Myopia affects more people worldwide than any other chronic condition, and it is increasing in all populations across the globe. It affects ∼25% of the U.S. general population between the ages of 12 and 54 years. The present study is a genetic investigation of X-linked high-grade myopia that maps to Xq28.
Abstract
Purpose.
Myopia is a common vision problem affecting almost one third of the world's population. It can occur as an isolated genetic condition or be associated with other anomalies and/or syndromes. Seventeen myopia loci have been identified on various chromosomes; however, no specific gene mutations have yet been identified.
Methods.
Two large multigeneration Asian Indian pedigrees (UR006 and UR077) with isolated, nonsyndromic myopia were studied, in which the condition appeared to segregate as an X-linked recessive trait (MYP1; MIM 310460). The degree of myopia was variable in both families, ranging from −6 to −23 D (mean, –8.48 D) with the majority >7.0 D. To map the myopia locus in these families, polymorphic microsatellite markers covering the entire X chromosome were used in linkage analyses performed on 42 genomic DNA samples (13 affected and 29 normal) from both families.
Results.
Marker DXYS154, which is located within the pseudoautosomal region in distal Xq28 (PAR2; pseudoautosomal region 2), gave a combined maximum LOD score of 5.3 at θ = 0 under an autosomal recessive model. Other markers in the region (near but not within the PAR2 region) that showed no recombination with the phenotype in both the families included DXS1108, DXS8087, and F8i13.
Conclusions.
Observation of recombination in family UR006 refined the disease locus to a ∼1.25-Mb region flanked by the proximal marker DXS1073 and distal marker DXYS154. Mutation search in exons and splice junctions of candidate genes CTAG2, GAB3, MPP1, F8Bver, FUNDC2, VBP1, RAB39B, CLIC2, TMLHE, SYBL, IL9R, SPRY3, and CXYorf1 did not detect a pathogenic or predisposing variant.
Myopia (MYP), generally known as near-sightedness, is the natural refractive state of having a focal point in front of the retina: Near objects are in focus, but distant objects appear out of focus. The condition is due to excessive elongation of the eyeball in the axial dimension.1 It is generally categorized as mild (0–1.5 D), moderate (1.5–6.0 D), and high-grade (>6.0 D). Pathologic myopia (>8.0 D) is often associated with retinal disease, cataract, glaucoma, and choroidal neovascularization, and the risk of threats to vision can also occur in patients with moderate- and/or high-grade myopia.2 There are a few factors that have been hypothesized as potential environmental risk factors for myopia, but to date, none have been conclusively proven, including night lighting,3,4 excessive nearwork,5 and certain occupations.6 The prevalence of myopia is often presumed to be much higher, in that exact population-based data are not available. In general, high-grade myopia has a prevalence of ∼2.8% to 9.1%7,8; however, it is specific to the age and sex of the individual ethnic population. The prevalence of X-linked myopia is uncertain, as there is a paucity of data due to a very low number of published reports of families with X-linked myopia.9–13
Several genome-wide linkage studies conducted using multiplex MYP families of different ethnic origins showed evidence of linkage at 17 loci for either high- or low-grade myopia (MYP) on Xq28 (MYP1 [MIM 310460]), 18p11.31 (MYP2 [MIM 160700]), 12q21-q23 (MYP3 [MIM 603221]), 17q21-q22 (MYP5 [MIM 608474]), 22q12 (MYP6 [MIM 608908]), 11p13 (MYP7 [MIM609256]), 3q26 (MYP8 [MIM 609257]), 4q12 (MYP9 [MIM 609258]), 8p23 (MYP10 [MIM 609259]), 4q22-q27 (MYP11 [MIM 609994]), 2q37.1 (MYP12 [MIM 609995]), Xq23-q25 (MYP13 [MIM 300613]), 1p36 (MYP14 [MIM 610320]), 10q21.1 (MYP15 [MIM 612717]), 5p15.33-p15.2 (MYP16 [MIM 612554]), 7p15 (MYP17 [MIM 608367]), and 14q22.1-q24.2 (MYP18; [MIM 613626]).9,10,14–25 In addition, three independent association and linkage studies of different populations have identified myopia loci at 11q24.1, 15q14, and 15q25, respectively.15,16,26–28 Increased incidence of gene polymorphism associated with pathologic myopia was reported recently16,17,29–31; however, no specific gene mutations or pathogenic causative genomic variations have yet been identified for these loci. The majority of these studies were performed on autosomal dominant MYP families. X-linked myopia may occur as an isolated genetic anomaly or it may be associated with various other anomalies, such as night blindness,32–35 cone–rod dysfunction,36 albinism,37 Aland Island eye disease,38,39 and blue cone monochromacy,40 combined with astigmatism, impaired vision, hypoplasia of the optic nerve heads, and deuteranopia (color blindness).9,12 Although most reports have associated X-linked myopia with other anomalies, a few have suggested that it is isolated.11,41,42
We report MYP1 locus homogeneity to the previously defined Xq28 region9 in two large multigeneration Asian Indian families with nonsyndromic high-grade myopia (UR006 and UR077), and the exclusion of potential candidate genes located in this region. Recombination in family UR006 refined the previously mapped MYP1 disease locus to ∼1.25 Mb flanked by the proximal marker DXS1073 and distal marker DXYS154. Thirteen positional candidate genes were excluded by direct sequence analysis of known exons and splice junctions. The present study represents the first confirmation of locus homogeneity of the MYP1 locus for nonsyndromic X-linked high-grade myopia without any associated anomalies to Xq28.
Materials and Methods
Family UR006
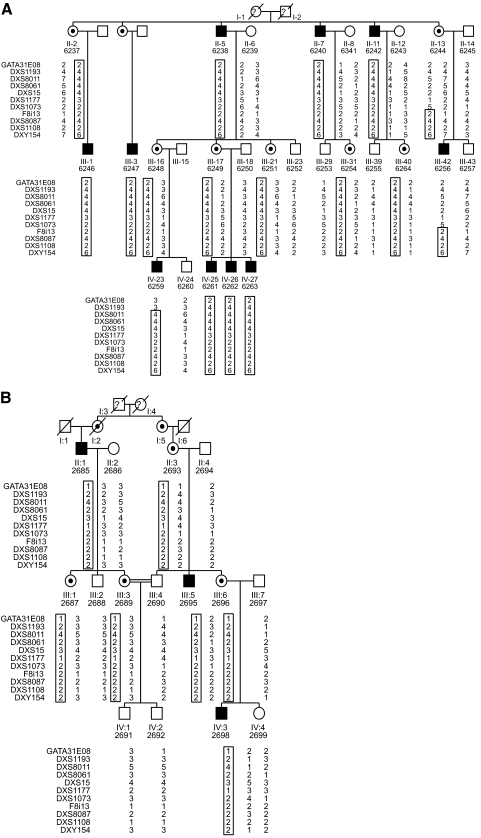
The six-generation pedigree UR006 with isolated, nonsyndromic, high-grade myopia was from Gujarat, in western India. The original pedigree consisted of 60 persons with 10 affected individuals (Fig. 1A). The age at onset ranged from 4 to 6 years, and the degree of myopia among affected family members was variable, ranging from −6 to −23 D (mean, −9.53 D), with the majority >8.0 D.
Figure 1.
Partial pedigrees of myopia families UR006 (A) and UR077 (B). Filled symbols: affected individuals; open symbols: normal individuals; question mark: deceased, data not available; center dot: obligate carrier females. The DNA samples used in the analysis are numbered under each symbol in the pedigrees. Common haplotype among the affected individual are boxed.
Family UR077
This five-generation pedigree UR077 is from the southern state of India, Andhra Pradesh, and consists of 22 individuals, with 3 male members affected with myopia (Fig. 1B). The age at onset was between 4 and 12 years, and the degree of myopia was between −6.00 to −9.5 D (mean, 7.43).
Fifty-four individuals from both the pedigrees who joined in the study were followed up for several years by RS and VVR. To exclude those with a known syndromic form of myopia, such as Marfan syndrome, Stickler syndrome, or Knobloch syndrome, a detailed physical examination was performed and medical and ophthalmic histories were acquired; those with other known ocular or systemic diseases were also excluded. Detailed ophthalmic examinations were conducted by experienced ophthalmologists (RS and VVR) for each patient included in the study, including visual acuity, slit lamp, intraocular pressure, axial length, color vision testing, and funduscopic examination. Electroretinogram (ERG) testing was performed in selected affected individuals. In all patients, individual eye refraction was performed (RT5100; Nidek, Gamagori, Japan; Fig. 2). For the genotyping analysis, the minimal requirements of phenotype for a subject was considered to be high-grade myopia if the refraction error in the either eye was −6 D or below and a maximum age at onset was 4 to 12 years. In both the pedigrees, only male members were affected and the degree of myopia increased with advance age. The affected subjects had high-grade myopia, and the carrier females had no functional or structural visual problems. No other associated visual anomalies, specifically color vision defect or night blindness, were present in any member of these pedigrees. No male-to-male transmission was observed in either family and none of the female members was affected, suggestive of an X-linked recessive mode of inheritance with full penetrance. The research adhered to the tenets of the Declaration of Helsinki. Informed consent was obtained from all subjects who participated in the study, and their blood samples were collected for the genetic studies. Both are unrelated families of different geographic regions, live in urban areas, and are of higher socioeconomic condition than the Indian average. None of the affected individuals had a history of occupational exposures none were involved in any hazardous occupations.
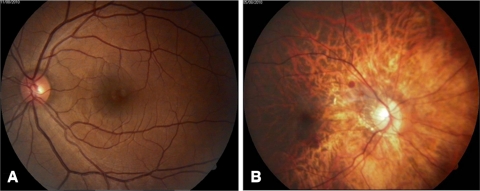
Figure 2.
Fundus photographs of individuals from family UR006: (A) unaffected individual (6253) with normal fundus and (B) affected individual (6247) with high-grade myopia (−12.5 D) with tessellation pattern.
Linkage Analysis
Genomic DNA was purified from peripheral blood samples with a DNA extraction kit (Gentra Systems, Inc., Minneapolis, MN). In the present study, we used 42 samples including 13 affected and 29 normal individuals from both the families, numbered under each individual in the pedigrees (Fig. 1). Initially, we presumed that the phenotype in both the pedigrees (UR006 and UR077) would be allelic to the known MYP1 locus on Xq28. To test this assumption, we performed linkage analysis with the 25 microsatellite markers previously used for genetic analysis of the candidate genomic regions, including the markers that covered the entire X-chromosome. These markers were selected from the Marshfield, Genethon, and CHLC collection (NIH/CEPH Collaborative Mapping Group, 1992).43,44 One oligonucleotide primer of each marker was labeled with γ[32P]ATP, with T4 polynucleotide kinase. PCR was performed from 120 ng of genomic DNA in a total volume of 15-μL mixture per reaction containing 0.4 pM of labeled forward primer, 2.6 pM of unlabeled reverse primer, 1.3 μM of each dNTP, and 0.25 U Taq polymerase (Pharmacia, San Diego, CA). PCR products were separated by electrophoresis in a 6% denaturing urea/polyacrylamide gel. Genotyping was performed as previously described.45
Family information and marker genotypes were stored in a pedigree program (Cyrillic 2.1; Cherwell Software, Colorado Spring, CO). Statistical analysis was performed on the basis of an X-linked recessive trait for X chromosome and recessive for PAR2 (pseudoautosomal region 2) with 100% penetrance. For all markers, the allele frequencies were kept equal, and the gene frequency was set to 0.00001. Maximum LOD scores (Zmax) were calculated for each marker by the use of the ILINK, MLINK, and LINKMAP programs of LINKAGE ver. 5.246 and FASTLINK ver. 3.0.47 Two-point and multipoint analysis for both X-chromosome and PAR2 were performed separately as described.48,49
Candidate Gene Eye Tissue Expression Studies
For reverse transcription–polymerase chain reaction (RT-PCR), total RNA from sclera, cornea, optic nerve, and retina was extracted from pooled human donor eyes (Pennsylvania Lions Eye Bank, Philadelphia) using an extraction reagent (TRIzol; Invitrogen, Carlsbad, CA). The eyes were treated by submersion in RNA stabilizer (RNAlater solution; Ambion, Inc., Austin, TX) within 2 to 12 hours postmortem. RT-PCR was performed using standard methods with random hexamers and reverse transcriptase (SuperScript II; Invitrogen). Subsequent gene-specific PCR for all tested candidate genes was performed using Taq polymerase (Platinum; Invitrogen) to validate gene expression in human ocular tissue. The resulting PCR amplicons were visualized on 2% agarose gels after electrophoresis and staining with ethidium bromide.
Sequencing
Genomic DNA from two affected and two normal individuals were PCR-amplified for regions covering the entire coding sequence and splice junctions of the genes CTAG2, GAB3, MPP1, F8Bver, FUNDC2, VBP1, RAB39B, CLIC2, TMLHE, SYBL, IL9R, SPRY3, and CXYorf1. Detailed PCR conditions for exons of each gene are available on request (Supplementary Table S1, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.10-6815/-/DCSupplemental). Amplified PCR products were purified with spin columns (QIAquick, Qiagen, Valencia, CA) and sequenced directly (BigDye Terminators Sequencing Kit; Applied Biosytems, Inc. [ABI], Foster City, CA) in both directions with an automated genetic analysis system (3100; ABI) and were analyzed with a commercial software program package (Sequencer 4.1; Gene Codes, Ann Arbor, MI).
Electronic Database Information
The following sites were accessed during the research for this study: The UCSC Human Genome Browser (http://genome.cse.ucsc.edu/, University of California Santa Cruz); MERLIN (http://www.sph.umich.edu/csg/abecasis/Merlin/, the University of Michigan, Ann Arbor, MI); National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/, National Institutes of Health [NIH], Bethesda, MD); and Online Mendelian Inheritance in Man (OMIM) (http://www.ncbi.nlm.nih.gov/Omim/, NCBI, NIH).
Results
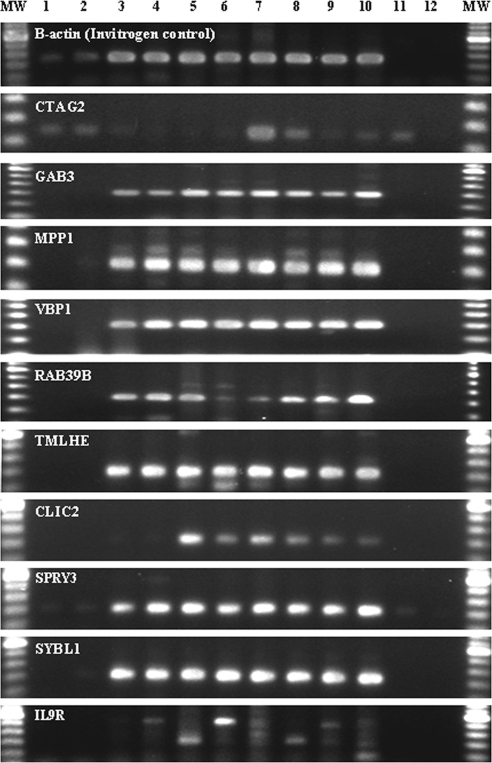
All myopia subjects included in the analysis had high-grade myopia in one or both the eyes (>6.0 D) without any associated anomalies, including color vision defect. All affected subjects developed high-grade myopia before they were of school age. None of the unrelated individuals in this family was observed with any form of myopia. The results of two-point linkage analyses of myopia families UR006 and UR077 and polymorphic microsatellite markers covering the Xq28 genomic region at various recombination fractions are shown in Table 1. A maximum combined two-point LOD score of 5.3 was obtained for marker DXYS154, which is located in the pseudoautosomal region of distal Xq28 (PAR2) at recombination fraction theta = 0. Other markers in the region (near but not within PAR2) that showed no recombination with the phenotype included DXS1108, DXS8087, and F8i13. However, recombination was observed between the phenotype and markers GATA31E08, DXS1193, DXS8011, DXS8061, DXS15, DXS1177, and DXS1073 for family UR006. Other X chromosome markers that showed negative LOD scores are not shown. RT-PCR results confirmed expression of all genes tested in human ocular tissue (Fig. 3). Sequence analysis of known exons and splice junctions of 13 positional candidate genes did not yield any mutations in the exons and intron–exon boundaries for the affected and normal individuals.
Table 1.
Combined Two-Point LOD Scores for MYP1 and Xq28 Markers for Families UR006 and UR077
| Markers | Family | Recombination Fractions |
Physical Position (Mb) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.01 | 0.05 | 0.10 | 0.20 | 0.30 | 0.40 | |||
| GATA31E08 | UR006 | −∞ | −2.87 | −1.55 | −1.04 | −0.59 | −0.35 | −0.17 | |
| UR077 | −∞ | −0.84 | −0.20 | −0.02 | 0.16 | 0.16 | 0.10 | ||
| Combined | −∞ | −3.71 | −1.75 | −1.02 | −0.43 | −0.19 | −0.07 | 140.062 | |
| DXS1193 | UR006 | −∞ | 0.15 | 0.69 | 0.80 | 0.70 | 0.49 | 0.25 | |
| UR077 | 1.70 | 1.71 | 1.59 | 1.44 | 1.11 | 0.76 | 0.39 | ||
| Combined | −∞ | 1.86 | 1.66 | 2.24 | 1.81 | 1.25 | 0.64 | 148.187 | |
| DXS8011 | UR006 | −∞ | 2.15 | 2.60 | 2.57 | 2.16 | 1.54 | 0.79 | |
| UR077 | 1.30 | 1.23 | 1.14 | 1.03 | 0.80 | 0.56 | 0.29 | ||
| Combined | −∞ | 3.38 | 3.74 | 3.60 | 2.96 | 2.10 | 1.08 | 149.617 | |
| DXS8061 | UR006 | −∞ | 1.56 | 2.04 | 2.06 | 1.75 | 1.25 | 0.63 | |
| UR077 | 2.10 | 2.03 | 1.89 | 1.71 | 1.33 | 0.92 | 0.47 | ||
| Combined | −∞ | 3.59 | 3.93 | 3.77 | 3.08 | 2.17 | 1.10 | 151.772 | |
| DXS15 | UR006 | −∞ | 2.15 | 2.60 | 2.57 | 2.16 | 1.54 | 0.79 | |
| UR077 | 1.70 | 1.71 | 1.59 | 1.44 | 1.11 | 0.76 | 0.38 | ||
| Combined | −∞ | 3.86 | 4.19 | 4.01 | 3.27 | 2.30 | 1.17 | 152.128 | |
| DXS1177 | UR006 | −∞ | 2.15 | 2.60 | 2.57 | 2.16 | 1.54 | 0.79 | |
| UR077 | 2.00 | 1.98 | 1.84 | 1.67 | 1.29 | 0.88 | 0.45 | ||
| Combined | −∞ | 4.13 | 4.44 | 4.24 | 3.45 | 2.42 | 1.24 | 152.285 | |
| DXS1073 | UR006 | −∞ | 1.38 | 1.87 | 1.89 | 1.60 | 1.13 | 0.58 | |
| UR077 | 2.00 | 2.01 | 1.87 | 1.69 | 1.31 | 0.90 | 0.46 | ||
| Combined | −∞ | 3.39 | 3.74 | 3.58 | 2.91 | 2.03 | 1.04 | 152.547 | |
| F8i13 | UR006 | 2.22 | 2.18 | 2.00 | 1.78 | 1.34 | 0.91 | 0.46 | |
| UR077 | 0.92 | 0.91 | 0.84 | 0.76 | 0.58 | 0.40 | 0.20 | ||
| Combined | 3.14 | 3.09 | 2.84 | 2.54 | 1.92 | 1.31 | 0.66 | 153.482 | |
| DXS8087 | UR006 | 1.15 | 1.12 | 1.01 | 0.87 | 0.61 | 0.38 | 0.18 | |
| UR077 | 1.07 | 1.05 | 0.97 | 0.86 | 0.65 | 0.42 | 0.19 | ||
| Combined | 2.22 | 2.17 | 1.98 | 1.73 | 1.26 | 0.80 | 0.37 | 153.8 | |
| DXS1108 | UR006 | 1.29 | 1.26 | 1.14 | 0.99 | 0.69 | 0.42 | 0.19 | |
| UR077 | 0.92 | 0.91 | 0.84 | 0.76 | 0.58 | 0.40 | 0.20 | ||
| Combined | 2.21 | 2.17 | 1.98 | 1.75 | 1.27 | 0.82 | 0.39 | 154.515 | |
| DXY154 | UR006 | 3.99 | 3.91 | 3.59 | 3.19 | 2.33 | 1.42 | 0.57 | |
| UR077 | 1.31 | 1.27 | 1.13 | 0.96 | 0.62 | 0.33 | 0.11 | ||
| Combined | 5.30 | 5.18 | 4.72 | 4.15 | 2.95 | 1.75 | 0.68 | 154.731 | |
Figure 3.
Gene-specific PCR for some tested candidate genes by reverse transcription-polymerase chain reaction. RT-PCR results: lane 1: sclera; lane 2: cornea; lane 3: optic nerve; lane 4: retina; lane 5: lung; lane 6: skeletal muscle; lane 7: heart; lane 8: trachea; lane 9: kidney (all Clontech, Inc., Temecula, CA); lane 10: brain (Ambion); lane 11: cRNA; lane 12: no RT (H2O).
Discussion
Myopia is a heterogeneous disorder and has a multifactorial etiology in which both genetics and environment play important roles. Seventeen loci50 with evidence of linkage for either high- or low-grade myopia have been identified on various chromosomes.9,10,13–23,50,51 However, to date no convincing candidate gene has been found to be mutated in myopia. In the only known X-linked MYP1 family, affected males and homozygous females exhibited myopia and color vision defect.9 The same family was revisited by Young et al.13 to include additional subjects and genealogic data using polymorphic microsatellite markers. All affected males had high-grade myopia with protanopia and mild cone dysfunction on electroretinogram testing, whereas carrier females had normal findings in functional and structural ophthalmic evaluations. Zhang et al.10 mapped another X-linked myopia locus (MYP13) to the Xq23-q25 region. In both studies,10,13 all affected individuals had associated anomalies; however, in the present two families (UR006 and UR077), all female carriers were normal, and no affected male members had any associated anomalies, including color vision defect. Linkage analysis showed that the disease locus maps to the previously identified region on Xq289; however, haplotype analysis (Fig. 1A) revealed informative recombination events in the affected individual III-42 (6256) of family UR006 reduced the previously defined genomic interval of 6.8 cM at Xq27.3-Xq2813 to ∼1.4 cM, by flanking the proximal marker at DXS1073 and the distal boundary at DXYS154. Marker DXYS154 gave the highest combined LOD score at θ = 0; however, no distal markers are available. Although families UR006 and UR077 are of Indian origin, they belong to different ethnic groups. Haplotype analysis from the Xq28-linked region of affected individuals did not find extensive common haplotypes shared between these two families, yet a few common identical alleles were observed for selected markers. The MYP13 locus was excluded by linkage and haplotype analysis for both the families.
This 1.25-Mb linked genomic region on Xq28 contains multiple candidate genes for high-grade myopia, including CTAG2, GAB3, MPP1, F8Bver, FUNDC2, VBP1, RAB39B, CLIC2, and TMLHE, proximal to PAR2, and SYBL, IL9R, and SPRY3CXYorf1 in PAR2. Sequence analysis indicated that none of these genes is involved in the pathogenesis of the MYP1 phenotype in both the pedigrees. The absence of pathogenic polymorphic variants in the coding regions of these genes indicates that the mutation may lie in a regulatory region of one of these candidate genes, in another gene, or in another functional element (conserved or not) within the MYP1 region. It is also possible that the mutation is a duplication or a deletion within this region. However, a deletion of the coding sequence of genes outside PAR2 is excluded, because we successfully amplified all sequences in the males. There are genes in the PAR2 region that are only expressed from the X-chromosome; thus, they behave as X-linked recessive. HSPRY3 and SYBL1 are both inactive on Y and are subject to X inactivation in humans.52 In contrast, IL9R and CXYorf1 are expressed on Y and are not subject to X inactivation.53,54 Identification of the mutant gene would potentially provide significant insights into the molecular mechanisms underlying the etiology of nonsyndromic myopia.
Supplementary Material
Acknowledgments
The authors thank all members of families UR006 and UR077 for their participation and donation of blood samples.
Footnotes
Supported by funds from the Green Cross Blood Bank, Ahmedabad, India, and partially by funds from the University and Cantonal Hospital of Geneva. TLY lab is supported by funds from Research to Prevent Blindness, Inc., and NIH NEI R01 EY014685.
Disclosure: U. Ratnamala, None; R. Lyle, None; R. Raval, None; R. Singh, None; S. Vishnupriya, None; P. Himabindu, None; V. Rao, None; S. Aggarwal, None; P. Paluru, None; L. Bartoloni, None; T.L. Young, None; A. Paoloni-Giacobino, None; M.A. Morris, None; S.K. Nath, None; S.E. Antonarakis, None; U. Radhakrishna, None
References
- 1. Curtin BJ. The Myopias: Basic Science and Clinical Management. New York: Harper and Row; 1985; 237– 245 [Google Scholar]
- 2. Fredrick DR. Myopia. BMJ. 2002; 324: 1195– 1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Quinn GE, Shin CH, Maguire MG, Stone RA. Myopia and ambient lighting at night. Nature. 1999; 399: 113– 114 [DOI] [PubMed] [Google Scholar]
- 4. Zadnik K, Jones LA, Irvin BC, et al. Myopia and ambient night-time lighting. CLEERE Study Group. Collaborative Longitudinal Evaluation of Ethnicity and Refractive Error. Nature. 2000; 404: 143– 144 [DOI] [PubMed] [Google Scholar]
- 5. McBrien NA, Adams DW. A longitudinal investigation of adult-onset and adult-progression of myopia in an occupational group: refractive and biometric findings. Invest Ophthalmol Vis Sci. 1997; 38: 321– 333 [PubMed] [Google Scholar]
- 6. Adams DW, McBrien NA. Prevalence of myopia and myopic progression in a population of clinical microscopists. Optom Vis Sci. 1992; 69: 467– 473 [DOI] [PubMed] [Google Scholar]
- 7. Kempen JH, Mitchell P, Lee KE, et al. The prevalence of refractive errors among adults in the United States, Western Europe, and Australia. Arch Ophthalmol. 2004; 122: 495– 505 [DOI] [PubMed] [Google Scholar]
- 8. Wong TY, Foster PJ, Hee J, et al. Prevalence and risk factors for refractive errors in adult Chinese in Singapore. Invest Ophthalmol Vis Sci. 2000; 41: 2486– 2494 [PubMed] [Google Scholar]
- 9. Schwartz M, Haim M, Skarsholm D. X-linked myopia: Bornholm eye disease: linkage to DNA markers on the distal part of Xq. Clin Genet. 1990; 38: 281– 286 [PubMed] [Google Scholar]
- 10. Zhang Q, Guo X, Xiao X, Jia X, Li S, Hejtmancik JF. Novel locus for X linked recessive high myopia maps to Xq23–q25 but outside MYP1. J Med Genet. 2006; 43: e20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bartsocas CS, Kastrantas AD. X-linked form of myopia. Hum Hered. 1981; 31: 199– 200 [DOI] [PubMed] [Google Scholar]
- 12. Haim M, Fledelius HC, Skarsholm X-linked myopia in Danish family. Acta Ophthalmol (Copenh). 1988; 66: 450– 456 [DOI] [PubMed] [Google Scholar]
- 13. Young TL, Deeb SS, Ronan SM, et al. X-linked high myopia associated with cone dysfunction. Arch Ophthalmol. 2004; 122: 897– 908 [DOI] [PubMed] [Google Scholar]
- 14. Young TL, Ronan SM, Drahozal LA, et al. Evidence that a locus for familial high myopia maps to chromosome 18p. Am J Hum Genet. 1998; 63: 109– 119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Young TL, Ronan SM, Alvear AB, et al. A second locus for familial high myopia maps to chromosome 12q. Am J Hum Genet. 1998; 63: 1419– 1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Paluru P, Ronan SM, Heon E, et al. New locus for autosomal dominant high myopia maps to the long arm of chromosome 17. Invest Ophthalmol Vis Sci. 2003; 44: 1830– 1836 [DOI] [PubMed] [Google Scholar]
- 17. Stambolian D, Ibay G, Reider L, et al. Genomewide linkage scan for myopia susceptibility loci among Ashkenazi Jewish families shows evidence of linkage on chromosome 22q12. Am J Hum Genet. 2004; 75: 448– 459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hammond CJ, Andrew T, Mak YT, Spector TD. A susceptibility locus for myopia in the normal population is linked to the PAX6 gene region on chromosome 11: a genomewide scan of dizygotic twins. Am J Hum Genet. 2004; 75: 294– 304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang Q, Guo X, Xiao X, Jia X, Li S, Hejtmancik JF. A new locus for autosomal dominant high myopia maps to 4q22–q27 between D4S1578 and D4S1612. Mol Vis. 2005; 11: 554– 560 [PubMed] [Google Scholar]
- 20. Paluru PC, Nallasamy S, Devoto M, Rappaport EF, Young TL. Identification of a novel locus on 2q for autosomal dominant high-grade myopia. Invest Ophthalmol Vis Sci. 2005; 46: 2300– 2307 [DOI] [PubMed] [Google Scholar]
- 21. Wojciechowski R, Moy C, Ciner E, et al. Genomewide scan in Ashkenazi Jewish families demonstrates evidence of linkage of ocular refraction to a QTL on chromosome 1p36. Hum Genet. 2006; 119: 389– 399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nallasamy S, Paluru PC, Devoto M, Wasserman NF, Zhou J, Young TL. Genetic linkage study of high-grade myopia in a Hutterite population from South Dakota. Mol Vis. 2007; 13: 229– 236 [PMC free article] [PubMed] [Google Scholar]
- 23. Lam CY, Tam PO, Fan DS, et al. A genome-wide scan maps a novel high myopia locus to 5p15. Invest Ophthalmol Vis Sci. 2008; 49: 3768– 3778 [DOI] [PubMed] [Google Scholar]
- 24. Ciner E, Wojciechowski R, Ibay G, Bailey-Wilson JE, Stambolian D. Genomewide scan of ocular refraction in African-American families shows significant linkage to chromosome 7p15. Genet Epidemiol. 2008; 32: 454– 463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang Z, Xiao X, Li S, Zhang Q. Clinical and linkage study on a consanguineous Chinese family with autosomal recessive high myopia. Mol Vis. 2009; 15: 312– 318 [PMC free article] [PubMed] [Google Scholar]
- 26. Hysi PG, Young TL, Mackey DA, et al. A genome-wide association study for myopia and refractive error identifies a susceptibility locus at 15q25. Nat Genet. 2010; 42: 902– 905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Solouki AM, Verhoeven VJ, van Duijn CM, et al. A genome-wide association study identifies a susceptibility locus for refractive errors and myopia at 15q14. Nat Genet. 2010; 42: 897– 901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nakanishi H, Yamada R, Gotoh N, et al. A genome-wide association analysis identified a novel susceptible locus for pathological myopia at 11q24.1. PLoS Genet. 2009; 5: e1000660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nakanishi H, Hayashi H, Yamada R, et al. Single-nucleotide polymorphisms in the promoter region of matrix metalloproteinase-1, -2, and -3 in Japanese with high myopia. Invest Ophthalmol Vis Sci. 2010; 51: 4432– 4436 [DOI] [PubMed] [Google Scholar]
- 30. Metlapally R, Ki CS, Li YJ, et al. Genetic association of insulin-like growth factor-1 polymorphisms with high-grade myopia in an international family cohort. Invest Ophthalmol Vis Sci. 2010; 51: 4476– 4479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang Y, Li X, Yan N, Cai S, Liu X. Myopia: a collagen disease? Med Hypotheses. 2009; 73: 485– 487 [DOI] [PubMed] [Google Scholar]
- 32. Price MJ, Judisch GF, Thompson HS. X-linked congenital stationary night blindness with myopia and nystagmus without clinical complaints of nyctalopia. J Pediatr Ophthalmol Strabismus. 1988; 25: 33– 36 [DOI] [PubMed] [Google Scholar]
- 33. Bech-Hansen NT, Naylor MJ, Maybaum TA, et al. Mutations in NYX, encoding the leucine-rich proteoglycan nyctalopin, cause X-linked complete congenital stationary night blindness. Nat Genet. 2000; 26: 319– 323 [DOI] [PubMed] [Google Scholar]
- 34. Gal A, Schinzel A, Orth U, et al. Gene of X-chromosomal congenital stationary night blindness is closely linked to DXS7 on Xp. Hum Genet. 1989; 81: 315– 318 [DOI] [PubMed] [Google Scholar]
- 35. Orth U, Schinzel A, Machler M, Gal A. X-chromosomal hereditary night blindness: detection of carriers by segregation analysis with linked DNA markers (in German). Klin Monatsbl Augenheilkd. 1990; 196: 269– 272 [DOI] [PubMed] [Google Scholar]
- 36. Glass IA, Good P, Coleman MP, et al. Genetic mapping of a cone and rod dysfunction (Aland Island eye disease) to the proximal short arm of the human X chromosome. J Med Genet. 1993; 30: 1044– 1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Forsius H, Eriksson AW. A new eye syndrome with X-chromosomal transmission: a family clan with fundus albinism, fovea hypoplasia, nystagmus, myopia, astigmatism and dyschromatopsia (in German). Klin Monatsbl Augenheilkd. 1964; 144: 447– 457 [PubMed] [Google Scholar]
- 38. Alitalo T, Kruse TA, Forsius H, Eriksson AW, de la Chapelle A. Localization of the Aland Island eye disease locus to the pericentromeric region of the X chromosome by linkage analysis. Am J Hum Genet. 1991; 48: 31– 38 [PMC free article] [PubMed] [Google Scholar]
- 39. Jalkanen R, Bech-Hansen NT, Tobias R, et al. A novel CACNA1F gene mutation causes Aland Island eye disease. Invest Ophthalmol Vis Sci. 2007; 48: 2498– 2502 [DOI] [PubMed] [Google Scholar]
- 40. Lewis RA, Holcomb JD, Bromley WC, Wilson MC, Roderick TH, Hejtmancik JF. Mapping X-linked ophthalmic diseases. III. Provisional assignment of the locus for blue cone monochromacy to Xq28. Arch Ophthalmol. 1987; 105: 1055– 1059 [DOI] [PubMed] [Google Scholar]
- 41. Wold KC. Hereditary myopia. Arch Ophthalmol. 1949; 42: 225– 237 [DOI] [PubMed] [Google Scholar]
- 42. Gregg FM, Feinberg EB. X-linked pathologic myopia. Ann Ophthalmol. 1992; 24: 310– 312 [PubMed] [Google Scholar]
- 43. Gyapay G, Morissette J, Vignal A, et al. The 1993–94 Genethon human genetic linkage map. Nat Genet. 1994; 7: 246– 339 [DOI] [PubMed] [Google Scholar]
- 44. Buetow KH, Weber JL, Ludwigsen S, et al. Integrated human genome-wide maps constructed using the CEPH reference panel. Nat Genet. 1994; 6: 391– 393 [DOI] [PubMed] [Google Scholar]
- 45. Radhakrishna U, Blouin JL, Mehenni H, et al. Mapping one form of autosomal dominant postaxial polydactyly type A to chromosome 7p15–q11.23 by linkage analysis. Am J Hum Genet. 1997; 60: 597– 604 [PMC free article] [PubMed] [Google Scholar]
- 46. Lathrop GM, Lalouel JM, Julier C, Ott J. Strategies for multilocus linkage analysis in humans. Proc Natl Acad Sci U S A. 1984; 81: 3443– 3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cottingham RW, Jr, Idury RM, Schaffer AA. Faster sequential genetic linkage computations. Am J Hum Genet. 1993; 53: 252– 263 [PMC free article] [PubMed] [Google Scholar]
- 48. Ott J. Y-linkage and pseudoautosomal linkage. Am J Hum Genet. 1986; 38: 891– 897 [PMC free article] [PubMed] [Google Scholar]
- 49. Auricchio A, Brancolini V, Casari G, et al. The locus for a novel syndromic form of neuronal intestinal pseudoobstruction maps to Xq28. Am J Hum Genet. 1996; 58: 743– 748 [PMC free article] [PubMed] [Google Scholar]
- 50. Naiglin L, Gazagne C, Dallongeville F, et al. A genome wide scan for familial high myopia suggests a novel locus on chromosome 7q36. J Med Genet. 2002; 39: 118– 124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Paget S, Julia S, Vitezica ZG, Soler V, Malecaze F, Calvas P. Linkage analysis of high myopia susceptibility locus in 26 families. Mol Vis. 2008; 14: 2566– 2574 [PMC free article] [PubMed] [Google Scholar]
- 52. Charchar FJ, Svartman M, El-Mogharbel N, et al. Complex events in the evolution of the human pseudoautosomal region 2 (PAR2). Genome Res. 2003; 13: 281– 286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ciccodicola A, D'Esposito M, Esposito T, et al. Differentially regulated and evolved genes in the fully sequenced Xq/Yq pseudoautosomal region. Hum Mol Genet. 2000; 9: 395– 401 [DOI] [PubMed] [Google Scholar]
- 54. Matarazzo MR, Cuccurese M, Strazzullo M, et al. Human and mouse SYBL1 gene structure and expression. Gene. 1999; 240: 233– 238 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.