This study examines temporal variability in RPGR isoform expression and tests whether the constitutive RPGRex1-19 variant can substitute for RPGR function in photoreceptors.
Abstract
Purpose.
Mutations in the retinitis pigmentosa GTPase regulator (RPGR) gene are a frequent cause of X-linked retinitis pigmentosa. The RPGR transcript undergoes complex alternative splicing to express both constitutive (Rpgrex1-19) and RpgrORF15 variants. Both variants localize to photoreceptor connecting cilia and are believed to play roles in ciliary function. This study examined variability in isoform expression and tested whether the constitutive variant could substitute for Rpgr function in photoreceptors.
Methods.
Rpgrex1-19 and RpgrORF15 expression during retinal development were compared using immunoblot analysis and immunohistochemistry, and ciliary affinity in adult photoreceptors was assessed by protein fractionation. Transgenic mice expressing either the full-length Rpgrex1-19 or RpgrORF15 variant were studied using light and electron microscopy and immunofluorescence imaging. The results were compared with those of wild-type and Rpgr−/− mice.
Results.
Rpgr expression undergoes dynamic temporal regulation during retinal development, and variants exhibit variability for ciliary localization in adult photoreceptors. Transgenic expression of both variants grossly exceeded endogenous Rpgr expression in photoreceptors. Although both variants exhibited normal ciliary localization, overexpression of the Rpgrex1-19 variant resulted in atypical accumulation of Rpgr in photoreceptor outer segments, abnormal photoreceptor morphology, and severe retinal degeneration.
Conclusions.
The Rpgr isoform ratio in the adult retina is critical to photoreceptor integrity. The utilization of distinct Rpgr variants at different stages of photoreceptor maturation suggests independent roles in photoreceptor function. Finally, misexpression of Rpgrex1-19 causes retinal degeneration that is considerably more severe than that caused by Rpgr knockout but photoreceptors tolerate overexpression of RpgrORF15 without evidence of degeneration.
X-linked retinitis pigmentosa (XLRP) represents a severe form of retinitis pigmentosa (RP), a group of inherited retinal dystrophies that result in photoreceptor cell death and the accumulation of intraretinal pigmentlike deposits.1,2 Symptoms include night blindness, progressive loss of peripheral visual fields and eventual loss of central vision.3 Mutations in the retinitis pigmentosa GTPase regulator (RPGR) gene account for more than 70% of XLRP and approximately 10% of all RP cases.1,2,4 Ablation of the Rpgr gene in mice5 and naturally occurring mutations in dogs6 also lead to photoreceptor cell degeneration, suggesting that Rpgr is essential for mammalian photoreceptor survival. In addition, both early cone photoreceptor defects and rod degeneration indicate that Rpgr is necessary for the survival of rods and cones.5,7,8
Rpgr transcripts undergo a complex splicing process using alternative splicing sites and polyadenylation signals to generate constitutive Rpgrex1-19 transcripts and highly variable RpgrORF15 transcripts (Fig. 1).2,9–11 The Rpgrex1-19 variants are widely expressed and contain exons 1-13 and 16-19, whereas numerous RpgrORF15 variants are preferentially expressed in the retina and contain exons 1-13 plus a large, alternatively spliced C-terminal exon 14/15.2,5 Although both Rpgrex1-19 and RpgrORF15 localize to the connecting cilia through interaction of their constitutive N-terminal domain with Rpgrip12–14 and evidence suggests that they regulate protein trafficking through the photoreceptor connecting cilia,5,9,15 little is known regarding the physiological significance of the expression of two distinct variants.
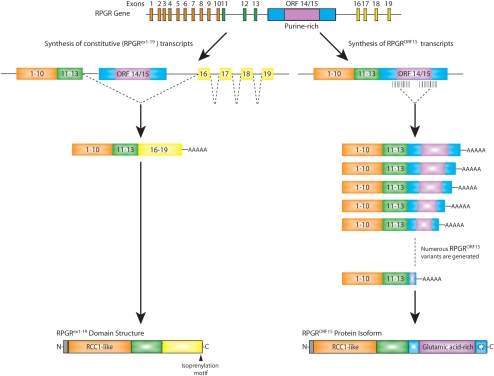
Figure 1.
The Rpgr gene structure and Rpgr expression in the retina. Alternative splicing leads to two groups of Rpgr transcripts. Rpgrex1-19 includes exons 1-13 and exons 16-19, and RpgrORF15 includes exons 1-13 plus a large, alternatively spliced ORF 14/15. Orange, exons encoding RCC1-like domain common to all Rpgr variants; green, remainder of exons common to all Rpgr variants; yellow, exons 16-19 encoding Rpgrex1-19 specific C-terminal domain, with isoprenylation motif; blue/purple, large exon (ORF 14/15) encoding C-terminal domain of RpgrORF15; purple, alternatively spliced region of ORF14/15 encoding glutamic acid-rich domain.
To further investigate the significance of variable variant expression in photoreceptors, we compared Rpgrex1-19 and RpgrORF15 expression during retinal development. Using immunoblot analysis and immunofluorescence microscopy, we observed dynamic temporal regulation of Rpgr expression during retinal development. Although Rpgrex1-19 is highly expressed in developing photoreceptors, expression is significantly downregulated in mature cells. Emergence of the RpgrORF15 variant, on the other hand, correlates with photoreceptor maturation. After examining transgenic lines expressing only Rpgrex1-19, we also report that an abundance of Rpgrex1-19 expression in mature photoreceptors results in abnormal accumulation of protein in the outer segments, disruption of outer segment morphology, and rapid retinal degeneration.
Methods
Animals
C57BL/6 wild-type mice were obtained from Harlan Laboratories (Houston, TX). The full-length Rpgrex1-19 was cloned from C57BL/6 wild-type retinal cDNA. Because of variable internal splicing of the ORF14/15 exon present in RpgrORF15 transcripts,9–11 a full-length RpgrORF15 transcript has never, to our knowledge, been successfully cloned. Thus, RpgrORF15 was cloned from a combination of genomic DNA and cDNA. Exons 1-13 were amplified from retinal cDNA and were joined with a full-length, unspliced ORF14/15 exon amplified from genomic DNA. Each clone was introduced into a pCBA vector between the cytomegalovirus (CMV) enhancer β-actin promoter (CBA), which has been shown to drive expression in both rods and cones, and a bovine growth hormone (BGH) polyadenylation sequence. N-terminal 3x-Myc tags were added to distinguish between native and transgenic Rpgr expression. Transgenic mice were generated by pronuclear injection of the described transgenic constructs (designated mRDef and mROrf) into C57BL/6 wild-type embryos. Founder mice were bred with C57BL/6 wild-type mice and Rpgr−/− mice, to generate transgenic mice on a wild-type and Rpgr null background, respectively. Rpgr−/− littermates were used as a control for assessment of retinal phenotype.
All animals were maintained on a 12-hour light–dark cycle, with food and water ad libitum and were handled in accordance with the institutional guidelines approved by the Texas A&M University IACUC (Institutional Animal Care and Use Committee).
Reverse Transcription (RT-PCR)
Total RNA was isolated from retina (TRIzol reagent; Invitrogen, Carlsbad, CA), and cDNA was generated with a PCR system (Superscript One-Step RT-PCR system; Invitrogen), according to the manufacturer's instructions.
Antibodies
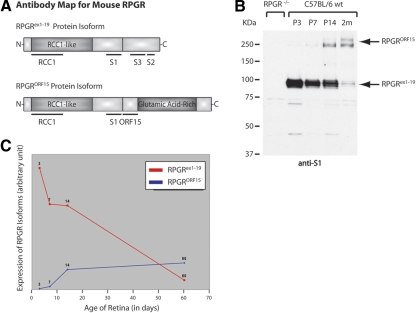
The polyclonal ORF15 antibody was generated in guinea pig and has been characterized.15 The other Rpgr antibodies have also been described.12,15 The locations of the Rpgr antibodies are shown in the antibody map in Figure 2A. Green cone opsin antibody (JH492) was provided by Jeremy Nathans (Johns Hopkins University School of Medicine) and was used as described.16 Anti-glial fibrillary acidic protein (GFAP) and acetylated α-tubulin (T6793) antibodies were obtained from Sigma-Aldrich (St. Louis, MO).
Figure 2.
Antibody map for mouse Rpgr and comparison of Rpgrex1-19 and RpgrORF15 expression in the developing retina. (A) Top: the structure of the Rpgrex1-19 variant and the location of domains used to generate our RCC1 and S1 polyclonal antibodies, which detect all Rpgr variants, and our S2 and S3, default specific polyclonal antibodies. Bottom: the structure of the Rpgr isoform structure and location of the common domains used to generate polyclonal antibodies against all Rpgr variants (RCC1 and S1) and the ORF15-specific domain used to generate our polyclonal ORF15 antibody. (B) Immunoblot analysis of retinal homogenate from wild-type mice at P3, P7, and P14 and at 2 months. The RpgrORF15 variants are approximately 250 kDa and the Rpgrex1-19 variants are roughly 100 kDa. Left: retinal homogenate from Rpgr−/− mice, illustrating antibody specificity. The faint smaller bands detected in the negative control are the result of antibody background and are not detected by any of our other antibodies against Rpgr. (C) Relative expression of Rpgrex1-19 and RpgrORF15 in the developing retina.
Goat anti-rabbit IgG-horseradish peroxidase conjugate and goat anti-mouse IgG-alkaline phosphatase conjugate (Pierce Biotech, Rockford, IL) were used as secondary antibodies. Alexa fluorochrome-conjugated secondary antibodies for immunostaining were used (Invitrogen-Molecular Probes, Inc., Eugene, OR).
Immunoblot Analyses
Tissues were homogenized in buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 0.5% NP40) containing a protease inhibitor cocktail (Sigma-Aldrich) and were centrifuged at 1000g for 2 minutes. For denaturing gel electrophoresis, samples were mixed with 4× SDS sample buffer with β-mercaptoethanol, separated on polyacrylamide gels, and then transferred to PVDF membranes (Immobilon-P; Millipore). After the membrane was blocked in 5% skim milk in PBS with 0.1% Tween, the proteins were detected by applying primary antibody overnight followed by the appropriate secondary antibody for 2 hours. Immunoreactive bands were quantitatively analyzed by using Image J (developed by Wayne Rasband, National Institutes of Health [NIH], Bethesda, MD; available at http://rsb.info.nih.gov/ij/index.html). Band densities were measured by using a simple graphic method that involves generating lane profile plots, drawing lines to enclose peaks of interest, and then measuring the peak areas (detailed description of method available at http://rsb.info.nih.gov/ij/docs/menus/analyze.html#gels/NIH). A marker (Precision Plus Prestained Standard; Bio-Rad, Hercules, CA) ranging from 10 to 250 to 25 kDa was used.
Cellular Fractionation
Four mouse retinas were dissected and kept on ice or 4°C for the remainder of the procedure, unless otherwise noted. The retinas were homogenized in tissue fractionation buffer (50 mm Tris [pH 7.4]; 150 mm NaCl, and protease inhibitor). The suspension was centrifuged at 500g for 2 minutes to remove large debris. The supernatant was centrifuged again at 35,000g for 30 minutes. The supernatant was transferred to a fresh tube and was designated the cytosolic fraction. The pellet was gently washed in fractionation buffer and was resuspended in NP40 buffer (50 mM Tris, [pH 7.4], 150 mM NaCl, and 1% Nonidet P-40). After incubating 30 minutes at room temperature, the samples were centrifuged at 35,000g for 30 minutes. The supernatant was collected and designated as the detergent-soluble fraction. The pellet was carefully washed in NP40 buffer, resuspended in tissue fractionation buffer, and designated the axoneme-enriched fraction.
Immunohistochemistry
Unfixed eyes were embedded in optimal cutting temperature (OCT) compound and were snap frozen in liquid nitrogen. Cryosections (10-μm) were cut and collected on pretreated glass slides (Superfrost Plus; Fisher Scientific, Pittsburgh, PA). Sections were stored at −20°C and used within 2 to 3 days. Immunofluorescence staining was performed as described elsewhere.5,12
Histology
Eyes were fixed in 4% formaldehyde and were embedded in OCT compound. Histology procedures were then performed as published.17
Dissociated Photoreceptors
Dissociated photoreceptor fragments were obtained by mechanical detachment from freshly dissected mouse retinas, as previously described.18 In brief, retinas were suspended in Ringer solution and were gently homogenized by five passes through a disposable transfer pipette. Cell fragments were allowed to adhere for 5 minutes to pretreated glass slides (Superfrost Plus Microscope Slides; Fisher Scientific). Adhered cell fragments were fixed for 5 minutes in ice-cold methanol, before proceeding with typical immunocytochemical staining as previously described.5,12
Electron Microscopy
After removal of the lens and vitreous, enucleated eyes were fixed in 2% formaldehyde and 2% glutaraldehyde in 0.1 M cacodylate buffer. Eyes were washed in 0.1 M cacodylate buffer for 3 days and were postfixed in 1% OsO4 for 1 hour at room temperature. They were then washed once in 0.1 M cacodylate buffer and once in water and were gradually dehydrated in 30%, 50%, 70%, 80%, 90%, and 95% cold ethanol for 15 minutes each. After warming to room temperature, the eyes were incubated in 100% ethanol (three times, 15 minutes each) followed by propylene oxide (PO). They were infiltrated with 1:2 epoxy (Epon Araldite with 1.5% DMP-30):PO for 1 hour, 1:1 epoxy:PO for 1 hour, 3:1 epoxy:PO for 1 hour, and then with 100% Epoxy. After transfer to flat molds, they were heat cured at 65°C for 2 days to polymerize.
Results
Rpgr Expression in the Developing Retina
Since the retina co-expresses both the Rpgrex1-19 and the RpgrORF15 variants, we examined whether expression of Rpgr variants is altered in a temporal manner. We previously showed that Rpgr immunoreactivity is first detectable at the apices of the developing photoreceptor layer at postnatal day (P)3, which correlates with the timing and location of connecting cilia formation.5 To further study the dynamics of Rpgr expression, we compared the expression of Rpgrex1-19 and RpgrORF15 at specific phases of retinal development. In the mouse, much of retinal development takes place in the 3 weeks after birth in a process very similar to third-trimester human retinal development.19 Proliferation, migration, and differentiation of neuronal precursor cells in the mouse retina is initiated at embryonic day (E)12 and continues through final neuronal differentiation and maturation at approximately P8.20 The final stages of neuronal differentiation and retinal vascular development occur when mice open their eyes and vision is initiated at around P14. Since many factors critical to establishing this visual pathway are regulated during the first 2 postnatal weeks,21 we analyzed Rpgr protein levels in retinal homogenates from P3, P7, P14, and adult (2-month) wild-type mice by using our anti-S1 antibody (Fig. 2A), which recognizes both Rpgr variants. Rpgrex1-19 migrates as a 95- to 100-kDa band on Western blot analysis. Protein expression was detected at times of neuronal differentiation in the retina and decreased with age, with robust expression at P3 compared to adult expression levels (Fig. 2B). In contrast, RpgrORF15 migrated at approximately 200 kDa and the emergence of the RpgrORF15 variants correlated with the maturation of photoreceptors (Fig. 2B; Fig. 3B). Relative intensities of the Rpgrex1-19 and RpgrOFR15 bands were quantified with the ImageJ software (Fig. 2C). Thus, our data show a correlation between changes in the Rpgr isoform ratio and photoreceptor development and maturation.
Figure 3.
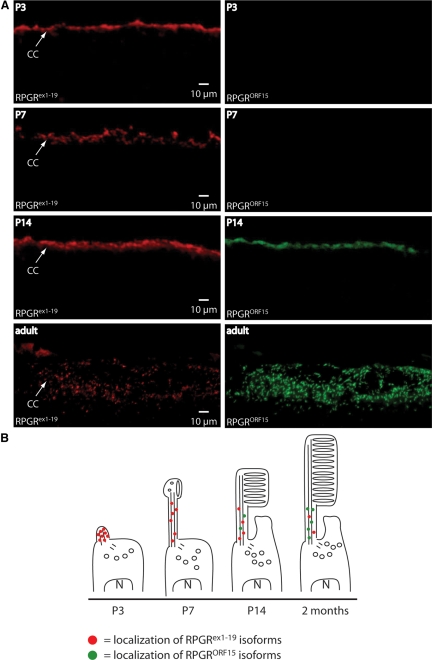
Immunohistochemical analysis of Rpgrex1-19 and RpgrORF15 expression and localization in the developing retina and a representation of photoreceptor development at the analyzed time points. (A) Double-immunostaining of retinal cryosections using our rabbit polyclonal anti-S2 antibody to detect Rpgrex1-19 specific variants (top row) and our guinea pig polyclonal anti-ORF15 antibody to detect RpgrORF15 specific variants (second row). Nuclear staining with DAPI (third row) and DIC images (bottom row) are shown to monitor the developmental progression of the retina. (B) Illustration representing the development of photoreceptor cells with representative expression and localization of Rpgrex1-19 and RpgrORF15 variants.
To compare the localization of the Rpgrex1-19 and RpgrORF15 variants during retinal development, we performed immunohistochemistry on wild-type P3, P7, P14, and adult retinas. Rpgrex1-19 and RpgrORF15 variants were detected with the anti-S3 and anti-Rpgr ORF15 antibodies, respectively (Fig. 3A; and antibody map, Fig. 2A). In the P3 retina, the Rpgrex1-19 was detected as a narrow band at the apex of the developing photoreceptor layer (Fig. 3A). The appearance of Rpgr at this time point is consistent with the appearance and location of the emerging photoreceptor connecting cilia (Fig. 3B). The well-defined band persisted through day 14 but severely diminished in the adult retina. RpgrORF15 was nearly undetectable until P14 and increased in intensity in the adult retina (Fig. 3A). These data are consistent with those in our isoform-specific protein level analysis shown in Figure 2B.
Subcellular Distribution of Rpgr Variants in Retina
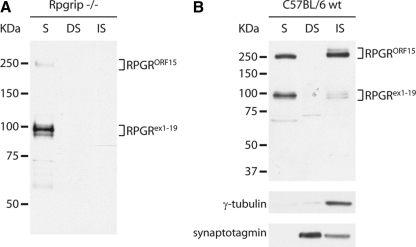
The N-terminal domain common to both Rpgrex1-19 and RpgrORF15 interacts with Rpgrip, a structural component of the ciliary axoneme.12 This interaction anchors Rpgr to the connecting cilia of rod and cone photoreceptors. On fractionation of retinal tissue, Rpgr is present in the insoluble, ciliary axoneme–enriched fraction in addition to the cytosolic fraction.12 Ciliary localization of all Rpgr variants was lost in Rpgrip−/− mice, hence eliminating Rpgr from the insoluble fraction (Fig. 4A).
Figure 4.
Fractionation of retinal homogenate illustrates ciliary localization of Rpgr variants. (A) Fractionation of retinal homogenate from Rpgrip−/− retina shows failure of both Rpgrex1-19 and RpgrORF15 variants to properly localize to the connecting cilia. S, soluble protein fraction; DS, NP40 detergent soluble fraction; IS, NP40 detergent insoluble fraction. (B) Immunoblot of fractionated retinal homogenate from C57BL/6 wild-type mice. Rpgr was normally distributed between the S fraction (unbound Rpgr) and the NP40 IS fraction (Rpgrip bound Rpgr) with a higher proportion of RpgrORF15 in the NP40 IS fraction.
To examine whether both Rpgr variants share equal affinity toward Rpgrip in the connecting cilia, we fractionated retinal homogenates from 2-month-old mice. The cytosolic fraction (soluble, S), the detergent soluble (DS) fraction (Nonidet P-40 soluble fraction), and insoluble (IS) fraction were analyzed by immunoblot analysis, using our polyclonal anti-S1 antibody (Fig. 4B). γ-Tubulin and synaptotagmin were used as quality controls to ensure that the detergent soluble and insoluble fractions were enriched for the membrane-bound and ciliary axoneme–bound proteins, respectively. Although the amounts of the Rpgrex1-19 and RpgrORF15 variants in the soluble fraction were approximately equal, a larger percentage of the total RpgrORF15 isoform population was found in the insoluble fraction. These results indicate that the two groups of variants do not share equal affinity for the ciliary fraction.
Expression of Rpgrex1-19 and RpgrORF15 in Transgenic Mice
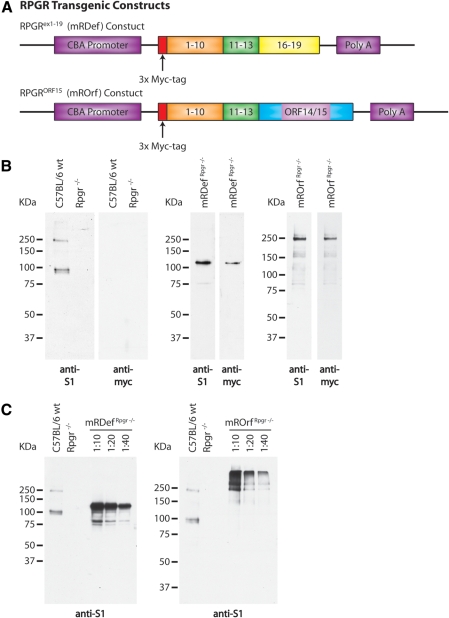
Rpgrex1-19 and RpgrORF15 variants both interact with Rpgrip and thus are likely to share some functional redundancy. However, the discrete C-terminal domains, evidence of developmental differences in isoform expression, and differential binding to the axoneme suggest that the two Rpgr variants also possess independent functions. To investigate the unique role of these two groups of variants in photoreceptor viability, we produced transgenic mice expressing only Rpgrex1-19 or RpgrORF15. The mRDef transgenic mice expressed a full-length Rpgrex1-19 transcript with an N-terminal 3x-Myc tag. This construct (Fig. 5A) is expressed from a CMV/β-actin promoter (CBA) that drives expression in both rods and cones22,23 (see the Methods section). This line was examined on both a wild-type and Rpgr null background, herein referred to as mRDefRpgr wt and mRDefRpgr−/−, respectively. The mROrf transgenic mice likewise express an RpgrORF15 construct from a CBA promoter that includes an N-terminal 3x-Myc tag (Fig. 5A). This line was examined on an Rpgr−/− background, herein referred to as mROrfRpgr−/−.
Figure 5.
Schematic illustration of transgenic constructs and confirmation of transgene expression. (A) Top: an Rpgrex1-19 transcript was cloned between the cytomegalovirus (CMV) enhancer β-actin promoter (CBA) and a bovine growth hormone (BGH) polyadenylation sequence. Bottom: a full-length RpgrORF15 transcript was cloned from a combination of genomic and cDNA. Exons 1-13 were cloned from wild-type retinal cDNA and the final exon, ORF14/15, was cloned from genomic cDNA. An N-terminal Myc tag was integrated in both transgenic constructs to allow for differentiation between transgenic and native Rpgr expression. (B) Left: immunoblot analysis of retinal homogenate from wild-type and Rpgr−/− mice using our polyclonal anti-S1 and monoclonal anti-myc antibodies. Middle: verification of transgene expression by immunoblot analysis of retinal homogenate from mRDefRpgr−/− transgenic mice. Right: verification of transgene expression by immunoblot analysis of retinal homogenate from mROrfRpgr−/− transgenic mice. (C) Comparison of transgenic expression levels with Rpgr expression in wild-type retina by immunoblot analysis with the anti-S1 antibody.
To confirm that the promoter was driving expression of the transgene in the retina, we analyzed retinal homogenate from mRDefRpgr−/− and mROrfRpgr−/− transgenic mice by immunoblot analysis. Expression was first confirmed by using the S1 antibody followed by a monoclonal anti-myc antibody. An immunoblot of retinal homogenate from wild-type and Rpgr−/− mice verified the specificity of our antibodies (Fig. 5B). The presence of the N-terminal 3x-Myc tag increased the size of the transgenic Rpgrex1-19 protein by an estimated 27 kDa. Although the transgenic RpgrORF15 protein has the same N-terminal tag, the increase in size is not evident due to decreased separation of proteins larger than ∼250 kDa on the gel.
To estimate the expression level of the transgenic protein, we compared serial dilutions of retinal homogenate from mRDefRpgr−/− and mROrfRpgr−/− transgenic mice with retinal homogenate from wild-type mice (Fig. 5C). By comparing the 10-, 20-, and 40-fold dilutions, we estimate there is approximately an 80-fold increase in Rpgrex1-19 expression and about a 40-fold increase in RpgrORF15 expression in our transgenic mice in comparison to wild-type Rpgr expression levels.
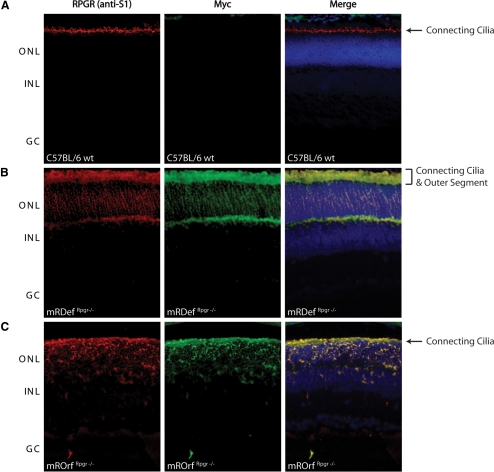
Localization of Transgenic Rpgr Protein Variants
Using immunofluorescence microscopy, we previously determined that Rpgr is concentrated in the connecting cilia of rod and cone photoreceptors,5 as is shown in Figure 6A. To examine the subcellular localization of transgenic Rpgr proteins, we compared frozen retinal cryosections from mRDefRpgr−/− and mROrfRpgr−/− mice with retinal sections from wild-type mice. All retinal sections were probed with both the S1 antibody and a monoclonal anti-myc antibody (Fig. 6). Unlike wild-type Rpgr staining, which localizes to the connecting cilium, Rpgr staining in the mRDefRpgr−/− transgenic mice not only labeled the connecting cilia but extended into the inner and outer segments (Fig. 6B). We observed considerable mislocalization of Rpgrex1-19 protein around the photoreceptor nuclei and synaptic regions, areas that are not associated with any cilia or basal body structures. In the mROrfRpgr−/− transgenic mice, Rpgr was observed in the connecting cilia and inner segment but not in the outer segment (Fig. 6C). Although Rpgr did mislocalize to the cell body in the mROrfRpgr−/− model, there appeared to be less mislocalization than in the mRDefRpgr−/− animals. The data confirm that both lines express transgenic Rpgr in the photoreceptors and that overexpression of different Rpgr variants results in protein mislocalization. Although localization of either Rpgr variant in the inner segment is atypical, it is likely to be the result of the abundance of protein production taking place in the biosynthetic inner segment. More important, however, only overexpression of Rpgrex1-19 resulted in protein accumulation in the outer segment.
Figure 6.
Comparison of native Rpgr localization with Rpgrex1-19 and RpgrORF15 transgenic expression by immunohistochemical analysis of frozen retinal cryosections. Double-staining of (A) wild-type, (B) transgenic Rpgrex1-19 expression in mRDefRpgr−/− retina, and (C) RpgrORF15 expression in mROrfRpgr−/− retina with our anti-S1 polyclonal antibody (red) and anti-myc monoclonal antibody (green).
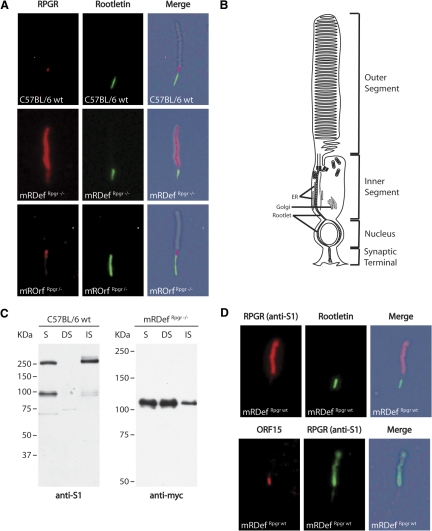
To gain better resolution of protein localization, we co-labeled mechanically dissociated photoreceptors for Rpgr and rootletin, a structural component of the inner segment. These preparations contained mostly shaken-off rod outer segments attached to the connecting cilia with a portion of the inner segments at the proximal end of the connecting cilia. Comparison of the immunofluorescence and DIC images of a wild-type photoreceptor confirmed Rpgr localization to the connecting cilium (Fig. 7A). A diagram of a photoreceptor cell is shown in Figure 7B to help illustrate the subcellular compartments. The staining pattern of RpgrORF15 in mROrfRpgr−/− dissociated photoreceptors strongly resembled labeling of Rpgr in wild-type photoreceptors. However, comparison of immunolabeled Rpgrex1-19 in mRDefRpgr−/− photoreceptors showed intense outer segment staining in addition to the normal ciliary staining (Fig. 7A). These data are consistent with our immunofluorescence staining of retinal sections (Fig. 6) and confirm that overexpression of Rpgrex1-19 results in an atypical accumulation of protein in the photoreceptor outer segments.
Figure 7.
Comparison of the subcellular distribution of Rpgr in photoreceptors from wild-type and transgenic retina. (A) Double-staining of a rod photoreceptor from a (top) wild-type, an (middle) mRDefRpgr−/− transgenic, and an (bottom) mROrfRpgr−/− transgenic retina with the anti-S1 and rootletin antibody. (B) Schematic representation of a rod photoreceptor illustrates subcellular compartments. (C) Fractionation of retinal homogenate from wild-type and mRDefRpgr−/− retinas shows accumulation of excess Rpgrex1-19 protein in the membrane-bound fraction. S, soluble protein fraction; NS, NP40 detergent soluble fraction; IS, NP40 detergent insoluble fraction. Left: immunoblot of fractionated retinal homogenate from wild-type mice. Rpgr is normally distributed between the soluble fraction (unbound Rpgr) and the NP40 insoluble fraction (Rpgrip bound Rpgr). Right: immunoblot of fractionated retinal homogenate from mRDefRpgr−/− mice shows accumulation of default protein in the membrane-bound, NP40 soluble fraction. (D) Double-staining of rod photoreceptors from mRDef transgenic mice on a wild-type background. Top: double-staining of all Rpgr variants (anti-S1) and rootletin (anti-Root6). Bottom: double-staining of only the RpgrORF15 variants (anti-ORF15) and rootletin.
The C terminus of the Rpgrex1-19 peptide contains an isoprenylation signal and is isoprenylated in tissue culture.1,9,22 As isoprenylation often facilitates binding of proteins to target membranes,24,25 we hypothesized that excess Rpgrex1-19 accumulates in the outer segments of mRDef photoreceptors due to interactions with the disc membranes. To test this hypothesis, we compared fractionated retinal homogenate from mRDefRpgr−/− and wild-type mice (Fig. 7C). In the wild-type retina, Rpgrex1-19 was primarily found in the cytosolic (S) and ciliary axoneme enriched (IS) fractions. In the mRDef retina, there was a moderate increase in the presence of Rpgrex1-19 in both the cytosolic (S) and ciliary axoneme–enriched (IS) fractions. However, the most significant change was the accrual of protein in the detergent soluble fraction (NS). Since Nonidet P-40 solubilizes membrane-bound proteins, we conclude that Rpgrex1-19 accumulates in the outer segments by interacting with the membranes.
Since Rpgrex1-19 and RpgrORF15 compete for interaction with Rpgrip in the photoreceptor connecting cilia, we also examined whether an increase in the Rpgrex1-19 concentration disrupts localization of RpgrORF15 to the connecting cilia. We labeled dissociated photoreceptors from transgenic mRDef mice on a wild-type Rpgr background for all Rpgr variants (anti-S1) and only RpgrORF15 (anti-ORF15; Fig. 7D). Photoreceptors were also labeled with anti-rootletin to confirm the location of subcellular compartments. Despite the overabundance and mislocalization of Rpgrex1-19, localization of native RpgrORF15 to the photoreceptor connecting cilia remained unaltered. If Rpgrex1-19 and RpgrORF15 share equal affinity for Rpgrip, then we would expect the excess Rpgrex1-19 protein to compete for Rpgrip and diminish the ciliary presence of RpgrORF15; however, this was not observed. This is consistent with our earlier finding that a larger proportion of RpgrORF15 than Rpgrex1-19 was found in the ciliary-enriched fraction (Fig. 4) and supports our conclusion that RpgrORF15 has a higher affinity for binding Rpgrip in the connecting cilia.
Retinal Disease in Rpgrex1-19 Transgenic Mice
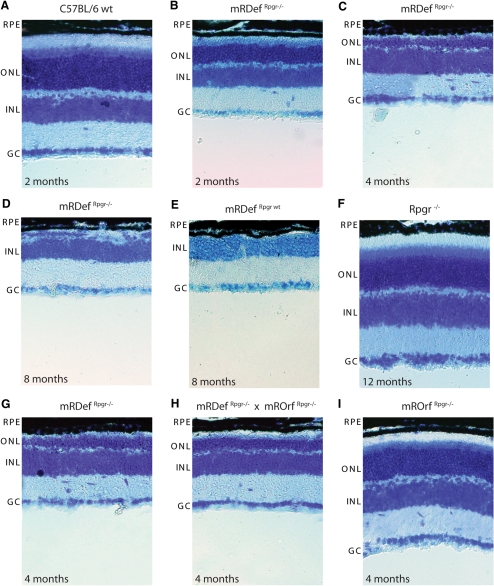
To evaluate the effects of transgene expression on photoreceptor cell survival, we examined retinal morphology in wild-type mice at 2 months of age and mRDefRpgr−/− mice from 2 to 8 months of age (Figs. 8A–D). Although the retinal morphology of mRDefRpgr−/− mice was comparable to that of the wild-type at the completion of retinal development (P21), retinal cell loss was apparent in young mRDefRpgr−/− retinas. By 2 months of age, the inner and outer segments of the mRDefRpgr−/− mice were shortened, and a decreased number of nuclei in the outer nuclear layer provided evidence of significant photoreceptor cell loss (Fig. 8B). Compared with Rpgr−/− mice (Fig. 8F), in which retinal cell loss is relatively slow,5 degeneration in the mRDef mice was rapid with complete loss of photoreceptors by 8 months (Fig. 8D). Since the rate of degeneration was similar on both the Rpgr−/− and wild-type backgrounds (Figs. 8D, 8E), we conclude that retinal cell loss results from the overexpression of the Rpgrex1-19 transgene.
Figure 8.
Phenotypic analysis of mRDef transgenic mice by light microscopy. Histologic sections of (A) wild-type retina at 2 months, (B–D) mRDefRpgr−/− at 2 to 8 months, and (E) mRDefRpgr wt at 8 months. (G–I) mRDefRpgr−/− mice were crossed with mROrfRpgr−/− mice, and the retinal phenotypes of 4-month-old single- and double-transgenic littermates were assessed by light microscopy at 4 months of age. RPE, retinal pigment epithelium; ONL, outer nuclear layer; INL, inner nuclear layer; GC, ganglion cell layer.
To investigate whether concurrent overexpression of RpgrORF15 would alter the mRDef transgenic phenotype, we bred mRDef and mROrf mice on an Rpgr−/− background. We compared the retinal morphology of mRDefRpgr−/−, mROrfRpgr−/−, and mRDef/mROrfRpgr−/− littermates at 4 months of age (Figs. 8G–I), and found that the severity of the mRDef transgenic phenotype was unaffected by the presence of RpgrORF15. 21 Importantly, overexpression of RpgrORF15 in the mROrfRpgr−/− mice did not result in measurable degeneration at 4 months of age (Fig. 8I). These results suggest that photoreceptors can tolerate overexpression of the RpgrORF15 variant but not the Rpgrex1-19 variant. Thus, we conclude that the retinal cell loss in mRDef transgenic mice results in a neomorphic phenotype independent of RpgrORF15 expression.
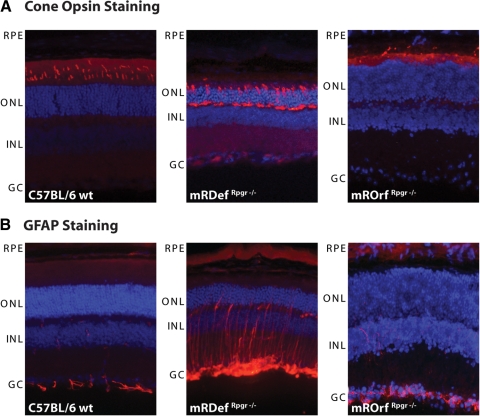
Mislocalization of cone opsins in cone photoreceptor cell bodies and synapses is a prominent phenotype in Rpgr−/− mouse retinas before retinal cell loss is even apparent.5 Both blue and green cone opsins, which normally localize in the outer segments, partially mislocalize to the inner segment, perinuclear area, and synaptic regions as early as postnatal day 20 in Rpgr−/− mice (data not shown).5 To observe whether expression of Rpgrex1-19 or RpgrORF15 rescues the phenotype in cone photoreceptors, we compared mRDefRpgr−/−, mROrfRpgr−/−, and wild-type control retinas by immunofluorescence for cone opsin (Fig. 9A). Like Rpgr−/− retinas,5 opsins in the mRDefRpgr−/− cone photoreceptors show mislocalization in the inner segment, perinuclear regions, and synaptic terminals. In contrast, cone opsin staining in the mROrfRpgr−/− was confined to the cone OS, as in the wild-type, indicating restoration of Rpgr function in cone cells. Thus, comparison of the number and integrity of cone cells between the two transgenic mice demonstrated that RpgrORF15 but not Rpgrex1-19 is able to rescue the Rpgr−/− phenotype.
Figure 9.
Mislocalization of opsins and upregulation of GFAP in mRDef transgenic mice. (A) Immunohistochemical analysis of opsin localization in retinal cryosections with a green cone opsin–specific antibody. (Left) Wild-type, (middle) mRDefRpgr−/− transgenic, and (right) mROrfRpgr−/− transgenic retina. (B) Upregulation of GFAP immunoreactivity in mRDefRpgr−/− retina. (Left) Wild-type, (middle) mRDefRpgr−/− transgenic, and (right) mROrfRpgr−/− transgenic retina. RPE, retinal pigment epithelium; ONL, outer nuclear layer; INL, inner nuclear layer; GC, ganglion cell layer.
The upregulation of GFAP expression in the retina is a nonspecific marker of retinal degeneration. In Rpgr−/− retinas, GFAP upregulation indicates degenerative changes before retinal cell loss.5 As an additional outcome measure for the mRDefRpgr−/− and mROrfRpgr−/− transgenic phenotypes, we examined GFAP expression in transgenic and control animals (Fig. 9B). As expected, GFAP was clearly unregulated in the mRDefRpgr−/− mice, with expansion of staining into the outer retina. Virtually no GFAP signal was detected in the wild-type and mROrfRpgr−/− retinas.
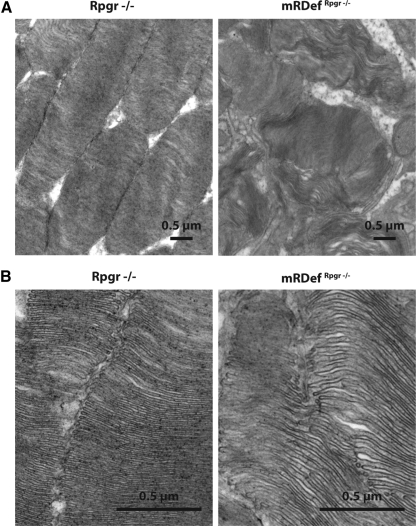
Because of the rate of degenerative changes and the aforementioned accumulation of Rpgrex1-19 protein in the outer segments of mRDefRpgr−/− transgenic mice, photoreceptor outer segment morphology was assessed by electron microscopy at 2 months of age (Fig. 10). The outer segments were notably disorganized in the transgenic mice with disruption of the conventional parallel arrangement of disc membranes and poorly defined outer segment morphologies. Perimeters were undefined and disc diameters were greatly expanded. Stacks of disc membranes were occasionally arranged parallel to the long axis of the outer segments instead of the normal perpendicular orientation. Although such defects are not seen in the Rpgr−/− mice, these observations are reminiscent of the abnormal disc morphology seen in the Rpgrip−/− mice.26
Figure 10.
Ultrastructural examination of photoreceptor outer segments in mRDef transgenic mice at two magnifications. TEM image of photoreceptor outer segments in 2-month-old Rpgr−/− (left) and mRDefRpgr−/− (right) retinas. Magnification: (A) ×11,000; (B) ×44,000.
Discussion
An important finding of this work is the differential regulation of Rpgr variant expression during photoreceptor development. Previous analyses of mice lacking Rpgr indicated that Rpgr is not essential for mammalian photoreceptor development.5 The robust expression of the Rpgrex1-19 variant during retinal development and subsequent attenuation of expression in mature photoreceptors is nonetheless suggestive of a functional, albeit redundant, role during development. These conclusions are consistent with a recent report identifying two Rpgr orthologues in zebrafish, which were also reported to have more widespread expression during development. Like many zebrafish orthologues of human genes, the two homologous RPGR genes reported (zfrpgr1 and zfrpgr2) are probably attributable to a genome duplication that occurred in teleosts. Unlike mammals, Rpgr knockdown in zebrafish results in developmental abnormalities, including failure to develop photoreceptor outer segments,27 suggesting that Rpgr is required for normal retinal development. In addition, our data also indicate that emergence of the RpgrORF15 variants follows a reciprocal expression pattern, coinciding with photoreceptor maturation. This supports the idea that RpgrORF15 plays a physiological role in the integrity of mature photoreceptors. Although it has been speculated that RpgrORF15 is the functionally significant variant in photoreceptors,23 the dynamics of Rpgr expression in emerging photoreceptors suggests that both the RpgrORF15 and Rpgrex1-19 variants retain some independent, isoform-specific functions.
In this study, we also addressed whether the Rpgrex1-19 variant alone is able to restore function to photoreceptors lacking endogenous Rpgr. Photoreceptors in Rpgrex1-19 transgenic mice exhibited atypical accumulation and interaction of Rpgrex1-19 protein with the membranous outer segments, severe histopathologic changes in the photoreceptor outer segments, mislocalization of cone opsin, and upregulation of GFAP. Expression of the Rpgrex1-19 transgene resulted in a substantially more severe phenotype than that of the previously reported Rpgr−/− mice5 with photoreceptor degeneration apparent from an early age.
Rpgrex1-19 expression in our transgenic mice exceeded wild-type endogenous Rpgrex1-19 expression by several fold. Thus, the observed phenotype may be a nonspecific consequence associated with the intense level of overexpression. However, since similar overexpression of the RpgrORF15 variant did not result in atypical accumulation of protein in the outer segment or in a degenerative retinal phenotype, we conclude that our report describes an Rpgrex1-19-specific phenomenon. Although endogenous Rpgrex1-19 expression in adult photoreceptors is minimal, further investigation of this acquired function, may nonetheless provide evidence of native Rpgrex1-19 function in developing and/or mature photoreceptors.
The Rpgrex1-19 variant differs from the RpgrORF15 variant by the presence of a C-terminal isoprenylation motif.1,9,28,29 By immunofluorescence, we observe that excess Rpgrex1-19 protein accumulates in photoreceptor outer segments. Such mislocalized accumulation is likely related to the membranous nature of the outer segment structure and the inherent properties of the isoprenylation signal. In general, isoprenylation motifs signal the addition of prenyl groups at carboxyl-terminal cysteine residues. The functional consequence of such posttranslational protein modification is anchorage of prenylated proteins to cell membranes.24,25
Given evidence of severely diminished Rpgrex1-19 expression in mature photoreceptors and the affinity of Rpgrex1-19 to tightly bind Rpgrip in the connecting cilia,12–14 the presence of Rpgrip is most likely sufficient to limit localization of endogenous Rpgr to the connecting cilia in mature photoreceptors. This probability suggests that if the concentration of Rpgrex1-19 exceeds the binding capacity of Rpgrip or if binding is otherwise interrupted, Rpgrex1-19 may begin to mislocalize and accumulate in the photoreceptor outer segments. By electron microscopy, it is clear that this accumulation of protein both functionally and morphologically disrupts the organized structure of the outer segments' membranous disks. In addition, isoprenylation of Rpgr may be necessary for some form of ciliary trafficking during very early stages of photoreceptor development before the appearance of the outer segments. Thus, the Rpgrex1-19 variants may possess different functions during early development, compared with the function of RpgrORF15 after ciliogenesis and outer segment maturation.
Although we have shown that expression of a single RpgrORF15 variant substantially rescues the Rpgr knockout phenotype,23 these more recent findings should also be taken into consideration when designing therapeutic treatment for RPGR patients. Unfortunately, there is a void in knowledge regarding protein expression in RPGR patients. Although a majority of RPGR mutations are in the ORF15 exon and thus only directly affect the integrity of the RpgrORF15 variants, there is some possibility of indirect effects on Rpgrex1-19 expression as well. Given that all RPGR variants are under the control of a single promoter, there is some possibility that Rpgrex1-19 expression is affected by endogenous attempts to compensate for the loss of functional RpgrORF15. This theory may explain some of the variability associated with human genotype–phenotype correlation, as well as, the surprisingly mild phenotype of Rpgr null mouse models when compared with XlRP3-affected humans and dogs. Furthermore, if this theory is upheld, then introduction of the RpgrORF15 variant by gene therapy also has the potential to affect endogenous Rpgr expression. In either case, upregulation of Rpgrex1-19 expression has the potential to be more detrimental to photoreceptor integrity and disease progression than the lack of functional RpgrORF15.
Footnotes
Supported by National Eye Institute Grants EY14188 (D-H.-H.) and EY017037 (B.P.).
Disclosure: R.N. Wright, None; D.-H. Hong, None; B. Perkins, None
References
- 1. Meindl A, Dry K, Herrmann K, et al. A gene (RPGR) with homology to the RCC1 guanine nucleotide exchange factor is mutated in X-linked retinitis pigmentosa (RP3). Nat Genet. 1996;13:35–42 [DOI] [PubMed] [Google Scholar]
- 2. Vervoort R, Lennon A, Bird AC, et al. Mutational hot spot within a new RPGR exon in X-linked retinitis pigmentosa. Nat Genet. 2000;25:462–466 [DOI] [PubMed] [Google Scholar]
- 3. Kennan A, Aherne A, Humphries P. Light in retinitis pigmentosa. Trends Genet. 2005;21:103–110 [DOI] [PubMed] [Google Scholar]
- 4. Bunker CH, Berson EL, Bromley WC, Hayes RP, Roderick TH. Prevalence of retinitis pigmentosa in Maine. Am J Ophthalmol. 1984;97:357–365 [DOI] [PubMed] [Google Scholar]
- 5. Hong DH, Pawlyk BS, Shang J, Sandberg MA, Berson EL, Li T. A retinitis pigmentosa GTPase regulator (RPGR)-deficient mouse model for X-linked retinitis pigmentosa (RP3). Proc Natl Acad Sci U S A. 2000;97:3649–3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang Q, Acland GM, Wu WX, et al. Different RPGR exon ORF15 mutations in Canids provide insights into photoreceptor cell degeneration. Hum Mol Genet. 2002;11:993–1003 [DOI] [PubMed] [Google Scholar]
- 7. Berson EL, Gouras P, Gunkel RD, Myrianthopoulos NC. Rod and cone responses in sex-linked retinitis pigmentosa. Arch Ophthalmol. 1969;81:215–225 [DOI] [PubMed] [Google Scholar]
- 8. Sharon D, Sandberg MA, Rabe VW, Stillberger M, Dryja TP, Berson EL. RP2 and RPGR mutations and clinical correlations in patients with X-linked retinitis pigmentosa. Am J Hum Genet. 2003;73:1131–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yan D, Swain PK, Breuer D, et al. Biochemical characterization and subcellular localization of the mouse retinitis pigmentosa GTPase regulator (mRpgr). J Biol Chem. 1998;273:19656–19663 [DOI] [PubMed] [Google Scholar]
- 10. Kirschner R, Rosenberg T, Schultz-Heienbrok R, et al. RPGR transcription studies in mouse and human tissues reveal a retina-specific isoform that is disrupted in a patient with X-linked retinitis pigmentosa. Hum Mol Genet. 1999;8:1571–1578 [DOI] [PubMed] [Google Scholar]
- 11. Hong DH, Li T. Complex expression pattern of RPGR reveals a role for purine-rich exonic splicing enhancers. Invest Ophthalmol Vis Sci. 2002;43:3373–3382 [PubMed] [Google Scholar]
- 12. Hong DH, Yue G, Adamian M, Li T. Retinitis pigmentosa GTPase regulator (RPGRr)-interacting protein is stably associated with the photoreceptor ciliary axoneme and anchors RPGR to the connecting cilium. J Biol Chem. 2001;276:12091–12099 [DOI] [PubMed] [Google Scholar]
- 13. Boylan JP, Wright AF. Identification of a novel protein interacting with RPGR. Hum Mol Genet. 2000;9:2085–2093 [DOI] [PubMed] [Google Scholar]
- 14. Roepman R, Bernoud-Hubac N, Schick DE, et al. The retinitis pigmentosa GTPase regulator (RPGR) interacts with novel transport-like proteins in the outer segments of rod photoreceptors. Hum Mol Genet. 2000;9:2095–2105 [DOI] [PubMed] [Google Scholar]
- 15. Hong DH, Pawlyk B, Sokolov M, et al. RPGR isoforms in photoreceptor connecting cilia and the transitional zone of motile cilia. Invest Ophthalmol Vis Sci. 2003;44:2413–2421 [DOI] [PubMed] [Google Scholar]
- 16. Chiu MI, Nathans J. A sequence upstream of the mouse blue visual pigment gene directs blue cone-specific transgene expression in mouse retinas. Vis Neurosci. 1994;11:773–780 [DOI] [PubMed] [Google Scholar]
- 17. Li T, Sandberg MA, Pawlyk BS, et al. Effect of vitamin A supplementation on rhodopsin mutants threonine-17 → methionine and proline-347 → serine in transgenic mice and in cell cultures. Proc Natl Acad Sci U S A. 1998;95:11933–11938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Muresan V, Joshi HC, Besharse JC. Gamma-tubulin in differentiated cell types: localization in the vicinity of basal bodies in retinal photoreceptors and ciliated epithelia. J Cell Sci. 1993;104:1229–1237 [DOI] [PubMed] [Google Scholar]
- 19. Connolly SE, Hores TA, Smith LE, D'Amore PA. Characterization of vascular development in the mouse retina. Microvasc Res. 1988;36:275–290 [DOI] [PubMed] [Google Scholar]
- 20. Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: lessons from the retina. Nat Rev Neurosci. 2001;2:109–118 [DOI] [PubMed] [Google Scholar]
- 21. Dorrell MI, Aguilar E, Weber C, Friedlander M. Global gene expression analysis of the developing postnatal mouse retina. Invest Ophthalmol Vis Sci. 2004;45:1009–1019 [DOI] [PubMed] [Google Scholar]
- 22. McGee Sanftner LH, Abel H, Hauswirth WW, Flannery JG. Glial cell line derived neurotrophic factor delays photoreceptor degeneration in a transgenic rat model of retinitis pigmentosa. Mol Ther. 2001;4:622–629 [DOI] [PubMed] [Google Scholar]
- 23. Hong DH, Pawlyk BS, Adamian M, Sandberg MA, Li T. A single, abbreviated RPGR-ORF15 variant reconstitutes RPGR function in vivo. Invest Ophthalmol Vis Sci. 2005;46:435–441 [DOI] [PubMed] [Google Scholar]
- 24. Zhang FL, Casey PJ. Protein prenylation: molecular mechanisms and functional consequences. Annu Rev Biochem. 1996;65:241–269 [DOI] [PubMed] [Google Scholar]
- 25. Clarke S. Protein isoprenylation and methylation at carboxyl-terminal cysteine residues. Annu Rev Biochem. 1992;61:355–386 [DOI] [PubMed] [Google Scholar]
- 26. Zhao Y, Hong DH, Pawlyk B, et al. The retinitis pigmentosa GTPase regulator (RPGR)-interacting protein: subserving RPGR function and participating in disk morphogenesis. Proc Natl Acad Sci U S A. 2003;100:3965–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shu X, Zeng Z, Gautier P, et al. Zebrafish Rpgr is required for normal retinal development and plays a role in dynein-based retrograde transport processes. Hum Mol Genet. 2010;19:657–670 [DOI] [PubMed] [Google Scholar]
- 28. Shu X, Black GC, Rice JM, et al. RPGR mutation analysis and disease: an update. Hum Mutat. 2007;28:322–328 [DOI] [PubMed] [Google Scholar]
- 29. Wright AF, Shu X. Focus on molecules: RPGR. Exp Eye Res. 2007;85:1–2 [DOI] [PubMed] [Google Scholar]