Abstract
Positive feedback is a ubiquitous signal transduction motif that allows systems to convert graded inputs into decisive, all-or-none outputs. Here we investigate why the positive feedback switches that regulate polarization of budding yeast, calcium signaling, Xenopus oocyte maturation, and various other processes use multiple interlinked loops rather than single positive feedback loops. Mathematical simulations revealed that linking fast and slow positive feedback loops creates a “dual-time” switch that is both rapidly inducible and resistant to noise in the upstream signaling system.
Studies in many biological systems have identified positive feedback as the key regulatory motif in the creation of switches with all-or-none “digital” output characteristics (1). Although a single positive feedback loop (A activates B and B activates A) or the equivalent double-negative feedback loop (A inhibits B and B inhibits A) can, under the proper circumstances, generate a bistable all-or-none switch (1–5), it is intriguing that many biological systems have not only a single but multiple positive feedback loops (Table 1). Three examples of positive feedback systems are shown in more detail in Fig. 1. Polarization in budding yeast depends on two positive feedback loops, a rapid loop involving activity cycling of the small guanosine triphosphatase Cdc42 and a slower loop that may involve actin-mediated transport of Cdc42 (Fig. 1A) (6). In many cell types, the induction of prolonged Ca2+ signals involves initial rapid positive feedback loops centered on Ca2+ release mediated by inositol 1,4,5-trisphosphate (IP3) combined with a much slower loop that induces Ca2+ influx mediated by the depletion of Ca2+ stores (7, 8) (Fig. 1B). Xenopus oocytes respond to maturation-inducing stimuli by activating a rapid phosphorylation/dephosphorylation–mediated positive feedback loop (between Cdc2, Myt1, and Cdc25) and a slower translational positive feedback loop [between Cdc2 and the the mitogen-activated protein kinase (MAPK or ERK) cascade, which includes Mos, MEK (MAPK kinase), and p42] (Fig. 1C).
Table 1.
Examples of interlinked positive feedback loops in biological regulation.
| System | Positive feedback loops | References |
|---|---|---|
| Mitotic trigger | Cdc2 → Cdc25 → Cdc2 Cdc2 -| Wee1 -| Cdc2 Cdc2 -| Myt1 -| Cdc2 |
(12, 13) |
| p53 regulation | p53 → PTEN -| Akt → Mdm-2 -| p53 p53 → p21 -| CDK2 -| Rb -|Mdm-2 -| p53 |
(14) |
| Xenopus oocyte maturation | Cdc2 → Mos → Cdc2 Cdc2 → Cdc25 → Cdc2 Cdc2 → Myt1 → Cdc2 |
(11) |
| Budding yeast traversal of START | Cdc28 → Cln transcription → Cdc28 Cdc28 -| Sic1 -| Cdc28 |
(15) |
| Budding yeast polarization | Cdc42 → Cdc24 → Cdc42 Cdc42 → actin → Cdc42 |
(6, 16, 17) |
| Eukaryotic chemotaxis | PIP3 → Rac/Cdc42 → PIP3 PIP3 → Rac/Cdc42 → actin → PIP3 |
(18) |
| Muscle cell fate specification | MyoD → MyoD Myogenin → myogenin MyoD → CDO → MyoD MyoD → Akt2 → MyoD |
(19–21) |
| B cell fate specification | IL-7 → EBF → IL-7 EBF -| Notch-1 -|E2A → EBF → Pax-5 -| Notch-1 -| E2A→ EBF |
(22, 23) |
| Notch/delta signaling | Notch (cell A) -| Delta (cell A) -| Notch (cell A) Notch (cell A) -| Delta (cell A) → Notch (cell B) -| Delta (cell B) → Notch (cell A) |
(24) |
| EGF receptor signaling | EGFR -| PTP -| EGFR Sos → Ras → Sos ERK2 → arachidonic acid → ERK2 EGFR → sheddases → EGFR |
(25–28) |
| S. cerevisiae galactose regulation | Gal2 → galactose -| Gal80 -| Gal2 Gal3 -| Gal80 -| Gal3 |
(29) |
| Blood clotting | thrombin → Xa:Va → thrombin XIIa → XIIa IXa:VIIIa → Xa → IXa:VIIIa |
(30) |
| Platelet activation | activation → ADP secretion → activation activation → 5-HT secretion → activation activation → TxA2 secretion → activation activation → aggregation → activation |
(31) |
| Ca2+ spikes/oscillations | Ca2+cyt → PLC → IP3 → Ca2+cyt Ca2+cyt → IP3R → Ca2+cyt Ca2+cyt → IP3R -| Ca2+ER -| SOC → Ca2+cyt |
(7, 8) |
ADP, adenosine 5′-diphosphate; CDK, cyclin-dependent kinase; cyt, cytochrome; CDO, a component of a cell surface receptor; EGFR, epidermal growth factor receptor; ER, endoplasmic reticulum; 5-HT, serotonin (5-hydroxytryptamine); IL-7, interleukin-7; IP3R, inositol 1,4,5-trisphosphate receptor; PIP3, phosphatidylinositol 3,4,5-trisphosphate; PLC, phospholipase C; PTEN, phosphatase and tensin homolog deleted on chromosome 10; PTP, protein tyrosine phosphatase; S. cerevisiae, Saccharomyces cerevisiae; TxA2, thromboxane A2.
Fig. 1.
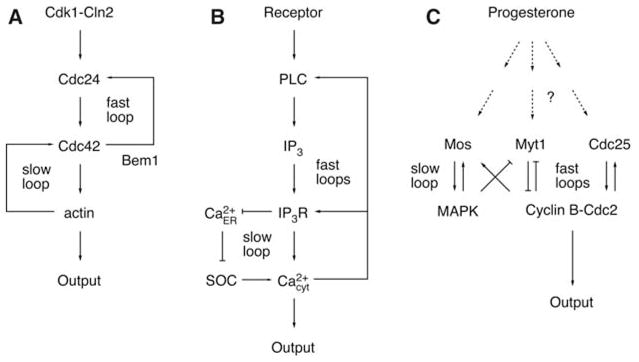
Schematic views of positive feedback loops in three systems. (A) Establishment of polarity in budding yeast. (B) Mammalian calcium signal transduction. (C) Xenopus oocyte maturation.
The presence of multiple interlinked positive loops raises the question of the performance advantage of the multiple-loop design. One clue is provided by recent studies of budding yeast polarization. When the slow positive feedback loop is selectively compromised by treatment with the actin-depolymerizing agent latrunculin, the result is rapid but unstable cell polarization (6). In contrast, cells lacking a functional fast loop (by deletion of Bem1) form stable poles, but with reduced speed (6). These experimental observations led us to hypothesize that the slow positive feedback loop is crucial for the stability of the polarized “on” state, whereas the fast loop is critical for the speed of the transition between the unpolarized “off” state and polarized on state.
To test this hypothesis computationally, we created models of positive feedback switches containing either a single positive feedback loop (Fig. 2A) or two interlinked loops (Fig. 2B). For the single-loop switch, we assumed either fast or slow kinetics for the activation and inactivation of loop component A. For the dual-loop switch, we assumed either fast kinetics for both the A and B loops, slow kinetics for both loops, or fast kinetics for the A loop and slow for the B loop (9).
Fig. 2.

Calculated responses of single and dual positive feedback loop switches to stimuli. (A) A one-loop switch. (B) A two-loop switch. (C to G) Feedback loop output (y axis) as a function of time (x axis) for single-loop and two-loop switches. (C) One slow loop. (D) Two slow loops. (E) One fast loop. (F) Two fast loops. (G) One slow loop and one fast loop. The curves on the left assume a noise-free stimulus; the curves on the right assume a noisy stimulus.
Each model switch responded to a noise-free stimulus (Fig. 2, C to G, left) and a noisy stimulus (Fig. 2, C to G, right) as shown. As expected, the single-slow-loop switch turned on and off slowly and filtered out noise (Fig. 2C). Adding a second slow loop produced a higher basal activity in the off state, a quicker switch from off to on, and a slower switch from on to off (Fig. 2D). The behavior of the two-slow-loop switch was exactly equivalent to that of a single-loop switch in which the concentration of B was doubled. Thus, adding a second loop with identical kinetic constants provides a backup in the event of gene deletion, but does not otherwise alter the behavior of the system beyond what could be achieved with a single loop.
The single-fast-loop switch turned on and off rapidly and was highly susceptible to noise in both the off and on states (Fig. 2E), and adding a second fast loop quickened the transition from off to on and delayed the transition from on to off (Fig. 2F). Thus, the fast-loop switch achieved more rapid responses, but at the cost of increased noise.
In contrast, the system in which a slow and a fast positive feedback loop are linked together introduces marked advantages over single-loop systems, as well as dual-loop systems with the same time constant. In this “dual-time” switch, the output turned on rapidly, as a consequence of the kinetic properties of the fast loop, and turned off slowly as a consequence of the kinetics of the slow loop (Fig. 2G). This allows for independent tuning of the activation and deactivation times. More important, although the dual-time switch exhibited high noise sensitivity when in the off state, as a result of the rapid responses of its fast loop, it became resistant to noise once it settled in its on state as a result of the properties of its slow loop. Thus, the dual-time switch provides the ability to transit rapidly from the off state to the on state together with robust stability of the on state (10).
These computational studies help understand the yeast phenotypes described above and provide a rationale for the existence of dual-time positive feedback systems in Ca2+ signaling, oocyte maturation, and other biological systems. In the case of Ca2+ signaling, the dual-time system enables rapid Ca2+ responses from IP3-induced Ca2+ release, while also enabling long-term robust Ca2+ signals once the store-operated Ca2+ influx is triggered. Although weak stimuli or noise have been shown to trigger IP3-mediated Ca2+ spikes, more persistent stimuli are needed to induce Ca2+ influx and prolonged Ca2+ responses (7). These long-term Ca2+ signals are required for T-cell activation and differentiation and many other cellular processes (7, 8). Xenopus oocyte maturation includes a period termed interkinesis, during which Cdc2 becomes partially deactivated (11). We conjecture that the slow positive feedback loop helps prevent a transition to the off state during this critical interkinesis period.
Our study suggests that many biological systems have evolved interlinked slow and fast positive feedback loops to create reliable all-or-none switches. These dual-time switches have separately adjustable activation and deactivation times. They combine the important features of a rapid response to stimuli and a marked resistance to noise in the upstream signaling pathway.
Acknowledgments
We thank R. Brandman, Y. Brandman, T. Galvez, R. S. Lewis, L. Milenkovic, D. Mochly-Rosen, M. P. Scott, P. M. Vitorino, and R. Wedlich-Soldner who provided helpful suggestions. This work was supported by an NSF predoctoral fellowship awarded to O.B., NIH grants GM46383 to J.E.F., GM057063 to R.L., and MH064801 and GM063702 to T.M.
References and Notes
- 1.Ferrell JE, Jr, Xiong W. Chaos. 2001;11:227. doi: 10.1063/1.1349894. [DOI] [PubMed] [Google Scholar]
- 2.Laurent M, Kellershohn N. Trends Biochem Sci. 1999;24:418. doi: 10.1016/s0968-0004(99)01473-5. [DOI] [PubMed] [Google Scholar]
- 3.Gardner TS, Cantor CR, Collins JJ. Nature. 2000;403:339. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 4.Becskei A, Seraphin B, Serrano L. EMBO J. 2001;20:2528. doi: 10.1093/emboj/20.10.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Isaacs FJ, Hasty J, Cantor CR, Collins JJ. Proc Natl Acad Sci USA. 2003;100:7714. doi: 10.1073/pnas.1332628100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wedlich-Soldner R, Wai SC, Schmidt T, Li R. J Cell Biol. 2004;166:889. doi: 10.1083/jcb.200405061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berridge MJ. Novartis Found Symp. 2001;239:52. [PubMed] [Google Scholar]
- 8.Lewis RS. Annu Rev Immunol. 2001;19:497. doi: 10.1146/annurev.immunol.19.1.497. [DOI] [PubMed] [Google Scholar]
-
9.The ordinary differential equations for the one- and two-loop positive feedback switches are
- One loop
-
Two loops
kout_on = 2, kout_off = 0.3, kout_min = 0.001, kmin = 0.01, n = 3, ec50 = 0.35. For a fast loop, τ = 0.5. For a slow loop, τ = 0.008. The equations were solved numerically with Matlab 7.0.
- 10.An interesting variation on this scheme can be envisioned by assuming that A and B have distinct effects on the output, and that both effects are required to activate the output. For example, A and B could phosphorylate different sites on the output protein, so that the protein is only activated when both sites are phosphorylated. The behavior of this dual-time AND switch is essentially the mirror image of the dual-time system shown in Fig. 2E: It turns on slowly, turns off rapidly, and acquires noise resistance when it has been in the off state for a period of time determined by the slow loop.
- 11.Abrieu A, Doree M, Fisher D. J Cell Sci. 2001;114:257. doi: 10.1242/jcs.114.2.257. [DOI] [PubMed] [Google Scholar]
- 12.Solomon MJ, Glotzer M, Lee TH, Philippe M, Kirschner MW. Cell. 1990;63:1013. doi: 10.1016/0092-8674(90)90504-8. [DOI] [PubMed] [Google Scholar]
- 13.Hoffmann I, Clarke PR, Marcote MJ, Karsenti E, Draetta G. EMBO J. 1993;12:53. doi: 10.1002/j.1460-2075.1993.tb05631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris SL, Levine AJ. Oncogene. 2005;24:2899. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 15.Levine K, Tinkelenberg AH, Cross F. Prog Cell Cycle Res. 1995;1:101. doi: 10.1007/978-1-4615-1809-9_8. [DOI] [PubMed] [Google Scholar]
- 16.Butty AC, et al. EMBO J. 2002;21:1565. doi: 10.1093/emboj/21.7.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wedlich-Soldner R, Altschuler S, Wu L, Li R. Science. 2003;299:1231. doi: 10.1126/science.1080944. [DOI] [PubMed] [Google Scholar]
- 18.Weiner OD, et al. Nat Cell Biol. 2002;4:509. doi: 10.1038/ncb811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thayer MJ, et al. Cell. 1989;58:241. doi: 10.1016/0092-8674(89)90838-6. [DOI] [PubMed] [Google Scholar]
- 20.Cole F, Zhang W, Geyra A, Kang JS, Krauss RS. Dev Cell. 2004;7:843. doi: 10.1016/j.devcel.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 21.Kaneko S, et al. J Biol Chem. 2002;277:23230. doi: 10.1074/jbc.M201733200. [DOI] [PubMed] [Google Scholar]
- 22.Singh H, Medina KL, Pongubala JM. Proc Natl Acad Sci USA. 2005;102:4949. doi: 10.1073/pnas.0500480102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Medina KL, et al. Dev Cell. 2004;7:607. doi: 10.1016/j.devcel.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 24.Lodish H, et al. Molecular Cell Biology. 5. Freeman; New York: 2004. [Google Scholar]
- 25.Reynolds AR, Tischer C, Verveer PJ, Rocks O, Bastiaens PI. Nat Cell Biol. 2003;5:447. doi: 10.1038/ncb981. [DOI] [PubMed] [Google Scholar]
- 26.Margarit SM, et al. Cell. 2003;112:685. doi: 10.1016/s0092-8674(03)00149-1. [DOI] [PubMed] [Google Scholar]
- 27.Bhalla US, Ram PT, Iyengar R. Science. 2002;297:1018. doi: 10.1126/science.1068873. [DOI] [PubMed] [Google Scholar]
- 28.Shvartsman SY, et al. Am J Physiol Cell Physiol. 2002;282:C545. doi: 10.1152/ajpcell.00260.2001. [DOI] [PubMed] [Google Scholar]
- 29.Acar M, Becskei A, van Oudenaarden A. Nature. 2005;435:228. doi: 10.1038/nature03524. [DOI] [PubMed] [Google Scholar]
- 30.Beltrami E, Jesty J. Proc Natl Acad Sci USA. 1995;92:8744. doi: 10.1073/pnas.92.19.8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holmsen H. Proc Natl Sci Counc Repub China B. 1991;15:147. [PubMed] [Google Scholar]


