Abstract
Human STIM (stromal interacting molecule) proteins are parts of elaborate eukaryotic Ca2+ signaling systems that include numerous plasma membrane (PM), endoplasmic reticulum (ER), and mitochondrial Ca2+ transporters, channels and regulators. STIM2 and STIM1 function as Ca2+ sensors with different sensitivities for ER Ca2+. They translocate to ER-PM junctions and open PM Orai Ca2+ influx channels when receptor-mediated Ca2+ release lowers ER Ca2+ levels. The resulting increase in cytosolic Ca2+ leads to the activation of numerous Ca2+ effector proteins that in turn regulate differentiation, cell contraction, secretion and other cell functions. In this review, we use an evolutionary perspective to survey molecular activation mechanisms in the Ca2+ signaling system with a particular focus on regulatory motifs and functions of the two STIM proteins. We discuss the presence and absence of STIM genes in different species, the order of appearance of STIM versus Orai, and the evolutionary addition of new signaling domains to STIM proteins.
Introduction
Our review sheds light on the uses, mechanisms, and origins of the two vertebrate STIM proteins by taking an evolutionary approach that considers STIM1 and STIM2 as components of integrated Ca2+ signaling systems with ancient origins. Primordial cells had to regulate the contents of their cytoplasm to create an environment that was favorable for the basic chemical reactions needed for life. Ca2+ ions, which are highly abundant in seawater and on land, interact with phosphate and can interfere with nucleic acid metabolism1. This makes Ca2+ a prominent and ancient hazard. Eukaryotes addressed this challenge by efficiently pumping and transporting Ca2+ out of the cytoplasm into the extracellular space or into membrane-enclosed intracellular stores. The resulting steep concentration gradients across the plasma membrane (PM) and internal membranes provided cells with an opportunity to use these gradients as the driving forces to regulate Ca2+ influx and release from internal stores as part of different signaling mechanisms. Not surprisingly, the origins of such Ca2+ signaling pathways are very old. Use of Ca2+ as a signaling molecule is ubiquitous in all eukaryotes, and it is used as a signaling molecule in some bacteria as well2.
Human cells use diverse sensors, Ca2+ buffering proteins, channels, pumps, and exchangers to regulate the transport of Ca2+ ions between the cytosol, internal organelles and the extracellular space (Figure 1). Years of study have uncovered the identities of many of these genes. Two of the most recently discovered proteins are the endoplasmic reticulum (ER) Ca2+ sensor, STIM3,4, and the PM Ca2+ channel regulated by STIM, Orai5–7. Together, they form the main components of a long hypothesized store-operated Ca2+ entry (SOCE) pathway8. This pathway includes a number of interesting new signaling concepts. First, it is one of only a few known "inside-out" signaling mechanisms. Specifically, it allows changes in luminal ER Ca2+ to regulate the opening of a PM Ca2+ channel. Second, STIM proteins have the interesting characteristic that they can directly bridge the ER to the PM at specialized junctions, the ER-PM junctions, that now appear to be ubiquitous in all eukaryotic cells. Third, the activation of STIM involves an oligomerization process that is different from other receptors that oligomerize, in that ligand dissociation (i.e., Ca2+ dissociation) rather than ligand binding triggers STIM oligomerization and activation. In this review, we survey existing knowledge about the evolutionary history of human Ca2+ signaling, with a particular emphasis on this intriguing STIM-controlled ER-to-PM Ca2+ signaling pathway.
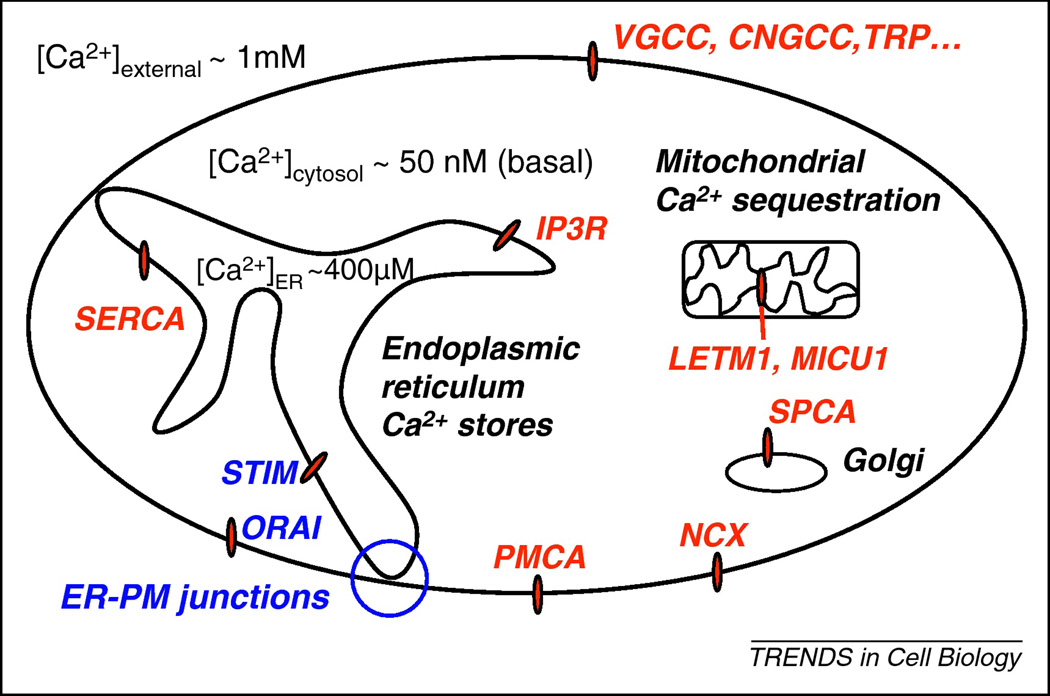
Figure 1.
Schematic representation of relevant Ca2+ regulators. Shown is a diagram of a eukaryotic cell with the localization of PM channels (VGCC, CNGCC, TRP, ORAI), Ca2+ pumps (PMCA, SERCA, SPCA), internal channels (IP3R), Ca2+ exchangers (NCX, LETM1), and key regulatory proteins (STIM, MICU1) indicated. Also indicated are approximate values for the Ca2+ concentration in the extracellular space (~1 mM), the cytoplasm (~50 nM under basal conditions), and the ER (~400 µM).
Conserved generators of transmembrane Ca2+ gradients
The most fundamental task in setting up the Ca2+ signaling system is to maintain low basal cytoplasmic Ca2+ concentration. Long term reduction of cytoplasmic Ca2+ is primarily carried out by extruding Ca2+ out of the cell via active Ca2+ pumping (PMCA proteins) and by Na+/Ca2+ exchange (NCX and NCKX proteins)1,9 (Figure 1 and see Glossary for abbreviations used). As might be expected, homologs of these pumps or exchangers are widely present in eukaryotes (see Figure 2). The best known exchangers are homologs of the cardiac Na+/Ca2+ exchanger NCX9. NCKX has an additional driving force from a co-transported K+ ion, allowing cytosolic Ca2+ in retinal photoreceptor and other cells to be more effectively lowered into the tens of nanomolar range10. A likely reason for the prevalence of both pump- and exchange-based Ca2+ extrusion is that exchange can rapidly lower high cytoplasmic Ca2+ concentrations due to a low affinity and high capacity of the exchangers, whereas the ATP-consuming PMCA pump has a higher affinity but lower capacity that allows it to operate effectively at low cytosolic Ca2+ concentrations.
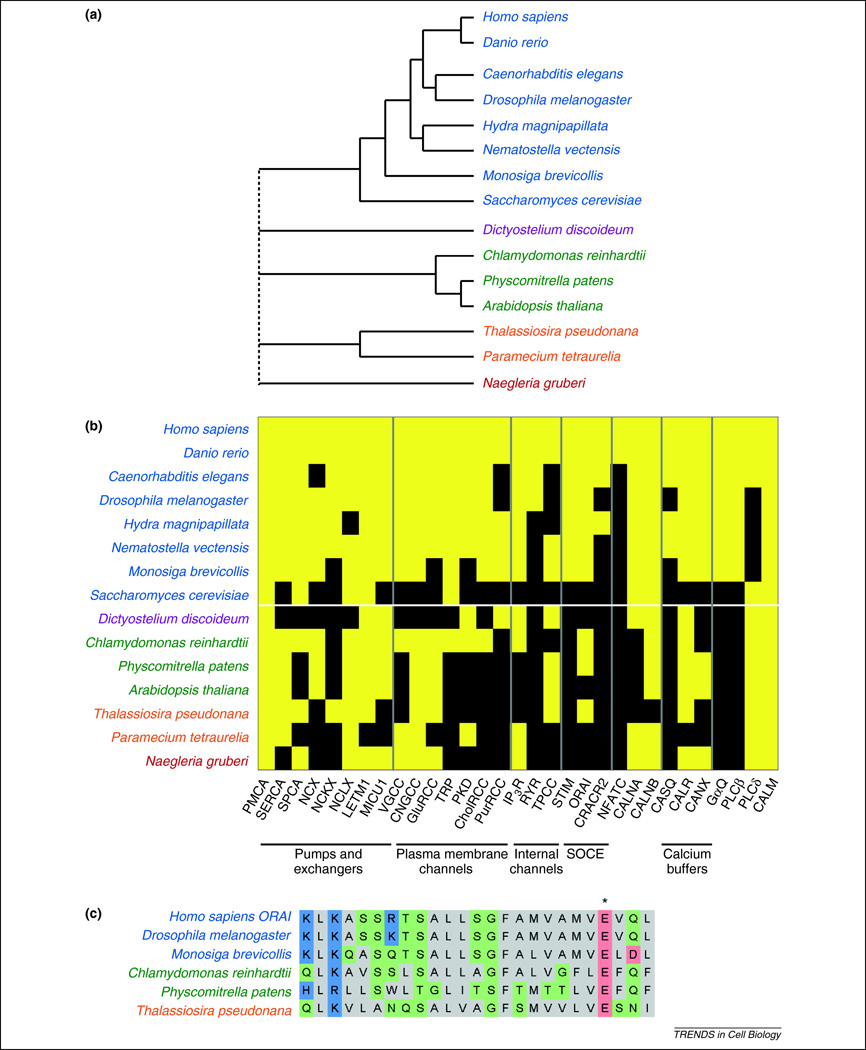
Figure 2.
Phylogenetic profiles of eukaryotic Ca2+ signal generating genes. (a) A consensus cladogram showing the topology (branch lengths are not intended to be to scale) of the phylogenetic relationships between the species shown in b22,96–98. Colored type is used to indicate membership in different major branches of the eukaryotic tree. The dotted line joining the major branches indicates an unclear phylogenetic relationship. (b) A representative set of species with sequenced genomes are labeled on the left. Human gene families are listed below (e.g., “STIM” includes the genes STIM1 and STIM2). Yellow indicates evidence for the presence of a homolog of a human gene family in a particular species, and black indicates the lack of an apparent homolog12,17,22–24,26,27,33,34,36,40,42,61,64,99,86,100–103,90,91,94. (c) Shown is a portion of a multiple sequence alignment of Orai1 homologs. The organism corresponding to each sequence is indicated to the left. Only one sequence is given for human Orai, because the sequences of Orai1, Orai2, and Orai3 are identical in this region. The E106 residue in human Orai1 is indicated with an asterisk. This residue has been implicated as a key determinant of ion specificity.
While long-term cytosolic Ca2+ levels are regulated at the PM, shorter term transient Ca2+ increases often result from release of Ca2+ stored inside the lumen of the ER11. The ER can typically release and reuptake Ca2+ ions on a time scale of seconds, often 10 to 100 times faster than extrusion out of the cell. This allows cells to terminate Ca2+ transients by rapidly pumping Ca2+ into ER stores with Ca2+ leaving the cell much more slowly. This transport from the cytoplasm into the ER is primarily mediated by SERCA Ca2+ pumps. A second related pump, the Ca2+ ATPase, SPCA12, is more ubiquitously present and pumps Ca2+ into the Golgi, secretory vesicles, endosomes, and likely other compartments. Although not every organism has a homolog of both SERCA and SPCA (e.g., Saccharomyces cerevisiae lacks a SERCA homolog and Arabidopsis thaliana lacks an SPCA homolog12), a homolog of at least one internal pump appears to be present in almost every eukaryote. As will be discussed below, the ER is not only a Ca2+ store, but Ca2+ also has important regulatory roles inside the ER.
Mitochondria handle Ca2+ very differently from the ER. They take up Ca2+ instead of releasing it upon cell stimulation13. This Ca2+ uptake involves a mitochondrial uniporter channel and an as of yet poorly understood process where Ca2+ released from the ER directly enters mitochondria14. Due to high Ca2+ cooperativity in uniporter activation, Ca2+ uptake into mitochondria also plays a safety role in limiting peak cytosolic Ca2+ levels. Inside mitochondria, Ca2+ is stored as a phosphate precipitate due to the high pH and high phosphate content in the mitochondrial inner matrix13,15. This precipitation limits mitochondrial free Ca2+ concentration to levels much lower than those in the ER. When cytosolic Ca2+ signals are terminated, mitochondria slowly transport Ca2+ back out into the cytosol. A primary role of this transient increase in mitochondrial Ca2+ is to link cell activation to an enhancement of ATP production and other metabolic activities16.
While the molecular identity of the uniporter is not yet known, a mitochondrial H+/Ca2+ antiporter (LETM1)17, a mitochondrial Na+/Ca2+ exchanger (NCLX)18, and a Ca2+ binding protein that regulates uniporter activity (MICU1) have all been recently discovered19. LETM1 may provide cells with a mechanism to link mitochondrial Ca2+ to the mitochondrial proton gradient and aerobic ATP production by participating in both Ca2+ uptake and extrusion17. All three of these genes are widely conserved in eukaryotes, suggesting that mitochondrial Ca2+ regulation became important in the earliest protozoans but was lost in some lineages such as fungi (Figure 2). Indeed, the presence of MICU1 homologs in distant protozoan lineages and absence of a homolog in Saccharomyces cerevisiae were key observations that allowed for its identification19.
Thus, the stage for Ca2+ signaling in humans is set by a conserved transport system that generates Ca2+ gradients across the PM as well as the ER, mitochondrial and other internal membranes, and provides a driving force for Ca2+ influx or release when channels are opened. Markedly, subsets of these ancient transporters were lost in multiple eukaryotic lineages12,19 (Figure 2) but each species has at least some of these Ca2+ gradient generators.
Ancient roots, but great plasticity of PM Ca2+ channels
Cytosolic Ca2+ signals are almost exclusively generated by regulated opening and closing of Ca2+ channels and many of these channels are in the PM1. Humans have diverse PM Ca2+ conducting channels to respond to an array of stimuli with a particular importance in highly specialized cells such as muscle cells and sensory neurons. Given that many of these specialized cells are unique to animals, it is remarkable that representatives of most human PM Ca2+ channels are found in distant eukaryotic lineages (Figure 2). This implies that single-celled eukaryotes, living over 1 billion years ago, already used influx of extracellular Ca2+ as a major signaling mechanism to respond to a variety of external cues.
Humans have gene families encoding PM Ca2+ channels that are gated by different mechanisms, including voltage-gated Ca2+ channels (VGCC) that are activated by PM depolarization1, transient receptor potential (TRP) channels that are often regulated by different sensory inputs such as heat and mechanical stress1,20, cyclic nucleotide-gated channels (CNGCC) that are typically opened by the intracellular second messengers cAMP or cGMP21, and glutamate receptor channels (GLURCC) that respond to the extracellular presence of the amino acid glutamate. The VGCC, CNGCC, and GLURCC families each have apparent homologs in the Naegleria gruberi genome22 and in either plants or algae23. The presence of these distant homologs (Figure 2) suggests that they were likely present in the earliest eukaryotes. The TRP family has homologs in Saccharomyces cerevisiae24 and in the algae Chlamydomonas reinhardtii23. The polycystic kidney disease (PKD) channels, which are a subtype of TRP channel often associated with specialized cell extensions called primary cilia25, are widely represented. These observations suggest that TRP channels also arose early. Finally, the ATP-regulated purinergic Ca2+ channels (PurRCC) and the cholinergic Ca2+ channels (ChoRCC), which have been largely implicated in cell-cell communication in higher eukaryotes, may be more recent inventions as homologs of these channels appear to be less widespread. However, PurRCC homologs have been identified in Dictyostelium discoideum26 and the Chlamydomonas reinhardtii genome contains a ligand-gated ion channel with similarity to ChoRCCs23,27. As a note of caution, the ion selectivity and subcellular localization of Ca2+ channel homologs is difficult to discern from sequence homology alone and some of these homologs may conduct other cations and may not act in the PM28. Nevertheless, evolutionary analyses argue that PM Ca2+ channels of many subtypes arose early in eukaryotic evolution, though particular subtypes were also often lost in different eukaryotic lineages.
Evolution of the intracellular Ca2+ release system
Humans have three major known routes for Ca2+ release from internal stores. The most prominent mechanism is based on inositol trisphosphate (IP3)-mediated opening of IP3 receptor (IP3R) Ca2+ channels29, a process that is further amplified by Ca2+ binding to IP3Rs themselves30. A second ER release mechanism is based on Ca2+-triggered opening of ryanodine receptor (RyR) Ca2+ channels31, a process that can be modulated by cyclic adenosine diphosphate (cADP)-ribose and, in the case of skeletal muscle, by a physical interaction with a PM-localized voltage-gated Ca2+ channel. A third release mechanism involves the opening of two-pore Ca2+ channels, endosome- and vacuole-localized proteins that can be regulated by nicotinic acid adenine dinucleotide phosphate (NAADP) but have otherwise largely unresolved functions32.
Both the IP3R and the two-pore channel have distant homologs in the plant kingdom and protozoan lineages while the RyR appears more recently23,33,34. It is also interesting that the most common regulation of human IP3 production, via G-protein (GαQ) activation of phospholipase C β (PLCβ)35, is likely to be a relatively recent phenomenon (appearing after the IP3R) while the Ca2+ regulated phospholipase C δ (PLCδ), which also produces IP3, likely arose earlier and is more broadly present than IP3R Ca2+ channels (Figure 2)36. This suggests that hydrolysis of phosphoinositide lipids by phospholipase C has older roles than triggering Ca2+ release, possibly in the production of IP6 and other higher phosphorylated inositol products35,37. It is then conceivable that the IP3R may have initially contributed to an indirect Ca2+ amplification process whereby IP3R-mediated release of Ca2+ increased PLCδ activity which in turn produced more IP3, more Ca2+ release and more polyinositol phosphates. However, there is also evidence for IP3-mediated Ca2+ signaling in organisms that lack IP3R homologs38,39, although the channels responsible have not been identified and the mechanism may be indirect. Thus, an alternative possibility is that IP3 generation was initially linked to Ca2+ release from internal stores by multiple mechanisms, and different mechanisms remain in different modern organisms.
While Ca2+ release from internal stores is an ancient signaling mechanism prominent in all human cells, each of these types of release channels has also been lost during the evolution of a number of modern organisms (Figure 2 and Box 2). For example, the IP3R is absent from the genome of the diatom Thalassiosira pseudonana40, and the two-pore channel was lost during the evolution of Caenorhabditis elegans41. A number of species, including the yeast Saccharomyces cerevisiae42, lack all three (IP3R, RyR, and two-pore) of these channel types. However, the Saccharomyces cerevisiae uses a TRP-family channel (YVC1) to release Ca2+ from vacuolar stores24. Thus, while internal Ca2+ release mechanisms are present in almost every eukaryote, the channels used, and presumably the current and activation properties of those channels, vary widely.
Box 2.
Brief description of model organisms
Xenopus laevis. African clawed frog.
Gallus gallus. Chicken.
Danio rerio. The zebrafish is a tropical freshwater fish and a common model organism for studying vertebrate development.
Caenorhabditis elegans. This is a millimeter-long roundworm that serves as a popular model for genetics and development.
Drosophila melanogater. Fruit fly.
Hydra magnipapillata. This organism is a small (millimeter-scale), radially symmetric freshwater invertebrate. It is also known as a freshwater polyp.
Nematostella vectensis. Like Hydra, the sea anemone belongs to the phylum Cnidaria which are among the simplest animals with neurons, muscle fibers, and epithelial cells.
Monosiga brevicollis. This aquatic protozoan is one of the closest known single-celled relatives of multicellular animals. It is also known as a type of choanoflagellate.
Saccharomyces cerevisiae. The common budding yeast is a popular model organism.
Dictyostelium discoideum. Also known as a slime mold, D. discoideum is a species of amoeba that has both single-celled and multicellular states, and serves as a model for motility, chemotaxis and differentiation.
Chlamydomonas reinhardtii. This single-celled green alga swims by the use of two flagella. It lives in both soil and fresh water.
Physcomitrella patens. This multicellular moss is used as a model system for plant genetics and development. Mosses are estimated to have had a common ancestor with flowering plants approximately 200–400 million years ago.
Arabidopsis thaliana. This flowering plant is a popular model for genetics, and its genome was the first plant genome to be sequenced.
Thalassiosira pseudonana. These are single-celled organisms commonly known as diatoms. They live in oceans and derive energy from photosynthesis.
Paramecium tetraurelia. Paramecium are single-celled protozoan that live in freshwater environments, move by use of cilia that cover the cell surface, and prey on other single-celled organisms such as bacteria and algae.
Naegleria gruberi. These are single-celled protozoans that are capable of transitioning between a state in which they move in an amoeboid process to a state in which they move through flagellar motion.
Placing STIM proteins into the eukaryotic evolutionary map
In addition to having cytosolic signaling roles, Ca2+ also plays an important role inside the ER where it regulates a host of Ca2+-dependent chaperones that assist protein folding43. A requirement of Ca2+ for protein folding and processing continues throughout the secretory system44,45. Several Ca2+-dependent chaperone proteins (such as calnexin and calreticulin) are highly conserved46 (Figure 2), and even in yeast where ER Ca2+ levels are lower (~10 µM) and the calnexin homolog does not appear to be Ca2+-dependent, depletion of secretory pathway Ca2+ results in defects in protein processing and secretion47. This raises the question of how cells can maintain sufficient ER Ca2+ concentration (~400 µM in humans) to prevent ER stress in spite of variable Ca2+ signaling activity. It is also not obvious how cells can generate long-term cytosolic Ca2+ signals since cells with elevated cytosolic Ca2+ are expected to slowly lose all Ca2+ from the ER and cytosol by PMCA-mediated extrusion from the cell.
STIM proteins solve this problem by providing a functional connection between the regulation of ER and cytosolic Ca2+. STIM1 was first identified3,4 as the elusive ER Ca2+ sensor that can control ER Ca2+ levels and enable long-term increases in cytosolic Ca2+upon receptor stimulation. Initial studies showed that ER-localized STIM1 proteins are kept inactive by binding of Ca2+ to an EF hand located in the ER luminal part of STIM13,48. Furthermore, Ca2+ dissociation induced by ER Ca2+ depletion led to the translocation of STIM1 to sites in the ER close to the PM without insertion of STIM1 into the PM3. Imaging of fluorescent protein conjugated STIM1 using total internal reflection fluorescence microscopy (TIRF), which allows monitoring specifically of events close to the PM, suggested that dedicated ER-PM junctions exist where STIM1 remains on the ER side in close proximity to the PM, enabling potential direct regulation of PM Ca2+ channels3. This provided evidence for the now well established model that STIM proteins have a direct ER to PM signaling role. While the existence of an ER store-operated Ca2+ influx process (SOCE) was hypothesized to exist in 19868, it took close to 20 years to identify STIM1 as the first molecular component of this SOCE pathway. A second component, the PM Ca2+ channel Orai5–7 was then identified and found to be directly activated by STIM at these ER-PM junctions36 and STIM2 was found to have similar roles as STIM1 in regulating Ca2+ influx49. Furthermore, accessory proteins were identified such as calmodulin (CALM)50, the Ca2+ regulated adaptor protein CRACR251 and possibly other STIM-regulated proteins such as voltage-gated Ca2+ channels52,53, Trp channels54,55 and adenylyl cyclases56.
The physiological roles of STIM proteins and SOCE have been extensively characterized in T-cells where they are critically required for allowing sustained Ca2+ signaling, and long-term activation of CALM, the protein phosphatase calcineurin (CALNA and CALNB) and the transcription factor NFAT, which in turn leads to T-cell differentiation and activation57. Recent studies further showed that mutations or deletion of STIM or Orai result in severe immunodeficiencies5,58. STIM and Orai have been found in every mammalian cell type examined thus far and in a number of model organisms, and regulate many additional processes such as neuronal excitability59 and muscle activity60, suggesting that they are likely core elements of all human Ca2+ signaling systems.
At what point along the trajectory of human evolution did this STIM signaling system evolve? One can imagine two scenarios for the ancient roles of the SOCE system. STIM-mediated signaling could initially have served to stabilize ER Ca2+ levels and regulate the function of ER chaperones. Alternatively, the main role of the ancient STIM pathway could have been to sustain cytosolic Ca2+ signals when receptor stimuli persistently increased IP3 levels. A survey of the evolutionary record provides some insights. First, STIM and Orai homologs are present in the genome of the single-celled choanoflagellate Monosiga brevicollis61 (see also62), which diverged from the human line prior to the apparent origins of the RyR, the muscle ER Ca2+ storage protein calsequestrin (CASQ)63, or the T-cell differentiation transcription factor NFAT (Figure 2b)57. A second notable observation is that a number of species that have the IP3R lack STIM and Orai. Examples include Paramecium tetraurelia33 and Naegleria gruberi22. Therefore, the presence of STIM and Orai is not a universal complement to IP3-mediated Ca2+ signaling.
Remarkably, distant but recognizable Orai homologs appear in the genomes of green algae (Chlamydomonas reinhardtii)27, moss (Physcomitrella patens)64, and of the diatom Thalassiosira pseudonana40. While it is impossible to determine by sequence analysis alone whether these homologs function as store-operated Ca2+ channels, it is interesting to note that putative homologs in each of these species have conserved the E106 residue (Figure 2c), which has been identified as the main Ca2+ binding site controlling ion selectivity in the pore of Orai165–67. Several of these species with Orai ancestors lack the IP3R (Physcomitrella patens is an example23,64). The distant Orai homologs are suggestive of the presence of SOCE without IP3-mediated signaling. This could imply an initial evolutionary role for SOCE in regulating ER Ca2+ levels before SOCE became critical for prolonging IP3-mediated Ca2+ signals. However, the co-appearance of clear STIM homologs with PLCβ and GαQ in Monosiga brevicollis and animals, supports the alternative model that STIM arose as a complement to receptor-operated IP3 signaling, and that this system co-opted an existing Orai channel for SOCE. Additionally, the appearance of Orai before STIM could indicate that the earliest Orai proteins were controlled by a different gating mechanism. Indeed, there is evidence that human Orai1 and Orai3 isoforms may have a second role as part of an arachidonic acid-gated Ca2+ channel68. Thus, this question remains until some of the functions of these ancestral homologs are better characterized. (See Box 1 for additional food for thought on why ER Ca2+ levels are below the extracellular levels and a putative alternative SOCE mechanism to regulate Ca2+ influx in fungi.)
Box 1.
ER Ca2+ homeostasis without STIM and Orai
Eukaryotic cells need to keep a high Ca2+ concentration (~400 µM) in the ER to regulate chaperones and enzymes that control protein folding, modification, and secretion. How do organisms that lack STIM and Orai regulate ER Ca2+? One mechanism has been uncovered in the yeast Saccharomyces cerevisiae. In this organism, depletion of Ca2+ from the secretory pathway results in Ca2+ influx across the plasma membrane through a voltage-gated Ca2+ channel homolog Cch1 (which has no homology to Orai)104 and this leads indirectly to refilling of ER Ca2+ stores. This regulatory process involves two stress response pathways. One is the unfolded protein response (UPR)105 that is triggered by low ER and Golgi Ca2+ which leads to incorrect folding, glycosylation, and sorting of secreted proteins. This in turns activates the cell wall integrity MAP kinase pathway which protects cells from lysis during polarized growth and in response to hypo-osmotic shock105,106. This suggests that yeast employ a STIM-independent ER Ca2+ homeostasis mechanism that is triggered indirectly by defects in the secretory system. While there is no evidence for a similar UPR- related Ca2+ influx mechanism in humans, a close link between ER Ca2+ homeostasis and ER protein folding and cell stress has also been found in humans and other animals107. It is therefore conceivable that STIM or other sensors may, in addition to their signaling and homeostatic roles, have roles related to the ER stress response.
If high ER Ca2+ levels prevent ER stress, this raises the question of why Ca2+ levels in the ER are not much higher and are instead relatively close to the threshold for triggering a stress response (basal ER Ca2+ is ~ 400 µM [Ca2+] compared to ~ 1.5 mM outside of the cell). A plausible reason is that ER Ca2+-regulated chaperones also have a regulatory role so that the level of ER Ca2+ may adjust the rate of secretion of selected proteins. Furthermore, the level of ER Ca2+ may have to be kept below that of the extracellular environment to reduce the chance that adhesion proteins that have weak Ca2+ binding sites108 inadvertently polymerize along the secretory path before reaching the outside of cells (Ca2+ is needed for many secreted proteins to bind to each other). Thus, several open questions remain concerning how ER Ca2+ levels are regulated, how STIM signaling is connected to protein folding and secretion and how these processes are related to ER stress and human disease.
Core regulatory motifs of human STIMs with common evolutionary roots
After STIM1 and STIM2 proteins were discovered in 2005, a burst of research led to the characterization of domains and sequence motifs that define the molecular signaling steps in the activation of these two luminal ER Ca2+ sensors (Figure 3a). Key parts of the activation mechanisms are shared between STIM1 and STIM249,69. In short, activation begins with Ca2+ dissociation from a conserved EF-SAM module in the lumen of the ER3,70 that triggers STIM oligomerization followed by translocation of the oligomers to ER-PM junctions where they directly interact with PM-localized Orai channels and induce Ca2+ influx.
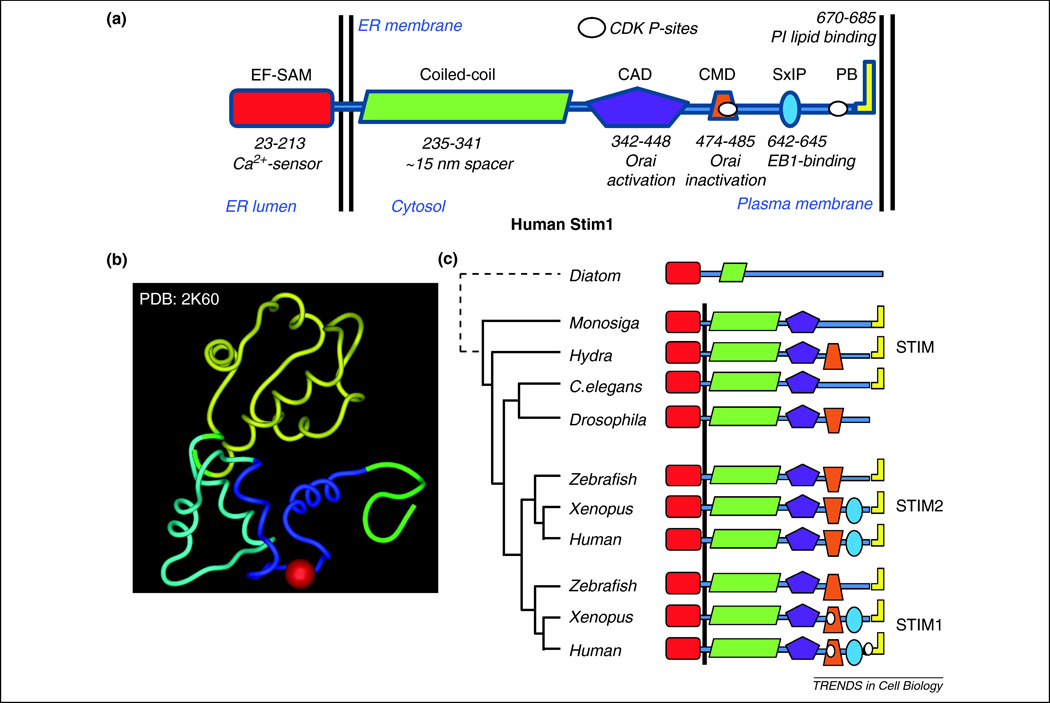
Figure 3.
Evolution of regulatory motifs in STIM proteins. (a) A diagram of the domain organization of human STIM1. The domains are annotated with residue numbers and brief functional descriptions. (b) High-resolution structure of the EF-SAM domain of STIM1 in Ca2+-bound form70. (c) Diagrams of the domain organization of STIM homologs in Monosiga brevicollis (Monosiga), Hydra magnipapillata (Hydra), Caenorhabditis elegans (C. elegans), Drosophila melanogaster (Drosophila), Danio rerio (Zebrafish), Xenopus laevis (Xenopus), and Homo sapiens (Human). Diagrams for both STIM1 and STIM2 are shown for the vertebrate organisms. Invertebrates and Monosiga brevicollis have only a single STIM homolog. Also included is a distant STIM-related protein in Thalassiosira pseudonana (Diatom). The tree indicates the topology of the phylogenetic relationships between the included STIM proteins.
A detailed description of this signaling process begins with the luminal EF-SAM module of both STIM1 and STIM2, which each bind a single Ca2+ ion (Figure 3b)70, suppressing their homo or hetero-oligomerization49,70–72. Several studies suggest that STIM may already be present as an inactive dimer in unstimulated cells73–75 and then undergo higher order oligomerization upon Ca2+ dissociation. Independent of the basal state, upon depletion of ER Ca2+ stores, STIM proteins undergo a rapid increase in their oligomeric state, and this increase triggers their translocation to the PM where they bind and activate Orai. Live cell fluorescence resonance energy transfer (FRET) experiments show that oligomerization occurs within seconds after a drop in ER Ca2+ while STIM is still in ER membranes away from ER-PM junctions71. In turn, this rapid oligomerization process is sufficient for STIM to translocate to ER-PM junctions and activate Orai76. Post-oligomerization, the translocation process likely involves a local passive diffusion of STIM proteins over a few micrometers71 to reach nearby ER-PM junctions.
The evolutionarily conserved minimal functional motifs of STIM1 and STIM2 (Figure 3c) consist of the EF-SAM Ca2+ sensor in the lumen of the ER, a single transmembrane spanning domain and a cytosolic coiled-coil domain that likely serves as a ~15 nm spacer (estimated as 0.15 nm per amino acid for an alpha helix77) to bridge the space between the ER and PM (in STIM homologs, this domain is often broken into two segments). A subsequent CRAC activation domain (CAD; also termed a STIM1 Orai activating region, SOAR) also contributes to the oligomerization73,78,79 and acts as the main mediator of Orai recruitment and activation. The formation of a CAD tetramer is sufficient for binding to Orai79 and, within the native protein, the same CAD is required to recruit and activate PM-localized Orai at ER-PM junctions.
Evolutionary and functional separation of STIM1 and STIM2
STIM1 and STIM2 proteins appeared by gene duplication from a common ancestral STIM protein during the evolution of vertebrates approximately 500 million years ago62 (Figure 3c). The conservation of both types of STIM proteins in all currently sequenced vertebrate species suggests that they have non-redundant important roles. A main molecular difference between STIM1 and STIM2 is an approximately 2-fold difference in their respective Ca2+ sensitivity. The ER Ca2+ level where translocation starts to be triggered is approximately 400 µM for STIM2 and 200 µM for STIM149. In cells where both proteins are present, this difference in Ca2+ affinity makes STIM2 more likely to be a main regulator of basal ER and cytosolic Ca2+ levels. In addition, STIM2 can also generate Ca2+ signals for weak receptor stimuli that cause only a small reduction in ER Ca2+. STIM1, which is typically present at much higher concentrations than STIM2, is only activated for stronger receptor stimuli. The activation of both STIM isoforms is very steep, changing cooperatively with ER Ca2+49. STIM2 has further been shown to have a lower relative activity than STIM1 in triggering Orai activation49,69, suggesting that it is well suited for basal Ca2+ homeostasis, which typically requires small Ca2+ fluxes. Nevertheless, in T-cells, STIM2 is strongly upregulated following stimulation and prolongs the long-term Ca2+ signals80, possibly allowing ER Ca2+ levels to remain sufficiently high to prevent an ER stress response while still driving long term SOCE Ca2+ influx. STIM2 also has important functions in neuronal and muscle Ca2+ homeostasis59,81, consistent with a ubiquitous role in long term Ca2+ homeostasis and signaling.
Evolution of additional regulatory features of STIM1 and STIM2
In addition to the four core domains, all vertebrates and some invertebrates, but not insects, have a polybasic charged motif at the C-terminus of STIM that is reminiscent of an electrostatic phosphoinositide lipid binding motif82. Indeed, this motif has been shown to bind to phosphoinositide lipids in vitro83, and phosphoinositide lipids are highly enriched in the PM. While the eight charges in a STIM1 monomer are too weak to induce PM binding on their own82, it is interesting that the translocation of oligomerized STIM1 to ER-PM junctions in the absence of Orai requires this motif71. The motif is not required to trigger Orai activation in cells where both proteins are overexpressed79. Since STIM and Orai are present physiologically at much lower levels, the polybasic region likely enhances the retention of STIM at ER-PM junctions and thereby increases the kinetics and cooperativity of STIM-mediated Orai binding and activation. The same motif may play a subsequent additional role by participating in the interaction with Orai84. The Orai-independent lipid interaction of STIM is likely a result of an oligomerization-mediated exposure of a large polybasic surface (consisting of the combined polybasic tails) that then becomes sufficiently strong to bind to phosphoinositide lipids in ER-PM junctions71,85. In other words, each polybasic C-terminal peptide may only contribute a weak binding activity that generates in the oligomer a high affinity phosphoinositide interaction and drives the translocation. While the polybasic motif is conserved in STIM proteins in vertebrates62 and in Monosiga brevicollis86, it is not always present. STIM proteins in Drosophila melanogaster do not have the polybasic motif62 and may instead rely on direct regulation of Orai by STIM without support by this intriguing lipid facilitation mechanism.
Some invertebrate and all vertebrate STIMs also have a stretch of negative charges referred to as a CRAC modulatory domain (CMD) or inactivation domain of STIM (IDSTIM) that mediates delayed Orai inactivation87,50. This domain appears to be absent in Monosiga brevicollis, and the Monosiga brevicollis genome86 lacks an apparent homolog of the recently characterized SOCE regulator CRACR2A or its paralog CRACR2B (grouped together as CRACR2 in Figure 2). CRACR2A is an EF-hand containing protein that interacts with both STIM and Orai. Upon ER Ca2+ release, CRACR2A first enhances STIM-Orai interaction in its Ca2+ free form and then suppresses STIM-Orai interaction following Ca2+ binding51. The absence of CMD domains and CRACR2 in M. brevicollis suggests that Ca2+-dependent feedback mechanisms to inactivate Orai may have been elaborations only advantageous in some multicellular organisms. Nevertheless, the role of this region in different multicellular species is at this point not yet fully understood.
STIM also has an [S/T]xIP sequence, a transport motif that mediates STIM binding to the microtubule plus-end tracking protein EB1 and directs STIM movements within the ER membrane88,89. An [S/T]xIP motif is present in STIM1 and STIM2 in many vertebrate genomes including those of Xenopus laevis and Gallus gallus, but it is absent in Danio rerio and in the invertebrates Drosophila melanogaster and Caenorhabditis elegans90–94. The EB1 binding interaction is apparently lost upon oligomerization88, suggesting that microtubule binding helps STIM proteins localize to regions in the ER closer to the periphery of cells without directly mediating translocation to ER-PM junctions. As discussed above, STIM oligomers may then translocate to ER-PM junctions over short distances by a diffusion and capture mechanism. In addition, the STIM-mediated coupling of ER and microtubules may also support, together with other coupling mechanisms, the well known role of microtubules in ER morphology and tubule formation88,89. Finally, phosphorylation sites matching a cyclin-dependent kinase consensus sequence have been identified in STIM195. Phosphorylation of these sites suppresses STIM1-mediated Ca2+ influx during mitosis95. They are specific to STIM1, and the predicted phosphorylation sites are only conserved in higher vertebrates. It is unclear whether a similar mechanism is used in other organisms. Thus, while the pathway includes a number of important modulators, the main function of STIM1 and STIM2 proteins is to monitor ER luminal Ca2+ levels and enhance Ca2+ influx across the PM when ER Ca2+ levels drop below critical respective thresholds.
The above discussion argues that STIM proteins are part of interconnected cytosolic, ER and mitochondrial Ca2+ signaling systems that differ between organisms and cell types. The purpose of such complex systems with Ca2+ as a single nodal point is likely to integrate multiple upstream receptor and sensory inputs that merge onto Ca2+, which then controls in parallel a large number of mediators that in turn coordinate processes such as cell contraction, secretion, translation, transcription and differentiation to generate an organized cell specific physiological response.
Conclusions
We have discussed STIM proteins using an evolutionary perspective that provides valuable molecular insights into the activation and role of STIM in the context of other Ca2+ channels, transporters and regulatory processes. This led to insights into the functions and origins of the SOCE signaling pathway. Our considerations suggest that the ER Ca2+ sensor STIM may have arisen in evolution after an existing Orai PM Ca2+ channel and co-opted Orai for two purposes: first, to stabilize ER Ca2+ levels and, second, to support increases in amplitude and duration of IP3R signaling. These increases in IP3R signaling may have coincided with the appearance of receptor coupling to PLCβ for IP3 production. Duplication of STIM in an early vertebrate ancestor allowed specialization of two regulatory functions with STIM2 specializing as a basal homeostatic Ca2+ regulator that also responds to weak receptor stimuli (sensing small decreases in ER Ca2+) and STIM1 playing a primary role in prolonging and increasing cytosolic Ca2+ signals following strong receptor stimulation that triggers larger drops in ER Ca2+.
Glossary
Important Ca2+-related transport and signaling proteins.
- CALM
Calmodulin. Most ubiquitous and important mediator of cytosolic Ca2+ signaling. Binds four Ca2+ ions.
- CANX
Calnexin. Ca2+-regulated ER chaperone.
- CALNA, CALNB
Calcineurin (catalytic and regulatory subunits, respectively). Ca2+/CALM-regulated Ser/Thr phosphatase.
- CALR
Calreticulin. Ca2+-regulated ER chaperone.
- CASQ
Calsequestrin. Ca2+-binding protein that enhances storage capacity of the ER.
- CholRCC
PM-localized cholinergic Ca2+ channels (e.g., acetylcholine receptors).
- CNGCC
PM cyclic nucleotide-gated Ca2+ channels.
- CRACR2
Ca2+-regulated adapter for Orai and STIM. CRACR2A has a characterized role in regulating SOCE51, but we also include here its less characterized paralog CRACR2B.
- EB1
Microtubule plus end binding protein.
- GluRCC
PM-localized glutamate-regulated Ca2+ channels (nicotinic).
- GαQ
G-protein alpha subunit that regulates PLCβ.
- IP3R
IP3-regulated ER-localized Ca2+ channel.
- LETM1
Mitochondrial Ca2+/H+ exchanger.
- MICU1
Ca2+ sensor that regulates the Ca2+ uniporter.
- NCKX
(SLC24A1–5) PM-localized Na+/K+/Ca2+ exchanger.
- NCLX
(SLC24A6). PM-localized Na+/Li+/Ca2+ exchanger.
- NCX
(SLC8A). PM-localized Na+/Ca2+ exchanger.
- NFATc
Nuclear factor for activated T-cell, key regulator of T-cell differentiation.
- Orai
PM Ca2+ channel regulated by the ER Ca2+ sensor STIM
- PKD
Polycystic kidney disease Ca2+ channel, TRP subfamily.
- PLCβ,δ
Phospholipase C, hydrolyzes phosphatidylinositol 4,5-bisphosphate, produces inositol 1,4,5-trisphosphate (IP3) and diacylglycerol.
- PMCA
(ATP2B). PM Ca2+ pump regulated by Ca2+/CALM.
- PurRCC
ATP-regulated purinergic PM Ca2+ channels.
- RyR
ER-localized Ca2+ channel regulated by Ca2+.
- SERCA
(ATP2A) ER-localized Ca2+ pump.
- SPCA
(ATP2C). Golgi- and endosome-localized Ca2+ pump.
- STIM
ER luminal Ca2+ sensor that controls store-operated Ca2+ influx into cells.
- TPCC
Two-pore channel. Internal endosomal NAADP-regulated Ca2+ channel.
- TRP
(TRPc, TRPv …) PM Ca2+ channels often involved in sensory signal transduction.
- VGCC
PM-localized voltage-gated Ca2+ channels.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Clapham DE. Calcium signaling. Cell. 2007;131:1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 2.Dominguez DC. Calcium signalling in bacteria. Mol. Microbiol. 2004;54:291–297. doi: 10.1111/j.1365-2958.2004.04276.x. [DOI] [PubMed] [Google Scholar]
- 3.Liou J, et al. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr. Biol. 2005;15:1235–1241. doi: 10.1016/j.cub.2005.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roos J, et al. STIM1, an essential and conserved component of store-operated Ca2+ channel function. J. Cell Biol. 2005;169:435–445. doi: 10.1083/jcb.200502019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feske S, et al. A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature. 2006;441:179–185. doi: 10.1038/nature04702. [DOI] [PubMed] [Google Scholar]
- 6.Zhang SL, et al. Genome-wide RNAi screen of Ca(2+) influx identifies genes that regulate Ca(2+) release-activated Ca(2+) channel activity. Proc. Natl. Acad. Sci. U.S.A. 2006;103:9357–9362. doi: 10.1073/pnas.0603161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vig M, et al. CRACM1 is a plasma membrane protein essential for store-operated Ca2+ entry. Science. 2006;312:1220–1223. doi: 10.1126/science.1127883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Putney JW. A model for receptor-regulated calcium entry. Cell Calcium. 1986;7:1–12. doi: 10.1016/0143-4160(86)90026-6. [DOI] [PubMed] [Google Scholar]
- 9.Lytton J. Na+/Ca2+ exchangers: three mammalian gene families control Ca2+ transport. Biochem. J. 2007;406:365–382. doi: 10.1042/BJ20070619. [DOI] [PubMed] [Google Scholar]
- 10.Schnetkamp PPM. The SLC24 Na+/Ca2+-K+ exchanger family: vision and beyond. Pflugers Arch. 2004;447:683–688. doi: 10.1007/s00424-003-1069-0. [DOI] [PubMed] [Google Scholar]
- 11.Meldolesi J, Pozzan T. The endoplasmic reticulum Ca2+ store: a view from the lumen. Trends Biochem. Sci. 1998;23:10–14. doi: 10.1016/s0968-0004(97)01143-2. [DOI] [PubMed] [Google Scholar]
- 12.Wuytack F, Raeymaekers L, Missiaen L. PMR1/SPCA Ca2+ pumps and the role of the Golgi apparatus as a Ca2+ store. Pflugers Arch. 2003;446:148–153. doi: 10.1007/s00424-003-1011-5. [DOI] [PubMed] [Google Scholar]
- 13.Rizzuto R, Bernardi P, Pozzan T. Mitochondria as all-round players of the calcium game. J. Physiol. (Lond.) 2000;529(Pt 1):37–47. doi: 10.1111/j.1469-7793.2000.00037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rizzuto R, et al. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science. 1998;280:1763–1766. doi: 10.1126/science.280.5370.1763. [DOI] [PubMed] [Google Scholar]
- 15.Spät A, Szanda G, Csordás G, Hajnóczky G. High- and low-calcium-dependent mechanisms of mitochondrial calcium signalling. Cell Calcium. 2008;44:51–63. doi: 10.1016/j.ceca.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rizzuto R, et al. Ca(2+) transfer from the ER to mitochondria: when, how and why. Biochim. Biophys. Acta. 2009;1787:1342–1351. doi: 10.1016/j.bbabio.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang D, Zhao L, Clapham DE. Genome-wide RNAi screen identifies Letm1 as a mitochondrial Ca2+/H+ antiporter. Science. 2009;326:144–147. doi: 10.1126/science.1175145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palty R, et al. NCLX is an essential component of mitochondrial Na+/Ca2+ exchange. Proc. Natl. Acad. Sci. U.S.A. 2010;107:436–441. doi: 10.1073/pnas.0908099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perocchi F, et al. MICU1 encodes a mitochondrial EF hand protein required for Ca(2+) uptake. Nature. 2010 doi: 10.1038/nature09358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montell C. The TRP superfamily of cation channels. Sci. STKE. 2005;2005:re3. doi: 10.1126/stke.2722005re3. [DOI] [PubMed] [Google Scholar]
- 21.Kaupp UB, Seifert R. Cyclic nucleotide-gated ion channels. Physiol. Rev. 2002;82:769–824. doi: 10.1152/physrev.00008.2002. [DOI] [PubMed] [Google Scholar]
- 22.Fritz-Laylin LK, et al. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell. 2010;140:631–642. doi: 10.1016/j.cell.2010.01.032. [DOI] [PubMed] [Google Scholar]
- 23.Wheeler GL, Brownlee C. Ca2+ signalling in plants and green algae--changing channels. Trends Plant Sci. 2008;13:506–514. doi: 10.1016/j.tplants.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 24.Denis V, Cyert MS. Internal Ca(2+) release in yeast is triggered by hypertonic shock and mediated by a TRP channel homologue. J. Cell Biol. 2002;156:29–34. doi: 10.1083/jcb.200111004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou J. Polycystins and primary cilia: primers for cell cycle progression. Annu. Rev. Physiol. 2009;71:83–113. doi: 10.1146/annurev.physiol.70.113006.100621. [DOI] [PubMed] [Google Scholar]
- 26.Ludlow MJ, Traynor D, Fisher PR, Ennion SJ. Purinergic-mediated Ca2+ influx in Dictyostelium discoideum. Cell Calcium. 2008;44:567–579. doi: 10.1016/j.ceca.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merchant SS, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor CW, Prole DL, Rahman T. Ca(2+) channels on the move. Biochemistry. 2009;48:12062–12080. doi: 10.1021/bi901739t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berridge MJ. Inositol trisphosphate and calcium signalling mechanisms. Biochim. Biophys. Acta. 2009;1793:933–940. doi: 10.1016/j.bbamcr.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 30.Bezprozvanny I, Watras J, Ehrlich BE. Bell-shaped calcium-response curves of Ins(1,4,5)P3- and calcium-gated channels from endoplasmic reticulum of cerebellum. Nature. 1991;351:751–754. doi: 10.1038/351751a0. [DOI] [PubMed] [Google Scholar]
- 31.Hamilton SL, Serysheva II. Ryanodine receptor structure: progress and challenges. J. Biol. Chem. 2009;284:4047–4051. doi: 10.1074/jbc.R800054200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu MX, et al. Calcium signaling via two-pore channels: local or global, that is the question. Am. J. Physiol., Cell Physiol. 2010;298:C430–C441. doi: 10.1152/ajpcell.00475.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ladenburger E, Korn I, Kasielke N, Wassmer T, Plattner H. An Ins(1,4,5)P3 receptor in Paramecium is associated with the osmoregulatory system. J. Cell. Sci. 2006;119:3705–3717. doi: 10.1242/jcs.03075. [DOI] [PubMed] [Google Scholar]
- 34.Wilczynska Z, et al. Release of Ca2+ from the endoplasmic reticulum contributes to Ca2+ signaling in Dictyostelium discoideum. Eukaryotic Cell. 2005;4:1513–1525. doi: 10.1128/EC.4.9.1513-1525.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rhee SG. Regulation of phosphoinositide-specific phospholipase C. Annu. Rev. Biochem. 2001;70:281–312. doi: 10.1146/annurev.biochem.70.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rebecchi MJ, Pentyala SN. Structure, function, and control of phosphoinositide-specific phospholipase C. Physiol. Rev. 2000;80:1291–1335. doi: 10.1152/physrev.2000.80.4.1291. [DOI] [PubMed] [Google Scholar]
- 37.Tsui MM, York JD. Roles of inositol phosphates and inositol pyrophosphates in development, cell signaling and nuclear processes. Adv. Enzyme Regul. 2010;50:324–337. doi: 10.1016/j.advenzreg.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krinke O, Novotná Z, Valentová O, Martinec J. Inositol trisphosphate receptor in higher plants: is it real? J. Exp. Bot. 2007;58:361–376. doi: 10.1093/jxb/erl220. [DOI] [PubMed] [Google Scholar]
- 39.Belde PJ, Vossen JH, Borst-Pauwels GW, Theuvenet AP. Inositol 1,4,5-trisphosphate releases Ca2+ from vacuolar membrane vesicles of Saccharomyces cerevisiae. FEBS Lett. 1993;323:113–118. doi: 10.1016/0014-5793(93)81460-h. [DOI] [PubMed] [Google Scholar]
- 40.Armbrust EV, et al. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science. 2004;306:79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- 41.Zhu MX, et al. Calcium signaling via two-pore channels: local or global, that is the question. Am. J. Physiol., Cell Physiol. 2010;298:C430–C441. doi: 10.1152/ajpcell.00475.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cherry JM, et al. Genetic and physical maps of Saccharomyces cerevisiae. Nature. 1997;387:67–73. [PMC free article] [PubMed] [Google Scholar]
- 43.Coe H, Michalak M. Calcium binding chaperones of the endoplasmic reticulum. Gen. Physiol. Biophys. 2009;28:F96–F103. Spec No Focus. [PubMed] [Google Scholar]
- 44.Ivessa NE, De Lemos-Chiarandini C, Gravotta D, Sabatini DD, Kreibich G. The Brefeldin A-induced retrograde transport from the Golgi apparatus to the endoplasmic reticulum depends on calcium sequestered to intracellular stores. J. Biol. Chem. 1995;270:25960–25967. doi: 10.1074/jbc.270.43.25960. [DOI] [PubMed] [Google Scholar]
- 45.Missiaen L, Dode L, Vanoevelen J, Raeymaekers L, Wuytack F. Calcium in the Golgi apparatus. Cell Calcium. 2007;41:405–416. doi: 10.1016/j.ceca.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 46.Navazio L, et al. Functional conservation of calreticulin in Euglena gracilis. J. Eukaryot. Microbiol. 1998;45:307–313. doi: 10.1111/j.1550-7408.1998.tb04541.x. [DOI] [PubMed] [Google Scholar]
- 47.Antebi A, Fink GR. The yeast Ca(2+)-ATPase homologue, PMR1, is required for normal Golgi function and localizes in a novel Golgi-like distribution. Mol. Biol. Cell. 1992;3:633–654. doi: 10.1091/mbc.3.6.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang SL, et al. STIM1 is a Ca2+ sensor that activates CRAC channels and migrates from the Ca2+ store to the plasma membrane. Nature. 2005;437:902–905. doi: 10.1038/nature04147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brandman O, Liou J, Park WS, Meyer T. STIM2 is a feedback regulator that stabilizes basal cytosolic and endoplasmic reticulum Ca2+ levels. Cell. 2007;131:1327–1339. doi: 10.1016/j.cell.2007.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mullins FM, Park CY, Dolmetsch RE, Lewis RS. STIM1 and calmodulin interact with Orai1 to induce Ca2+-dependent inactivation of CRAC channels. Proc. Natl. Acad. Sci. U.S.A. 2009;106:15495–15500. doi: 10.1073/pnas.0906781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Srikanth S, et al. A novel EF-hand protein, CRACR2A, is a cytosolic Ca2+ sensor that stabilizes CRAC channels in T cells. Nat. Cell Biol. 2010;12:436–446. doi: 10.1038/ncb2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Y, et al. The calcium store sensor, STIM1, reciprocally controls Orai and CaV1.2 channels. Science. 2010;330:105–109. doi: 10.1126/science.1191086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park CY, Shcheglovitov A, Dolmetsch R. The CRAC channel activator STIM1 binds and inhibits L-type voltage-gated calcium channels. Science. 2010;330:101–105. doi: 10.1126/science.1191027. [DOI] [PubMed] [Google Scholar]
- 54.Yuan JP, et al. TRPC channels as STIM1-regulated SOCs. Channels (Austin) 2009;3:221–225. doi: 10.4161/chan.3.4.9198. [DOI] [PubMed] [Google Scholar]
- 55.Ng LC, Airey JA, Hume JR. The contribution of TRPC1 and STIM1 to capacitative Ca(2+) entry in pulmonary artery. Adv. Exp. Med. Biol. 2010;661:123–135. doi: 10.1007/978-1-60761-500-2_8. [DOI] [PubMed] [Google Scholar]
- 56.Lefkimmiatis K, et al. Store-operated cyclic AMP signalling mediated by STIM1. Nat. Cell Biol. 2009;11:433–442. doi: 10.1038/ncb1850. [DOI] [PubMed] [Google Scholar]
- 57.Crabtree GR, Olson EN. NFAT signaling: choreographing the social lives of cells. Cell. 2002;109 Suppl:S67–S79. doi: 10.1016/s0092-8674(02)00699-2. [DOI] [PubMed] [Google Scholar]
- 58.Picard C, et al. STIM1 mutation associated with a syndrome of immunodeficiency and autoimmunity. N. Engl. J. Med. 2009;360:1971–1980. doi: 10.1056/NEJMoa0900082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Berna-Erro A, et al. STIM2 regulates capacitive Ca2+ entry in neurons and plays a key role in hypoxic neuronal cell death. Sci Signal. 2009;2:ra67. doi: 10.1126/scisignal.2000522. [DOI] [PubMed] [Google Scholar]
- 60.Launikonis BS, Murphy RM, Edwards JN. Toward the roles of store-operated Ca(2+) entry in skeletal muscle. Pflugers Arch. 2010 doi: 10.1007/s00424-010-0856-7. [DOI] [PubMed] [Google Scholar]
- 61.Cai X. Unicellular Ca2+ signaling 'toolkit' at the origin of metazoa. Mol. Biol. Evol. 2008;25:1357–1361. doi: 10.1093/molbev/msn077. [DOI] [PubMed] [Google Scholar]
- 62.Cai X. Molecular evolution and functional divergence of the Ca(2+) sensor protein in store-operated Ca(2+) entry: stromal interaction molecule. PLoS ONE. 2007;2:e609. doi: 10.1371/journal.pone.0000609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Royer L, Ríos E. Deconstructing calsequestrin. Complex buffering in the calcium store of skeletal muscle. J. Physiol. (Lond.) 2009;587:3101–3111. doi: 10.1113/jphysiol.2009.171934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rensing SA, et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- 65.Zhou Y, Ramachandran S, Oh-Hora M, Rao A, Hogan PG. Pore architecture of the ORAI1 store-operated calcium channel. Proc. Natl. Acad. Sci. U.S.A. 2010;107:4896–4901. doi: 10.1073/pnas.1001169107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yeromin AV, et al. Molecular identification of the CRAC channel by altered ion selectivity in a mutant of Orai. Nature. 2006;443:226–229. doi: 10.1038/nature05108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Prakriya M, et al. Orai1 is an essential pore subunit of the CRAC channel. Nature. 2006;443:230–233. doi: 10.1038/nature05122. [DOI] [PubMed] [Google Scholar]
- 68.Shuttleworth TJ. Arachidonic acid, ARC channels, and Orai proteins. Cell Calcium. 2009;45:602–610. doi: 10.1016/j.ceca.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Parvez S, et al. STIM2 protein mediates distinct store-dependent and store-independent modes of CRAC channel activation. FASEB J. 2008;22:752–761. doi: 10.1096/fj.07-9449com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stathopulos PB, Zheng L, Li G, Plevin MJ, Ikura M. Structural and mechanistic insights into STIM1-mediated initiation of store-operated calcium entry. Cell. 2008;135:110–122. doi: 10.1016/j.cell.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 71.Liou J, Fivaz M, Inoue T, Meyer T. Live-cell imaging reveals sequential oligomerization and local plasma membrane targeting of stromal interaction molecule 1 after Ca2+ store depletion. Proc. Natl. Acad. Sci. U.S.A. 2007;104:9301–9306. doi: 10.1073/pnas.0702866104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stathopulos PB, Zheng L, Ikura M. Stromal interaction molecule (STIM) 1 and STIM2 calcium sensing regions exhibit distinct unfolding and oligomerization kinetics. J. Biol. Chem. 2009;284:728–732. doi: 10.1074/jbc.C800178200. [DOI] [PubMed] [Google Scholar]
- 73.Covington ED, Wu MM, Lewis RS. Essential role for the CRAC activation domain in store-dependent oligomerization of STIM1. Mol. Biol. Cell. 2010;21:1897–1907. doi: 10.1091/mbc.E10-02-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Baba Y, et al. Coupling of STIM1 to store-operated Ca2+ entry through its constitutive and inducible movement in the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 2006;103:16704–16709. doi: 10.1073/pnas.0608358103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Penna A, et al. The CRAC channel consists of a tetramer formed by Stim-induced dimerization of Orai dimers. Nature. 2008;456:116–120. doi: 10.1038/nature07338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Luik RM, Wang B, Prakriya M, Wu MM, Lewis RS. Oligomerization of STIM1 couples ER calcium depletion to CRAC channel activation. Nature. 2008;454:538–542. doi: 10.1038/nature07065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Berg JM, Tymoczko JL, Stryer L. Biochemistry. W. H. Freeman; 2007. [Google Scholar]
- 78.Yuan JP, et al. SOAR and the polybasic STIM1 domains gate and regulate Orai channels. Nat. Cell Biol. 2009;11:337–343. doi: 10.1038/ncb1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Park CY, et al. STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to Orai1. Cell. 2009;136:876–890. doi: 10.1016/j.cell.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Oh-Hora M, et al. Dual functions for the endoplasmic reticulum calcium sensors STIM1 and STIM2 in T cell activation and tolerance. Nat. Immunol. 2008;9:432–443. doi: 10.1038/ni1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim JA, et al. Electromechanical modulation of catabolic and anabolic pathways in chronically inactive, but neurally intact, muscles. Muscle Nerve. 2010 doi: 10.1002/mus.21720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Heo WD, et al. PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science. 2006;314:1458–1461. doi: 10.1126/science.1134389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ercan E, et al. A conserved, lipid-mediated sorting mechanism of yeast Ist2 and mammalian STIM proteins to the peripheral ER. Traffic. 2009;10:1802–1818. doi: 10.1111/j.1600-0854.2009.00995.x. [DOI] [PubMed] [Google Scholar]
- 84.Calloway N, Holowka D, Baird B. A basic sequence in STIM1 promotes Ca2+ influx by interacting with the C-terminal acidic coiled coil of Orai1. Biochemistry. 2010;49:1067–1071. doi: 10.1021/bi901936q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Walsh CM, et al. Role of phosphoinositides in STIM1 dynamics and store-operated calcium entry. Biochem. J. 2010;425:159–168. doi: 10.1042/BJ20090884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.King N, et al. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Derler I, et al. A Ca2(+)release-activated Ca2(+) (CRAC) modulatory domain (CMD) within STIM1 mediates fast Ca2(+)-dependent inactivation of ORAI1 channels. J. Biol. Chem. 2009;284:24933–24938. doi: 10.1074/jbc.C109.024083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Grigoriev I, et al. STIM1 is a MT-plus-end-tracking protein involved in remodeling of the ER. Curr. Biol. 2008;18:177–182. doi: 10.1016/j.cub.2007.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Honnappa S, et al. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 2009;138:366–376. doi: 10.1016/j.cell.2009.04.065. [DOI] [PubMed] [Google Scholar]
- 90.Adams MD, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 91.C. elegans Sequencing Consortium Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 92.International Chicken Genome Sequencing Consortium Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 93.Klein SL, et al. Genetic and genomic tools for Xenopus research: The NIH Xenopus initiative. Dev. Dyn. 2002;225:384–391. doi: 10.1002/dvdy.10174. [DOI] [PubMed] [Google Scholar]
- 94.Jekosch K. The zebrafish genome project: sequence analysis and annotation. Methods Cell Biol. 2004;77:225–239. doi: 10.1016/s0091-679x(04)77012-0. [DOI] [PubMed] [Google Scholar]
- 95.Smyth JT, et al. Phosphorylation of STIM1 underlies suppression of store-operated calcium entry during mitosis. Nat. Cell Biol. 2009;11:1465–1472. doi: 10.1038/ncb1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rodríguez-Ezpeleta N, et al. Toward resolving the eukaryotic tree: the phylogenetic positions of jakobids and cercozoans. Curr. Biol. 2007;17:1420–1425. doi: 10.1016/j.cub.2007.07.036. [DOI] [PubMed] [Google Scholar]
- 97.Schierwater B, et al. Concatenated analysis sheds light on early metazoan evolution and fuels a modern "urmetazoon" hypothesis. PLoS Biol. 2009;7:e20. doi: 10.1371/journal.pbio.1000020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Longhorn SJ, Foster PG, Vogler AP. The nematode-arthropod clade revisited: phylogenomic analyses from ribosomal protein genes misled by shared evolutionary biases. Cladistics. 2007;23:130–144. doi: 10.1111/j.1096-0031.2006.00132.x. [DOI] [PubMed] [Google Scholar]
- 99.Aury J, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- 100.Chapman JA, et al. The dynamic genome of Hydra. Nature. 2010;464:592–596. doi: 10.1038/nature08830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Eichinger L, et al. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Putnam NH, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 103.Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 104.Locke EG, Bonilla M, Liang L, Takita Y, Cunningham KW. A homolog of voltage-gated Ca(2+) channels stimulated by depletion of secretory Ca(2+) in yeast. Mol. Cell. Biol. 2000;20:6686–6694. doi: 10.1128/mcb.20.18.6686-6694.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bonilla M, Cunningham KW. Mitogen-activated protein kinase stimulation of Ca(2+) signaling is required for survival of endoplasmic reticulum stress in yeast. Mol. Biol. Cell. 2003;14:4296–4305. doi: 10.1091/mbc.E03-02-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Levin DE. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2005;69:262–291. doi: 10.1128/MMBR.69.2.262-291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Malhotra JD, Kaufman RJ. The endoplasmic reticulum and the unfolded protein response. Semin. Cell Dev. Biol. 2007;18:716–731. doi: 10.1016/j.semcdb.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Leckband D, Prakasam A. Mechanism and dynamics of cadherin adhesion. Annu Rev Biomed Eng. 2006;8:259–287. doi: 10.1146/annurev.bioeng.8.061505.095753. [DOI] [PubMed] [Google Scholar]