Abstract
Epidermal growth factor receptor (EGFR) mutation is frequently observed in human cancer and contributes to the growth, survival and therapeutic resistance of tumors. EGFRvIII is an oncogenic EGFR mutant resulting from the deletion of exons 2–7 and is the most common EGFR mutant observed in glioblastoma multiforme, an aggressive brain tumor. EGFRvIII is constitutively active but poorly ubiquitinated, leading to inefficient receptor trafficking to lysosomes and unattenuated oncogenic signaling. The mechanism by which EGFRvIII evades downregulation is not fully understood although recent studies suggest that its interaction with the ubiquitin ligase Cbl may be compromised. In this study, we examine the regulation of EGFRvIII by the recently identified negative regulator, LRIG1, which targets EGFR through recognition of its extracellular domain. Here, we determine whether the extracellular domain deletion in EGFRvIII renders it refractory to LRIG1 regulation. We find that EGFRvIII retains interaction with LRIG1 and is in fact more sensitive to LRIG1 action than wild-type receptor. We demonstrate that LRIG1 regulation of EGFRvIII is distinct from the only other known mechanism of EGFR regulation, Cbl-mediated degradation. Ectopic expression of LRIG1 in EGFRvIII(+) glioblastoma cells opposes EGFRvIII-driven tumor cell proliferation, survival, motility and invasion. Finally, RNAi-mediated silencing of LRIG1 alters EGFRvIII intracellular trafficking and leads to enhanced EGFRvIII expression, suggesting that loss of LRIG1 in tumors may contribute to a permissive environment for EGFRvIII overexpression, contributing to EGFRvIII oncogenesis.
Keywords: EGFRvIII, LRIG1, negative regulator, glioblastoma
Introduction
Aberrant activation of the ErbB signaling network occurs in cancer via autocrine production of ligand, receptor overexpression, gene amplification and mutation. In the case of the epidermal growth factor receptor (EGFR), each of these mechanisms is operative (Sibilia et al., 2007). Overexpression of EGFR has been documented in a variety of solid tumor types and predicts reduced recurrence-free and overall patient survival. Amplification of the EGFR gene occurs in a high proportion of glioblastomas and is often accompanied by gene rearrangement (Nicholas et al., 2006). The most common of these rearrangements leads to the deletion of exons 2–7, yielding a receptor that lacks amino acids 6–273 of the extracellular domain, known as EGFR variant III (EGFRvIII). This deletion renders EGFRvIII unable to bind ligand, resulting in a receptor with weak but constitutive signaling activity, leading to enhanced tumor cell growth and survival (Rosell et al., 2006). Expression of EGFRvIII is a negative prognostic indicator in glioblastoma and mediates resistance to EGFR-targeted tyrosine kinase inhibitors (TKIs) through drug-resistant activation of Akt (Learn et al., 2004). As such, coexpression of the tumor suppressor phosphatase and tensin homologue, which opposes Akt activation, with EGFRvIII, is associated with response to EGFR-targeted TKIs both in cultured cell experiments and in a clinical setting (Mellinghoff et al., 2005).
In normal cells, ligand-stimulated activation of receptor tyrosine kinases (RTKs) is followed by attenuation of receptor signaling through internalization, ubiquitination and degradation in lysosomes. A tight coupling of these opposing processes assures the maintenance of signaling within a narrow window commensurate with normal physiology. The importance of this regulatory mechanism is underscored by the fact that a noninternalizing EGFR mutant elicits enhanced mitogenesis and cellular transformation (Wells et al., 1990). EGFRvIII is very inefficiently degraded compared to wild-type EGFR and expression of EGFRvIII is sufficient for cellular transformation (Davies et al., 2006).
The mechanism by which EGFRvIII evades downregulation, has been intensely studied yet remains controversial. A recent study found that inefficient internalization accompanied by efficient recycling to the plasma membrane contributes to the long half-life of EGFRvIII (Grandal et al., 2007). The ubiquitin ligase Cbl plays an essential role in EGFR degradation and is recruited to the receptor following ligand stimulation via two mechanisms: through a direct interaction with Y1045 of the receptor and/or indirectly through interaction with the adaptor protein Grb2 (Waterman et al., 2002). Hypophosphorylation of EGFRvIII on Y1045 has been reported to attenuate Cbl interaction with the receptor, promoting EGFRvIII stability (Han et al., 2006). However, an opposing study found that EGFRvIII interaction with Cbl remained intact and that EGFRvIII was subject to Cbl regulation in a tyrosine phosphorylation-dependent manner (Davies et al., 2006). Further study is needed to resolve whether EGFRvIII eludes regulation by Cbl.
Leucine rich repeat and immunoglobulin-like domain protein-1 (LRIG1), a recently identified negative regulator of ErbB and Met RTKs, enhances both basal and ligand-stimulated receptor ubiquitination and degradation. With respect to the EGF receptor, LRIG1 has been reported to function by augmenting Cbl recruitment to the receptor (Gur et al., 2004). However, the requirement of Cbl for LRIG1 action is not universal as LRIG1-mediated Met degradation is independent of Cbl (Shattuck et al., 2007). LRIG1 opposes tumor cell proliferation, motility and invasion in vitro and has been proposed to function as a tumor suppressor (Hedman et al., 2002). Although genetic evidence of LRIG1’s tumor suppressor activity is currently lacking, LRIG1 null mice develop a psoriasiform epidermal hyperplasia, consistent with a role for LRIG1 as a growth suppressor (Suzuki et al., 2002). In addition, LRIG1 is underexpressed in a number of tumor types (Thomasson et al., 2003; Tanemura et al., 2005; Lindström et al., 2008; Miller et al., under revision).
LRIG1 interaction with ErbB receptors is specified by a mutual interaction amongst extracellular domains (Gur et al., 2004). EGFRvIII lacks 267 amino acids of the receptor extracellular domain comprising the dimerization arm and a portion of the ligand-binding domain. This large deletion could conceivably render EGFRvIII refractory to LRIG1 action by disrupting complex formation and in turn could contribute to the long half-life of the receptor. In this manuscript we examine whether EGFRvIII escapes regulation by LRIG1. We find that EGFRvIII retains the ability to interact with LRIG1 and provide evidence that LRIG1 opposes EGFRvIII oncogenic activity.
Results
LRIG1 interacts with and destabilizes EGFRvIII
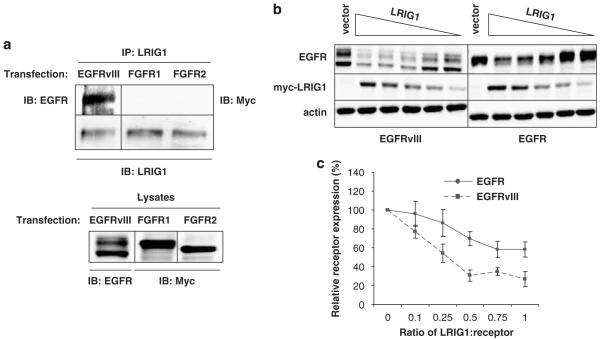
LRIG1 has recently been found to enhance ErbB and Met degradation (Gur et al., 2004; Laederich et al., 2004; Shattuck et al., 2007). Recognition of members of the ErbB family by LRIG1 requires the receptor extracellular domain, although the specific regions required are unknown (Gur et al., 2004). To examine whether the deletion in EGFRvIII affects recognition by LRIG1, a co-immunoprecipitation experiment was performed. EGFRvIII, Myc-FGFR1 or Myc-FGFR2 were transfected into HEK-293T cells, which express moderate levels of endogenous LRIG1. EGFRvIII precipitated with endogenous LRIG1 (Figure 1a) but FGFR1 and FGFR2 did not, indicating that endogenous LRIG1 and EGFRvIII form a specific complex in cells.
Figure 1.
Leucine rich repeat and immunoglobulin-like domain protein-1 (LRIG1) interacts with and destabilizes epidermal growth factor receptor variant III (EGFRvIII). (a) HEK-293T cells were transfected with EGFRvIII, myc-tagged FGFR1 or myc-tagged FGFR2. Lysates were immunoprecipitated with anti-LRIG1-151 antibody and blotted with EGFR, myc (FGFR1/2) and LRIG1 antibodies. (b) HEK-293T cells were transfected with EGFR or EGFRvIII and decreasing concentrations of LRIG1. Lysates were collected and blotted with EGFR, myc and actin antibodies. Image is representative of three independent experiments. (c) Percent of relative receptor present in (b) was analysed via densitometry and graphed for three independent experiments with standard error shown. Fractions on the x axis represent the ratio of LRIG1 cDNA transfected to EGFRvIII cDNA transfected. Western blots are representative of at least three experiments.
To examine destabilization further, LRIG1 and EGFRvIII were cotransfected into HEK-293T cells and the expression of EGFRvIII was examined by western blotting. As shown in Figure 1b (left panel), EGFRvIII expression was diminished when cotransfected with an equivalent amount of LRIG1 vector. As the LRIG1 level was decreased, EGFRvIII expression was recovered. This was compared to the efficiency with which LRIG1 destabilized wild-type EGFR to determine if the deletion in EGFRvIII protects it from LRIG1-mediated degradation. As shown in Figure 1b (right panel), a 1:1 ratio of LRIG1 to wild-type EGFR vector similarly resulted in decreased EGFR expression. As observed with EGFRvIII, as the LRIG1 level was decreased, EGFR expression was recovered. Plots of the recovery curves from several independent experiments (Figure 1c) demonstrated that EGFRvIII was more sensitive to LRIG1 action than wild-type EGFR. These data indicate that the deletion in EGFRvIII does not render this receptor refractory to LRIG1 action but may instead sensitize the receptor to LRIG1-mediated degradation.
LRIG1 destabilizes EGFRvIII in a Cbl-independent manner
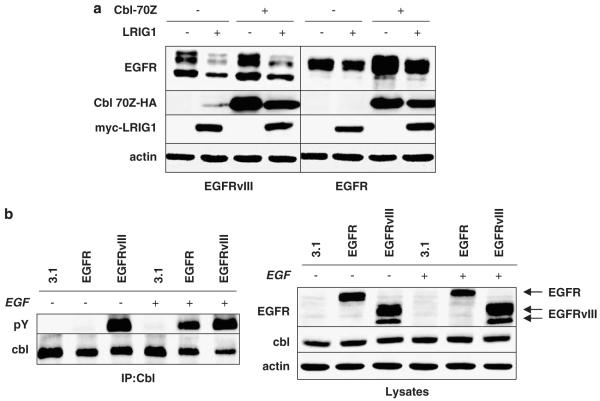
Previous studies of LRIG1-mediated degradation of EGFR found that dominant negative Cbl (Cbl-70Z) interfered with LRIG1-enhanced EGF-stimulated receptor ubiquitination, suggesting that Cbl is required for LRIG1 function (Gur et al., 2004). To determine whether Cbl is involved in LRIG1-mediated EGFRvIII degradation, EGFRvIII expression was examined in HEK-293T cells. EGFRvIII was cotransfected with vector control or myc-tagged LRIG1 in the absence or presence of hemagglutinin (HA)-tagged Cbl-70Z. As shown in Figure 2a (left panel), expression of Cbl-70Z led to a modest stabilization of EGFRvIII indicating that Cbl-70Z functions in a dominant-negative fashion with respect to this receptor. However, Cbl-70Z had no effect on the degree to which LRIG1 destabilized EGFRvIII, indicating that LRIG1 acts in a Cbl-independent manner with respect to this receptor. This finding is somewhat unexpected given the reported requirement of Cbl for LRIG1-mediated ligand-stimulated degradation of wild-type EGFR. To address this issue, a similar experiment was performed with wild-type EGFR (right panel of Figure 2a). Expression of Cbl-70Z led to the dramatic stabilization of EGFR, indicating that Cbl-70Z acts in a dominant-negative fashion with respect to this receptor, as expected. However, Cbl-70Z had no significant effect on the degree to which LRIG1 destabilized EGFR (compare the difference in lanes 1 and 2 to that in lanes 3 and 4). These results were consistently observed in a number of independent experiments (n > 10) and in several cell types (data not shown) and strongly suggest that LRIG1 does not employ Cbl to mediate ligand-independent degradation of the EGFR.
Figure 2.
Leucine rich repeat and immunoglobulin-like domain protein-1 (LRIG1)’s ability to suppress epidermal growth factor receptor variant III (EGFRvIII) is Cbl independent. (a) HEK-293T cells were cotransfected with specified combinations of EGFR, EGFRvIII, Cbl-70Z-HA, LRIG1-myc and pcDNA3.1 empty vector control. Lysates were blotted with indicated antibodies. (b) HEK-293T cells transfected with pcDNA3.1, EGFR or EGFRvIII were serum starved overnight then left untreated or treated with 3 nM EGF for 10 min. Cbl was immunoprecipitated from cleared lysates with Cbl antibody and blotted with phosphotyrosine (pY) and Cbl antibody. Western blots are representative of at least three experiments.
As Cbl tyrosine phosphorylation is a prerequisite for its action as an ubiquitin ligase (Peschard et al., 2007), we examined whether Cbl was tyrosine phosphorylated and therefore competent to participate in LRIG1-mediated EGFRvIII degradation. Endogenous Cbl was immunoprecipitated and blotted with an anti-phosphotyrosine antibody. As shown in Figure 2b (left panel), EGFRvIII expression in HEK-293T cells led to robust Cbl tyrosine phosphorylation indicating that the lack of Cbl involvement in LRIG1-mediated EGFRvIII degradation is not due to the absence of Cbl tyrosine phosphorylation. Lysates corresponding to Figure 2a are shown in Figure 2b, right panel.
LRIG1 destabilizes EGFRvIII in glioblastoma cells
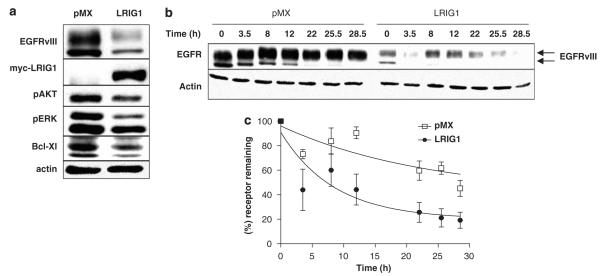
To extend our observations to a clinically relevant cell line, the impact of LRIG1 on EGFRvIII expression in glioblastoma cells was determined. U87MG-EGFRvIII cells have been engineered to express a high level of EGFRvIII and are a frequently used model of EGFRvIII(+) glioblastoma (Wang et al., 2006). These cells express a low but detectable level of endogenous LRIG1 making them an appropriate system for examining the impact of ectopic expression of LRIG1. To determine whether LRIG1 was capable of destabilizing EGFRvIII in glioblastoma cells, U87MG-EGFRvIII cells were transduced with the pMX-pie retroviral system. Cells were transduced with either empty retrovirus (pMX) or with retrovirus containing myc-tagged LRIG1. Puromycin-resistant clones were pooled to avoid artifacts due to clonal variation. As shown in Figure 3a, expression of LRIG1 in U87MG-EGFRvIII cells led to a dramatic loss of EGFRvIII expression. Coincident with the loss of EGFRvIII was a significant decrease in the constitutive phosphorylation of Akt and p42/44 MAP kinases as well as the expression of the anti-apoptotic protein, Bcl-XL, likely as a consequence of the decrease in receptor level.
Figure 3.
Leucine rich repeat and immunoglobulin-like domain protein-1 (LRIG1) destabilizes epidermal growth factor receptor variant III (EGFRvIII) in glioblastoma cells and decreases the half-life of EGFRvIII. U87MG-EGFRvIII cells were transduced with empty virus (pMX) or LRIG1-myc virus. (a) Cells were serum starved for 24 h then collected and blotted with phospho-AKT (S473), phospho-Erk (42/44), Bcl-XL, EGFR, myc and actin antibodies. For (b) and (c) U87MG-EGFRvIII cells transduced with either pMX or LRIG1-myc were incubated with media containing 100 μg/ml cycloheximide and time points were collected at 0, 3.5, 8, 12, 22, 22.5 and 28.5 h. (b) Western blot of EGFRvIII levels with cycloheximide time points. (c) The amount of receptor from six independent experiments was quantified using densitometry and graphed using a nonlinear regression line with one phase decay and a β value of 20% for LRIG1 and 40% for pMX. The β values were obtained from the average of six independent experiments by measuring the amount of receptor that was refractory to degradation. Western blots are representative of at least three experiments.
EGFRvIII is a long-lived protein due to a combination of inefficient internalization coupled with efficient recycling to the plasma membrane (Grandal et al., 2007). To determine whether LRIG1 decreases EGFRvIII protein stability, receptor half-life was measured in U87MG-EGFRvIII-pMX and –LRIG1 cells. Cells were treated with cycloheximide to halt new protein synthesis and the half-life of existing EGFRvIII was measured by examining its decay over time. This experiment was repeated six times and a representative western blot is shown in Figure 3b. Quantification of the data is shown in Figure 3c. Expression of LRIG1 led to a marked decrease in EGFRvIII half-life, from 16.4 to 5.9 h.
Ectopic expression of LRIG1 decreases the aggressive behavior of EGFRvIII(+) glioblastoma cells
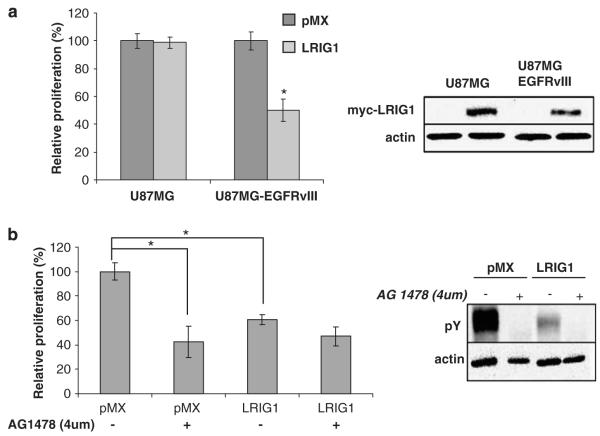
To examine whether LRIG1 impacts the growth of U87MG-EGFRvIII cells, a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was performed. An equivalent number of –pMX and –LRIG1 cells were seeded into wells and 48 h later, the total number of viable cells was measured, as shown in Figure 4a. After 48 h, there were approximately twofold more –pMX cells as compared to –LRIG1 cells indicating that LRIG1-expressing cells are at a growth disadvantage. Conversely, LRIG1 expression had no significant effect on the growth of the U87MG parental cells despite equivalent LRIG1 expression (right panel).
Figure 4.
Leucine rich repeat and immunoglobulin-like domain protein-1 (LRIG1) decreases the proliferation of epidermal growth factor receptor variant III (EGFRvIII)(+) glioblastoma cells. (a) Left panel: Cells were grown in complete media for 48 h. An 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was then conducted to measure cellular proliferation. Graph shown is a representative sample of at least three independent experiments with at least five individual readings per experiment. Bars represent standard error. Right panel: U87MG and U87MG-EGFRvIII cells were transduced with either pMX empty virus or pMX-LRIG1 virus and stable clones were pooled to avoid clonal variation. Total cell lysates were blotted with anti-LRIG1 and anti-actin antibodies. (b) U87MG-EGFRvIII pMX and LRIG1 cells were analysed via western blot after treatment for 48 h with AG1478 (right panel). An MTT of these cells was conducted as described in (a, left panel). The asterisk (*) represents P<0.05 from a Student’s t-test. Western blots are representative of at least three experiments.
The effect of LRIG1 on U87MG-EGFRvIII cell growth was comparable to AG1478, a specific EGF receptor TKI (Figure 4b). In this experiment, cells were treated with a concentration of AG1478 (4 μM) that completely inhibited EGFRvIII tyrosine phosphorylation (right panel) and the viable cell number was measured after 48 h. At this concentration, AG1478 inhibited cell growth by approximately 50%, whereas LRIG1 expression led to ~40% decline in cell growth. The modest difference in the efficiencies of cell growth inhibition between AG1478 treatment and LRIG1 expression is likely due to residual EGFRvIII tyrosine phosphorylation in –LRIG1 cells (right panel) because AG1478 treatment of –LRIG1 cells neutralized this difference. These results suggest that LRIG1 inhibits U87MG-EGFRvIII cell growth mainly through its effects on EGFRvIII expression. These results are in agreement with the observation that expression of LRIG1 had no significant effect on parental U87MG cell growth (Figure 4a).
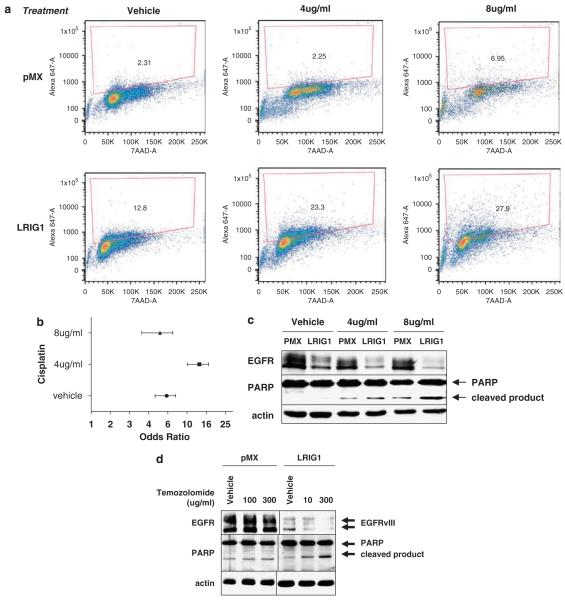
Viable cell number is an outcome of the balance between proliferation and apoptosis and EGFRvIII is known to suppress apoptosis through the activation of phosphatidylinositol-3 kinase (PI-3) kinase and the upregulation of Bcl-XL (Moscatello et al., 1998; Nagane et al., 1998). EGFRvIII(+) glioblastomas are highly invasive and infiltration of the surrounding tissue renders complete surgical resection improbable. This necessitates the use of adjuvant therapies such as the DNA-damaging agent cisplatin (cis-diamminedichloro-platinum(II)). EGFRvIII(+) glioblastoma cells are resistant to cisplatin but this resistance can be attenuated by the addition of AG1478 (Nagane et al., 1998, 2001). To examine whether LRIG1 expression sensitizes cells to cisplatin treatment, U87MG-EGFRvIII-pMX and –LRIG1 cells were treated with either vehicle control or cisplatin, and terminal transferase dUTP nick-end labeling (TUNEL) staining was measured by flow cytometry. A representative experiment chosen from three independent experiments is shown in Figure 5a. Interestingly, LRIG1-expressing cells are significantly more apoptotic both basally and following cisplatin treatment (Figure 5b). This is paralleled in a western blot of poly-ADP ribose polymerase (PARP) cleavage, a hallmark of apoptosis (Figure 5c).
Figure 5.
Leucine rich repeat and immunoglobulin-like domain protein-1 (LRIG1) sensitizes epidermal growth factor receptor variant III (EGFRvIII)(+) glioblastoma cells to drug-induced apoptosis. For (a) through (c), U87MG-EGFRvIII pMX and LRIG1 cells were treated with cisplatin or vehicle control for 48 h. Cells were collected and terminal transferase dUTP nick-end labeling (TUNEL) staining was analysed by flow cytometry. Shown is a representative example of three independent experiments. (a) Shown are scatter plots of DNA content (7AAD) vs TUNEL signal (Alexa 647) with gated region representing percent of TUNEL positive cells. (b) Forest plot graphical representation of odds ratio calculations with bars representing 95% confidence intervals. χ2-Analysis revealed P<0.001 for pMX vs LRIG1 for all three conditions. (c) Cells treated as described in (a) were analysed by western blotting with anti-poly-ADP ribose polymerase (PARP), anti-EGFR and anti-actin antibodies. (d) U87MG-EGFRvIII pMX and LRIG1 cells were treated with temozolomide or vehicle control for 48 h in serum-free media and analysed by western blotting with anti-PARP, anti-EGFR, and anti-actin antibodies. Western blots are representative of at least three experiments.
The oral alkylating agent temozolomide is used concurrently with radiotherapy followed by adjuvant administration and is the standard of care for newly diagnosed glioblastoma (Aoki et al., 2007). To examine whether LRIG1 expression sensitizes cells to temozolomide, PARP cleavage was visualized by western blotting. pMX cells did not demonstrate a significant increase in PARP cleavage compared to vehicle control whereas the increase was evident in LRIG1 cells at both doses of temozolomide. These results indicate that ectopic LRIG1 expression ‘tips the balance’ of EGFRvIII(+) glioblastoma cells towards apoptosis and sensitizes cells to both cisplatin and temozolomide.
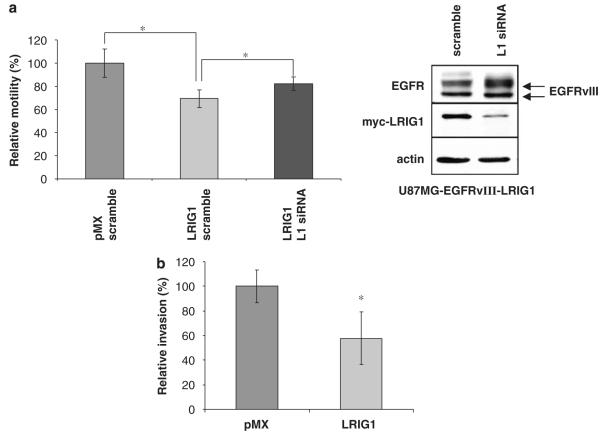
We next examined whether ectopic expression of LRIG1 could attenuate the motility and invasion of U87MG-EGFRvIII cells. As shown in Figures 6a and b, LRIG1-expressing cells were significantly less motile (a) and invasive (b) than control cells. To determine whether the differences between pMX and LRIG1 cells were due to LRIG1 expression, LRIG1-transduced cells were treated with either scramble control or LRIG1-specific siRNA (Figure 6a, left panel). Depletion of LRIG1 in LRIG1-transduced cells led to an increase in EGFRvIII expression (Figure 6a, right panel) and a restoration of motility. However, motility was not completely restored, most likely due to residual LRIG1 observed in these cells following knockdown. These data suggest that loss or depletion of LRIG1 could contribute to the aggressive nature of EGFRvIII(+) tumors.
Figure 6.
Leucine rich repeat and immunoglobulin-like domain protein-1 (LRIG1) decreases the motility and invasion of epidermal growth factor receptor variant III (EGFRvIII)(+) glioblastoma cells. For both motility and invasion assays, differences in cell growth between –pMX and –LRIG1 cells during the time period of the assay were taken into account and results were normalized so as to reflect true differences in motility or invasion rather than cell growth. (a) Left panel: Cell motility was measured using a Boyden chamber assay with the U87MG-EGFRvIII pMX and LRIG1 cells treated with either LRIG1 siRNA (black bars) or scramble control (light and dark gray bars). The assay was performed in triplicate with the average of 3, 20 × objective images per chamber shown with error bars representing standard error. Experiment shown is representative of three independent experiments. Right panel: Myc-LRIG1 levels and EGFRvIII levels were evaluated by western blot. (b) Matrigel invasion chambers were used to measure cellular invasion. The assay was performed in triplicate with the average of 3, 20 × objective images per chamber shown with error bars representing standard error. Experiment shown is representative of three independent experiments. The asterisk (*) represents P<0.05 from a Student’s t-test.
Loss of LRIG1 contributes to enhanced EGFRvIII expression
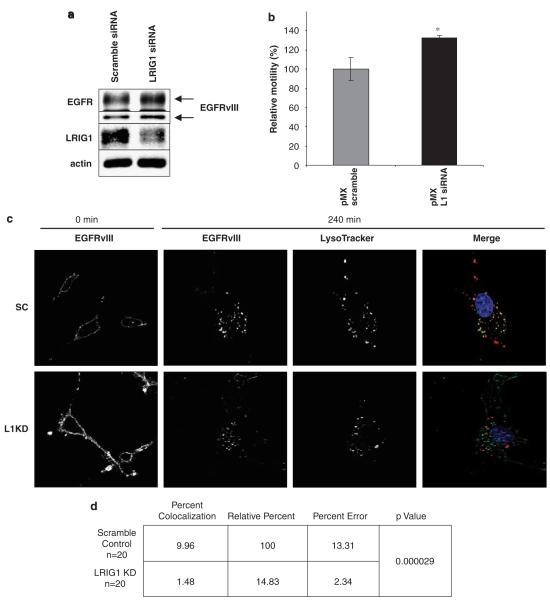
U87MG-EGFRvIII cells express ~1.5 million EGFRvIII receptors per cell along with a modest level of endogenous LRIG1. To determine whether depletion of LRIG1 could enhance the expression of EGFRvIII in this setting, endogenous LRIG1 was depleted by siRNA. LRIG1 knockdown led to a modest but reproducible increase in total EGFRvIII expression (Figure 7a), indicating that loss of LRIG1 is sufficient to enhance EGFRvIII expression, even in this setting of substantial receptor overexpression. Densitometry analysis of EGFRvIII levels from six independent experiments revealed ~40% increase in EGFRvIII expression following LRIG1 knockdown (1.4 fold ± 0.07s.e.). Interestingly, loss of LRIG1 resulted in an increase in tumor cell motility (Figure 7b). These data indicate that LRIG1 acts to oppose EGFRvIII signaling by destabilizing this receptor and that loss of LRIG1 creates a ‘permissive environment’ for receptor overexpression.
Figure 7.
Leucine rich repeat and immunoglobulin-like domain protein-1 (LRIG1) knockdown stabilizes epidermal growth factor receptor variant III (EGFRvIII) and alters EGFRvIII trafficking patterns. (a) Endogenous LRIG1 was knocked down using siRNA targeted against LRIG1 or scrambled control in U87MG-EGFRvIII cells. After 72 h, whole cell lysates were collected and immunoblotted for EGFR, LRIG1 and actin. (b) Differences in migration following LRIG1 siRNA were analysed by the Boyden chamber assay. The assay was performed in triplicate with the average of 3, 20 × objective images per chamber shown with error bars representing standard error. Experiment shown is representative of three independent experiments. The asterisk (*) represents P<0.05 from a Student’s t-test. (c) U87MG-EGFRvIII cells were transfected with either scrambled control (top row) or LRIG1 knockdown (bottom row) RNAi oligos. Trafficking of EGFRvIII was analysed by surface labeling EGFRvIII and following internalization by confocal scanning immunofluorescence microscopy. Lysotracker is used to visualize lysosomes. Single channels are represented for EGFRvIII at 0 and 240 min as well as lysotracker at 240 min. The merged image demonstrates colocalization (yellow staining) of EGFRvIII and lysosomes. (d) Olympus Fluoview software was used to analyse the percent of pixel colocalization for the region of interest within 20 different cells from the experiment described in (c). Percent colocalization represents the percentage of pixels within the region of interest that have both EGFRvIII and lysotracker staining. Relative percentage compares the amount of colocalization in the scramble control compared to LRIG1 knockdown. P-value was computed using a two-tailed t-test.
Recycling of RTKs after internalization prolongs their effective plasma membrane residency time and promotes sustained signaling. EGFRvIII evades downregulation through a combination of inefficient internalization and efficient recycling. To examine the mechanism by which LRIG1 depletion enhances EGFRvIII expression, the impact of LRIG1 depletion on of EGFRvIII trafficking was investigated. Endogenous LRIG1 was depleted in U87MG-EGFRvIII-pMX cells and the trafficking of cell-surface resident EGFRvIII was examined by immunofluorescence. As shown in Figure 7c, when LRIG1 was intact, cell surface EGFRvIII internalized and colocalized with lysosomes indicating that despite its propensity to evade downregulation, EGFRvIII does undergo some trafficking to lysosomes for degradation. In contrast, when LRIG1 was depleted, the colocalization of EGFRvIII with lysosomes was minimal, indicating that LRIG1 loss prolongs EGFRvIII half-life by promoting its escape from lysosomal degradation. It should be noted that increased expression of surface-localized EGFRvIII was noted with LRIG1 knockdown, in agreement with the results observed with western blotting. Scrambled control and LRIG1 knockdown ‘0 min’ panels are shown at the same exposure in Figure 7c to illustrate this difference. Quantification of the differences in lysosomal colocalization is shown in Figure 7d. Collectively, our results uncover a new mechanism of EGFRvIII regulation and describe for the first time a Cbl-independent mechanism of EGFR degradation.
Discussion
Aberrant or unopposed signaling from RTKs is a recurrent theme in human diseases, most notably in cancer. Receptor activation in healthy cells is balanced by receptor deactivation, mediated by a variety of negative regulatory molecules. These molecules fall into several categories, including phosphatases and ubiquitin ligases, and impinge upon both the magnitude and duration of RTK signaling (Sweeney et al., 2006; Kirisits et al., 2007). Evasion of downregulation is a powerful mechanism by which RTKs can tip the balance in the favor of uncontrolled growth and eventual tumor formation. Indeed, EGFRvIII internalization is significantly impaired compared to wild-type receptor and when coupled with efficient recycling, provides a mechanism by which growth and survival signals are amplified by this receptor (Grandal et al., 1997; Huang et al., 2007).
In this study, we considered whether EGFRvIII eludes regulation by the negative regulatory protein, LRIG1. The phenotype of the LRIG1 knockout mouse, a psoriatic-like epidermal hyperplasia, supports its role as a growth suppressor and negative regulator of EGFR signaling because transgenic mice overexpressing EGFR ligands (Cook et al., 1997; Vassar and Fuchs, 1991) display a phenotype similar to LRIG1−/− mice. Interestingly, loss of a distinct negative regulator of ErbB receptors, Mig6/RALT, also manifests in an epidermal hyperplasia, indicating that the skin is a sensitive reporter of ErbB oversignaling (Ferby et al., 2006). Deletion analysis of LRIG1 and ErbB receptors found that the extracellular domains of both proteins are necessary and sufficient for interaction (Gur et al., 2004). LRIG1 represents the first example of a protein that selects EGFR for degradation through recognition of its extracellular domain, raising the possibility that oncogenic receptors may escape regulation through deletion of LRIG1 recognition elements.
In this study, we find that the deletion in EGFRvIII does not render this receptor refractory to regulation by LRIG1. We demonstrate that EGFRvIII retains interaction with LRIG1 by co-immunoprecipitation analysis and that EGFRvIII is destabilized by LRIG1 coexpression as efficiently, if not more efficiently, than wild-type receptor. Despite a previous report that LRIG1 acts in a Cbl-dependent manner with respect to EGFR degradation, we find that Cbl activity is dispensable for LRIG1-mediated ligand-independent degradation of EGFRvIII, representing the first example of a Cbl-independent means of EGFR degradation. Presumably, LRIG1 acts by recruiting novel components of the protein degradation machinery to its targets and studies are ongoing in our lab to identify these components.
EGFRvIII(+) tumors are highly aggressive and refractory to conventional therapeutic approaches. In this study, we find that ectopic expression of LRIG1 potently inhibits tumor cell behaviors that impact clinical outcome, including growth, motility and invasion. EGFRvIII(+) glioblastomas are highly invasive and readily infiltrate surrounding brain tissue, forming a mesh-like network of tumor, making complete surgical removal impossible. Chemoresistance of remaining tumor tissue presents a significant problem, leading to recurrence following surgical resection in nearly all patients (Raizer, 2005). Treatment of U87MG-EGFRvIII cells with AG1478, a specific EGFR TKI, sensitizes these cells to cisplatin, presumably through an attenuation of EGFRvIII-mediated survival signals (Huang et al., 2007). LRIG1, by virtue of its effects on EGFRvIII stability, attenuates signaling through the pro-survival PI-3 kinase pathway and leads to a sensitization of U87MG-EGFRvIII cells to both cisplatin and the oral alkylating agent, temozolomide. These results suggest that restoration of LRIG1 to EGFRvIII(+) tumors could have far-reaching effects on several key parameters of aggressive tumor cell behavior and could perhaps represent an alternative approach for tumors that develop resistance to TKIs. However, it should be emphasized that restoration of a large transmembrane protein such as LRIG1 to tumor cells is currently not feasible and represents a technical hurdle. Use of a soluble version of LRIG1 or peptides derived from LRIG1 (Goldoni et al., 2007) may hold promise for future therapies.
Mechanisms of receptor overexpression have been the subject of intense study for some years due to their clear link to the oncogenic phenotype. An interesting theme that has emerged from recent work suggests that the loss of negative regulatory molecules creates a ‘permissive environment’ for receptor overexpression. Negative regulatory molecules continue to antagonize receptor signaling, even in the face of overwhelming receptor overexpression, and silencing or loss of these molecules further promotes tumor cell growth and survival. This is evidenced by recent work demonstrating that reconstitution of the negative regulator Mig6/RALT in ErbB2-amplified breast cancer cells inhibits ErbB2-dependent mitogenesis and reverses Herceptin resistance (Anastasi et al., 2005). These findings suggest that loss of RALT and other negative regulatory molecules such as LRIG1 may cooperate with receptor amplification to drive full oncogenic signaling. In agreement with this, depletion of LRIG1 from U87MG-EGFRvIII cells leads to a further increase in EGFRvIII expression through decreased receptor trafficking to lysosomes. These results indicate that although EGFRvIII is not refractory to LRIG1 by ‘design’, depletion or loss of LRIG1 in EGFRvIII(+) tumors could contribute to EGFRvIII overexpression and drive the oncogenic phenotype.
Materials and methods
Reagents and cell culture
AG 1478 and Cisplatin were purchased from Calbiochem (San Diego, CA, USA). Temozolomide was purchased from Sigma (St Louis, MO, USA). HEK-293T cells were purchased from American Type Culture Collection (Manassas, VA, USA). All U87 cell lines were provided by Dr Webster Cavenee and are described in Huang et al. (1997). All cell lines were cultured in Dulbecco’s modified Eagle’s medium (Mediatech, Washington, DC, USA)/10% fetal calf serum (FCS) at 37 °C and 10% CO2. Serum-starved media contains 0.1% FCS. Antibodies used include Cbl C-15 and EGFR/EGFRvIII 1005 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), phosphotyrosine (pY) 4G10 (Upstate, Billerica, MA, USA), myc (Invitrogen, Carlsbad, CA, USA), actin AC-15 (Sigma), HA (Zymed Laboratories, Carlsbad, CA, USA), pAKT (S473) and p-Erk (Cell Signaling Technologies, Danvers, MA, USA), PARP and Bcl-XL (Cell Signaling Technologies), and LRIG1-151 (Agrisera, Vännäs, Sweden). Transfections were performed using FuGENE 6 (Roche Applied Science, Indianapolis, IN, USA) according to the manufacturer’s instructions. U87MG-EGFRvIII cells were transduced with pMX-pie (pMX) retrovirus containing LRIG1-myc or control empty virus as previously described (Shattuck et al., 2007) and selected with 0.75 μg/ml puromycin. Puromycin resistant clones were pooled to avoid artifact due to clonal variation.
Immunoprecipitation and western blot analysis
Cells were lysed in co-immunoprecipitation buffer (20 mM Tris, pH 7.4, 137 mM NaCl, 0.1% Nonidet P-40, 100 μM 4-(2-Aminoethyl) benzenesulfonyl fluoride hydrochloride, and 4 μg/ml each of aprotinin, leupeptin and pepstatin), and cleared lysates were precipitated with 2 μg of antibody or control antibody. Precipitates were resolved by 8% SDS–polyacrylamide gel electrophoresis (PAGE), transferred to nitrocellulose, and blotted with indicated antibodies. Detection of antibodies was carried out using horseradish peroxidase-conjugated secondary antibodies (Zymed Laboratories), followed by development with SuperSignal West chemicals (Pierce, Rockford, IL, USA). An Alpha Innotech imaging station with FluorChem software was used to capture images.
Migration and invasion assays
For the migration assay, cells transduced with LRIG1-myc or control pMX were plated (2 × 104 per well) onto 24-well Boyden chambers (Corning, Lowell, MA, USA) with 8-μm pore polycarbonate membranes in complete medium. After 24 h, filters were fixed and stained according to the manufacturer’s instructions. Filters were air-dried and photographed using a 20 × objective. Three fields of view for each well were counted, and the results were averaged among three chambers for each cell line. For the Matrigel invasion assay, transduced U87MG-EGFRvIII cells were plated (2 × 104 per well) onto 24-well Matrigel invasion chambers (BD Biosciences, Bedford, MA, USA). After 24 h in the invasion chambers, cells were fixed and stained according to the manufacturer’s instructions. Chambers were photographed and quantified as described above.
Half-life analysis
Cells were seeded in 35 mm plates and allowed to adhere overnight before addition of 100 μg/ml cyclohexamide (Sigma). Time points were collected in sample buffer and resolved by SDS–PAGE. Densitometry was performed using FluorChem software.
Apoptosis analysis
Cisplatin was dissolved in dimethyl sulfoxide (DMSO) at 2 μg/ml then diluted into media just before use. Temozolomide was dissolved in 1 part DMSO and 3 parts phosphate-buffered saline (PBS) at 16.67 μg/ml. For TUNEL analysis, 5 × 105 cells were seeded in 6-cm dishes and allowed to adhere overnight. Media containing cisplatin or vehicle (DMSO) control was then added, and after 48 h, cells were fixed in 1% paraformaldehyde in PBS and permeabilized with 70% ethanol before proceeding with the TUNEL protocol. APO-BRDU kit with Alexa 647 antibody (Phoenix Flow Systems, San Diego, CA, USA) was used according to the manufacturer’s protocol. Cells were analysed by flow cytometry on a Becton Dickson LSR II and data were analysed using FlowJo software. For PARP analysis of cisplatin and temozolomide-treated cells, cells were treated as described for TUNEL and PARP cleavage was evaluated by western blotting. Temozolomide treatment was performed in serum-free media.
Growth analysis
Cells were seeded (2.5 × 104 cells per well) overnight in 24-well plates, and then placed in conditioned media for 48 h with MTT (Sigma) added to the media for the last 3–5 h of growth. Results were quantified by dissolving the MTT crystals in acidic isopropanol and recording absorption at 570 nm with a baseline subtraction at 655 nm. At least five points were averaged for each condition, and the experiment was repeated at least three times with a representative experiment selected.
RNAi experiments
LRIG1 was depleted in U87-EGFRvIII cells using Dharmacon ON-TARGETplus SMARTpool (Dharmacon, Lafayette, CO, USA). ON-TARGETplus siControl nontargeting pool was used as a control. Cells were transfected with 100 nM siRNA using DharmaFect1, per the manufacturer’s instructions. The medium was replaced after 24 h, and cells were left to incubate for an additional 24–48 h. Lysates were collected and analysed as described.
Immunofluorescence and confocal microscopy
Glass cover slips were placed in six-well plates with 2 × 104 U87MG-EGFRvIII cells per well. After settling overnight, cells were transfected with RNAi as specified above. After 48 h, cells were serum starved overnight. For surface labeling, cells were placed in ice-cold serum-free media containing 1 μg/ml anti-EGFR Ab-1 (clone 528; Lab Vision, Fremont, CA, USA) for 30 min. After washing briefly in PBS, cells were placed in prewarmed serum-starved media containing 50 nM Lysotracker (Invitrogen), and incubated at 37 °C for the indicated times. Cells were fixed with 4% paraformaldehyde and then washed two times with PBS. After incubating overnight in blocking solution (PBS containing 1% bovine serum albumin, 0.01% sodium azide, 2% NP-40, 5% goat serum), cells were treated with blocking solution containing 2 μg/ml Alexafluor anti-mouse 488 (Invitrogen) for 1 h, washed two times with PBS and mounted onto slides. Images were acquired on an Olympus FV1000 Spectral Scan Confocal microscope and images were analysed with Olympus Fluoview software (Olympus, Center Valley, PA, USA). Regions of interest (ROI) were drawn to contain the entire cytosol of cells. ROI were then subjected to colocalization analysis and computed in threshold mode.
Acknowledgements
We thank Dr Webster Cavenee from the Ludwig Institute for Cancer Research, University of California at San Diego, La Jolla, California for the U87MG parental and EGFRvIII expressing glioblastoma cells. We thank Dr Laurel Beckett, Chief of the Division of Biostatistics at UC Davis School of Medicine, for statistical consultation. We thank Carol Oxford, Manager of the UC Davis Optical Biology Core, for her assistance in the flow cytometry experiments. This work was supported by NIH grants CA118384 (CS) and GM068994 (KLC). DLS is a recipient of a DOD BCRP predoctoral fellowship Award no. W81XWH-06-1-0772.
References
- Anastasi S, Sala G, Huiping C, Caprini E, Russo G, Iacovelli S, et al. Loss of RALT/MIG-6 expression in ERBB2-amplified breast carcinomas enhances ErbB-2 oncogenic potency and favors resistance to Herceptin. Oncogene. 2005;24:4540–4548. doi: 10.1038/sj.onc.1208658. [DOI] [PubMed] [Google Scholar]
- Aoki T, Hashimoto N, Matsutani M. Management of glioblastoma. Expert Opin Pharmacother. 2007;8:3133–3146. doi: 10.1517/14656566.8.18.3133. [DOI] [PubMed] [Google Scholar]
- Cook PW, Piepkorn M, Clegg CH, Plowman GD, DeMay JM, Brown JR, et al. Transgenic expression of the human amphiregulin gene induces a psoriasis-like phenotype. J Clin Invest. 1997;100:2286–2294. doi: 10.1172/JCI119766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies GC, Ryan PE, Rahman L, Zajac-Kaye M, Lipkowitz S. EGFRvIII undergoes activation-dependent downregulation mediated by the Cbl proteins. Oncogene. 2006;25:6497–6509. doi: 10.1038/sj.onc.1209662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferby I, Reschke M, Kudlacek O, Knyazev P, Panté G, Amann K, et al. Mig6 is a negative regulator of EGF receptor-mediated skin morphogenesis and tumor formation. Nat Med. 2006;12:568–573. doi: 10.1038/nm1401. [DOI] [PubMed] [Google Scholar]
- Goldoni S, Iozzo RA, Kay P, Campbell S, McQuillan A, Agnew C, et al. A soluble ectodomain of LRIG1 inhibits cancer cell growth by attenuating basal and ligand-dependent EGFR activity. Oncogene. 2007;26:368–381. doi: 10.1038/sj.onc.1209803. [DOI] [PubMed] [Google Scholar]
- Grandal MV, Zandi R, Pedersen MW, Willumsen BM, van Deurs B, Poulsen HS. EGFRvIII escapes down-regulation due to impaired internalization and sorting to lysosomes. Carcinogenesis. 2007;7:1408–1417. doi: 10.1093/carcin/bgm058. [DOI] [PubMed] [Google Scholar]
- Gur G, Rubin C, Katz M, Amit I, Citri A, Nilsson J, et al. LRIG1 restricts growth factor signaling by enhancing receptor ubiquitylation and degradation. EMBO J. 2004;23:3270–3281. doi: 10.1038/sj.emboj.7600342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W, Zhang T, Yu H, Foulke JG, Tang CK. Hypophosphorylation of residue Y1045 leads to defective downregulation of EGFRvIII. Cancer Biol Ther. 2006;5:1361–1368. doi: 10.4161/cbt.5.10.3226. [DOI] [PubMed] [Google Scholar]
- Hedman H, Nilsson J, Guo D, Henriksson R. Is LRIG1 a tumour suppressor gene at chromosome 3p14.3? Acta Oncol. 2002;41:352–354. doi: 10.1080/028418602760169398. [DOI] [PubMed] [Google Scholar]
- Huang HS, Nagane M, Klingbeil CK, Lin H, Nishikawa R, Ji XD, et al. The enhanced tumorigenic activity of a mutant epidermal growth factor receptor common in human cancers is mediated by threshold levels of constitutive tyrosine phosphorylation and unattenuated signaling. J Biol Chem. 1997;272:2927–2935. doi: 10.1074/jbc.272.5.2927. [DOI] [PubMed] [Google Scholar]
- Huang PH, Mukasa A, Bonavia R, Flynn RA, Brewer ZE, Cavenee WK, et al. Quantitative analysis of EGFRvIII cellular signaling networks reveals a combinatorial therapeutic strategy for glioblastoma. Proc Natl Acad Sci USA. 2007;104:12867–12872. doi: 10.1073/pnas.0705158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisits A, Pils D, Krainer M. Epidermal growth factor receptor degradation: an alternative view of oncogenic pathways. Int J Biochem Cell Biol. 2007;39:2173–2182. doi: 10.1016/j.biocel.2007.07.012. [DOI] [PubMed] [Google Scholar]
- Laederich MB, Funes-Duran M, Yen L, Ingalla E, Wu X, Carraway KL, III, et al. The leucine-rich repeat protein LRIG1 is a negative regulator of ErbB family receptor tyrosine kinases. J Biol Chem. 2004;279:47050–47056. doi: 10.1074/jbc.M409703200. [DOI] [PubMed] [Google Scholar]
- Learn CA, Hartzell TL, Wikstrand CJ, Archer GE, Rich JN, Friedman AH, et al. Resistance to tyrosine kinase inhibition by mutant epidermal growth factor receptor variant III contributes to the neoplastic phenotype of glioblastoma multiforme. Clin Cancer Res. 2004;10:3216–3224. doi: 10.1158/1078-0432.ccr-03-0521. [DOI] [PubMed] [Google Scholar]
- Lindström AK, Ekman K, Stendahl U, Tot T, Henriksson R, Hedman H, et al. LRIG1 and squamous epithelial uterine cervical cancer: correlation to prognosis, other tumor markers, sex steroid hormones, and smoking. Int J Gynecol Cancer. 2008;18:312–317. doi: 10.1111/j.1525-1438.2007.01021.x. [DOI] [PubMed] [Google Scholar]
- Mellinghoff IK, Wang MY, Vivanco I, Haas-Kogan DA, Zhu S, Dia EQ, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353:2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- Moscatello DK, Holgado-Madruga M, Emlet DR, Montgomery RB, Wong AJ. Constitutive activation of phosphatidylinositol 3-kinase by a naturally occurring mutant epidermal growth factor receptor. J Biol Chem. 1998;273:200–206. doi: 10.1074/jbc.273.1.200. [DOI] [PubMed] [Google Scholar]
- Nagane M, Levitzki A, Gazit A, Cavenee WK, Huang HJ. Drug resistance of human glioblastoma cells conferred by a tumor-specific mutant epidermal growth factor receptor through modulation of Bcl-XL and caspase-3-like proteases. Proc Natl Acad Sci USA. 1998;95:5724–5729. doi: 10.1073/pnas.95.10.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagane M, Narita Y, Mishima K, Levitzki A, Burgess AW, Cavenee WK, et al. Human glioblastoma xenografts overexpressing a tumor-specific mutant epidermal growth factor receptor sensitized to cisplatin by the AG1478 tyrosine kinase inhibitor. J Neurosurg. 2001;95:472–479. doi: 10.3171/jns.2001.95.3.0472. [DOI] [PubMed] [Google Scholar]
- Nicholas MK, Lukas RV, Jafri NF, Faoro L, Salgia R. Epidermal growth factor receptor-mediated signal transduction in the development and therapy of gliomas. Clin Cancer Res. 2006;12:7261–7270. doi: 10.1158/1078-0432.CCR-06-0874. [DOI] [PubMed] [Google Scholar]
- Peschard P, Kozlov G, Lin T, Mirza IA, Berghuis AM, Lipkowitz S, et al. Structural basis for ubiquitin-mediated dimerization and activation of the ubiquitin protein ligase Cbl-b. Mol Cell. 2007;27:474–485. doi: 10.1016/j.molcel.2007.06.023. [DOI] [PubMed] [Google Scholar]
- Raizer JJ. HER1/EGFR tyrosine kinase inhibitors for the treatment of glioblastoma multiforme. J Neurooncol. 2005;74:77–86. doi: 10.1007/s11060-005-0603-7. [DOI] [PubMed] [Google Scholar]
- Rosell R, Taron M, Reguart N, Isla D, Moran T. Epidermal growth factor receptor activation: how exon 19 and 21 mutations changed our understanding of the pathway. Clin Cancer Res. 2006;12:7222–7231. doi: 10.1158/1078-0432.CCR-06-0627. [DOI] [PubMed] [Google Scholar]
- Shattuck DL, Miller JK, Laederich M, Funes M, Petersen H, Carraway KL, III, et al. LRIG1 is a novel negative regulator of the Met receptor and opposes Met and Her2 synergy. Mol Cell Biol. 2007;27:1934–1946. doi: 10.1128/MCB.00757-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibilia M, Kroismayr R, Lichtenberger BM, Natarajan A, Hecking M, Holcmann M. The epidermal growth factor receptor: from development to tumorigenesis. Differentiation. 2007;75:770–787. doi: 10.1111/j.1432-0436.2007.00238.x. [DOI] [PubMed] [Google Scholar]
- Sweeney C, Miller JK, Shattuck D, Carraway KL., III ErbB receptor negative regulatory mechanisms: implications in cancer. J Mammary Gland Biol Neoplasia. 2006;11:89–99. doi: 10.1007/s10911-006-9015-3. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Miura H, Tanemura A, Kobayashi K, Kondoh G, Sano S, et al. Targeted disruption of LIG-1 gene results in psoriasiform epidermal hyperplasia. FEBS Lett. 2002;521:67–71. doi: 10.1016/s0014-5793(02)02824-7. [DOI] [PubMed] [Google Scholar]
- Tanemura A, Nagasawa T, Inui S, Itami S. LRIG-1 provides a novel prognostic predictor in squamous cell carcinoma of the skin: immunohistochemical analysis for 38 cases. Dermatol Surg. 2005;31:423–430. doi: 10.1111/j.1524-4725.2005.31108. [DOI] [PubMed] [Google Scholar]
- Thomasson M, Hedman H, Guo D, Ljungberg B, Henriksson R. LRIG1 and epidermal growth factor receptor in renal cell carcinoma: a quantitative RT–PCR and immunohistochemical analysis. Br J Cancer. 2003;89:1285–1289. doi: 10.1038/sj.bjc.6601208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassar R, Fuchs E. Transgenic mice provide new insights into the role of TGF-alpha during epidermal development and differentiation. Genes Dev. 1991;5:714–727. doi: 10.1101/gad.5.5.714. [DOI] [PubMed] [Google Scholar]
- Wang MY, Lu KV, Zhu S, Dia EQ, Vivanco I, Shackleford GM, et al. Mammalian target of rapamycin inhibition promotes responses to epidermal growth factor receptor kinase inhibitors in PTEN-deficient and PTEN-intact glioblasma cells. Cancer Research. 2006;66:7864–7869. doi: 10.1158/0008-5472.CAN-04-4392. [DOI] [PubMed] [Google Scholar]
- Waterman H, Katz M, Rubin C, Shtiegman K, Lavi S, Elson A, et al. A mutant EGF-receptor defective in ubiquitylation and endocytosis unveils a role for Grb2 in negative signaling. EMBO J. 2002;21:303–313. doi: 10.1093/emboj/21.3.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells A, Welsh JB, Lazar CS, Wiley HS, Gill GN, Rosenfeld MG. Ligand-induced transformation by a noninternalizing epidermal growth factor receptor. Science. 1990;247:962–964. doi: 10.1126/science.2305263. [DOI] [PubMed] [Google Scholar]