Based on transcriptome responses of chick retina/RPE to unilateral spectacle lens wear, plus or minus lenses induce markedly different retinal responses, with initial responses quite different from those when growth patterns are well established. The lists of altered genes identify promising signaling candidates and regulatory pathways for future study, including the hypothesis that retinal circadian and clock genes may participate in the mechanisms governing refractive development.
Abstract
Purpose.
Because of the retina's role in refractive development, this study was conducted to analyze the retinal transcriptome in chicks wearing a spectacle lens, a well-established means of inducing refractive errors, to identify gene expression alterations and to develop novel mechanistic hypotheses about refractive development.
Methods.
One-week-old white Leghorn chicks wore a unilateral spectacle lens of +15 or −15 D for 6 hours or 3 days. With total RNA from the retina/(retinal pigment epithelium, RPE), chicken gene microarrays were used to compare gene expression levels between lens-wearing and contralateral control eyes (n = 6 chicks for each condition). Normalized microarray signal intensities were evaluated by analysis of variance, using a false discovery rate of <10% as the statistical criterion. Selected differentially expressed genes were validated by qPCR.
Results.
Very few retina/RPE transcripts were differentially expressed after plus lens wear. In contrast, approximately 1300 transcripts were differentially expressed under each of the minus lens conditions, with minimal overlap. For each condition, low fold-changes typified the altered transcriptome. Differentially regulated genes under the minus lens conditions included many potentially informative signaling molecules and genes whose protein products have roles in intrinsic retinal circadian rhythms.
Conclusions.
Plus or minus lens wear induce markedly different, not opposite, alterations in retina/RPE gene expression. The initial retinal responses to defocus are quite different from those when the eye growth patterns are well established, suggesting that different mechanisms govern the initiation and persistence or progression of refractive errors. The gene lists identify promising signaling candidates and regulatory pathways for future study, including a potential role for circadian rhythms in refractive development.
The molecular mechanisms governing normal refractive development or underlying the development of refractive errors are poorly understood. Genetic factors have been implicated in both myopia1 and hyperopia.2 It has been long maintained that myopia in particular arises from complex interactions of both environmental and genetic influences, but the relative roles of environment versus genes remain to be fully defined.3–5 Contemporary laboratory and clinical research indicates that the visual image modulates refractive development because image blur or defocus alters eye growth; the retina in large part governs these processes.6–9 Much laboratory research in this area has used one of two experimental approaches: (1) form-deprivation myopia, in which wearing an image-diffusing goggle or lid suturing blurs the retinal image and induces ipsilateral myopia; and (2) wearing defocusing spectacle lenses to shift the image plane in front of or behind the retina, inducing compensating changes in eye growth to reposition the retina at the image location. With spectacle lens wear, plus (i.e., convex or [+]) spectacle lenses shift the visual image forward and slow eye growth to permit the anatomic adjustments for distant images to focus in the retina; minus (i.e., concave or [−]) spectacle lenses shift the visual image farther back and accelerate eye growth, also permitting distant images to focus on the retina. These image-related adjustments in ocular growth include not only changed scleral growth but also altered choroidal thickness: choroidal thinning with goggle or (−) lens wear that stimulate eye growth and choroidal thickening after (+) lens wear.10 The early kinetics of choroidal thickness responses vary somewhat between these eye growth models in chick.11 With spectacle lens removal, eyes previously beneath a (+) spectacle lens are hyperopic; those previously beneath a (−) spectacle lens are myopic.9 Form-deprivation myopia occurs in children,12 and the eyes of young adults adjust axial dimensions to acute defocus.13 The extent to which the signaling and molecular mechanisms that underlie the responses to goggles or spectacle lens wear account for clinical ametropia is unknown7; spectacle lens wear may be most useful for studying emmetropization mechanisms,9,14 although it is unclear whether emmetropia is the physiologic end point of human refractive development.
Through such experimental approaches, numerous neurotransmitters or other signaling molecules have been implicated in the pathway(s) linking visual input and refractive development.7 However, an organized framework is not available for the retinal signaling that underlies mechanisms for either emmetropization or the development of refractive errors. Visual stimuli that alter eye growth induce changes in gene expression at the level of the transcriptome (molecular signatures) in the retina.15 We hypothesized that these molecular signatures not only can identify important retinal mediators of refractive development but also can identify signaling pathways or networks, which may provide leads for novel approaches to understand laboratory and clinical conditions.
Paralleling a protocol we had used previously to study form-deprivation myopia,16 we profiled gene expression in the combined retina and retinal pigment epithelium (RPE) of chicks wearing a unilateral spectacle lens of (+) or (−) power. We conducted these assays after either 6 hours or 3 days of lens wear. The shorter duration activates a retina-to-sclera signaling pathway that subsequently modulates scleral biochemistry, indicating initiation of at least some aspect(s) of the eye growth signaling pathway; the longer duration corresponds to a time when changes in refraction and eye size are manifest.11 Because of their links to perturbed eye growth, rapid alterations in retinal dopamine physiology17 and the immediate early gene EGR118–21 after experimentally altered visual input also support the notion that the 6-hour sampling time reflects early activation of a retina-to-sclera signaling pathway. Although minor contralateral refractive effects develop when chicks wear a unilateral spectacle lens,10 potential individual differences in gene expression complicate statistical approaches to interbird comparisons, and our bioinformatics approach accordingly emphasized the experimental-to-contralateral control eye comparison in individual birds.
Methods
One-day-old white Leghorn chicks (Moyer's Chick, Inc., Quakertown, PA) were maintained under a 12-hour light–dark cycle, under incandescent lighting (General Electric, Fairfield, CT) with irradiance of approximately 1600 μW/cm2 at chick eye level. They received food (Purina Chick Chow, Indianapolis, IN) and water ad libitum. When the chicks were 1 week of age and under inhalation ether anesthesia, a ring-shaped piece of Velcro was secured to the periorbital feathers of one eye with cyanoacrylate glue; experimental eyes were alternated between right and left in each series of chicks. Within 1 hour of the onset of the light phase on the next day and without sedation, either a +15- or –15-D clear 12-mm diameter PMMA (polymethyl methacrylate) contact lens (ABB CONCISE Optical Group LLC, Marshfield, MA) was secured to the experimental eye with complementary ring-shaped Velcro; the contralateral eye was not fitted with a lens and served as the within-subject control. After 6 hours or 3 days of spectacle lens wear (n = 8 for each time and for either [+] or [−] lens wear), the chicks were killed by decapitation. For 6 hours of lens wear, the chicks were killed at 6 to 7 hours into the light phase, the lenses having been applied at the onset of the light phase. For 3 days of lens wear, the chicks were killed at 2 to 3 hours into the light phase. The ocular effects of spectacle lens wear by chicks are well characterized,9 and ocular refractions and eye measurements were not obtained to avoid potential anesthesia effects and to minimize postmortem mRNA degradation. As quickly as possible, the eyes were enucleated and opened at the equator; the retina/RPE was dissected together from the lens-wearing and contralateral control eyes. The tissues were individually frozen and stored in liquid nitrogen until processed. The microarray targets (as cDNA) were prepared from total RNA from each eye separately without pooling, using six chicks from each of the four experimental groups.
To verify the refractive responses, two additional groups of day-old white Leghorn chicks (Charles River Laboratories, Preston, CT; n = 6/cohort) were reared in identical conditions. After 3 days of unilateral +15- or −15-D lens wear, they were anesthetized with a mixture of ketamine (20 mg/kg) and xylazine (5 mg/kg), and both eyes were measured by refractometry and ultrasound, as described elsewhere.22 While still under anesthesia, the chicks were killed by decapitation. The experiments conformed both to the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research and to the University of Pennsylvania Institutional Animal Care and Use Committee approval.
RNA Isolation
RNA was isolated from each preparation (Trizol reagent; Invitrogen, Carlsbad, CA) followed by purification and DNase treatment on RNeasy columns (Qiagen, Inc., Valencia, CA). To quantify the RNA and determine its purity, we measured the samples on a spectrophotometer (ND-1000 UV-Vis; NanoDrop Technologies, Wilmington, DE), with 260/280 nm absorbance ratios between 1.8 and 2.1. To evaluate RNA integrity further, an aliquot of each RNA sample was loaded onto an RNA chip (6000 Nano Laboratory-Chip) and placed in a bioanalyzer (model 2100; Agilent Technologies, Santa Clara, CA). RNA integrity was verified by electropherograms and gel image analysis to visualize the intact ribosomal bands using the system software. Aliquots of the RNA samples were stored individually at −80°C.
Microarray Target Preparation and Hybridization
Microarray services were provided by the Penn Microarray Facility of the University of Pennsylvania School of Medicine. All protocols were conducted as described in the manufacturers' manuals (Ovation Manual, NuGen Technologies, Inc., San Carlos, CA; GeneChip Expression Analysis Technical Manual, Affymetrix Inc., Santa Clara CA). Briefly, 100 ng of total RNA was converted to first-strand cDNA using reverse transcriptase primed by a poly(T) oligomer that incorporated a synthetic RNA sequence. Second-strand cDNA synthesis was followed by ribo-SPIA (Single Primer Isothermal Amplification, NuGEN Ovation kit) for linear amplification of each transcript, and the resulting cDNA was fragmented, assessed with the bioanalyzer, and biotinylated. cDNA yields ranged from 7.1 to 10.2 μg, and 3.75 μg was added to hybridization cocktails (Affymetrix), heated at 99°C for 2 minutes, and hybridized for 16 hours at 45°C to chicken gene microarrays (Chicken Genome GeneChips; Affymetrix) (http://www.osa.sunysb.edu/udmf/ArraySheets/chicken_datasheet.pdf). The microarrays were then washed at low (6× SSPE) and high (100 mM MES and 0.1 M NaCl) stringency and stained with streptavidin-phycoerythrin. Fluorescence was amplified by adding biotinylated anti-streptavidin and an additional aliquot of streptavidin-phycoerythrin stain. A confocal scanner was used to collect fluorescence signal after excitation at 570 nm.
Bioinformatics Analyses
Hybridization signals were quantified (Command Console and Expression Console; Affymetrix) for each probe; default values provided by Affymetrix were applied to all analysis parameters. Border pixels were removed, and the average intensity of pixels within the 75th percentile was computed for each probe. Probe intensities were exported in .cel file format (Affymetrix).
The .cel files were imported into genomics software (Genomics Suite, ver. 6.4; Partek Inc., St. Louis, MO). To permit comparison between arrays, RMA (robust multiarray average) was applied to yield background-adjusted, normalized, log2-transformed signal intensities. We performed a three-way, mixed-model analysis of variance (ANOVA; factors: time, [6 hours, 3 days]; lens, −, 0, + [i.e., minus, no or plus lens]; chick ID, [random effect], with an interaction term [time × lens]). With the ANOVA, four pairwise comparisons were also calculated: 6 hours, + vs. 0; 6 hours, − vs. 0; 3 days, + vs. 0; and 3 days, − vs. 0. All resulting P values were corrected for multiple comparisons using the Benjamini-Hochberg step-up method to yield false discovery rates (FDRs), as implemented in the genomics software (Partek). An FDR of <10% was considered significant and was used as the primary cutoff criterion for identifying differentially expressed transcripts. For the pairwise comparisons, fold-change in gene expression was also calculated, comparing the lens-wearing to contralateral control eye.
Using Affymetrix probeset identifiers, two web-based tools were used to evaluate the differentially expressed transcripts meeting the above <10% FDR statistical criterion. VENNY (http://bioinfogpcnbcsices/tools/venny/indexhtml) was used to generate the Venn diagram of the overlapping transcripts between the two lens conditions and two times (Fig. 1). To evaluate potential networks and pathways implicated by the differentially expressed transcripts, we used pathway analysis (IPA ver. 8.6-3003, build 92475; Ingenuity Systems, Redwood, CA; http://www.ingenuity.com/) with the following general analysis settings: chicken genome array as the reference set, direct and indirect relationships, endogenous chemicals included, 35 molecules/network, 25 networks, all data sources and species, nervous system tissues/CNS (central nervous system) cell lines with a relaxed filter on the molecules, and their relationships. For conformity in data reporting, we used the Affymetrix notations for gene symbols throughout.
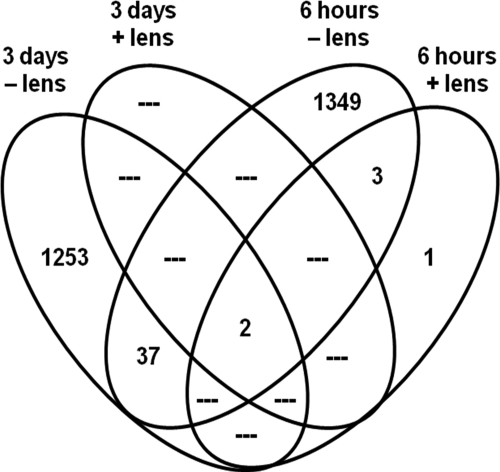
Figure 1.
Distinct and overlapping differentially expressed transcripts. Venn diagram for the distinct and overlapping differentially expressed retina/RPE transcripts for chicks wearing a unilateral +15- or –15-D spectacle lens for 6 hours or 3 days. The number of transcripts is shown in each cell for genes meeting the statistical criteria described in the Methods (i.e., ANOVA corrected for multiple comparisons with FDR <10%). See Supplementary Tables S1 to S4 (http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.10-6727/-/DCSupplemental) for complete gene lists.
Real-Time Quantitative RT-PCR
To ascertain the reliability of the microarray results, we conducted both biological and technical validations of the expression profiling for selected known genes, using real-time quantitative reverse transcription-polymerase chain reaction (qPCR). Biological replicates are assays on animals different from those subjected to microarray analysis but reared and processed contemporaneously; here, we used the retina/RPE from the two other chicks reared contemporaneously under each of the four conditions. Technical validations are measurements of aliquots from the same biological specimen with a different measuring technique; here, for technical validations, we used residual RNA from the retina/RPE assayed by microarray. For both types of qPCR validations, 1 μg of retina/RPE RNA was converted into cDNA (Superscript III First Strand cDNA Synthesis Supermix for qPCR; Invitrogen).
For the biological validations, we selected transcripts of known genes with a fold-change of approximately ≥1.4 in either the up- or downregulated direction, including some genes that were differentially expressed under more than one condition. Within that group, we included vasoactive intestinal peptide (VIP), noggin (NOG), and bone morphogenetic protein 2 (BMP2), because of their relation to findings in our prior profiling of the retina/RPE in form-deprivation myopia.16 For technical validations, four genes/conditions with comparatively high microarray fold-changes were selected from among the genes validated in the biologically independent samples; and qPCR assays were conducted with cDNA from both retinas of all six chicks studied by microarray with the same primers (see Results for genes/conditions). For the technical validations, the normalized expression value for each gene in the experimental retina/RPE was compared to that of its contralateral control using a paired t-test.
For qPCR, primer sets optimized for chicken sequences were purchased (Quantitect Primer Assays; Qiagen) for BMP2 (NM_204358); dual-specificity phosphatase 4 (DUSP4; NM_204838); glyceraldehyde-3-phosphate dehydrogenase (GAPDH; NM_204305); myosin, heavy chain 13, skeletal muscle (MYH13; XM_001231455); NOG (NM_204123); oxysterol binding protein 6 (OSBPL; XM_421982; XM_001233035); phosphodiesterase 3A, cGMP-inhibited (PDE3A;XM_416416); urotensin 2 domain containing (UTS2D; NM_206989; VIP (NM_205366). The primer sets are designed to amplify across intron–exon boundaries; details of size and position of amplicons on each gene of interest are available online from the manufacturer (Qiagen; www.qiagen.com/GeneGlobe).
Using a real-time PCR system (model 7300; Applied Biosystems, Inc., [ABI], Foster City, CA) and 96-well plates, triplicate 30 μL PCR reactions were performed for each gene of interest using cDNA (8 ng/reaction), the gene-specific primers and master mix (QuantiFast SYBR Green RT-PCR; Qiagen). The PCR reaction comprised 40 cycles at 95°C for 15 seconds and 60°C for 32 seconds. The analyses were performed using the system software (model 7300; ABI) and followed directions provided by the manufacturer (Guide to Performing Relative Quantification of Gene Expression RT-qPCR) using the comparative ΔΔCt relative quantification method.23 Before quantitative analysis, the efficiencies of the reference gene GAPDH and the genes of interest were determined to be the same; GAPDH expression was verified as unaltered across experimental conditions.
Results
Refractive Responses
As described,11 spectacle lens wear altered ocular growth to shift the retina toward the image plane of distant objects; at 3 days, the optical compensation was somewhat more advanced for +15-D lens wear (+11.8 ± 1.3 D; mean ± SEM) than for −15-D lens wear (−8.9 ± 0.7 D), although eye growth had not yet completely compensated for lenses of either sign. Eyes beneath the (+) lens were shorter than contralateral eyes, because of a shorter vitreous chamber (by 0.47 ± 0.05 mm); eyes beneath the (−) lens were longer than their contralateral eyes, because of a longer vitreous chamber (by 0.33 ± 0.08 mm).
Alterations in Retina/RPE Gene Expression
With the chicken gene microarrays (Affymetrix), we profiled gene expression in the retina/RPE of chicks wearing a unilateral +15- or −15-D spectacle lens for 6 hours or 3 days, using the contralateral eye as the control. With a conservative 10% FDR, most identified transcripts were differentially expressed at a relatively low fold-change compared with the contralateral eyes (Table 1). In fact, all identified transcripts were differentially expressed at less than a 2.3 fold-change, with the exception of 22 of the 603 differentially expressed transcripts after 3 days of (−) lens wear (Table 1, Supplementary Tables S1–S3, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.10-6727/-/DCSupplemental).
Table 1.
Differentially Expressed Retina/RPE Transcripts
| +15-D Lens |
−15-D Lens |
|||||||
|---|---|---|---|---|---|---|---|---|
| 6 h |
3 d |
6 h |
3 d |
|||||
| Down-regulated | Up-regulated | Down-regulated | Up-regulated | Down-regulated | Up-regulated | Down-regulated | Up-regulated | |
| Transcript number | ||||||||
| Total number | 6 | 2 | 1391 | 1292 | ||||
| Down- vs. up-regulated | 6 | — | 2 | — | 1252 | 139 | 662 | 630 |
| Fold-change | ||||||||
| Mean (±SD) | −1.30 ± 0.15 | — | −1.69 ± 0.07 | — | −1.29 ± 0.12 | +1.17 ± 0.12 | −1.26 ± 0.13 | +1.28 ± 0.47 |
| Maximum | −1.44 | — | −1.74 | — | −1.92 | +2.20 | −2.25 | +5.47 |
| Minimum | −1.13 | — | −1.64 | — | −1.05 | +1.03 | −1.06 | +1.04 |
The data represent those differentially expressed transcripts comparing the lens-wearing to contralateral control eyes and meeting the statistical criteria described in the Methods (i.e., ANOVA corrected for multiple comparisons with FDR <10%).
Remarkable differences occurred between both lens sign and time of image defocus (Table 1, Fig. 1). Surprisingly few transcripts were affected by (+) lens wear: 6 transcripts at 6 hours and 2 transcripts at 3 days. All altered transcripts after (+) lens wear were downregulated, and the two transcripts identified after 3 days of lens wear also were downregulated at the earlier time (Supplementary Tables S1, S4, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.10-6727/-/DCSupplemental). In striking contrast, (−) lens wear resulted in some 1300 differentially expressed genes at each time (Table 1, Fig. 1). At 6 hours of (−) lens wear, most altered transcripts were downregulated; at 3 days, there was a more even balance in the number of downregulated and upregulated transcripts.
A surprisingly small number of differentially expressed transcripts were common to two or more different conditions (Fig. 1; Supplementary Table S4, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.10-6727/-/DCSupplemental). The microarrays identified only two transcripts as differentially expressed in all four conditions, DUSP4 (dual specificity phosphatase 4) and a nonannotated transcript; the retinal/RPE expression of both transcripts was downregulated under each condition. Three transcripts were altered after 6 hours of either (+) or (−) lens wear: RSPO2 (R-spondin 2 homolog, Xenopus laevis), PDE3A (phosphodiesterase 3A, cGMP-inhibited), and another transcript not currently annotated; all three were downregulated under either lens. Comparing the (−) lens-wearing conditions, only 39 transcripts common to both times were altered based on the probeset identification (Affymetrix ID) number (Fig. 1). Because individual genes are represented in the chicken microarray by multiple transcripts with different probeset ID numbers, we also conducted a manual search of the lists of differentially expressed genes for each (−) lens condition. Based on the gene symbols, we found 31 additional named genes that were differentially expressed after both 6 hours and 3 days of (−) lens wear; these 31 additional genes were identified through different probeset Affymetrix ID numbers at the two times (Supplementary Table S4, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.10-6727/-/DCSupplemental).
Validations by qPCR
For the genes/conditions selected for biological validations, qPCR was performed on retina/RPE RNA from two chicks not used for microarray profiling. Of these genes, seven were confirmed by qPCR and six were not confirmed (Table 2). The technical validations used the same six RNA samples as were assayed by microarray for four genes/conditions in Table 2; all four confirmed the profiling results. For the technical validations, the mean (±SEM) fold-changes with statistical significances were: BMP2 minus lens, 6 hours: −3.25 ± 2.41, P = 0.03; BMP2 minus lens, 3 days: −3.26 ± 1.42, P = 0.05; VIP minus lens, 6 hours: −1.84 ± 0.20, P = 0.01; DUSP4 minus lens, 3 days: −2.00 ± 0.31, P = 0.03.
Table 2.
Selected Differentially Expressed Genes Studied by qPCR as Biological Validations
| Probeset ID* | Gene Title (Gene Symbol) | Lens Sign | Wearing Time | Fold-Change by Microarray† | Fold-Change by qPCR‡ |
|---|---|---|---|---|---|
| Gga.385.1.S1_at | Dual specificity phosphatase 4 (DUSP4) | + | 3 d | −1.64 | −1.29 (−1.23, −1.35) |
| Gga.3950.1.S1_at | Bone morphogenetic protein 2 (BMP2) | − | 6 h | −1.78 | −2.82 (−2.38, −3.26) |
| Gga.3950.1.S1_at | Bone morphogenetic protein 2 (BMP2) | − | 3 d | −2.25 | −2.42 (−2.71, −3.13) |
| Gga.666.1.S1_a_at | Vasoactive intestinal peptide (VIP) | − | 6 h | −1.68 | −2.39 (−1.28, −3.50) |
| Gga.9482.1.S1_at | Urotensin 2 domain containing (UTS2D) | − | 3 d | −1.46 | −2.15 (−1.90, −2.40) |
| Gga.449.1.S1_at | Noggin (NOG) | − | 6 h | +1.64 | +1.48 (+1.45, +1.51) |
| Gga.449.1.S1_at | Noggin (NOG) | − | 3 d | +1.38 | +1.35 (+1.14, +1.55) |
| Gga.9482.1.S1_at | Urotensin 2 domain containing (UTS2D) | − | 6 h | −1.44 | Not confirmed (−1.00, −1.59) |
| Gga.385.1.S1_at | Dual specificity phosphatase 4 (DUSP4) | + | 6 h | −1.44 | Not confirmed (−1.10, +1.27) |
| GgaAffx.511.1.S1_at | Myosin,heavy chain 13, skeletal muscle (MYH13) | − | 3 d | +2.78 | Not confirmed (−1.03, +1.00) |
| GgaAffx.22824.1.S1_s_at | Oxysterol binding protein-like 6 (OSBPL6) | − | 6 h | −1.34 | Not confirmed (+1.01, +1.05) |
| GgaAffx.22824.1.S1_s_at | Oxysterol binding protein-like 6 (OSBPL6) | − | 3 d | −1.42 | Not confirmed (+1.03, +1.60) |
| Gga.14256.1.S1_s_at | Phosphodiesterase 3A, cGMP-inhibited (PDE3A) | − | 6 h | −1.47 | Not confirmed (−1.42, +1.07) |
For all fold-change representations, negative numbers represent downregulation of a transcript in the retina/RPE of lens-wearing eyes compared with its expression in contralateral control eyes; positive numbers, upregulation of a transcript relative to contralateral control eyes.
Affymetrix, Santa Clara, CA.
Fold-change of gene expression by microarray for lens-wearing versus contralateral control eye for selected genes meeting the statistical criteria described in the Methods (i.e., ANOVA corrected for multiple comparisons with FDR <10%).
Mean fold-change of confirmed genes from the qPCR (quantitative RT-PCR) of two independent biological replicates; individual qPCR values in parentheses; lens-wearing vs. contralateral control eye.
Analysis of Patterns of Altered Gene Expression of Individual Genes and Patterns of Genes
To organize the long lists of differentially expressed transcripts under the two (−) lens conditions, we used both pathways analysis (Ingenuity Systems) and a manual evaluation based on the authors' familiarity with laboratory and clinical studies of refractive mechanisms. We sought both to identify novel signaling molecules and to group known genes into categories potentially informative for mechanisms governing the accelerated eye growth and myopic refraction developing under these (−) lens conditions. We did not perform such analyses for the (+) lens conditions because so few genes were differentially expressed.
We used a database of biological and functional data (IPA, Ingenuity Systems) to generate de novo networks of interactions among the differentially expressed genes at both 6 hours and 3 days of (−) lens wear (Supplementary Table S5, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.10-6727/-/DCSupplemental). At each experimental time point, the proportion of network molecules from the experimentally identified gene lists fell considerably after the first few networks; and accordingly, only the top five networks are shown for each condition (Supplementary Table S5, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.10-6727/-/DCSupplemental). The database also contains directionally informative canonical pathways based on previously established biological and functional models of relationships between genes and gene products. From this database's assessment of the statistical significance between its database of canonical pathways and the lists of differentially expressed genes at either 6 hours or 3 days (IPA, Ingenuity Systems), Supplementary Tables S6A and S6B (http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.10-6727/-/DCSupplemental) provide those canonical pathways for which P < 0.05.
We also manually generated a list of potentially informative differentially expressed genes (Table 3) based on gene products related to the extensive published pharmacology or prior reports on retinal gene expression in models of refractive development, areas that have been reviewed recently.7,15 These include genes related to amino acid neurotransmitters, acetylcholine, neuropeptides, proteins, and clock/circadian rhythms. We also manually generated a list of differentially expressed genes common to both the current investigation and prior reports on retinal gene expression in refractive development (Supplementary Table 7, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.10-6727/-/DCSupplemental).
Table 3.
Differential Expression of Selected Genes by Microarray Profiling after Minus Lens Wear for 6 Hours or 3 day
| Gene Symbol | Gene Title | Fold-Change*† |
|
|---|---|---|---|
| 6 h | 3 d | ||
| Amino Acid Signaling Genes | |||
| Glutamate-related genes | |||
| GLS | Glutamase | −1.56 | |
| GRM5 | Glutamate receptor, metabotropic 5 | −1.49, −1.44, −1.30 | — |
| GRIN2A | Glutamate receptor, ionotropic, N-methyl D-aspartate 2A | −1.35 | — |
| GRIA4 | Glutamate receptor, ionotrophic, AMPA4 | −1.28 | — |
| GRIK2 | Glutamate receptor, ionotropic, kainate 2 | −1.23 | — |
| GRID2 | Glutamate receptor, ionotropic, delta 2 | −1.23 | −1.23 |
| GRID1 | Glutamate receptor, ionotropic, delta 1 | −1.19 | — |
| GRIA3 | Glutamate receptor, ionotrophic, AMPA 3 | −1.18 | — |
| GRIP1 | Glutamate receptor interacting protein 1 | −1.15 | — |
| NARG1L | NMDA receptor regulated 1-like | — | −1.15 |
| GRINL1B | Glutamate receptor, ionotropic, N-methyl D-aspartate-like 1B | — | −1.10 |
| GABA-related genes | |||
| GABRR3 | Gamma-aminobutyric acid (GABA) receptor, rho 3 | — | −1.62 |
| SLC6A1 | Solute carrier family 6 (neurotransmitter transporter, GABA), member 1 | — | −1.48 |
| ABAT | 4-aminobutyrate aminotransferase | — | −1.23 |
| LOC428967 | Similar to gamma-aminobutyric acid (GABA) A receptor, pi | — | −1.20 |
| GABRG2 | Gamma-aminobutyric acid (GABA) A receptor, gamma 2 | — | −1.19 |
| GAD2 | Glutamate decarboxylase 2 (pancreatic islets and brain, 65kda) | — | −1.12 |
| Glycine-related gene | |||
| GLRA2 | Glycine receptor, alpha 2 | — | −1.44 |
| Acetylcholine-Related Genes | |||
| CHRM2 | Cholinergic receptor, muscarinic 2 | −1.21 | — |
| ACHE | Acetylcholinesterase | — | +1.24, +1.26 |
| Miscellaneous Peptide, Protein and Other Genes | |||
| VIP | Vasoactive intestinal peptide | −1.68 | — |
| BMP2 | Bone morphogenetic protein 2 | −1.78 | −2.25 |
| NOG | Noggin | +1.65 | +1.38 |
| FST | Follistatin | — | −1.22 |
| ADMP | Anti-dorsalizing morphogenetic protein | — | +1.26 |
| GCG | Glucagon | — | −1.40 |
| BDNF | Brain-derived neurotrophic factor | −1.31, −1.21 | −1.21 |
| FGFBP2 | Fibroblast growth factor binding protein 2 | −1.14 | — |
| NGFB | Nerve growth factor, beta polypeptide | — | −1.14 |
| NPY7R | Neuropeptide Y7 receptor | — | +1.29 |
| OGFRL1 | Opioid growth factor receptor-like 1 | — | +1.33 |
| NRG3 | Neuregulin 3 | −1.23 | — |
| PREP | Prolyl endopeptidase | — | −1.23 |
| NAALADL2 | N-acetylated alpha-linked acidic dipeptidase-like 2 | −1.40, −1.30 | — |
| NOS2 | Nitric oxide synthase 2, inducible | −1.23 | — |
| HMOX2 | Heme oxygenase (decycling) 2 | — | +1.17 |
| CNR1 | Cannabinoid receptor 1 (brain) | — | −1.33 |
| CTR BETA 2 | Thyroid hormone receptor beta 2 | −1.53 | — |
| DIO2 | Deiodinase, iodothyronine, type II | — | −1.13 |
| TRIP4 | Thyroid hormone receptor interactor 4 | — | +1.18 |
| ITPR1 | Inositol 1,4,5-triphosphate receptor, type 1 | −1.41, −1.38, −1.32 | — |
| EGR1 | Early growth response 1 | — | −1.57, −1.29 |
| Clock and Circadian Genes | |||
| OPN4 | Opsin 4 (melanopsin) | −1.33 | — |
| LOC395334 | Photopigment melanopsin-like | −1.29 | −1.41 |
| LOC424283 | Similar to peripheral clock protein 2 | −1.27 | — |
| PER3 | Period homolog 3 (Drosophila) | −1.26, −1.21 | — |
| MTNR1A | Melatonin receptor 1A | −1.25 | — |
| CLOCK | Clock homolog (mouse) | −1.18 | — |
| NPAS2 | Neuronal PAS domain protein 2 | −1.14 | −1.29 |
| CRY1 | Cryptochrome 1 (photolyase-like) | — | −1.35 |
Fold-change of gene expression by microarray for lens-wearing vs. contralateral control eye for selected genes meeting the statistical criteria described in the Methods (i.e., ANOVA corrected for multiple comparisons with FDR <10%). Negative numbers represent downregulation of a transcript in the retina/RPE of lens-wearing eyes compared with its expression in contralateral control eyes; positive numbers, upregulation of a transcript relative to contralateral control eyes.
When a gene was identified as differentially expressed by more than one transcript, all fold-change values are listed.
All primary data have been deposited in the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/, National Center for Biotechnology Information, Bethesda, MD) database (accession number GSE24641).
Discussion
Affymetrix Chicken GeneChips contain 32,773 chicken transcripts, representing over 28,000 genes (http://www.osa.sunysb.edu/udmf/ArraySheets/chicken_datasheet.pdf). We used these microarrays to profile the retina/RPE transcriptome in chick eyes beneath either a +15- or −15-D spectacle lens for either 6 hours or 3 days. Spectacle lens wear is a well-established technique for modifying eye growth and refractive development in young chicks and mammals, and it is widely used to study developmental mechanisms for emmetropia and refractive errors. Given the current lack of a cohesive biological understanding of the regulatory mechanism(s) for either emmetropization or refractive errors, we approached these profiles with a view toward generating novel, mechanistic, conceptual hypotheses about refractive development as well as exploring individual genes and potential definable regulatory networks.
Low Fold-Changes in Altered Genes
Similar to prior reports using chicken genome-wide microarrays in whole retina with binocular (+) lens wear24 or in retina/RPE with form-deprivation myopia,16 low fold-changes characterize most differentially expressed transcripts induced by either (+) or (−) lens wear for either duration (Table 1). As before,16 we included a large number of chicks for microarray profiling without pooling of tissues, as a strategy to increase the statistical likelihood of identifying differentially expressed genes with fold-changes ratios while minimizing the prospect of confounding by outliers. Evidently, the robust developmental response of chick eyes to image defocus develops without major alteration in the expression level of retinal genes, at least those genes included in the microarray platform. To enrich the list of dysregulated genes and both identify affected gene networks and generate novel hypotheses, we identified differentially expressed genes on statistical significance and not arbitrary fold-change levels.
qPCR Validations
We assayed a large number of chicks (n = 6/cohort), analyzing each eye by a separate microarray to bolster the statistical validity of the core profiling results. How to best validate microarray data is an open question.25 Here, we conducted biological validations with qPCR on the retina/RPE from two chicks in each cohort reared contemporaneously with the chicks whose retinas were evaluated by microarrays. The correlation between microarray platforms and qPCR is 0.70 to 0.89, suggesting reasonably good (though not necessarily excellent) agreement between the two methods; the agreement between microarrays and qPCR deteriorates with low fold-changes in differential gene expression,26–29 as generally seen here. We found that 7 (54%) of 13 of the qPCR assessments validated the fold-change direction of the microarrays. Although the utility of validating microarrays with technical replicates has been questioned,25 we also validated the microarrays with technical replicates for four genes/conditions for thoroughness. Considering our 10% FDR and the low fold-changes in differentially expressed genes, this proportion of altered genes validated by qPCR on biologically independent birds and the confirming technical replicates preclude a major systematic error in the entire microarray procedure. Together, these validations indicate that the profiling can be interpreted in terms of its primary statistical analysis.
The results of the microarray analyses form the basis for the considerations below. Any future study of individual genes and/or biological pathways will require independent assessments because of the low fold-changes and the inherent statistical nature of microarray profiling analysis.
Myopic versus Hyperopic Defocus
Comparing (+) lens wear to (−) lens wear, marked differences developed in the altered retinal transcriptome. Myopic defocus from (+) lens wear caused differential expression of very few transcripts at either time. Two other available microarray studies assessed the response of whole retina to myopic defocus. One assayed retina after bilateral (+) lens wear for 24 hours,24 and the other assayed retina/RPE/choroid after 1 or 4 days of recovery from myopia induced by a unilateral goggle.30 Each, particularly the former, identified more differentially expressed genes than the current investigation; but differences in experimental design and analytical methods limit direct comparison between these studies.15 For instance, bilateral lens wear, different assay times and use of interbird, not intrabird, controls in the other available study of retinal gene expression after (+) lens wear24 are major experimental differences that could account for the differences from the present investigation.
In contrast to the few retina/RPE genes we identified after myopic defocus, the expression levels of many transcripts were affected by hyperopic defocus from (−) lens wear (Table 1, Fig. 1). Despite their opposite effects on refraction and eye growth, (+) or (−) lens wear exerted nonreciprocal effects on the retinal transcriptome. Choroidal compensation per se does not provide a simple explanation for these differences in gene expression because choroidal thickness changes rapidly in each condition, although in different directions with different time courses.11 Refractive compensation to either lens is incomplete by 3 days, so that defocus persists with lens wear of either sign. Further, the limited number of genes common to (+) or (−) lens wear were each downregulated under these conditions and not altered in opposite directions. In other direct comparisons of the effects of (+) and (−) lens wear on the retinal transcriptome, a limited number of common differentially expressed genes and nonreciprocal effects similarly were seen after 24 hours in the whole retina with binocular treatments24 and in the amacrine cell layer with monocular treatments.31 These results suggest that distinctive and not opposite mechanisms may underlie eye growth acceleration from (−) lens wear and eye growth arrest from (+) lens wear.
Onset versus Persistence and/or Progression of Refractive Changes
Pronounced differences with limited overlap occurred in the altered retinal/RPE transcriptome comparing 6 hours to 3 days of image defocus, particularly for (−) lens wear (Table 1, Fig. 1). Similarly, marked differences with limited overlap in the retinal/RPE transcriptome developed in form-deprivation myopia at these same two experimental time points.16 The 6-hour time point should reflect the retinal gene changes occurring at the onset of the visually driven growth change. Transcriptome changes after 3 days of lens wear reflect a time when the altered growth patterns are well established but before lens compensation is completed.11 In another study, none of the altered retinal genes identified by microarray after 24 hours of (+) lens wear were affected at 4 hours when assayed by qPCR.24 These findings suggest a hypothesis that mechanisms initiating a change in eye growth may differ from those underlying the persistence and/or progression of altered eye growth. A few, initial clinical observations support this hypothesis: an increase in axial growth develops before the onset of myopia32; outdoor activity may relate to myopia onset, not progression, while visual nearwork may relate to myopia progression rather than its onset (Mutti DO, et al. IOVS 2010;51:ARVO E-Abstract 2968).33 Children with ametropia are typically older developmentally than the young laboratory animals investigated in most mechanistic studies. Firmly establishing whether the mechanisms responsible for myopia onset differ from those responsible for its persistence and/or progression in children would seem fundamental to understanding myopia's pathogenesis and to developing future therapies.
Hyperopic Defocus versus Form-Deprivation Myopia
The wearing of (−) spectacle lenses or image-diffusing goggles each cause ocular elongation and a myopic refraction, but they are known to differ in a variety of other parameters, such as the time course of the biochemical response in sclera and in the electroretinogram.9 The methods in the present study were selected to closely parallel those in our prior investigation of the altered retinal transcriptome after unilateral goggle wear, and comparing the profiling results substantiates differences between lens-induced and form-deprivation myopia.16 While also revealing a transcriptome difference between the onset and progression of form-deprivation myopia, the 15 altered retina/RPE genes at 6 hours and 280 altered genes at 3 days of goggle wear16 differ dramatically from the more extensive response to (−) spectacle lens wear (Table 1, Fig. 1). The somewhat higher FDR of 13% in the goggle study is less restrictive than the 10% rate in the present study and presumably would bias toward longer, not shorter, gene lists. Potentially contributing to shorter gene lists, though, we did use a ≥1.2 fold-change filter in the bioinformatics analysis of the goggle study.16 Nevertheless, after (−) lens wear, 1015 genes after 6 hours and 608 genes after 3 days were differentially expressed at a ≥1.2 fold-change (Supplementary Tables S2, S3, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.10-6727/-/DCSupplemental). Further, very few differentially regulated genes were common to the two conditions (Supplementary Table S7, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.10-6727/-/DCSupplemental). Whether related to the differences in the time course of the axial or choroidal compensations, to the nature of the altered visual input or to some other yet to be defined parameter, the retina/RPE transcriptome response to (−) lens wear differs considerably from the response to goggle wear despite the growth and refractive similarities between the two conditions.
Networks and Pathways after Minus Lens Wear
For the (−) lens conditions, the networks and canonical pathways generated (IPA, Ingenuity Systems; Supplementary Tables S5, S6, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.10-6727/-/DCSupplemental) each differed between the two experimental time points.
After 6 hours of (−) lens wear, the top network functions generated by pathway analysis related to nervous system development/function, cell signaling, small molecule biochemistry, the cell cycle, and gene expression. After 3 days of (−) lens wear, the top functions included gene expression, cellular growth/proliferation, cell death, cell morphology, cellular assembly/organization, cell-to-cell signaling/interaction, cellular movement, and metabolism of amino acids and carbohydrates (Supplementary Table 5, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.10-6727/-/DCSupplemental). As one hypothesis from these general functions, the differentially expressed genes at 6 hours more prominently include signaling and developmental phenomena, and the differentially expressed genes at 3 days may also incorporate mechanisms by which the retina adjusts its area anatomically to the increased expansion of the vitreous chamber.
Like the networks, the archived canonical pathways to which the pathway analysis assigned differentially expressed genes were largely different between the two times (Supplementary Tables S6A, S6B, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.10-6727/-/DCSupplemental). The most statistically significant association occurred for the glutamate receptor signaling pathway, discussed further below. Many of the assigned canonical pathways related generally to receptor signaling, as might occur in neurons. Understanding how, or whether, these canonical pathways relate to either image defocus or the resultant accelerated eye growth response requires future study.
Selected Differentially Expressed Genes
Amino Acid Signaling Genes.
A marked disparity in differentially regulated genes related to amino acid signaling developed between the two (−) lens wear times (Table 3). Transcriptome alterations related to the excitatory amino acid glutamate occurred chiefly at the 6-hour time point. In fact, glutamate receptor signaling comprised the most significant canonical pathway identified at 6 hours of (−) lens wear (Supplementary Table S6A, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.10-6727/-/DCSupplemental). Supporting a role for glutamate signaling in refraction, local administration of excitatory neurotoxins and drugs interacting with excitatory amino acid receptors influence ocular growth and refractive development in the chick.34–38 A major excitatory neurotransmitter for vertical transmission through the retina, glutamate signaling also impacts the ON and OFF pathways previously implicated in eye growth of the chick.39–41
In contrast to the altered expression of glutamate-related genes mostly at 6 hours, altered expression of genes related to the inhibitory amino acids GABA and glycine occurred only at the 3-day time point (Table 3). Daily administration of drugs for 4 or 4.5 days that interact with GABA receptors alters the growth of chick eyes with either intact or altered visual input.42,43 These dosage schedules extend beyond the 3-day profiling time, when GABA genes were measured, and no data are available for shorter drug dosing times. Whether from adaptation to altered contrast44 or another mechanism, this shift in amino acid signaling genes dramatically underscores the 6-hour versus 3-day differences in retinal/RPE transcriptome alterations.
The changes in amino acid signaling genes may be particularly relevant to the role of retinal dopamine in regulating eye growth.7 Dopaminergic amacrine cells receive excitatory glutamatergic input from ON bipolar cells,45–47 and mice with an ON bipolar cell defect have low levels of dopamine and increased sensitivity to form-deprivation myopia.48 Conversely, dopamine amacrine cells are tonically inhibited by GABA.49–51
Immediate-Early Gene: EGR1.
The retinal expression of the immediate-early gene EGR1 (or ZENK) by RT-PCR or its gene product by immunohistochemistry is diminished in conditions stimulating eye growth (e.g., [−] lens or goggle wear) and increased in conditions inhibiting eye growth (e.g., [+] lens wear or myopia recovery). Accordingly, it has been suggested that the early activity of EGR1 may mediate the direction of eye growth.18,20,21,52 Among chick retinal neurons immunoreactive for this transcription factor are amacrine cells expressing glucagon20 (discussed below). Available data on EGR1 expression within the first few hours of altered unilateral visual input differ on whether the altered retinal gene expression occurs bilaterally21 or ipsilaterally.53 We did not detect differential retinal/RPE expression of EGR1 after 6 hours of minus lens wear (Table 3). Our analysis compared gene expression to the contralateral eye, however; and parallel EGR1 effects in both eyes after 6 hours of lens wear would have masked changes in expression as a result of our experimental design. With longer periods of (−) lens wear, available data suggest that ERG1 expression differences between defocused and contralateral eyes may increase,53 and we did find EGR1 expression to be downregulated in experimental eyes relative to contralateral eyes at 3 days in the (−) lens-wearing cohort (Table 3). Although more work is required to resolve detailed differences between specific studies, much evidence implicates EGR1 in the mechanism signaling the direction of eye growth18,20,21,52,53 including our profiling results after 3 days of lens wear.
Acetylcholine-Related Genes.
Acetylcholine has long been implicated in refractive development, but much is uncertain about the underlying mechanisms.7 Evidence of a role for acetylcholine has been the antimyopia activity of atropine and other muscarinic acetylcholine receptor antagonists in children54,55 and experimental animals,56,57 including chick.58 Whether these drugs inhibit myopia by acting at muscarinic acetylcholine receptors, though, has been questioned.52,59 Besides antimuscarinic drugs, nicotinic acetylcholine receptors may also have a role in refractive development; but only a few potentially pertinent laboratory and clinical epidemiologic studies are available.60–62 Organophosphate insecticides inhibit acetylcholinesterase (ACHE) and could elevate synaptic acetylcholine levels; environmental exposure has been associated with increasing myopia prevalence.63 In contrast, ACHE inhibitors suppress form-deprivation myopia in chick.64,65
Despite the evidence implicating the retina in refractive control,6–9 it is uncertain whether acetylcholine influences refractive development by action at the retina or through action elsewhere.7,18,52,64,66,67 Our profiling results suggest potential involvement of cholinergic signaling in the retina (Table 3): the chick cholinergic receptor CHRM2 transcript was downregulated at 6 hours, and two transcripts representing the gene for ACHE were upregulated at 3 days. Despite extensive interest in acetylcholine signaling, understanding the questions and contradictory results about the role of acetylcholine in refractive development requires future research.
Biologically Active Peptides, Proteins, and Other Genes.
We identified altered expression of genes for biologically active peptides, proteins, and other signaling molecules, some but not all of which have been identified in refractive research (Table 3). Two—vasoactive intestinal peptide (VIP) and glucagon—have generated interest as potential mediators of refractive development.7 The gene for VIP, downregulated only after 6 hours of (−) lens wear, is also downregulated in retina/RPE after 3 days of goggle wear.16 The retinal level of VIP is elevated in monkeys but not in chicks with form-deprivation myopia.7,68 Form-deprivation myopia in chicks, however, is fully inhibited by VIP antagonists but partially blocked by VIP.68 While consistent with a role for retinal VIP in eye growth control, research is needed to clarify its role in refractive development.
For chick retinal glucagon mRNA, (+) or (−) lens wear of up to 24 hours is known to affect its expression compared with that in untreated animals, but no differences between lens-wearing and contralateral control eyes have been seen for either lens sign at 6 hours,69 the common time with the current investigation. Based on growth responses to exogenous glucagon and several glucagon analogs activating or inhibiting its receptor in form-deprivation myopia or lens wear, retinal glucagon and/or a related peptide may provide an endogenous signal for inhibiting ocular elongation in chick.70–73 The reduced glucagon gene expression found here after 3 days of (−) lens wear (Table 3) conforms to that notion, assuming that the reduced mRNA results in diminished retinal glucagon release and lessening of a growth inhibitory signal.
The gene for bone morphogenetic protein 2 in retina/RPE has been found to be downregulated after both 6 hours and 3 days in form-deprivation myopia,16 the same alterations identified here after (−) lens wear (Table 3). In addition, genes for endogenous regulators of bone morphogenetic protein 2, noggin, and follistatin, were altered in (−) lens wear, suggesting a complex pathway involving bone morphogenetic protein 2 in either eye growth regulation and/or in the retinal response to blur and hyperopic defocus. Among other differentially expressed genes, transcripts for brain-derived neurotrophic factor were downregulated at each time. These findings are the first to implicate brain-derived neurotrophic factor in some aspect of refractive development, despite prior negative results.74,75
A number of other potentially informative molecular signaling candidates were identified in the present study. For example, the expression of retinal/RPE genes to the receptors for thyroid hormone, cannabinoids, neuropeptide Y, and opioids were affected by (−) lens wear (Table 3). Prior work has implicated nitric oxide synthase, the synthetic enzyme for nitric oxide, in refractive development7; the 3-day results also found differential expression of the gene for heme oxygenase 2, the biosynthetic enzyme for carbon monoxide (Table 3). In general, future work is needed to understand potential roles of these signaling molecules in refractive development.
Intrinsic Retinal Clock and Circadian Genes
Perhaps the most intriguing differentially expressed genes identified here are those involved in the intrinsic retinal clock and retinal circadian rhythms (Table 3). The retina has many intrinsic circadian rhythms, such as disc shedding and melatonin levels. Of potential pertinence to the role of the retina's rhythms in modulating eye growth, the axial growth of a chick eye follows a diurnal pattern, with elongation occurring chiefly during the daytime; this growth rhythm is disrupted by wearing a unilateral goggle or a (−) spectacle lens.76–79 The axial dimensions of rabbit,80 marmoset,81 and human82,83 eyes with nonrestricted vision also fluctuate during the day. Although findings in chick suggest that growth rhythms may relate to refractive development, no data on growth rhythms are yet available for laboratory mammals or children developing ametropias.
Like other tissues, the retina needs a circadian clock to match its daily rhythms to the 24-hour day. Biological clocks comprise interconnected transcriptional–translational feedback loops using the clock genes and their protein products.84 The retinal circadian clock exerts broad effects within the retina; an intact retinal circadian clock even seems essential for proper processing of visual input.85 Hyperopic defocus, through (−) lens wear, alters the expression of a significant proportion of intrinsic clock genes found in chick retina86 and one of the receptors to melatonin, a major retinal output of the circadian clock (Table 3). These changes occur mostly, but not exclusively, at the early time point.
Retinal dopamine may be important for the effects of defocus on circadian gene expression. Dopaminergic neurons express clock genes at comparatively high levels.87 Dopamine affects the circadian phase of Per2::Luc reporter gene expression,88 the expression patterns of clock and clock-controlled genes,89,90 and the circadian release of melatonin91 in retina.
Hyperopic defocus also affects the expression level of the gene for melanopsin, a light-sensitive pigment expressed in nonphotoreceptors of the vertebrate retina. In mammals, it is present in a subpopulation of ganglion cells that project to brain centers controlling circadian rhythms and pupil size.92,93 Circadian regulation of melanopsin expression depends in part on dopamine.94 Significantly, melanopsin-containing ganglion cells may also provide centrifugal input to dopaminergic amacrine cells and presumably influence their diurnal activity,95 although this is controversial.96 In the chick retina, melanopsin exists in two forms97 and is expressed by horizontal and bipolar cells in addition to ganglion cells.98 The diurnal physiologic activity of dopaminergic amacrine cells is believed to modulate refractive development in both experimental birds and mammals,7 and the effect of hyperopic defocus to alter the expression of clock genes and the genes for a melatonin receptor and melanopsin may provide mechanistic links between image clarity, dopaminergic amacrine cell activity, and refractive development.
Although the circadian time for harvesting retinal tissues differed by a few hours for the 6-hour and 3-day samples, our controls were contralateral eyes; and both eyes of individual chicks would be under the same circadian phase. Thus, unilateral alterations with (−) lens wear indicate an effect of defocus on circadian/clock genes, the specificity of which is further supported by absence of an effect on these genes by (+) lens wear or goggle wear with a similar protocol.16 Whether (+) lens or goggle wear alter circadian/clock genes at different times or under different conditions requires future study.
Ambient Lighting and Refractive Development
One means of assessing circadian influences on eye development could be to examine the refractive effects of external parameters already known to modify circadian rhythms in other systems. The altered light exposure patterns that influence refractive development experimentally or associate with refractive errors clinically often conform to the types of light exposure patterns used in entrainment models to study circadian rhythms.99
As extensive study has shown in the chick, disrupting the daily light–dark cycle perturbs the ordered growth of the eye, especially with rearing under constant light or constant dark,22,100–102 but also with variable length or interrupted photoperiods.11,102–104 Constant-light rearing of chicks also alters the responses to goggle or spectacle lens wear.22,105–107 Rearing rhesus monkeys under constant light, with or without a spectacle lens, also influences refraction, but with considerably less robust effects than in chick.108,109 Varying light intensity also modulates eye development in chick. Bright illumination comparable to outdoor sunlight alters the response to constant-light rearing,102,110 impairs the development of form-deprivation myopia111 and changes the time course of the compensation to spectacle lens wear.112 Long-term rearing of chicks in low-intensity light enlarges the vitreous chamber and induces myopia.113
Clinically, some feature of lighting has long been hypothesized to influence human refractive development.3 Lighting intensity, other photoperiod characteristics, and the age at which lighting may act have generated most recent clinical interest.
An antimyopia effect of outdoor activity is repeatedly noted,33,114,115 and may relate to time spent outdoors rather than outdoor physical activity per se.114,115 Higher intensity of outdoor than indoor lighting is one hypothesized mechanism for this effect.115 A role for specific indoor activities (e.g., nearwork) remains uncertain.33,115
A role for other photoperiod properties is more controversial. For instance, some116–118 but not all119–121 cross-sectional surveys associate later ametropia with disrupting the dark period in early childhood by ambient lighting at night. The only study to include ultrasound, while not reporting an overall effect on refraction, did find more high myopia and longer axial lengths with nighttime lighting exposure.122 Several reports have associated refraction with birthdate,123–125 suggesting among other hypotheses that perinatal daytime length is a myopia risk factor.123,124
Although it may seem contradictory that both more light (e.g., from a shorter or disrupted dark period) and indoor activities (e.g., resulting in less light during the day) each associate with myopia, circadian biology may link these diverse laboratory and clinical observations. Altering the light–dark photoperiod is well known to influence circadian rhythms.126 Dim lighting affects circadian rhythms in complex ways127; indoor lighting may be of inadequate intensity for proper clock entrainment.128 Based on our finding that hyperopic defocus alters intrinsic retinal clock and circadian gene expression while inducing myopia, addressing a role for circadian rhythms in refractive development may provide a unifying framework for understanding these disparate findings about rhythms, lighting, and refraction.
Comparison to Other Gene Expression Studies
Comparing gene expression between studies of refractive development is complicated by differences in species, experimental models, study duration, methods for detecting gene alterations, and the tissue assayed.15 On tissue in particular, broad sampling of a complex tissue such as the retina/RPE risks masking small effects in a limited number of discrete cells. Altered gene levels in a subtype or limited subtypes of retinal neurons may be obscured by stable or reciprocal changes or by high variability in other neurons. In this regard, higher levels of altered gene expression developed after 24 hours of the wearing of spectacle lens wear of +7 or −7 D in the amacrine cell layer isolated by laser capture microdissection.31 Using gene screening criteria of P ≤ 0.05 and ≥1.5 fold-change, (+) lens treatment altered the expression of 58 genes (30 upregulated, 28 downregulated, with a range of +7.7 to −5.7 fold-change); and (−) lens treatment altered the expression of 128 genes (63 upregulated, 65 downregulated, with a range of +8.0 to −27.4 fold-change).31 Restricting sampling to the amacrine cell layer, as well as differences in lens power, lens wear time or analytical approach, could have accounted for the differences from the present study.
Consistent with these qualifications, most overlap in genes between the present study and prior publications tends to be limited in number, with no universally identified genes (Supplementary Table S7, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.10-6727/-/DCSupplemental). From the perspective of intercellular signaling, some of the recurring genes are those encoding bone morphogenetic protein 2, glucagon, inducible nitric oxide synthase and vasoactive intestinal peptide. For understanding the mechanisms of refractive development, microarray platforms or other approaches to gene expression are designed to identify changes in RNA abundance, not protein products or posttranscriptional protein modifications. While posttranslational regulatory mechanisms most likely affect refractive development, RNA assays as performed in this study can provide important leads and generate potentially useful hypotheses for future study.
Overall Conclusion
In the present study, we identified novel genes and sorted differentially regulated genes into pathways and signaling networks, only some of which are explicitly discussed above. Perhaps because of the experimental design and informatics approaches, the present study and our prior report on form-deprivation myopia16 conform to some mechanistic hypotheses that could lead to a general, broad biological framework to understand the mechanism(s) that regulate emmetropization and cause refractive errors. These include the ideas that distinctive, not opposite, processes govern the upregulation and downregulation of eye growth and that the mechanisms precipitating changes in eye growth may differ from those responsible for persistence and/or progression of ametropias. The altered retinal/RPE clock and circadian genes after hyperopic defocus substantiates a potential role for endogenous retinal rhythms in refractive development. As a testable hypothesis, disrupted endogenous ocular clock or circadian rhythms may provide a framework for understanding the extensive clinical and laboratory literature on the potential role of lighting on refractive development. Establishing any of these concepts, though, will require future laboratory and clinical investigation. Significantly, many of these hypotheses can be addressed now in children while parallel investigations progress independently in the laboratory.
Supplementary Material
Footnotes
Supported by National Institutes of Health Grants R01-EY018838 (RAS), R01-EY013862 (TSK), R01-EY004864 (PMI), and P30 EY06360 (PMI); the Paul and Evanina Bell Mackall Foundation Trust (RAS); Research to Prevent Blindness (RAS, PMI). PMI is a recipient of the Research to Prevent Blindness Senior Scientific Investigator Award.
Disclosure: R.A. Stone, None; A.M. McGlinn, None; D.A. Baldwin, None; J.W. Tobias, None; P.M. Iuvone, None; T.S. Khurana, None
References
- 1. Hornbeak DM, Young TL. Myopia genetics: a review of current research and emerging trends. Curr Opin Ophthalmol. 2009;20:356–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wojciechowski R, Cogdon N, Bowie H, Munoz G, Gilbert D, West S. Familial aggregation of hyperopia in an elderly population of siblings in Salisbury, Maryland. Ophthalmology. 2005;112:78–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Curtin BJ. The Myopias. Philadelphia: Harper & Row, Publishers; 1985 [Google Scholar]
- 4. Goldschmidt E. The mystery of myopia. Acta Ophthalmol Scand. 2003;81:431–436 [DOI] [PubMed] [Google Scholar]
- 5. Morgan I, Rose K. How genetic is school myopia? Prog Retin Eye Res. 2005;24:1–38 [DOI] [PubMed] [Google Scholar]
- 6. Norton TT. Animal models of myopia: learning how vision controls the size of the eye. ILAR J. 1999;40:59–77 [DOI] [PubMed] [Google Scholar]
- 7. Stone RA. Myopia pharmacology: etiologic clues, therapeutic potential. In: Yorio T, Clark A, Wax M. eds. Ocular Therapeutics: an Eye on New Discoveries. New York: Elsevier/Academic Press; 2008:167–196 [Google Scholar]
- 8. Wallman J. Retinal control of eye growth and refraction. Prog Retin Eye Res. 1993;12:133–153 [Google Scholar]
- 9. Wallman J, Winawer J. Homeostasis of eye growth and the question of myopia. Neuron. 2004;43:447–468 [DOI] [PubMed] [Google Scholar]
- 10. Wildsoet C, Wallman J. Choroidal and scleral mechanisms of compensation for spectacle lenses in chicks. Vision Res. 1995;35:1175–1194 [DOI] [PubMed] [Google Scholar]
- 11. Kee C-s, Marzani D, Wallman J. Differences in time course and visual requirements of ocular responses to lenses and diffusers. Invest Ophthalmol Vis Sci. 2001;42:575–583 [PubMed] [Google Scholar]
- 12. Nathan JN, Kiely PM, Crewther SG, Crewther DP. Disease-associated visual image degradation and spherical refractive errors in children. Am J Optom Physiol Optics. 1985;62:680–688 [DOI] [PubMed] [Google Scholar]
- 13. Read SA, Collins MJ, Sander BP. Human optical axial length and defocus. Invest Ophthalmol Vis Sci. 2010;51:6262–6269 [DOI] [PubMed] [Google Scholar]
- 14. Norton TT, Siegwart JT. Animal models of emmetropization: matching axial length to focal plane. J Am Optom Assoc. 1995;66:405–414 [PubMed] [Google Scholar]
- 15. Stone RA, Khurana TS. Gene profiling in experimental models of eye growth: clues to myopia pathogenesis. Vision Res. 2010;50:2322–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McGlinn AM, Baldwin DA, Tobias JW, Budak MT, Khurana TS, Stone RA. Form deprivation myopia in chick induces limited changes in retinal gene expression. Invest Ophthalmol Vis Sci. 2007;48:3430–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Megaw PL, Morgan IG, Boelen MK. Dopaminergic behaviour in chicken retina and the effect of form deprivation. Aust N Z J Ophthalmol. 1997;25(suppl 1):S76–S78 [DOI] [PubMed] [Google Scholar]
- 18. Ashby R, McCarthy CS, Maleszka R, Megaw P, Morgan IG. A muscarinic cholinergic antagonist and a dopamine agonist rapidly increase ZENK mRNA expression in the form-deprived chicken retina. Exp Eye Res. 2007;85:15–22 [DOI] [PubMed] [Google Scholar]
- 19. Bitzer M, Schaeffel F. Defocus-induced changes in ZENK expression in the chicken retina. Invest Ophthalmol Vis Sci. 2002;43:246–252 [PubMed] [Google Scholar]
- 20. Fischer AJ, McGuire JJ, Schaeffel F, Stell WK. Light- and focus-dependent expression of the transcription factor ZENK in the chick retina. Nat Neurosci. 1999;2:706–712 [DOI] [PubMed] [Google Scholar]
- 21. Simon P, Feldkaemper M, Bitzer M, Ohngemach S, Schaeffel F. Early transcriptional changes in retinal and choroidal TGFβ-2, RALDH-2 and ZENK following imposed positive and negative defocus in chickens. Mol Vis. 2004;10:588–597 [PubMed] [Google Scholar]
- 22. Stone RA, Lin T, Desai D, Capehart C. Photoperiod, early post-natal eye growth, and visual deprivation. Vision Res. 1995;35:1195–1202 [DOI] [PubMed] [Google Scholar]
- 23. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408 [DOI] [PubMed] [Google Scholar]
- 24. Schippert R, Schaeffel F, Feldkaemper MP. Microarray analysis of retinal gene expression in chicks during imposed myopic defocus. Mol Vis. 2008;14:1589–1599 [PMC free article] [PubMed] [Google Scholar]
- 25. Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis: from disarray to consolidation of results. Nat Rev Genet. 2006;7:55–65 [DOI] [PubMed] [Google Scholar]
- 26. Dallas PB, Gottardo NG, Firth MJ, et al. Gene expression levels assessed by oligonucleotide microarray analysis and quantitative real-time RT-PCR: how well do they correlate? BMC Genomics. 2005;6:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kuo WP, Liu F, Trimarchi J, et al. A sequence-oriented comparison of gene expression measurements across different hybridization-based technologies. Nat Biotechnol. 2006;24:832–840 [DOI] [PubMed] [Google Scholar]
- 28. Morey JS, Ryan JC, Van Dolah FM. Microarray validation: factors influencing correlation between oligonucleotide microarrays and real-time PCR. Biol Proced Online. 2006;8:175–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. White CA, Salamonsen LA. A guide to issues in microarray analysis: application to endometrial biology. Reproduction. 2005;130:1–13 [DOI] [PubMed] [Google Scholar]
- 30. Summers Rada JA, Wiechmann AF. Ocular expression of avian thymic hormone: changes during the recovery from induced myopia. Mol Vis. 2009;15:778–792 [PMC free article] [PubMed] [Google Scholar]
- 31. Ashby RS, Feldkaemper MP. Gene expression within the amacrine cell layer of chicks after myopic and hyperopic defocus. Invest Ophthalmol Vis Sci. 2010;51:3726–3735 [DOI] [PubMed] [Google Scholar]
- 32. Thorn F, Gwiazda J, Held R. Myopia progression is specified by a double exponential growth function. Optom Vis Sci. 2005;82:286–297 [DOI] [PubMed] [Google Scholar]
- 33. Jones LA, Sinnott LT, Mutti DO, Mitchell GL, Moeschberger ML, Zadnik K. Parental history of myopia, sports and outdoor activities, and future myopia. Invest Ophthalmol Vis Sci. 2007;48:3524–3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Barrington M, Sattayasai J, Zappia J, Ehrilich D. Excitatory amino acids interfere with normal eye growth in posthatch chick. Curr Eye Res. 1989;8:781–792 [DOI] [PubMed] [Google Scholar]
- 35. Ehrlich D, Sattayasai J, Zappia J, Barrington M. Effects of selective neurotoxins on eye growth in the young chick. In: Bock GR, Widdows K. eds. Ciba Foundation Symposium 155: Myopia and the control of eye growth. Chichester: John Wiley & Sons; 1990:63–88 [DOI] [PubMed] [Google Scholar]
- 36. Fischer AJ, Pickett Seltner RL, Poon J, Stell WK. Immunocytochemical characterization of quisqualic acid- and N-methyl-D-aspartate-induced excitotoxicity in the retina of chicks. J Comp Neurol. 1998;393:1–15 [PubMed] [Google Scholar]
- 37. Fischer AJ, Seltner RL, Stell WK. Opiate and N-methyl-D-aspartate receptors in form-deprivation myopia. Vis Neurosci. 1998;15:1089–1096 [DOI] [PubMed] [Google Scholar]
- 38. Wildsoet CF, Pettigrew JD. Kainic acid-induced eye enlargement in chickens: differential effects on anterior and posterior segments. Invest Ophthalmol Vis Sci. 1988;29:311–319 [PubMed] [Google Scholar]
- 39. Crewther DP. The role of photoreceptors in the control of refractive state. Prog Retin Eye Res. 2000;19:421–457 [DOI] [PubMed] [Google Scholar]
- 40. Crewther DP, Crewther SG, Xie RZ. Changes in eye growth produced by drugs which affect retinal ON or OFF responses to light. J Ocul Pharmacol Ther. 1996;12:193–208 [DOI] [PubMed] [Google Scholar]
- 41. Crewther SG, Crewther DP. Inhibition of retinal ON/OFF systems differentially affects refractive compensation to defocus. Neuroreport. 2003;14:1233–1237 [DOI] [PubMed] [Google Scholar]
- 42. Chebib M, Hinton T, Schmid KL, et al. Novel, potent, and selective GABAC antagonists inhibit myopia development and facilitate learning and memory. J Pharmacol Exp Ther. 2009;328:448–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stone RA, Liu J, Sugimoto R, Capehart C, Zhu X, Pendrak K. GABA, experimental myopia and ocular growth in chick. Invest Ophthalmol Vis Sci. 2003;44:3933–3946 [DOI] [PubMed] [Google Scholar]
- 44. Demb JB. Functional circuitry of visual adaptation in the retina. J Physiol. 2008;586:4377–4384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Boatright JH, Gordon JR, Iuvone PM. Inhibition of endogenous dopamine release in amphibian retina by L-2-amino-4-phosphonobutyric acid (L-AP4) and trans- 2-aminocyclopentane-1,3-dicarboxylate (ACPD). Brain Res. 1994;649:339–342 [DOI] [PubMed] [Google Scholar]
- 46. Boelen MK, Boelen MG, Marshak DW. Light-stimulated release of dopamine from the primate retina is blocked by l-2-amino-4-phosphonobutyric acid (APB). Vis Neurosci. 1998;15:97–103 [DOI] [PubMed] [Google Scholar]
- 47. Dumitrescu ON, Pucci FG, Wong KY, Berson DM. Ectopic retinal ON bipolar cell synapses in the OFF inner plexiform layer: Contacts with dopaminergic amacrine cells and melanopsin ganglion cells. J Comp Neurol. 2009;517:226–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pardue MT, Faulkner AE, Fernandes A, et al. High susceptibility to experimental myopia in a mouse model with a retinal on pathway defect. Invest Ophthalmol Vis Sci. 2008;49:706–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Boatright JH, Rubim NM, Iuvone PM. Regulation of endogenous dopamine release in amphibian retina by gamma-aminobutyric acid and glycine. Vis Neurosci. 1994;11:1003–1012 [DOI] [PubMed] [Google Scholar]
- 50. Feigenspan A, Gustincich S, Raviola E. Pharmacology of GABA(A) receptors of retinal dopaminergic neurons. 84, J Neurophysiol. 2000;84:1697–1707 [DOI] [PubMed] [Google Scholar]
- 51. Marshburn PB, Iuvone PM. The role of GABA in the regulation of the dopamine/tyrosine hydroxylase-containing neurons of the rat retina. Brain Res. 1981;214:335–347 [DOI] [PubMed] [Google Scholar]
- 52. Bitzer M, Kovacs B, Feldkaemper M, Schaeffel F. Effects of muscarinic antagonists on ZENK expression in the chicken retina. Exp Eye Res. 2006;82:379–388 [DOI] [PubMed] [Google Scholar]
- 53. Ashby R, Kozulin P, Megaw PL, Morgan IG. Alterations in ZENK and glucagon RNA transcript expression during increased ocular growth in chickens. Mol Vis. 2010;16:639–649 [PMC free article] [PubMed] [Google Scholar]
- 54. Kennedy RH. Progression of myopia. Trans Am Ophthalmol Soc. 1995;93:755–800 [PMC free article] [PubMed] [Google Scholar]
- 55. Siatkowski RM, Cotter SA, Crockett RS, et al. Two-year multicenter, randomized double-masked, placebo-controlled, parallel safety and efficacy study of 2% pirenzepine ophthalmic gel in children with myopia. J AAPOS. 2008;12:332–339 [DOI] [PubMed] [Google Scholar]
- 56. Raviola E, Wiesel TN. An animal model of myopia. N Engl J Med. 1985;312:1609–1615 [DOI] [PubMed] [Google Scholar]
- 57. Tigges M, Iuvone PM, Fernandes A, et al. Effects of muscarinic cholinergic receptor antagonists on postnatal eye growth of rhesus monkeys. Optom Vis Sci. 1999;76:397–407 [DOI] [PubMed] [Google Scholar]
- 58. Stone RA, Lin T, Laties AM. Muscarinic antagonist effects on experimental chick myopia. Exp Eye Res. 1991;52:755–758 [DOI] [PubMed] [Google Scholar]
- 59. Luft WA, Ming Y, Stell WK. Variable effects of previously untested muscarinic receptor antagonists on experimental myopia. Invest Ophthalmol Vis Sci. 2003;44:1330–1338 [DOI] [PubMed] [Google Scholar]
- 60. Rahi JS, Cumberland PM, Peckham CS. Myopia over the lifecourse: prevalence and early life influences in the 1958 British birth cohort. Ophthalmology. 2011;118:797–804 [DOI] [PubMed] [Google Scholar]
- 61. Stone RA, Sugimoto R, Gill AS, Liu J, Capehart C, Lindstrom JM. Effects of nicotinic antagonists on ocular growth and experimental myopia. Invest Ophthalmol Vis Sci. 2001;42:557–565 [PubMed] [Google Scholar]
- 62. Stone RA, Wilson LB, Ying G-s, et al. Associations between childhood refraction and parental smoking. Invest Ophthalmol Vis Sci. 2006;47:4277–4287 [DOI] [PubMed] [Google Scholar]
- 63. Ishikawa S, Miyata M. Development of myopia following chronic organophosphate pesticide intoxication: an epidemiological and experimental study. In: Merigan WH, Weiss B. eds. Neurotoxicity of the Visual System. New York: Raven Press; 1980:233–254 [Google Scholar]
- 64. Cottriall CL, Brew J, Vessey KA, McBrien NA. Diisopropylfluorophosphate alters retinal neurotransmitter levels and reduces experimentally-induced myopia. Naunyn-Schmiedebergs Arch Pharmacol. 2001;364:372–382 [DOI] [PubMed] [Google Scholar]
- 65. Geller AM, Abdel-Rahman AA, Peiffer RL, Abou-Donia MB, Boyes WK. The organophosphate pesticide chlorpyrifos affects form deprivation myopia. Invest Ophthalmol Vis Sci. 1998;39:1290–1294 [PubMed] [Google Scholar]
- 66. Fischer AJ, Miethke P, Morgan IG, Stell WK. Cholinergic amacrine cells are not required for the progression and atropine-mediated suppression of form-deprivation myopia. Brain Res. 1998;794:48–60 [DOI] [PubMed] [Google Scholar]
- 67. Pendrak K, Lin T, Stone RA. Ciliary ganglion choline acetyltransferase activity in avian macrophthalmos. Exp Eye Res. 1995;60:237–243 [DOI] [PubMed] [Google Scholar]
- 68. Pickett Seltner RL, Stell WK. The effect of vasoactive intestinal peptide on development of form deprivation myopia in the chick: a pharmacological and immunocytochemical study. Vision Res. 1995;35:1265–1270 [DOI] [PubMed] [Google Scholar]
- 69. Buck C, Schaeffel F, Simon P, Feldkaemper M. Effects of positive and negative lens treatment on retinal and choroidal glucagon and glucagon receptor mRNA levels in the chicken. Invest Ophthalmol Vis Sci. 2004;45:402–409 [DOI] [PubMed] [Google Scholar]
- 70. Feldkaemper M, Schaeffel F. Evidence for a potential role of glucagon during eye growth regulation in chicks. Vis Neurosci. 2002;19:755–766 [DOI] [PubMed] [Google Scholar]
- 71. Vessey KA, Lencses KA, Rushforth DA, Hruby VJ, Stell WK. Glucagon receptor agonists and antagonists affect the growth of the chick eye: a role for glucagonergic regulation of emmetropization? Invest Ophthalmol Vis Sci. 2005;46:3922–3931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Vessey KA, Rushforth DA, Stell WK. Glucagon- and secretin-related peptides differentially alter ocular growth and the development of form-deprivation myopia in chicks. Invest Ophthalmol Vis Sci. 2005;46:3932–3942 [DOI] [PubMed] [Google Scholar]
- 73. Zhu X, Wallman J. Opposite effects of glucagon and insulin on compensation for spectacle lenses in chicks. Invest Ophthalmol Vis Sci. 2009;50:24–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Akamatsu S, Fujii S, Escaño MFT, Ishibashi K, Sekiya Y, Yamamoto M. Altered expression of genes in experimentally induced myopic chick eyes. Jpn J Ophthalmol. 2001;45:137–143 [DOI] [PubMed] [Google Scholar]
- 75. Mutti DO, Cooper ME, O'Brien S, et al. Candidate gene and locus analysis of myopia. Mol Vis. 2007;13:1012–1019 [PMC free article] [PubMed] [Google Scholar]
- 76. Nickla DL. The phase relationships between the diurnal rhythms in axial length and choroidal thickness and the association with ocular growth rate in chicks. J Comp Physiol A. 2006;192:399–407 [DOI] [PubMed] [Google Scholar]
- 77. Nickla DL, Wildsoet C, Wallman J. Visual influences on diurnal rhythms in ocular length and choroidal thickness in chick eyes. Exp Eye Res. 1998;66:163–181 [DOI] [PubMed] [Google Scholar]
- 78. Papastergiou GI, Schmid GF, Riva CE, Mendel MJ, Stone RA, Laties AM. Ocular axial length and choroidal thickness in newly hatched chicks and one-year-old chickens fluctuate in a diurnal pattern that is influenced by visual experience and intraocular pressure changes. Exp Eye Res. 1998;66:195–205 [DOI] [PubMed] [Google Scholar]
- 79. Weiss S, Schaeffel F. Diurnal growth rhythms in the chicken eye: relation to myopia development and retinal dopamine levels. J Comp Physiol A. 1993;172:263–270 [DOI] [PubMed] [Google Scholar]
- 80. Liu JHK, Farid H. Twenty-four-hour change in axial length in the rabbit eye. Invest Ophthalmol Vis Sci. 1998;39:2796–2799 [PubMed] [Google Scholar]
- 81. Nickla DL, Wildsoet C, Troilo D. Diurnal rhythms in intraocular pressure, axial length, and choroidal thickness in a primate model of eye growth, the common marmoset. Invest Ophthalmol Vis Sci. 2002;43:2519–2528 [PubMed] [Google Scholar]
- 82. Read SA, Collins MJ, Iskander DR. Diurnal variation of axial length, intraocular pressure, and anterior eye biometrics. Invest Ophthalmol Vis Sci. 2008;49:2911–2918 [DOI] [PubMed] [Google Scholar]
- 83. Stone RA, Quinn GE, Francis EL, et al. Diurnal axial length fluctuations in human eyes. Invest Ophthalmol Vis Sci. 2004;45:63–70 [DOI] [PubMed] [Google Scholar]
- 84. Tosini G, Pozdeyev N, Sakamoto K, Iuvone PM. The circadian clock system in the mammalian retina. BioEssays. 2008;30:624–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Storch K-F, Paz C, Signorovitch J, et al. Intrinsic circadian clock of the mammalian retina: importance for retinal processing of visual information. Cell. 2007;130:730–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Haque R, Ali RG, Biscoglia R, et al. CLOCK and NPAS2 have overlapping roles in the circadian oscillation of arylalkylamine N-acetyltransferase mRNA in chicken cone photoreceptors. J Neurochem. 2010;113:1296–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ruan G-X, Zhang D-Q, Shou T, Yamazaki S, McMahon DG. Circadian organization of the mammalian retina. Proc Natl Acad Sci USA. 2006;103:9703–9708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ruan GX, Allen GC, Yamazaki S, McMahon DG. An autonomous circadian clock in the inner mouse retina regulated by dopamine and GABA. PLoS Biology. 2008;6:e249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Jackson CR, Chaurasia SS, Hwang CR, Iuvone PM. Dopamine D4 receptor activation controls circadian timing of the adenylyl cyclase 1/cyclic AMP signaling system in mouse retina. Eur J Neurosci. In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Yujnovsky I, Hirayama J, Doi M, Borrelli E, Sassone-Corsi P. Signaling mediated by the dopamine D2 receptor potentiates circadian regulation by CLOCK:BMAL1. Proc Natl Acad Sci USA. 2006;103:6386–6391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Cahill GM, Besharse JC. Resetting the circadian clock in cultured Xenopus eyecups: Regulation of retinal melatonin rhythms by light and D2 dopamine receptors. J Neurosci. 1991;11:2959–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Bailes HJ, Lucas RJ. Melanopsin and inner retinal photoreception. Cell Mol Life Sci. 2010;67:99–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Paul KN, Saafir TB, Tosini G. The role of retinal photoreceptors in the regulation of circadian rhythms. Rev Endocr Metab Disord. 2009;10:271–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Sakamoto K, Liu C, Kasamatsu M, Pozdeyev NV, Iuvone PM, Tosini G. Dopamine regulates melanopsin mRNA expression in intrinsically photosensitive retinal ganglion cells. Eur J Neurosci. 2005;22:3129–3136 [DOI] [PubMed] [Google Scholar]
- 95. Zhang D-Q, Wong KY, Sollars PJ, Berson DM, Pickard GE, McMahon DG. Intraretinal signaling by ganglion cell photoreceptors to dopaminergic amacrine neurons. Proc Natl Acad Sci USA. 2008;105:14181–14186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Cameron MA, Pozdeyev N, Vugler AA, Cooper H, Iuvone PM, Lucas RJ. Light regulation of retinal dopamine that is independent of melanopsin phototransduction. Eur J Neurosci. 2009;29:761–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Bellingham J, Chaurasia SS, Melyan Z, et al. Evolution of melanopsin photoreceptors: discovery and characterization of a new melanopsin in nonmammalian vertebrates. PLoS Biol. 2006;4:1334–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Tomonari S, Takagi A, Akamatsu S, Noji S, Ohuchi H. A non-canonical photopigment, melanopsin, is expressed in the differentiating ganglion, horizontal, and bipolar cells of the chicken retina. Dev Dyn. 2005;234:783–790 [DOI] [PubMed] [Google Scholar]
- 99. Johnson CH, Elliott JA, Foster R. Entrainment of circadian programs. Chronobiol Int. 2003;20:741–774 [DOI] [PubMed] [Google Scholar]
- 100. Jensen LS, Matson WE. Enlargement of avian eye by subjecting chicks to continuous incandescent illumination. Science. 1957;125:741. [DOI] [PubMed] [Google Scholar]
- 101. Li T, Troilo D, Glasser A, Howland HC. Constant light produces severe corneal flattening and hyperopia in chickens. Vision Res. 1995;35:1203–1209 [DOI] [PubMed] [Google Scholar]
- 102. Oishi T, Murakami Y. Effects of duration and intensity of illumination on several parameters of the chick eye. Comp Biochem Physiol. 1985;81A:319–323 [DOI] [PubMed] [Google Scholar]
- 103. Li T, Howland HC, Troilo D. Diurnal illumination patterns affect the development of the chick eye. Vision Res. 2000;40:2387–2393 [DOI] [PubMed] [Google Scholar]
- 104. Osol G, Schwartz B, Foss DC. The effects of photoperiod and lid suture on eye growth in chickens. Invest Ophthalmol Vis Sci. 1986;27:255–260 [PubMed] [Google Scholar]
- 105. Bartmann M, Schaeffel F, Hagel G, Zrenner E. Constant light affects retinal dopamine levels and blocks deprivation myopia but not lens-induced refractive errors in chickens. Vis Neurosci. 1994;11:199–208 [DOI] [PubMed] [Google Scholar]
- 106. Guo SS, Sivak JG, Callender MG, Herbert KL. Effects of continuous light on experimental refractive errors in chicks. Ophthalmic Physiol Opt. 1996;16:486–490 [PubMed] [Google Scholar]
- 107. Padmanabhan V, Shih J, Wildsoet CF. Constant light rearing disrupts compensation to imposed- but not induced-hyperopia and facilitates compensation to imposed myopia in chicks. Vision Res. 2007;47:1855–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Smith EL, Bradley DV, Fernandes A, Hung L-F, Boothe RG. Continuous ambient lighting and eye growth in primates. Invest Ophthalmol Vis Sci. 2001;42:1146–1152 [PubMed] [Google Scholar]
- 109. Smith EL, III, Hung L-F, Kee C-S, Qiao-Grider Y, Ramamirtham R. Continuous ambient lighting and lens compensation in infant monkeys. Optom Vis Sci. 2003;80:374–382 [DOI] [PubMed] [Google Scholar]
- 110. Cohen Y, Belkin M, Yehezkel O, Avni I, Polat U. Light intensity modulated corneal power and refraction in the chick eye exposed to continuous light. Vision Res. 2008;48:2329–2335 [DOI] [PubMed] [Google Scholar]
- 111. Ashby R, Ohlendorf A, Schaeffel F. The effect of ambient illuminance on the development of deprivation myopia in chicks. Invest Ophthalmol Vis Sci. 2009;50:5348–5354 [DOI] [PubMed] [Google Scholar]
- 112. Ashby RS, Schaeffel F. The effect of bright light on lens compensation in chicks. Invest Ophthalmol Vis Sci. 2010;51:5247–5253 [DOI] [PubMed] [Google Scholar]
- 113. Cohen Y, Belkin M, Yehezkel O, Solomon AS, Polat U. Dependency between light intensity and refractive development under light-dark cycles. Exp Eye Res. 2011;92:40–46 [DOI] [PubMed] [Google Scholar]
- 114. Dirani M, Tong L, Gazzard G, et al. Outdoor activity and myopia in Singapore teenage children. B J Ophthalmol. 2009;93:997–1000 [DOI] [PubMed] [Google Scholar]
- 115. Rose KA, Morgan IG, Ip J, et al. Outdoor activity reduces the prevalence of myopia in children. Ophthalmology. 2008;115:1279–1285 [DOI] [PubMed] [Google Scholar]
- 116. Czepita D, Goslawski W, Mojsa A. Refractive errors among students occupying rooms lighted with incandescent or fluorescent lamps. Ann Acad Med Stetin. 2004;50:51–54 [PubMed] [Google Scholar]
- 117. Czepita D, Goslawski W, Mojsa A. Occurrence of refractive errors among students who before the age of two grew up under the influence of light emitted by incandescent or fluorescent lamps. Ann Acad Med Stetin. 2005;51:33–36 [PubMed] [Google Scholar]
- 118. Quinn GE, Shin CH, Maguire MG, Stone RA. Myopia and ambient lighting at night. Nature. 1999;399:113–114 [DOI] [PubMed] [Google Scholar]
- 119. Guggenheim JA, Hill C, Yam T-F. Myopia, genetics, and ambient lighting at night in a UK sample. Br J Ophthalmol. 2003;87:580–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Gwiazda J, Ong E, Held R, Thorn F. Myopia and ambient night-time lighting. Nature. 2000;404:144. [DOI] [PubMed] [Google Scholar]
- 121. Zadnik K, Jones LA, Irvin B, et al. Myopia and ambient night-time lighting. CLEERE Study Group: collaborative longitudinal evaluation of ethnicity and refractive error. Nature. 2000;404:143–144 [DOI] [PubMed] [Google Scholar]
- 122. Saw S-M, Zhang M-Z, Hong R-Z, Fu Z-F, Pang M-H, Tan DTH. Near-work activity, night-lights, and myopia in the Singapore-China study. Arch Ophthalmol. 2002;120:620–627 [DOI] [PubMed] [Google Scholar]
- 123. Deng L, Gwiazda J. Birth season, photoperiod, and infancy refraction. Optom Vis Sci. 2011;88:383–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Mandel Y, Grotto I, El-Yaniv R, et al. Season of birth, natural light, and myopia. Ophthalmology. 2008;115:686–692 [DOI] [PubMed] [Google Scholar]
- 125. McMahon G, Zayats T, Chen YP, Prashar A, Williams C, Guggenheim JA. Season of birth, daylight hours at birth, and high myopia. Ophthalmology. 2009;116:468–473 [DOI] [PubMed] [Google Scholar]
- 126. Golombek DA, Rosenstein RE. Physiology of circadian entrainment. Physiol Rev. 2010;90:1063–1102 [DOI] [PubMed] [Google Scholar]
- 127. Duffy JF, Czeisler CA. Effect of light on human circadian physiology. Sleep Med Clin. 2009;4:165–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Turner PL, Mainster MA. Circadian photoreception: ageing and the eye's important role in systemic health. Br J Ophthalmol. 2008;92:1439–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.