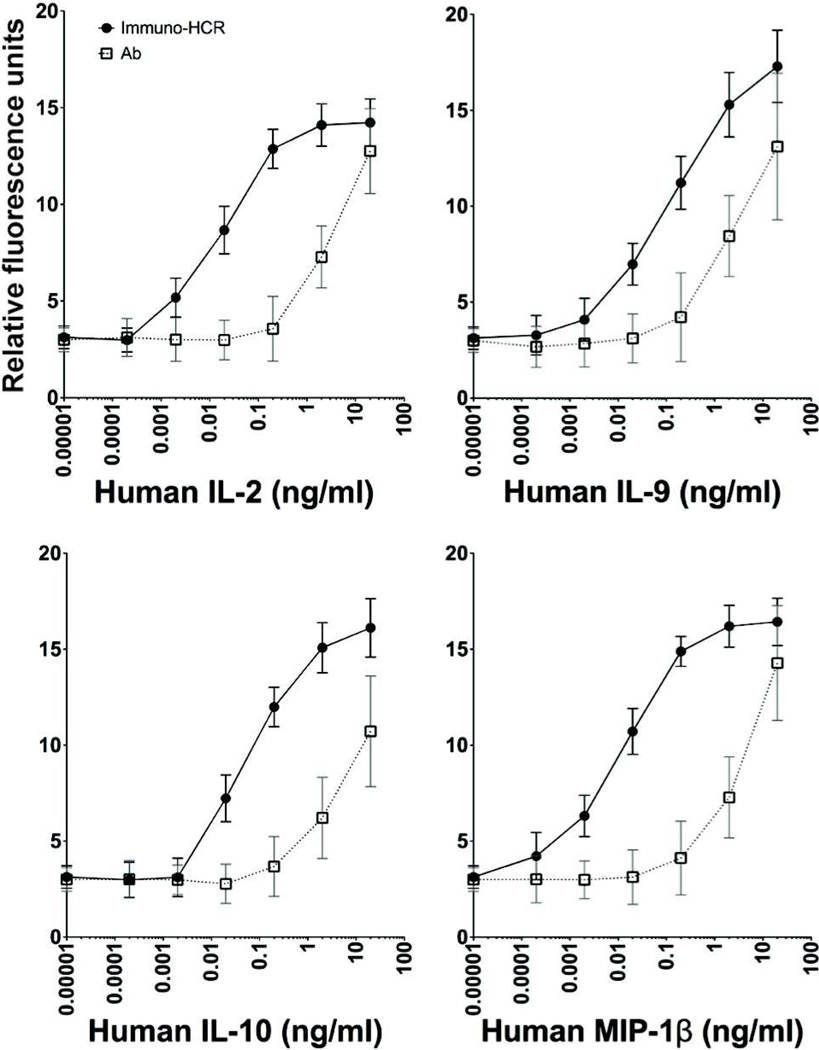
The initiator-antibody conjugates were then used to compare both the limit of detection and sensitivity of immuno-HCR-mediated detection of cytokines relative to the standard immunoassay used for cytokine microarrays in which the antibodies are directly conjugated with fluorescent dyes. We measured the mean fluorescent intensities resulting from a series of different concentrations of cytokines captured in a microarray using both directly-labeled antibodies and immune-HCR (Figure 3). The lower limit of detection (LOD) and sensitivity for all four analytes improved by an average of 200-fold using immuno-HCR (Table S-2). These improvements are similar to those previously reported for enzymatic amplifications based on rolling-circle amplification.15 The number of additional fluorophores incorporated per detection event amplified by immuno-HCR likely depends on several factors, including the time of the extension reaction and steric hindrances with growing chains. Nonetheless, comparing the signals measured using both methods here at concentrations within the overlapping linear ranges, we estimate that the number of fluorophores incorporated during immuno-HCR on the surface of the glass substrate is at least 4–5-fold greater than the number on directly-labeled detection antibodies (3–4 fluors).
Figure 3.
Immuno-HCR improves limits of detection and sensitivity relative to direct detection with fluorescently-labeled antibodies. Capture antibodies for four representative cytokines were spotted on a glass slide, and exposed to a range of concentrations of cytokines (as indicated). The captured cytokines were detected in a sandwich assay by antibodies conjugated to fluorescent dyes directly (open squares) or by immuno-HCR (filled circles). Data were collected from parallel experiments using the same imaging conditions for each one to allow direct comparisons of the resulting signals.
To confirm that the varying degrees in enhancement observed were not dependent on the specific pair of hairpins used for HCR, we used all four sets of initiators and hairpins to determine the limit of detection for the same analyte (human IL-9). Each initiator was conjugated separately to the detection antibody for IL-9 and was used to measure the mean fluorescent intensities resulting from immuno-HCR across a series of concentrations of IL-9. All four conjugates promoted immuno-HCR in a similar manner, and the lower limit of detection ranged from 0.01 to 0.02 ng/mL (Figure S-5). These data show the specific combination of initiator and hairpins used does not significantly affect the sensitivity, and suggest that variations in enhancement observed likely result from differences in the relative affinities of the antibodies used for each analyte.
Application of immuno-HCR to detect cytokines from single cells
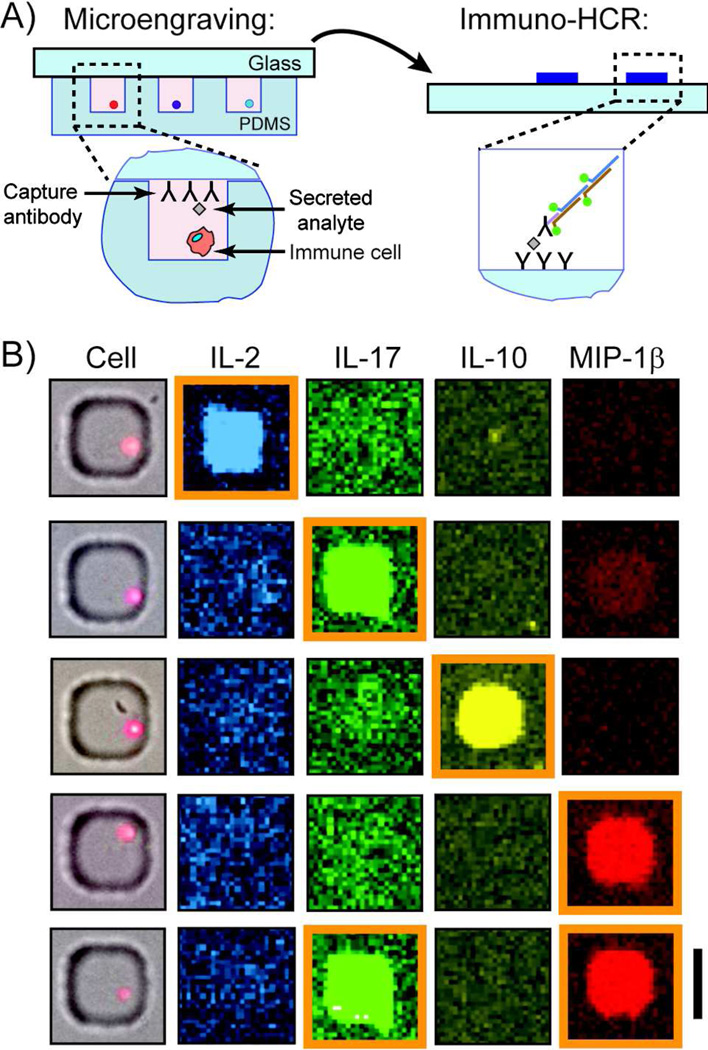
We next evaluated the utility of immuno-HCR to improve the detection of the number of single cells secreting cytokines by combining the method with microengraving (Figure 4A). Microengraving is a soft-lithographic technique for capturing proteins secreted by single cells in a paracrine manner,5 and can be integrated with other analytical operations to correlate other phenotypic traits, such as cell-specific lineage and gene expression.16, 17 Peripheral blood mononuclear cells (PBMCs) were stimulated for 24 h with CPG, LPS and tetanus toxoid, dispersed into an array of microwells, and then sealed in the wells with a glass slide that was pre-coated with capture antibodies for the cytokines of interest. After 1 h, the glass slide was removed and incubated with a mixture of antibodies specific for the captured analytes of interest (IL-2, IL-17, IL-10, and MIP-1β); each antibody bore a unique oligonucleotide for initiating HCR. Next, a mixture of four pairs of fluorescently-labeled hairpins was added to the surface of the printed microarray to amplify the signal from bound oligonucleotide-modified antibodies. Detection of the fluorescent signals resulting from oligonucleotide chains formed by HCR, and correlation of these signals with data collected from each microwell by image-based cytometry, allowed us to identify specific individual cells secreting each protein, or combinations of them (Figure 4B).
Figure 4.
Immuno-HCR allows detection of human cytokines and chemokines secreted from single stimulated PBMCs. A) Schematic illustration for application of immuno-HCR after microengraving to enable the detection of secreted products from single cells. Cells are deposited onto an array of microwells at a density of ~1 cell per well and the array is sealed with a glass slide bearing a mixture of capture antibodies. After incubation, the glass is removed, and immuno-HCR used to amplify the signal related to each capture event. (Imaging cytometry is used to score number of cells in each well—not shown). B) Representative composite micrographs of wells containing single PBMCs stimulated with CPG, LPS and tetanus toxoid for 24 h and their matching fluorescent micrographs for secreted proteins following microengraving and immuno-HCR. Positive secretion events for a particular cytokine are those with a median fluorescence intensity (MFI) greater than the background fluorescence plus three times the standard deviation (orange boxes). Scale bar is 50 µm.
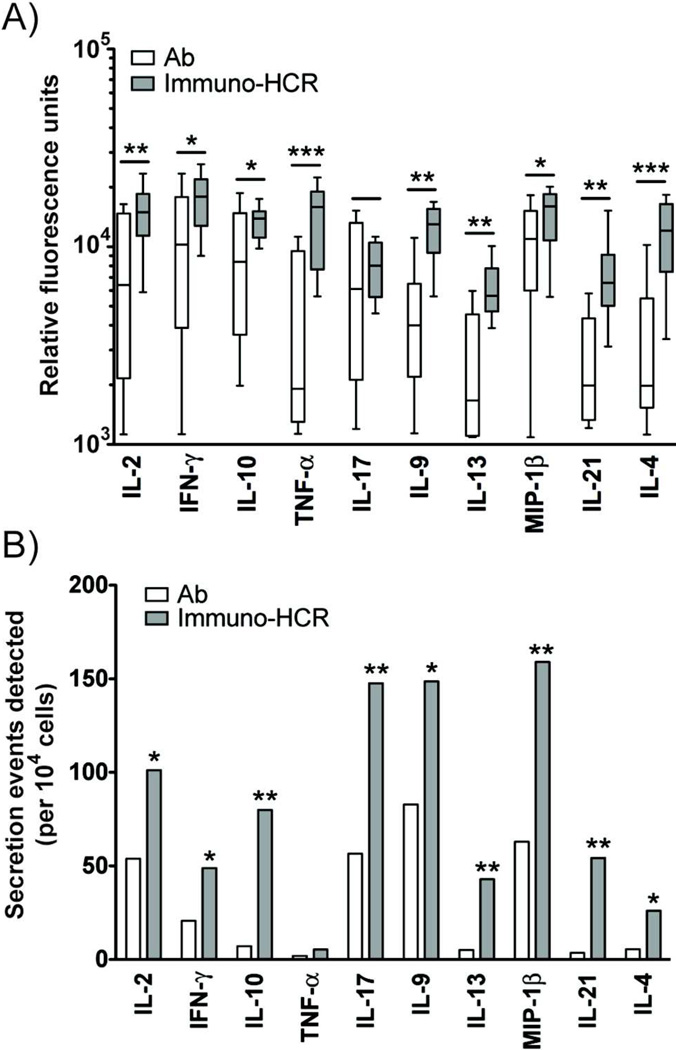
To assess the broad utility of immuno-HCR to improve the identification of cytokine-secreting cells for a range of analytes, we measured the relative fluorescence and number of events detected from stimulated PBMCs for ten different proteins (IL-2, IL-17, IFN-γ, IL-9, IL-10, IL-21, IL-13, TNF-α, IL-4 and MIP-1β). Both measures were enhanced using immuno-HCR relative to directly-labeled antibodies for detection (Figure 5). The average number of events detected per analyte increased 1.5-fold. These results, along with the LODs calculated from the calibration curves for each individual cytokine (Figure 3, Figure S-6, Table S-2), show that immuno-HCR increases the sensitivity for detecting minute amounts of secretions (<1 ng/mL) from single activated immune cells by microengraving.
Figure 5.
Amplification of signals by immuno-HCR is superior to detection by dye-conjugated antibodies for ten different human cytokines and chemokines released by single cells. A) Box plot comparing relative fluorescence units detected using directly labeled, fluorescent antibodies (white bars) and immuno-HCR (grey bars) for ten different proteins secreted from single cells. Statistical significance was determined by Student’s t-test. (*p< 0.05, **p< 0.01, ***p<0.005) B) Comparison of the numbers of positive events detected per 10,000 cells for ten proteins using directly labeled, fluorescent antibodies (white bars) and immuno-HCR (grey bars). Statistical significance was determined by Fisher’s exact test. (*p< 0.05, **p< 0.01, ***p<0.005). The data represent composite measurements from three separate microarrays generated in parallel by microengraving using divided aliquots from the same pool of stimulated cells. Each array captured up to four of the analytes.
CONCLUSIONS
Immuno-HCR provides an alternative, enzyme-free method for the amplification of fluorescence signals from micro-scale spots (~50 µm × 50 µm) of surface-captured proteins such as human cytokines and chemokines. This method lowers the limits of detection and improves the sensitivity for individual analytes in sandwich immunoassays on microarrays. Our data suggest that the degree of enhancement depends on the quality of the specific antibodies available for the assays, consistent with previous reports for protein microarrays, but nonetheless, this simple method enhanced the detection of secretions from individual PBMCs for all analytes evaluated.
The advantages of the method are that it is easy to integrate with standard protocols for processing microarrays including semi-automated washing and staining, it is isothermal, it is extensible to increase the degree of multiplexing, and it yields a convenient biomolecular scaffold on which a range of selective chemical modifications can be used (photocleavable linkers phosphorylation, biotinylation, and click chemistry18–21). One disadvantage is that the number of fluorophores associated with a given protein may vary in a non-linear manner. Since the number of unique events for individual analytes can vary from 1 in 10,000 cells to 1 in 100, the degree of polymerization of the extended HCR products may vary from assay to assay, depending on the abundance of specific populations of cytokine-secreting cells in a sample. Variations in the number of initiators and extent of conjugation among different antibody pairs may also contribute some additional inconsistencies that would impact the ability to quantify the amount of captured analytes in the current embodiment. One strategy to overcome these variations may be to create HCR products of finite size by engineering specific components that limit the extent of reaction.22 Augmenting microengraving with immuno-HCR should reduce the time required to detect secreted proteins from single cells, and provide new resolution of the kinetic variances of the secretion of cytokines and chemokines.
Supplementary Material
ACKNOWLEDGMENTS
The project described was supported in part by Award Number RC1AI086152 from the National Institute of Allergy and Infectious Diseases. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute Of Allergy And Infectious Diseases or the National Institutes of Health. The research was also supported in part by the Ragon Institute of MGH, MIT, and Harvard and the Charles A. Dana Foundation. Y.G. is supported by an NIGMS/NIH Biotechnology Training Predoctoral fellowship. J.C.L. is a Latham Family Career Development Professor.
Footnotes
SUPPORTING INFORMATION PARAGRAPH
Additional information as noted in the text. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Pantaleo G, Harari A. Nat Rev Immunol. 2006;6:417–423. doi: 10.1038/nri1840. [DOI] [PubMed] [Google Scholar]
- 2.Czerkinsky CC, Nilsson LA, Nygren H, Ouchterlony O, Tarkowski A. J Immunol Methods. 1983;65:109–121. doi: 10.1016/0022-1759(83)90308-3. [DOI] [PubMed] [Google Scholar]
- 3.Jones CN, Lee JY, Zhu J, Stybayeva G, Ramanculov E, Zern MA, Revzin A. Anal Chem. 2008;80:6351–6357. doi: 10.1021/ac8007626. [DOI] [PubMed] [Google Scholar]
- 4.Jin A, Ozawa T, Tajiri K, Obata T, Kondo S, Kinoshita K, Kadowaki S, Takahashi K, Sugiyama T, Kishi H, Muraguchi A. Nat Med. 2009;15:1088–1092. doi: 10.1038/nm.1966. [DOI] [PubMed] [Google Scholar]
- 5.Han Q, Bradshaw EM, Nilsson B, Hafler DA, Love JC. Lab Chip. 2010;10:1391–1400. doi: 10.1039/b926849a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schweitzer B, Roberts S, Grimwade B, Shao W, Wang M, Fu Q, Shu Q, Laroche I, Zhou Z, Tchernev VT, Christiansen J, Velleca M, Kingsmore SF. Nat Biotechnol. 2002;20:359–365. doi: 10.1038/nbt0402-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nallur G, Luo C, Fang L, Cooley S, Dave V, Lambert J, Kukanskis K, Kingsmore S, Lasken R, Schweitzer B. Nucleic Acids Res. 2001;29:E118. doi: 10.1093/nar/29.23.e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Venkataraman S, Dirks RM, Rothemund PW, Winfree E, Pierce NA. Nat Nanotechnol. 2007;2:490–494. doi: 10.1038/nnano.2007.225. [DOI] [PubMed] [Google Scholar]
- 9.Dirks RM, Pierce NA. Proc Natl Acad Sci U S A. 2004;101:15275–15278. doi: 10.1073/pnas.0407024101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zadeh JN, Steenberg CD, Bois JS, Wolfe BR, Pierce MB, Khan AR, Dirks RM, Pierce NA. J Comput Chem. 2011;32:170–173. doi: 10.1002/jcc.21596. [DOI] [PubMed] [Google Scholar]
- 11.Ogunniyi AO, Story CM, Papa E, Guillen E, Love JC. Nat Protoc. 2009;4:767–782. doi: 10.1038/nprot.2009.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartmann M, Roeraade J, Stoll D, Templin MF, Joos TO. Anal Bioanal Chem. 2009;393:1407–1416. doi: 10.1007/s00216-008-2379-z. [DOI] [PubMed] [Google Scholar]
- 13.Kersten B, Wanker EE, Hoheisel JD, Angenendt P. Expert Rev Proteomics. 2005;2:499–510. doi: 10.1586/14789450.2.4.499. [DOI] [PubMed] [Google Scholar]
- 14.Zhou H, Bouwman K, Schotanus M, Verweij C, Marrero JA, Dillon D, Costa J, Lizardi P, Haab BB. Genome Biol. 2004;5:R28.1–R28.12. doi: 10.1186/gb-2004-5-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mullenix MC, Sivakamasundari R, Feaver WJ, Krishna RM, Sorette MP, Datta HJ, Morosan DM, Piccoli SP. Clin Chem. 2002;48:1855–1858. [Google Scholar]
- 16.Story CM, Papa E, Hu CC, Ronan JL, Herlihy K, Ploegh HL, Love JC. Proc Natl Acad Sci U S A. 2008;105:17902–17907. doi: 10.1073/pnas.0805470105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gong Y, Ogunniyi AO, Love JC. Lab Chip. 2010;10:2334–2337. doi: 10.1039/c004847j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Armitage B. Chem Rev. 1998;98:1171–1200. doi: 10.1021/cr960428+. [DOI] [PubMed] [Google Scholar]
- 19.El-Sagheer AH, Brown T. Chem Soc Rev. 2010;39:1388–1405. doi: 10.1039/b901971p. [DOI] [PubMed] [Google Scholar]
- 20.Weisbrod SH, Marx A. Chem Commun (Camb) 2008:5675–5685. doi: 10.1039/b809528k. [DOI] [PubMed] [Google Scholar]
- 21.Cobb AJ. Org Biomol Chem. 2007;5:3260–3275. doi: 10.1039/b709797m. [DOI] [PubMed] [Google Scholar]
- 22.Choi HM, Chang JY, Trinh le A, Padilla JE, Fraser SE, Pierce NA. Nat Biotechnol. 2010;28:1208–1212. doi: 10.1038/nbt.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.