This study illustrates the discovery of a MERTK model with a slowly progressive form of retinal degeneration. These mice also demonstrate protective accumulation of TNF in the retina.
Abstract
Purpose.
To determine the basis and to characterize the phenotype of a chemically induced mutation in a mouse model of retinal degeneration.
Methods.
Screening by indirect ophthalmoscopy identified a line of N-ethyl-N-nitrosourea (ENU) mutagenized mice demonstrating retinal patches. Longitudinal studies of retinal histologic sections showed photoreceptors in the peripheral retina undergoing slow, progressive degeneration. The mutation was named neuroscience mutagenesis facility 12 (nmf12), and mapping localized the critical region to Chromosome 2.
Results.
Sequencing of nmf12 DNA revealed a point mutation in the c-mer tyrosine kinase gene, designated Mertknmf12. We detected elevated levels of tumor necrosis factor (Tnf, previously Tnfa) in retinas of Mertknmf12 homozygotes relative to wild-type controls and investigated whether the increase of TNF, an inflammatory cytokine produced by macrophages/monocytes that signals intracellularly to cause necrosis or apoptosis, could underlie the retinal degeneration observed in Mertknmf12 homozygotes. Mertknmf12 homozygous mice were mated to mice lacking the entire Tnf gene and partial coding sequences of the Lta (Tnfb) and Ltb (Tnfc) genes.2 B6.129P2-Ltb/Tnf/Ltatm1Dvk/J homozygotes did not exhibit a retinal degeneration phenotype and will, hereafter, be referred to as Tnfabc−/− mice. Surprisingly, mice homozygous for both the Mertknmf12 and the Ltb/Tnf/Ltatm1Dvk allele (Tnfabc−/−) demonstrated an increase in the rate of retinal degeneration.
Conclusions.
These findings illustrate that a mutation in the Mertk gene leads to a significantly slower progressive retinal degeneration compared with other alleles of Mertk. These results demonstrate that TNF family members play a role in protecting photoreceptors of Mertknmf12 homozygotes from cell death.
Proper function of cells in the retinal pigment epithelium (RPE) is required for normal vision and for the survival of photoreceptor cells. Its importance is underscored by the large number of diseases associated with improper function of the RPE, such as macular edema,1 Best's vitelliform macular degeneration,2–4 autosomal recessive macular degeneration,5 Leber congenital amaurosis,6–8 and Stargardt disease.9 Multiple instances of retinitis pigmentosa or allied disorders caused by mutations in the MERTK gene have been reported.10–14 The Royal College of Surgeons (RCS) rat is a classic model for assessing photoreceptor-RPE interaction.15–17 Mapping efforts determined that a deletion within the Mertk gene (Mertkrdy) led to a frameshift and a termination signal 20 codons after the start codon in RCS rats.18,19 Additionally, a targeted mutation of an exon encoding a portion of the Mer tyrosine kinase domain in Mertktm1Gkm mice has been reported.20 This murine allele closely recapitulates the RCS rat phenotype of rapid photoreceptor degeneration.21 Photoreceptor death in these Mertk mutants is associated with an inability of the RPE to properly phagocytize shed photoreceptor outer segment tips.22,23
The work presented here is the first to describe a missense mutation in the murine Mertk gene (Mertknmf12) associated with retinal degeneration. This mutation exhibits several hallmarks of degeneration reported in other rodent models of Mertk-mediated disease; however, the rate of degeneration is much slower in Mertknmf12 homozygotes than in RCS rats or Mertktm1Gkm homozygous mice. This slower rate of degeneration offers opportunities to better study the progression of photoreceptor loss under conditions of abnormal MERTK function. Additionally, our studies of the Mertknmf12 mutants suggest that TNF family members help to protect homozygous Mertknmf12 retinas from degeneration, providing novel insight into mechanisms affecting the rate of photoreceptor degeneration in MERTK-mediated photoreceptor cell death.
Materials and Methods
Mice were bred and housed in standardized conditions in the Research Animal Facility at The Jackson Laboratory and were monitored regularly to maintain a pathogen-free environment. Mice were provided free access to NIH 6% fat chow and acidified water in a vivarium with a 12-hour light/12-hour dark cycle. All mice were treated in accordance with protocols approved by the Animal Care and Use Committee at The Jackson Laboratory, and in compliance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research.
Mouse Production and Mapping
A C57BL/6J-nmf12/J × DBA/2J F2 intercross was generated by the Speed Expansion IVF (http://jaxmice.jax.org/services /speed expansion.html) service at The Jackson Laboratory. DNA isolated from tail tips of 13 affected and 13 unaffected mice were pooled and subjected to a genomewide scan using 92 SSLP markers distributed throughout the genome. Samples used in the DNA pools were tested individually to confirm the map location. The critical region was established by genotyping additional recombinant F2 mice with commercially available MIT markers and single nucleotide polymorphisms (SNPs) and was further refined through the generation of additional mapping markers.
Sequencing
Three pairs of eyes isolated from adult Mertknmf12 homozygous and wild-type mice were homogenized in a tissue homogenizer (VirTishear; VirTis, Gardiner, NY) for 2 minutes on ice with buffer (Trizol; Invitrogen, Carlsbad, CA). RNA was isolated according to the manufacturer's directions, and the total RNA quality was assessed by running a 5-μL aliquot of each RNA sample on a 1% agarose gel. RNA samples were reverse transcribed into cDNA (RETROscript; Ambion, Austin, TX). The Jackson Laboratory's Allele Typing and Sequencing Facility performed DNA sequencing (Big Dye Terminator chemistry). Data were analyzed with bioinformatics software (MacVector 7.2.3 [MacVector, Inc., Cary, NC] and Sequencher 4.2 [Gene Codes Corporation, Ann Arbor, MI]).
Electroretinography
Full-field ERG recordings were performed as previously described.24 Briefly, anesthesia was induced by intraperitoneal injection of a mixture of ketamine (70 mg/kg) and xylazine (15 mg/kg), and body temperature (36.5°C-37°C) was maintained with a temperature-controlled heating pad. A gold loop electrode was placed on the surface of the cornea and referenced to a gold wire placed in the mouth. A needle electrode in the tail served as ground. Signals were amplified (×10,000; CP511 AC amplifier; Grass Instruments, Quincy, MA), sampled at 0.8-ms intervals, and averaged. Rod-mediated ERGs were recorded from wild-type and mutant mice that were adapted to darkness overnight and exposed to short-wavelength flashes of light in a Ganzfeld dome (LKC Technologies, Inc., Gaithersburg, MD). Light intensities varied over a 4.0 log unit range up to the maximum allowable by the photopic stimulator (PS33 Plus; Grass Instruments). Cone-mediated responses were obtained with flashes of white light on a rod-saturating background after 10 minutes of light adaptation. Responses were averaged at all intensities, and up to 50 records were averaged for the weakest signals. A signal-rejection protocol was used to eliminate electrical artifacts produced by blinking and eye movements.
Microscopy and Immunohistochemistry
Retinas were fixed in 37.5% methanol/12.5% glacial acetic acid in (1×) phosphate-buffered saline, embedded in paraffin, and sectioned at 6-μm intervals. The sections were subsequently mounted on slides (SuperFrost Plus; Fisher Scientific, Pittsburgh, PA) and were stained with hematoxylin and eosin. Light microscopy and fluorescence microscopy were performed (DMR microscope; Leica, Wetzlar, Germany), and images were collected (FireCam; Leitz). Brightness and contrast of images compared between wild-type and mutant mice were equally optimized (Photoshop 7.0; Adobe, Mountain View, CA). Primary antibodies were used at the concentration of 1:200. Appropriate secondary antibodies conjugated to either CY3 (Jackson Immunochemicals, West Grove, PA) or Alexa-488 (Molecular Probes, Eugene, OR) were applied at a dilution of 1:500.
Western Blot Analysis
Six eyes each from C57BL/6J and C57BL/6J-Mertknmf12/J homozygous mice were homogenized in 300 μL RadioImmuno Precipitation Assay (RIPA) buffer containing protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN). Protein concentrations were determined using the Bio-Rad Protein Assay (Bio-Rad Laboratories, Inc., Hercules, CA), and 40 μg total protein for each sample was loaded with sample buffer (NuPAGE LDS; Invitrogen) and sample reducing agent (NuPAGE; Invitrogen). Samples were run on a 10% Bis-Tris gel with MOPS/SDS running buffer (NuPAGE; Invitrogen). Samples were transferred overnight onto a polyvinylidene difluoride (PVDF) membrane (Roche Diagnostics), and the membrane was blocked for 1 hour in Tris-buffered saline plus 5% milk powder before being probed with a polyclonal antibody against MERTK (af591) (R&D Systems, Minneapolis, MN). After washing, the membrane was probed with peroxidase-conjugated donkey anti-rabbit IgG (1:40,000; AffiniPure; Jackson ImmunoResearch, Inc.), and signal was detected with Western Lightning Chemiluminescent Reagent (Perkin Elmer Life Sciences, Inc., Waltham, MA).
Results
Mapping and Identification of the Mertknmf12 Mutation
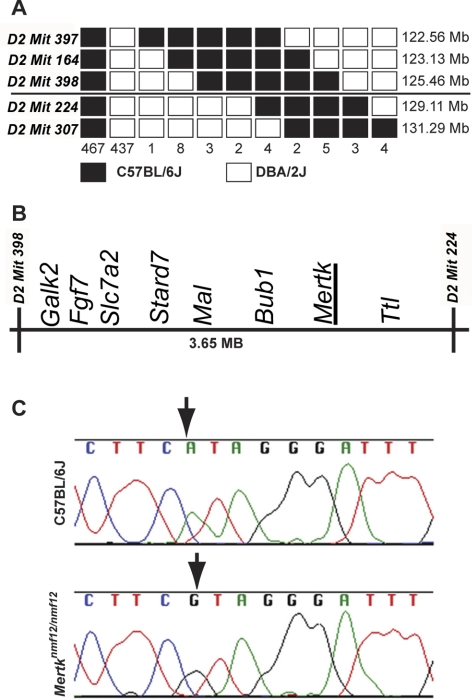
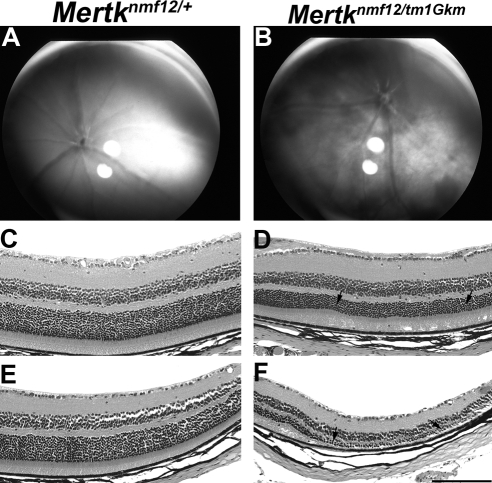
By in vitro fertilization, we generated a C57BL/6J-nmf12/nmf12 X DBA/2J F1 intercross, simultaneously producing 468 mice (representing 936 meioses) for mapping. A total of 32 F2 mice were identified with genetic crossovers between flanking markers D2Mit397 (122.56 Mb) and D2Mit307 (130.10 Mb). These 32 crossover mice were genotyped with a combination of MIT markers and SNPs (Fig. 1A), and the critical region was reduced to a 3.65-Mb region between markers D2Mit398 (125.46 Mb) and D2Mit224 (129.11 Mb; Fig. 1B). Within this critical region were 56 annotated transcripts, one of which encodes the Mertk gene. Sequencing of Mertk cDNA from Mertknmf12 homozygotes revealed a G-to-A substitution at base pair 2237 (Ensembl build 42: OTTMUSP0000016359; Fig. 1C). As further confirmation that the mutation in Mertk was causative of the observed retinal degenerative phenotype in Mertknmf12 homozygotes, we performed a complementation test between homozygous Mertknmf12 mice and mice carrying a targeted deletion (Mertktm1Gkm) in the Mertk gene.20 Mertknmf12 is not capable of complementing Mertktm1Gkm (Fig. 2), offering strong evidence that the two mutations are allelic.
Figure 1.
Mapping and cloning of the nmf12 mutation. (A) A haplotype map is shown, representing the location and number of crossovers in 936 meioses observed in 468 affected and unaffected progeny-tested mice. The numbers at the bottom of the graph represent the number of chromosomes bearing each depicted crossover. The critical interval spans 3.65 Mb between markers D2Mit398 (125.46 Mb) and D2Mit224 (129.11 Mb). Within this critical region (B) lies Mertk, a gene in which mutations have been shown to cause retinal degeneration. (C) Sequence chromatograms from control, C57BL/6J, and Mertknmf12 homozygotes. Analysis of these sequences reveals an A-to-G transition at base pair 2237 (A2237G).
Figure 2.
Complementation testing between Mertknmf12 and Mertktm1Gkm. Indirect ophthalmoscopy shows that at P45, the retinas of Mertknmf12/Mertktm1Gkm compound heterozygous mice demonstrate pan-retinal fundus patches (B), whereas the Mertknmf12/+ heterozygous littermates (as well as Mertknmf12 homozygotes) demonstrate a normal appearance in the central retina (A). Histologic examination of central and peripheral sections from the Mertknmf12/+ heterozygous littermates (C and E, respectively) appears normal. In contrast, the central retina of Mertknmf12/tm1Gkm compound heterozygotes (D) demonstrates slight thinning of the ONL (arrows), whereas the peripheral retina (F) demonstrates severe photoreceptor loss (arrows). Failure of the Mertknmf12/Mertktm1Gkm compound heterozygous mice to show a normal phenotype provides strong evidence that Mertknmf12 and Mertktm1Gkm do not complement and are likely to be allelic mutations. Scale bar, 50 μm (C–F).
Analysis of Mertknmf12 Protein Expression
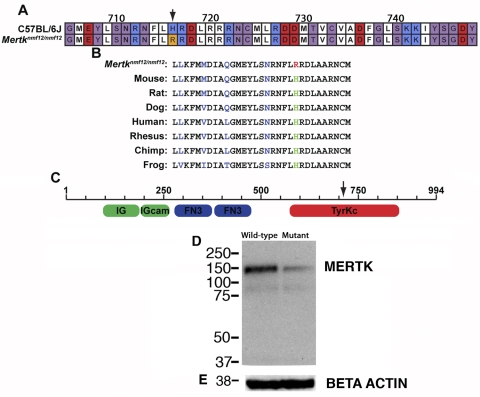
The missense mutation found in Mertknmf12 mutants causes a substitution at amino acid position 716 (Fig. 3A) and is predicted to alter a highly conserved histidine residue to arginine (H716R; Fig. 3B). This residue is located within the tyrosine kinase domain of the MERTK protein (Fig. 3C). Immunohistochemical studies showed that MERTK protein was present and properly localized to the RPE of Mertknmf12 homozygotes (data not shown). To determine whether this mutation could destabilize the MERTK protein and lead to its early degradation, Western blot analysis was performed. A 2.5-fold reduction in MERTK protein was evident in retinas of Mertknmf12 homozygotes (Fig. 3D).
Figure 3.
Analysis of Mertknmf12 protein expression and function. (A) Depiction of protein alignment between C57BL/6J and Mertknmf12 homozygotes (amino acids presented in randomly selected colors). A substitution of arginine for histidine is evident at amino acid 716 (H716R; arrow). This residue is conserved across species (B). (C) Schematic showing that the mutated amino acid falls within the tyrosine kinase domain of the Mertk gene. Western blot analysis shows a 2.5-fold reduction of MERTK protein in Mertknmf12 homozygous retinas (D). The blot was stripped and probed again with an antibody against β-actin as a loading control (E).
Analysis of Mertknmf12 Retinal Phenotype
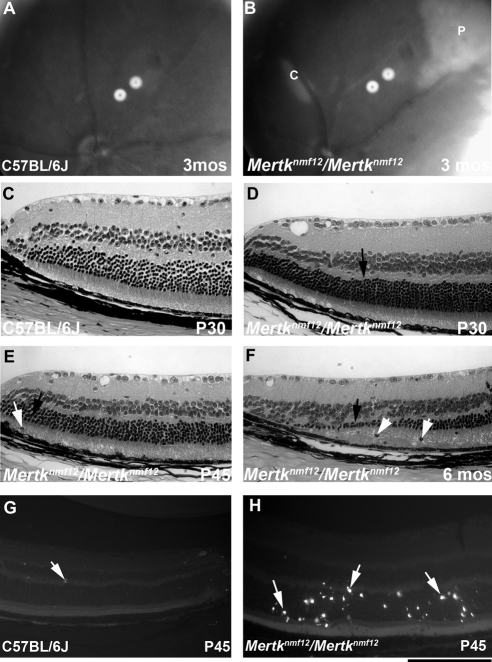
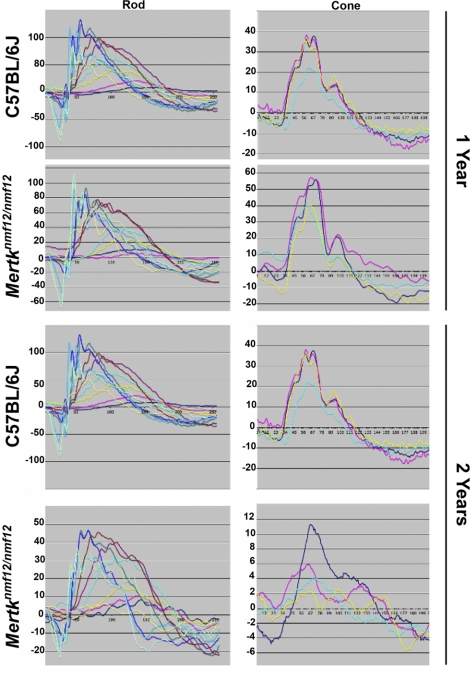
Mertknmf12 mutants were first identified by screening ENU-mutagenized mice by indirect ophthalmoscopy. The peripheral retina of Mertknmf12 homozygotes, when viewed by indirect ophthalmoscopy, appeared depigmented as early as postnatal day (P) 30 (data not shown). As the Mertknmf12 homozygotes age, patches become evident in the central portion of the retina in addition to those observed in the periphery (Figs. 4A, 4B). When viewed by histology at P30, wild-type mice have normal, well-structured retinas (Fig. 4C). Mertknmf12 homozygous retinas, when viewed at P30, demonstrate thinning of the peripheral outer nuclear layer (ONL; Fig. 4D). As time progresses, photoreceptor cell death increases and the ONL becomes increasingly thinner (Fig. 4E). As the majority of photoreceptors die, a single layer of photoreceptor nuclei is left adjacent to the RPE in the peripheral retina (Fig. 4F). Although the death of photoreceptor nuclei is most dramatic in the peripheral retina, presence of apoptotic nuclei is evident throughout the mutant retina at P45 (Figs. 4G, 4H). Most photoreceptors in the central retina, however, are spared (data not shown), and though electroretinographic (ERG) recordings do become progressively attenuated, ERG a- and b-waves are detectable in Mertknmf12 homozygotes as old as 2 years of age (Fig. 5).
Figure 4.
Analysis of the Mertknmf12 retinal phenotype. Fundus photographs of 3-month-old C57BL/6J (A) or homozygous Mertknmf12 (B) mice. Note the abnormal whitening of the peripheral (P) and central (C) retina that is likely to correspond to areas of ONL degeneration. Histologic sections of the peripheral retina from a P30 C57BL/6J mouse (C). The ONL in retinas of Mertknmf12 homozygotes aged P30, P45, and 6 months are shown (D–F, respectively). (D, arrow) Beginning of ONL cell thinning. This ONL thinning progresses over time. (E, F, black arrows) Thinning ONL. Pyknotic nuclei are present (E, F, white arrows) suggesting that cell death is taking place. Few TUNEL-positive cells (white arrow) are present in the retinas of P45 C57BL/6J mice (G), whereas many TUNEL-positive cells (white arrows) are present throughout the ONL in retinas of Mertknmf12 homozygotes (H). Scale bar, 50 μm (C–H).
Figure 5.
Analysis of the ERG function in eyes of Mertknmf12 mutants. Although dying cells are present throughout the retinas of Mertknmf12 homozygotes, the majority of photoreceptor cells in the central portion of the retina of Mertknmf12 homozygotes are preserved. This preservation is reflected in the ERG waveforms. Although the amplitude of the waveforms recorded from homozygous Mertknmf12 retina are progressively reduced over time compared with those of C57BL/6J mice, ERG waveforms are still detectable from eyes of Mertknmf12 homozygotes aged up to 2 years.
Analysis of TNF Expression in Mertknmf12 Eyes
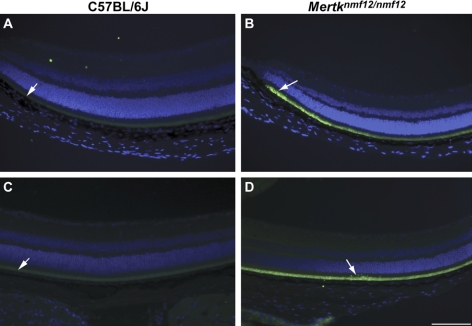
Studies involving Mertktm1Gkm homozygotes have demonstrated that serum levels of TNF are increased because of an inability of macrophages to phagocytize apoptotic nuclei.20,25 The increased TNF levels are thought to cause cellular damage and lethal tissue injury, and the small intestines of Mertktm1Gkm homozygotes were shown to be damaged as a result of excess TNF. Therefore we investigated whether TNF levels were increased in retinas of Mertknmf12 homozygotes. Immunohistochemistry with an antibody directed against TNF (ab9739; Abcam) was performed on wild-type (Figs. 6A, 6C) and Mertknmf12 homozygous (Figs. 6B, 6D) retinas. A marked increase in staining was observed in retinal sections from the Mertknmf12 homozygotes compared with controls.
Figure 6.
Immunohistochemical analysis of TNF expression in eyes of Mertknmf12 mutants. TNF is faintly present in the outer retina in peripheral (A) and central (C) retinas of C57BL/6J mice (white arrows). In comparison, TNF fluorescence is much more intense in the outer retina in the peripheral (B) and central (D) regions of Mertknmf12 homozygous retinas (white arrows). Scale bar, 50 μm (A–D).
Genetic Manipulation of Tumor Necrosis Factor Levels in Mertknmf12 Mutant Eyes
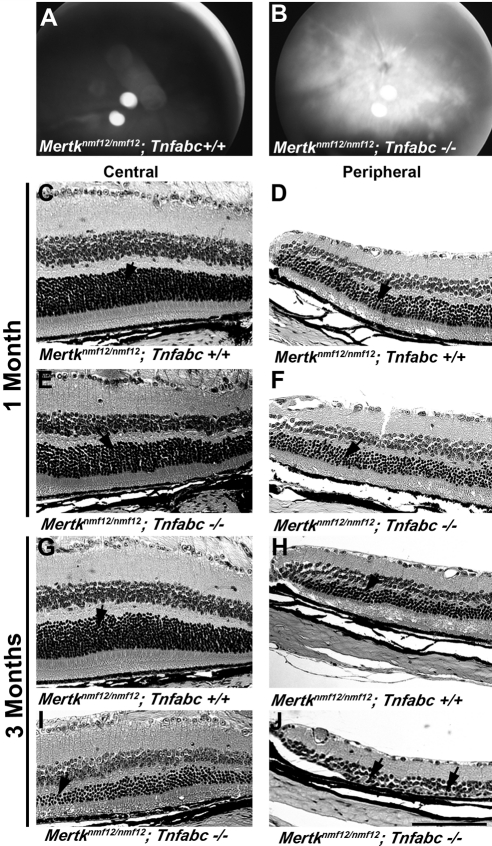
To ascertain the effect of increased TNF levels in the retinas of Mertknmf12 homozygotes, we performed a genetic experiment in which Mertknmf12 homozygotes were crossed to Tnfabc−/− homozygotes, and the resultant F1 offspring were intercrossed. Mertknmf12/Mertknmf12; Tnfabc−/− double homozygous mice exhibited an increased rate of photoreceptor cell degeneration compared with littermate mice homozygous for the Mertknmf12 mutation alone. At P45, when examined by indirect ophthalmoscopy, the central retinas of Mertknmf12/Mertknmf12; Tnfabc−/− double homozygotes (Fig. 7B) demonstrated an abnormal appearance across the entire retina, whereas the central retinas of homozygous Mertknmf12 mice appeared normal (Fig. 7A). Histologically, at 1 month of age, the thicknesses of Mertknmf12 homozygous ONL in the central (Fig 7C) and peripheral (Fig 7D) retina were similar to those observed for Mertknmf12/Mertknmf12; Tnfabc−/− mice (Figs. 7E, 7F). At 3 months of age, the central retina of Mertknmf12 homozygotes appeared relatively normal (Fig. 7G), whereas slight thinning of the ONL was evident in the peripheral retina (Fig. 7H). Demonstrating an accelerated rate of degeneration, the ONL of Mertknmf12/Mertknmf12; Tnfabc−/− double mutant mice was markedly thinned pan-retinally at 3 months of age (Figs. 7I, 7J), with an additional loss of cells in the inner nuclear layer in the peripheral retina.
Figure 7.
Genetic manipulation of tumor necrosis factor levels in eyes of Mertknmf12 mutants. Fundus photographs of P45 Mertknmf12/Mertknmf12; Tnfabc+/+ (A) and Mertknmf12/Mertknmf12; Tnfabc−/− (B) mice are presented. Note the pan-retinal patches in the latter. Histologic sections are of central (C) and peripheral (D) retinas of a 1-month-old Mertknmf12/Mertknmf12; Tnfabc+/+ retina and central (E) and peripheral (F) retinas of an age-matched Mertknmf12/Mertknmf12; Tnfabc−/− mouse. The thickness of the ONL is similar between Mertknmf12/Mertknmf12; Tnfabc+/+ and Mertknmf12/Mertknmf12; Tnfabc−/− mice (arrows). Central (G) and peripheral (H) retinas of a 3-month-old Mertknmf12/Mertknmf12; Tnfabc+/+ retina and central (I) and peripheral (J) retinas of an age-matched Mertknmf12/Mertknmf12; Tnfabc−/− retina are presented. Although the ONL of Mertknmf12/Mertknmf12; Tnfabc+/+ retinas remains relatively intact (G, H, arrows), extensive thinning of the ONL is evident in the central and peripheral retina (I, J, arrows, respectively) of the Mertknmf12/Mertknmf12; Tnfabc−/− mice. Scale bar, 50 μm (C–J).
Discussion
We have demonstrated that the nmf12 mutation maps to Chromosome 2 and that the observed phenotype segregates with a point mutation in the Mertk gene. This mutation leads to the replacement of a conserved histidine residue, within the MERTK tyrosine kinase domain, with arginine. The failure to complement the Mertktm1Gkm allele and the reduced levels of MERTK protein in Mertknmf12 homozygotes provide strong evidence that the missense mutation detected in the Mertk gene causes the observed retinal phenotype.
The ONL of Mertktm1Gkm homozygotes is almost completely normal at P20.21 Soon afterward (P25), the ONL becomes significantly thinned throughout the entire retina. Most photoreceptor nuclei are missing by P45, and no ERG activity is readily discernible after P40.21 In contrast, retinal thinning proceeds slowly from the periphery toward the central retina in Mertknmf12 homozygotes, and normal ONL thickness is preserved in the central retina of Mertknmf12 homozygotes as old as 2 years of age. Because of this conservation of the central retina, ERG measurements are also recordable up to 2 years of age in Mertknmf12 homozygotes.
The Mertktm1Gkm allele was generated by targeting an exon that encodes essential portions of the tyrosine kinase domain of MERTK.20 Whether the difference in the rate of retinal degeneration was due to allelic variance or genetic background is uncertain because the nmf12 allele arose on a mutagenized C57BL/6J background, whereas the Mertktm1Gkm mutant allele was targeted in a 129/Sv background and was maintained on a mixed genetic background of C57BL/6J and 129/Sv. It is interesting to note as well that the Mertktm1Gkm allele was not intended to be a null allele.20 Given that the rate of retinal degeneration in Mertktm1Gkm mutant mice is rapid and comparable to that seen in the rat Mertkrdy null allele and that no truncated protein is detectable in kidney or retina samples from Mertktm1Gkm mutant mice, the Mertktm1Gkm allele does appear to be null.21 The Mertknmf12 allele also affects the tyrosine kinase domain of MERTK but is a point mutation. In contrast to what is seen in the Mertktm1Gkm mutant mice, MERTK protein is detectable in retinal samples of Mertknmf12 mutant mice, though at a reduced amount compared with wild-type controls. Thus, the Mertknmf12 mutant protein may retain some functional activity and thereby cause a slower rate of photoreceptor cell loss than that observed in previously described models.
After the work of Camenisch et al.,20 who reported increased serum TNF levels in Mertktm1Gkm and associated this accumulation with tissue damage in the small intestine, we undertook to determine whether TNF levels were increased in the retinas of Mertknmf12 homozygous mice. Our immunohistochemical studies suggest that TNF levels are increased in the retinas of Mertknmf12 homozygous mice compared with those of wild-type controls. TNF is a cytokine that produces paradoxical effects in various instances. The role of TNF in uveoretinitis has been studied in depth, with increased TNF levels most often preceding tissue damage.26 Similarly, in many other types of retinal disease, increased levels of TNF are associated with increased retinal degeneration. Because of this, TNF inhibitory antibodies (infliximab, etanercept) have been developed and are being used in trials to treat human diseases such as age-related macular degeneration,27 Eales disease,28 neurosarcoidosis,29 various forms of ocular inflammation,30 diffuse subretinal fibrosis syndrome,31 and glaucoma.32
As has been noted33 and as our present study underscores, there are instances in which TNF plays a poorly understood protective role in retinal degeneration. It has been posited that in retinas of Mertkrdy rats, the RPE secretes a factor that might attract phagocytic cells, such as microglia, to help restore an environment more conducive to photoreceptor survival.34 Our studies suggest that TNF may be this predicted factor. That the loss of TNF function could potentially lead to deleterious effects should certainly be kept in mind when administering TNF inhibitors to human patients, and Mertknmf12 mutant mice offer a powerful tool to better illuminate the protective role TNF can play in various forms of retinal degeneration.
Acknowledgments
The authors thank Jeanie Hansen for excellent technical assistance and The Jackson Laboratory's Fine Mapping Service for performing the initial steps in the mapping process.
Footnotes
Supported by National Eye Institute Grants EY11996 and EY016501 (PMN), EY016313-01 (DMM), a JAX institutional core grant CA34196 (PMN), and the Foundation Fighting Blindness (MML, DV).
Disclosure: D.M. Maddox, None; W.L. Hicks, None; D. Vollrath, None; M.M. LaVail, None; J.K. Naggert, None; P.M. Nishina, None
References
- 1. Marmor MF. Mechanisms of fluid accumulation in retinal edema. Doc Ophthalmol. 1993;97:179–203 [DOI] [PubMed] [Google Scholar]
- 2. Allikmets R, Seddon JM, Bernstein PS, et al. Evaluation of the Best disease gene in patients with age-related macular degeneration and other maculopathies. Hum Genet. 1999;104:449–453 [DOI] [PubMed] [Google Scholar]
- 3. Bakall B, Marknell T, Ingvast S, et al. The mutation spectrum of the bestrophin protein- functional implications. Hum Genet. 1999;104:338–389 [DOI] [PubMed] [Google Scholar]
- 4. Caldwell GM, Kakuk LE, Griesinger IB, et al. Bestrophin gene mutations in patients with Best vitelliform macular dystrophy. Genomics. 1999;58:98–101 [DOI] [PubMed] [Google Scholar]
- 5. Ambati J, Ambati BK, Yoo SH, Ianchulev S, Adamis AP. Age-related macular degeneration: etiology, pathogenesis and and therapeutic strategies. Surv Ophthalmol. 2003;48:257–293 [DOI] [PubMed] [Google Scholar]
- 6. Hamel CP, Tsilou E, Pfeffer BA, Hooks JJ, Detrick B, Redmond TM. Molecular cloning and expression of RPE65, a novel retinal pigment epithelium-specific microsomal protein that is post-transcriptionally regulated in vitro. J Biol Chem. 1993;268:15751–15757 [PubMed] [Google Scholar]
- 7. Hamel CP, Jenkins NA, Gilbert DJ, Copeland NG, Redmond TM. The gene for the retinal pigment epithelium-specific protein, RPE65, is localized to human 1p31 and mouse 3. Genomics. 1994;20:509–512 [DOI] [PubMed] [Google Scholar]
- 8. Hamel CP, Griffion JM, Lasquellec L, Bazalgette C, Arnaud B. Retinal dystrophies caused by mutations in RPE65: assessments of visual functions. Br J Ophthalmol. 2001;85:424–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Simonelli F, Testa F, de Creechio G, et al. New ABCR mutations and clinical phenotype in Italian patients with Stargardt disease. Invest Ophthalmol Vis Sci. 2000;41:892–897 [PubMed] [Google Scholar]
- 10. Gal A, Li Y, Thompson DA, et al. Mutation in MERTK, the human orthologue of the RCS rat retinal dystrophy gene, cause retinitis pigmentosa. Nature. 2000;26:270–271 [DOI] [PubMed] [Google Scholar]
- 11. Thompson DA, McHenry CL, Li Y, et al. Retinal dystrophy due to paternal isodiomy for chromosome 1 or chromosome 2, with homoallelism for mutations in RPE65 or MERTK, respectively. Am J Hum Genet. 2002;70:224–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McHenry CL, Li Y, Feng W, et al. MERTK arginine-844-cysteine in a patient with severe rod-cone dystrophy: loss of mutant protein function in transfected cells. Invest Ophthalmol Vis Sci. 2004;45:1456–1462 [DOI] [PubMed] [Google Scholar]
- 13. Tschernutter A, Jenkins SA, Waseem NH, et al. Clinical characterization of a family with retinal dystrophy caused by a mutation in the Mertk gene. Br J Ophthalmol. 2007;90:718–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brea-Fernandez AJ, Pomares E, Brion MJ, et al. Novel splice donor site mutation in MERTK gene associated with retinitis pigmentosa. Br J Ophthalmol. 2008;92:1419–1423 [DOI] [PubMed] [Google Scholar]
- 15. Bourne MC, Campbell DA, Tansley K. Hereditary degeneration of the rat retina. Br J Ophthalmol. 1938;22:613–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dowling JC, Sidman RL. Inherited retinal dystrophy in the rat. J Cell Biol. 1962;14:73–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mullen RJ, LaVail MM. Inherited retinal dystrophy: primary defect in pigment epithelium determined with experimental rat chimeras. Science. 1976;192:799–801 [DOI] [PubMed] [Google Scholar]
- 18. D'Cruz PM, Yasumura D, Weir J, et al. Mutation of the receptor tyrosine kinase gene Mertk in the retinal dystrophic RCS rat. Hum Mol Genet. 2000;4:645–651 [DOI] [PubMed] [Google Scholar]
- 19. Vollrath D, Feng W, Duncan JL, et al. Correction of the retinal dystrophy phenotype of the RCS rat by viral gene transfer of Mertk. Proc Natl Acad Sci U S A. 2001;26:12584–12589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Camenisch TD, Koller BH, Earp HS, Matsushima GK. A novel receptor tyrosine kinase, Mer, inhibits TNF-a production and lipopolysaccharide-induced endotoxic shock. J Immunol. 1999;162:3498–3503 [PubMed] [Google Scholar]
- 21. Duncan JL, LaVail MM, Yasumura D, et al. An RCS-like retinal dystrophy phenotype in Mer knockout mice. Invest Ophthalmol Vis Sci. 2003;44:826–838 [DOI] [PubMed] [Google Scholar]
- 22. Feng W, Yasumura D, Matthes MT, LaVail MM, Vollrath D. Mertk triggers uptake of photoreceptor outer segments during phagocytosis by cultured retinal pigment epithelial cells. J Biol Chem. 2002;19:17016–17022 [DOI] [PubMed] [Google Scholar]
- 23. Hall MO, Obin MS, Heeb MJ, Burgess BL, Abrams TA. Both protein s and gas6 stimulate outer segment phagocytosis by cultured rat retinal pigment epithelial cells. Exp Eye Res. 2005;81:581–591 [DOI] [PubMed] [Google Scholar]
- 24. Collin GB, Cyr E, Bronson R, et al. Alms1-disrupted mice recapitulate human Alstrom syndrome. Hum Mol Genet. 2005;14:2323–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Scott RS, McMahon EJ, Pops SM, et al. Phagocytosis and clearance of apoptotic cells is mediated by Mer. Nature. 2001;411:207–211 [DOI] [PubMed] [Google Scholar]
- 26. Dick AD, Forrester JV, Liversidge J, Cope AP. The role of tumour necrosis factor (TNF-alpha) in experimental autoimmune uveoretinitis (EAU). Prog Retin Eye Res. 2004;23:617–637 [DOI] [PubMed] [Google Scholar]
- 27. Theodossiadis PG, Liarakos VS, Sfikakis PP, Vergados IA, Theodossiadis GP. Intravitreal administration of the anti-tumor necrosis factor agent infliximab for neovascular age-related macular degeneration. Am J Ophthalmol. 2009;147:825–830, 830.e1 [DOI] [PubMed] [Google Scholar]
- 28. Saxena S, Pant AB, Khanna VK, et al. Interleukin-1 and tumor necrosis factor-alpha: novel targets for immunotherapy in Eales disease. Ocul Immunol Inflamm. 2009;17:201–206 [DOI] [PubMed] [Google Scholar]
- 29. Salama B, Gicquel JJ, Lenoble P, Dighiero PL. Optic neuropathy in refractory neurosarcoidosis treated with TNF-alpha antagonist. Can J Ophthalmol. 2006;41:766–768 [DOI] [PubMed] [Google Scholar]
- 30. Theodossiadis PG, Markomichelakis NN, Sfikakis PP. Tumor necrosis factor antagonists: preliminary evidence for an emerging approach in the treatment of ocular inflammation. Retina. 2007;27:399–413 [DOI] [PubMed] [Google Scholar]
- 31. Adán A, Sanmarti R, Burés A, Casaroli-Marano RP. Successful treatment with infliximab in patients with diffus subretinal fibrosis syndrome. Am J Ophthalmol. 2007;143: 533–534 [DOI] [PubMed] [Google Scholar]
- 32. Nakazawa T, Nakazawa C, Matsubara A, et al. Tumor necrosis factor-alpha mediates oligodendrocyte death and delayed retinal ganglion cell loss in a mouse model of glaucoma. J Neurosci. 2006;26:12633–12641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. LaVail MM, Unoki K, Yasumura D, Matthes MT, Yancopoulos GD, Steinberg RH. Multiple growth factors, cytokines and neurotrophins rescue photoreceptors from the damaging effects of constant light. Proc Natl Acad Sci U S A. 1992;89:11249–11253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thanos S. Sick photoreceptors attract activated microglia from the ganglion cell layer: a model to study the inflammatory cascades in rats with inherited retinal dystrophy. Brain Res. 1992;588:21–28 [DOI] [PubMed] [Google Scholar]