Abstract
FACT (FAcilitates Chromatin Transcription/Transactions) plays a central role in transcription and replication in eukaryotes by both establishing and overcoming the repressive properties of chromatin. FACT promotes these opposing goals by interconverting nucleosomes between the canonical form and a more open reorganized form. In the forward direction, reorganization destabilizes nucleosomes, while the reverse reaction promotes nucleosome assembly. Nucleosome destabilization involves disrupting contacts among histone H2A-H2B dimers, (H3-H4)2 tetramers, and DNA. Here we show that mutations that weaken the dimer:tetramer interface in nucleosomes suppress defects caused by FACT deficiency in vivo in the yeast Saccharomyces cerevisiae. Mutating the gene that encodes the Spt16 subunit of FACT causes phenotypes associated with defects in transcription and replication, and we identify histone mutants that selectively suppress those associated with replication. Analysis of purified components suggests that the defective version of FACT is unable to maintain the reorganized nucleosome state efficiently, whereas nucleosomes with mutant histones are reorganized more easily than normal. The genetic suppression observed when the FACT defect is combined with the histone defect therefore reveals the importance of the dynamic reorganization of contacts within nucleosomes to the function of FACT in vivo, especially to FACT’s apparent role in promoting progression of DNA replication complexes. We also show that an H2B mutation causes different phenotypes, depending on which of the two similar genes that encode this protein are altered, revealing unexpected functional differences between these duplicated genes and calling into question the practice of examining the effects of histone mutants by expressing them from a single plasmid-borne allele.
FACT (FAcilitates Chromatin Transcription/Transactions) is a highly conserved histone chaperone with roles in both transcription and DNA replication (Reinberg and Sims 2006; Formosa 2008). In Saccharomyces cerevisiae, FACT is an Spt16-Pob3 heterodimer whose activity is supported by the High Mobility Group B (HMGB)-like DNA-binding protein Nhp6 (Brewster et al. 2001; Formosa et al. 2001). Susceptibility of nucleosomal DNA to digestion by some restriction endonucleases in vitro increases dramatically in the presence of FACT (Xin et al. 2009), indicating that FACT either induces a structural change or stabilizes an existing alternative nucleosomal structure (Winkler and Luger 2011). We have called this activity “nucleosome reorganization” (Formosa 2008). FACT enhances binding of TATA sequences within nucleosomes by TATA-binding protein (TBP/Spt15) (Biswas et al. 2005), suggesting that increasing access to DNA through reorganization is a physiologically important role of FACT.
FACT can induce displacement of H2A-H2B dimers from nucleosomes under some conditions in vitro (Belotserkovskaya et al. 2003; Xin et al. 2009). However, increased nuclease sensitivity does not require H2A-H2B loss (Xin et al. 2009). Reorganized nucleosomes instead appear to have the same composition as canonical nucleosomes, but the components are associated with one another in a substantially different manner. This change in structure increases the probability of H2A-H2B dimer loss, but dimer loss appears to be just one possible result of reorganization, not its mechanism. The effect of FACT on nucleosomes is not common to other histone chaperones, and FACT-mediated reorganization does not require ATP hydrolysis so it is unlike ATP-dependent chromatin remodeling (Orphanides et al. 1998; Clapier and Cairns 2009). FACT is essential for viability (Formosa 2008; Lolas et al. 2010), but the detailed nature of reorganized nucleosomes, the role of FACT in forming and resolving alternative nucleosome forms, and the importance of H2A-H2B dimer displacement to FACT activity in vivo remain poorly understood.
Partial loss-of-function mutations in the genes encoding FACT subunits cause a range of defects in transcription, replication, and other processes (Lycan et al. 1994; O’Donnell et al. 2004, 2009; Formosa 2008). Some of these defects can be enhanced by decreasing histone acetylation (Formosa et al. 2002), by blocking histone H3-K4 methylation (Biswas et al. 2006), by mutating histone genes themselves (Vandemark et al. 2008), or by inactivating the Hir/Hpc complex that is involved in regulating histone gene expression and depositing nucleosomes outside of S phase (Formosa et al. 2002). Some features of chromatin therefore support FACT activity in vivo, and their loss makes FACT defects more difficult to tolerate. In contrast, other chromatin factors oppose FACT activity, as the phenotypes caused by some FACT gene mutations can be suppressed by preventing methylation of H3-K36 (Biswas et al. 2006) or by inactivating the chromodomain-helicase Chd1 (Biswas et al. 2008). FACT gene mutations can also either enhance or suppress defects caused by mutating other chromatin factors such as the histone H3 or the Swi/Snf remodeling complex (Malone et al. 1991; Duina et al. 2007). Furthermore, phenotypes caused by FACT gene mutations can be either enhanced or suppressed by altering the ratio of expression of H2A-H2B relative to H3-H4 (Formosa et al. 2002). The properties of chromatin therefore affect the efficiency of FACT function in vivo, but it remains unclear how these properties influence the central activity of FACT.
To examine the relationship between FACT and nucleosome reorganization in vivo, we sought histone gene mutations that could compensate for FACT defects. We reasoned that deficiency for FACT activity could be counterbalanced by mutating the histones in a way that makes nucleosomes easier to reorganize. Such mutations would provide insight into both the functions of FACT in vivo and the nature of nucleosome reorganization. Here we report that the temperature sensitivity (Ts) and hydroxyurea sensitivity caused by the spt16-11 allele of FACT can be suppressed by weakening the H2A-H2B dimer interface with (H3-H4)2 tetramers within nucleosomes, but this does not significantly affect the Spt− phenotype also caused by spt16-11. Purified nucleosomes with these mutated histones display elevated rates of spontaneous reorganization using nuclease sensitivity as an assay, and this partially compensates for Spt16-11 protein’s defect in achieving stable reorganization in vitro. These results provide new insights into the mechanism of FACT activity in vivo, supporting an important role for the stability of the histone dimer:tetramer interface in nucleosome reorganization and revealing a potential role for reorganization in replication fork progression.
Materials and Methods
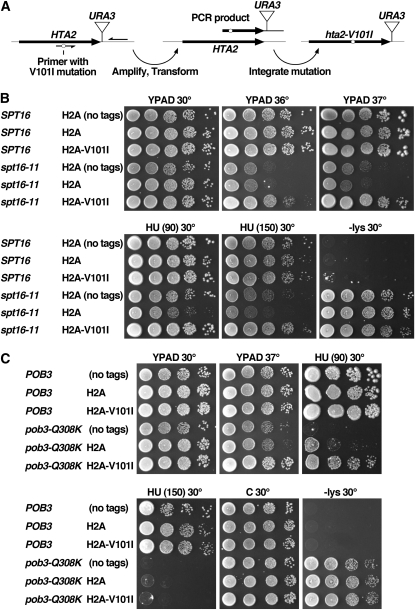
Strains are listed in Table 1 and in the supporting information, Table S1. W303 strains were derived from MSY1905 (kindly provided by M. Mitchell Smith, University of Virginia) by conversion of the KanMX marker replacing HHT1-HHF1 with HIS3 followed by standard crosses within the W303 background to introduce other mutations. A364a strains were constructed by integrating markers downstream of the histone genes and then amplifying the marked genomic locus by PCR using an upstream primer containing the desired mutation ∼30 nucleotides from the 5′ end and ∼25 nucleotides from the 3′ end and a downstream primer ∼200 bp distal to the marker (the strategy is outlined in Figure 3; the primers used are listed in Table S2; Toulmay and Schneiter 2006). The PCR product was used to transform a wild-type strain, selecting for transfer of the marker and then screening for cotransfer of the mutation by sequencing the entire histone gene. Standard crosses within the A364a background were then performed to obtain combinations of mutations.
Table 1 . Strains used.
| Strain | Genotype |
|---|---|
| Figure 1 | W303 background |
| DY10003 | MATaade2 can1 his3 leu2 trp1 ura3 spt16-11 hht1-hhf1-Δ(::HIS3) hht2-hhf2-∆(::KanMX) hta1-htb1-∆(::NatMX) hta2-htb2-∆(::HphMX) pJH33 (YCp URA3 HHT2-HHF2, HTA1-HTB1) |
| DY10004 | MATα ade2 can1 his3 leu2 trp1 ura3 spt16-11 hht1-hhf1-∆(::HIS3) hht2-hhf2-∆(::KanMX) hta1-htb1-∆(::NatMX) hta2-htb2-∆(::HphMX) pJH33 (YCp URA3 HHT2-HHF2, HTA1-HTB1) |
| Figure 3 | A364a background |
| 8127-7-4 | MATaura3-∆0 leu2-∆0 trp1-∆2 his3 lys2-128∂ |
| 8500-10-2 | MATaura3-∆0 leu2-∆0 trp1-∆2 his3 lys2-128∂ HTA1(220, His3MX) HTA2(30, URA3) |
| 8541-3-2 | MATaura3 leu2 trp1 his3 lys2-128∂ hta1-V101I(220, His3MX) hta2-V101I(30, URA3) |
| 8262-11-4 | MATaura3 leu2 trp1 his3 lys2-128∂ spt16-11 |
| 8554-5-3 | MATaura3 leu2 trp1 his3 lys2-128∂ spt16-11 HTA1(220, His3MX) HTA2(30, URA3) |
| 8541-4-2 | MATaura3 leu2 trp1 his3 lys2-128∂ hta1-V101I(220, His3MX) hta2-V101I(30, URA3) spt16-11 |
| 8127-7-4 | MATaura3-∆0 leu2-∆0 trp1-∆2 his3 lys2-128∂ |
| 8500-10-2 | MATaura3-∆0 leu2-∆0 trp1-∆2 his3 lys2-128∂ HTA1(220, His3MX) HTA2(30, URA3) |
| 8541-3-2 | MATaura3 leu2 trp1 his3 lys2-128∂ hta1-V101I(220, His3MX) hta2-V101I(30, URA3) |
| 8324-2-2 | MATaura3-∆0 leu2-∆0 trp1-∆2 his3 lys2-128∂ pob3-Q308K(LEU2) |
| 8500-2-2 | MATaura3-∆0 leu2-∆0 trp1-∆2 his3 lys2-128∂ HTA1(220, His3MX) HTA2(30, URA3) pob3-Q308K(LEU2) |
| 8555-4-2 | MATaura3 leu2 trp1 his3 lys2-128∂ hta1-V101I(220, His3MX) hta2-V101I(30, URA3) pob3-Q308K(LEU2) |
| Figure 4 | A364a background |
| 8483-9-1 | MATaura3-∆0 leu2-∆0 trp1-∆2 his3 lys2-128∂ HTB1(30, URA3) HTB2(30, His3MX) |
| 8606-3-4 | MATaura3 leu2 trp1 his3 lys2-128∂ htb1-A84D(30, URA3) HTB2(30, His3MX) |
| 8868-5-1 | MATaura3-∆0 leu2-∆0 trp1-∆2 his3 lys2-128∂ HTB1(30, URA3) htb2-A84D(30, His3MX) |
| 8442-4-3 | MATaura3-∆0 leu2-∆0 trp1-∆2 his3 lys2-128∂ htb1-A84D(30, URA3) htb2-A84D(30, His3MX) |
| 8482-5-3 | MATaura3 leu2 trp1 his3 lys2-128∂ HTB1(30, URA3) HTB2(30, His3MX) spt16-11 |
| 8606-2-1 | MATaura3 leu2 trp1 his3 lys2-128∂ htb1-A84D(30, URA3) HTB2(30, His3MX) spt16-11 |
| 8607-1-1 | MATaura3 leu2 trp1 his3 lys2-128∂ HTB1(30, URA3) htb2-A84D(30, His3MX) spt16-11 |
| 8455-8-4 | MATaura3 leu2 trp1 his3 lys2-128∂ htb1-A84D(30, URA3) htb2-A84D(30, His3MX) spt16-11 |
| 8625-3-4 | MATaura3 leu2 trp1 his3 lys2-128∂ HTB1(30, URA3) hta2-htb2-∆(::HIS3) spt16-11 |
| 8608-5-4 | MATaura3 leu2 trp1 his3 lys2-128∂ htb1-A84D(30, URA3) hta2-htb2-∆(::KanMX) spt16-11 |
| Table 2 | W303 background |
| DY9999 | MATaade2 can1 his3 leu2 trp1 ura3 hht1-hhf1-∆(::HIS3) hht2-hhf2-∆(::KanMX3) hta1-htb1-∆(::NatMX) hta2-htb2-∆(::HphMX) pJH33 (YCp URA3 HHT2-HHF2, HTA1-HTB1) |
| 8264-17-3 | MATaade2 can1 his3 leu2 trp1 ura3 pob3-Q308K hht1-hhf1-∆(::HIS3) hht2-hhf2-∆(::KanMX3) hta1-htb1-∆(::NatMX) hta2-htb2-∆(::HphMX) pJH33 (YCp URA3 HHT2-HHF2, HTA1-HTB1) |
Figure 3 .
H2A-V101I suppresses some phenotypes caused by two distinct FACT gene mutations. (A) Schematic showing the method used for integrating the hta2-V101I mutation into the genome (strategies for other genes shown in Figure S2 and Table S2). (B and C) Cultures of strains isogenic with the A364a genetic background and the additional genotypes shown (see Table 1 for the full genotypes) were tested as in Figure 1). “C” is complete synthetic medium; HU (90) and HU (150) are YPAD with 90 or 150 mm hydroxyurea. Growth on medium lacking lysine (−lys) reveals the Spt− phenotype in the strains with the lys2-128∂ allele (Simchen et al. 1984). Unless noted, all strains have marker genes (tags) inserted adjacent to the normal or mutated histone genes (Figure S2). Strains labeled “H2A-V101I” express this mutant protein from both HTA1 and HTA2 loci. Note that the severity of the Ts− and hydroxyurea sensitivity phenotypes caused by spt16-11 here is lower than in Figure 1 because the initial screen (Figure 1) was performed in the W303 genetic background where FACT gene mutations routinely cause stronger defects, whereas the integrants shown here were constructed in the A364a genetic background.
The S. cerevisiae genome contains two copies of each of the genes that encode the four core histones (Osley 1991). Plasmids pJH33, M4958, and M4959 carrying the genes HTA1-HTB1 and HHT2-HHF2 encoding histones H2A, H2B, H3, and H4 in vectors pRS316 (URA3), pRS315 (LEU2), or pRS314 (TRP1), respectively (Sikorski and Hieter 1989), were kindly provided by M. Mitchell Smith (University of Virginia) (Ahn et al. 2005). Mutagenized histone genes were obtained by amplifying M4958 using primers that align ∼400 bp into the vector sequences flanking the histone gene insert, resulting in a 5279-bp product with ∼400 bp of homology at each end to pRS414 or pRS415 (Figure 1). PCR was performed under standard conditions (100-µl reactions containing 20 pmol of each primer, 10 ng template, 0.2 mm each deoxynucleoside triphosphate (dNTP), 10 mm Tris–Cl pH 8.3, 1.5 mm MgCl2, 50 mm KCl; 30 cycles of 1 min at 94°, 1 min at 54°, 5 min at 72°) using Pfu polymerase (a small number of candidates were generated with a 1:200 mixture of Pfu:Taq polymerases, but most of these contained multiple mutations). Yeast strains DY10003 and DY10004 with the spt16-11 allele (Table 1) and carrying pJH33 were transformed with this PCR product mixed with vectors pRS414 (TRP1) or pRS415 (LEU2) (Christianson et al. 1992) that had been linearized with BamHI and HindIII. This yielded pLM04 (TRP1) and pTF238 (LEU2) derivatives by recombination in vivo. About 70,000 transformants were obtained and replica-plated to medium containing 5′-FOA to select for cells lacking the original wild-type histone plasmid (Boeke et al. 1987), thus demanding that the mutagenized plasmids expressed histones adequate for supporting viability. Strains surviving with only the mutagenized plasmids were replica-plated to 37° or to medium containing 120 mm hydroxyurea (HU), conditions nonpermissive for growth of the parent strains. About 2000 candidates were chosen for retesting, yielding several hundred suppressed strains. Plasmids were recovered from ∼100 of the strongest candidates, screened for the expected restriction digestion pattern, and used to transform DY10004 pJH33 to establish linkage of the suppressing mutation with the plasmid. Twenty-six of these plasmids were found to contain suppressor mutations and were sequenced using four primers to fully cover all four histone genes on each plasmid (Table S2). Eighteen of the plasmids had single mutations affecting 1 of 10 residues in H2A or H2B, distributed as shown in Table 2. Some of the 8 plasmids with multiple mutations also affected these same residues, and all plasmids had at least one mutation in H2A or H2B that mapped to the interface between H2A-H2B and H3-H4, but plasmids with complex mutations were not analyzed further. The screens for suppression of temperature sensitivity and HU sensitivity yielded overlapping results, so the resulting plasmids were combined for further analysis (Figure 1B).
Figure 1 .
Histone gene mutations can suppress defects caused by an spt16-11 mutation. (A) Scheme for mutagenizing histone genes. A plasmid carrying wild-type HHT2-HHF2 (H3-H4) and HTA1-HTB1 (H2A-H2B) was used as the template for PCR using primers TF04-25 and TF05-28 in the vector sequence flanking the histone gene insert (Table S2). The product was mixed with linearized vector DNA and used to transform DY10003 or DY10004 (Table 1). Recombination in vivo produced mutagenized histone gene plasmids. (B) Candidate plasmids with the single mutations indicated were recovered and used to transform strain DY10004 (Table 1), and then isolates lacking the wild-type histone gene plasmid were derived. Strains with only the mutated plasmid were grown to saturation, and aliquots of 10-fold serial dilutions were tested on YPAD (rich medium; Yeast Extract, Peptone, Adenine, Dextrose) at 25° or 34° or on HU (60) (YPAD with 60 mm hydroxyurea) at 25°.
Table 2 . Effects of spt16-11 suppressors in wild-type and pob3-Q308K strains.
| Histone | Mutation | No. of isolates | Phenotypes with pob3-Q308K | Phenotypes in wild type |
|---|---|---|---|---|
| H2A | hta1-A87T | 2 | ||
| H2A | hta1-R89G | 2 | Ts− HUs (enhanced defect) | Slight Ts−, Slight HUs, NaCl-s |
| H2A | hta1-N100D | 1 | Ts+ HUr (weak suppression) | |
| H2A | hta1-V101I | 1 | Ts+ HUr (weak suppression) | Slight Fmd-s |
| H2B | htb1-T55S | 2 | Fmd-s | |
| H2B | htb1-N66S | 1 | Fmd-s | |
| H2B | htb1-F68S | 2 | Ts− HUs (enhanced defect) | |
| H2B | htb1-T78P | 1 | Ts− HUs (enhanced defect) | |
| H2B | htb1-A80V | 2 | Ts− HUs (enhanced defect) | |
| H2B | htb1-A84V | 4 | Ts+ HUr (weak suppression) | |
| H2B | htb1-A84D | 1 | Ts− HUs (enhanced defect) | Mild Spt− |
| H2B | htb1-Y86C | 1 | Mixed weak effects | Slight Fmd-s, Slight HUs |
| H2B | htb1-I104F | 1 | Ts+ HUr (weak suppression) |
Strains 8264-17-3 (pob3-Q308K) and DY9999 (wild type) were transformed with LEU2 plasmids carrying the mutation indicated, and then transformants lacking the wild-type histone gene plasmid were screened for growth at elevated temperatures or on media containing 30–150 mm hydroxyurea, 1.2 M NaCl, 3% formamide, 10 mm caffeine, or 10 µg/ml camptothecin. The pob3-Q308K mutation causes Ts− and HUs phenotypes, and these were unaffected, enhanced, or suppressed as indicated. In all cases, the effects were small compared to other synthetic interactions observed with this allele. Moderate formamide sensitivity was observed where indicated (Fmd-s), with all other effects in the wild-type strain being weak. The number of times each mutation was isolated in the original screen for spt16-11 suppression is indicated as “No. of isolates.” HUs, hydroxyurea sensitive; HUr, hydroxyurea resistant; NaCl-s, NaCl sensitive.
Nucleosomes were reconstituted in vitro with recombinant histones, labeled with fluorescent dyes, and tested for binding affinity with FACT, sensitivity to DraI digestion, and retention of H2A-H2B dimers as described previously (Ruone et al. 2003; Rhoades et al. 2004; Xin et al. 2009). The spt16-11 (T828I, P859S) (Formosa et al. 2002), hta1-V101I, and htb1-A84D mutations were introduced into expression constructs using the Quikchange strategy (Stratagene), and the proteins were purified as previously described (Ruone et al. 2003; Rhoades et al. 2004; Xin et al. 2009).
Results
Defects caused by FACT mutants can be suppressed by histone mutants
The spt16-11 allele causes sensitivity to elevated temperatures, sensitivity to the replication toxin HU, and the Spt− phenotype (Formosa et al. 2001). The amount of Spt16 protein in an spt16-11 mutant drops about fivefold after a 3-hr incubation at 37° (Vandemark et al. 2008), so the Ts− phenotype probably reflects simple loss of FACT activity. HU inhibits ribonucleotide reductase (RNR), causing DNA replication to stall due to the shortage of dNTPs, but also leading to increased transcription of the genes encoding RNR. At least some FACT mutants retain normal induction of RNR gene transcription (Biswas et al. 2008; Formosa 2008), so HU sensitivity often reflects a defect in DNA replication, but indirect effects due to flawed transcription are also possible. The Spt− phenotype results from inappropriate transcription initiation start-site selection (Clark-Adams et al. 1988), indicating that spt16-11 causes a defect in transcription even at temperatures permissive for growth where Spt16 protein levels are normal. spt16-11 was chosen for the suppressor analysis described here because it causes a broad range of phenotypes associated with transcription and replication.
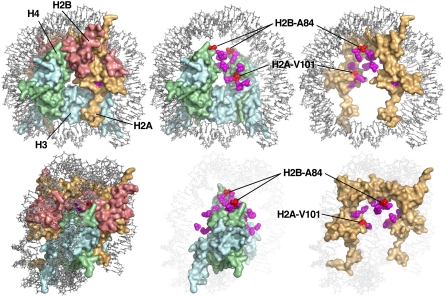
To detect histone gene mutations that can compensate for the defects caused by spt16-11, we constructed a strain with this mutation that lacked genomic versions of histone genes but instead carried a single copy of the genes encoding H2A, H2B, H3, and H4 on a plasmid (Materials and Methods). PCR amplification of the insert containing all four histone genes was used to introduce random mutations, and alleles capable of suppressing the Ts− or HU sensitivity phenotypes caused by spt16-11 were identified. This approach yielded 26 suppressing plasmids, 18 of which carried single mutations that altered 1 of 10 residues in H2A or H2B (Table 2, Figure 1B). H2B-A84 mutants were isolated five times (A84D four times and A84V once), but mutations affecting other residues were recovered only once or twice. The screen was therefore not saturated, but all of the mutated residues matched the same physical profile in that they affected residues that are buried within the histone octamer core on or near the surface of the H2A-H2B dimer that contacts the (H3-H4)2 tetramer (indicated in Figure 2 as either magenta or red residues). These results therefore strongly suggest that FACT activity in vivo involves disruption of the dimer:tetramer interface, that the spt16-11 mutation causes a defect in this function, and that weakening the interface by mutating residues in the interface reduces the requirement for efficient FACT. It should also be possible to interfere with this interface by mutating H3 or H4, so it is puzzling that no such mutations were identified, but it is possible that mutating the interface from the more highly conserved H3 or H4 protein side leads to inviability.
Figure 2 .
Histone gene mutations that suppress spt16-11 map to the dimer:tetramer interface. Histone residues identified in the suppressor screen are shown within the structure of a yeast nucleosome (PDB 1ID3) (White et al. 2001) as rendered in MacPyMOL (DeLano Scientific). Two orientations are shown (top and bottom panels) with a full nucleosome (left), H2A-H2B removed (center), or only H2A shown (right) to reveal buried sites. Sites of suppressor mutants are shown in magenta or red, with the latter indicating residues H2A-V101 and H2B-A84 that were chosen for further analysis.
Suppressors were isolated for the ability to reverse either the Ts− or the HU sensitivity caused by spt16-11, but each suppressor was found to suppress both phenotypes to variable but similar extents (Figure 1B). This suggests that both phenotypes have a common underlying defect. Suppression of spt16-11 was partially allele-specific, as some of the mutants weakly suppressed a pob3-Q308K mutation affecting the Pob3 subunit of FACT, but others had no effect and some even enhanced the defects caused by pob3-Q308K (Table 2, Figure S1A, Figure S4, and Figure 3). spt16-11 and pob3-Q308K therefore cause distinct defects in FACT function, and weakening the dimer:tetramer interface is strongly beneficial only to the cells with the spt16-11 deficiency. The histone gene mutations had little or no effect in an SPT16 wild-type strain (Table 2, Figure S1B). Altering the histones therefore caused a significant enough effect on nucleosome structure that an spt16-11 mutation was strongly suppressed, but the change was not sufficient to cause an obvious defect in an otherwise normal cell.
Integration of histone gene mutations reveals differences among expression contexts
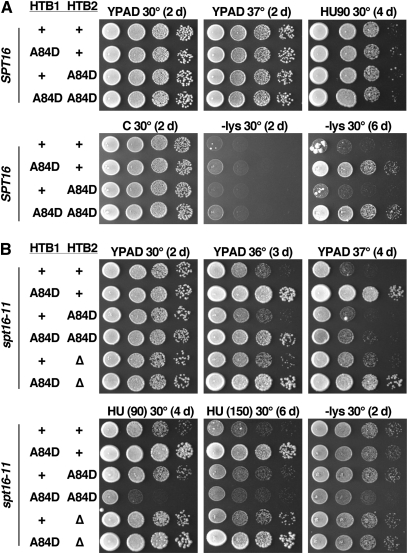
FACT mutants are sensitive to histone gene copy-number variation (Formosa et al. 2002), and it is difficult to maintain plasmids at uniform copy number throughout a population of cells. Furthermore, HTA1-HTB1 and HTA2-HTB2 encode slightly different amino acid sequences (Figure S3), transcription of each gene is regulated by different factors (Osley and Lycan 1987; Xu et al. 1992; Dollard et al. 1994; Hess and Winston 2005), and HTA1-HTB1 is essential for viability but HTA2-HTB2 is not (Formosa et al. 2002; Libuda and Winston 2006). Either the sequence differences or their transcription profiles under different conditions are therefore functionally important. To test the effects of the suppressing mutations in a more native context, we integrated two of the mutations into the genome at the endogenous loci. We chose H2A-V101I and H2B-A84D for this as they were two of the strongest suppressors of spt16-11 but they had opposite effects on the pob3-Q308K allele (Table 2).
We inserted selectable markers downstream of each gene that encodes H2A or H2B in separate wild-type strains (Figure S2). We then used genomic DNA from these strains to amplify each gene with the targeted mutation incorporated into one PCR primer (Figure 3A) (Toulmay and Schneiter 2006). The product carrying the desired mutation was used to transform a wild-type yeast strain, and transformants were screened by sequencing the entire histone gene to find strains in which the desired mutation but no unexpected additional mutation had been integrated into the genome along with the selectable marker. Finally, standard crosses were performed to obtain combinations of mutations.
Figure 3B shows the results obtained with the H2A-V101I expressed from both HTA1 and HTA2. Neither the integrated markers themselves (tags) nor the histone mutations affected the phenotypes tested in an SPT16 strain, but the tags themselves slightly enhanced the HU sensitivity and Spt− phenotypes (but not the Ts−) caused by spt16-11 (Figure 3B; compare rows 4 and 5: the Spt− phenotype is revealed by growth of these lys2-128∂ strains on medium lacking lysine). Marking the wild-type H2A genes therefore causes a minor defect in their expression; as noted previously and consistent with these results, a decrease in H2A-H2B expression is detrimental to some spt16 mutants (Formosa et al. 2002). To control for this effect, subsequent experiments always include marked versions of the H2A genes, whether wild type or mutant. This ensures that all comparisons are between strains that are as genetically matched as possible, although it remains possible that the markers could have a differential effect on mutant and wild-type alleles.
The integrated H2A-V101I mutation suppressed both the Ts− and hydroxyurea sensitivity phenotypes caused by spt16-11, but had only a slight effect on the Spt− phenotype (Figure 3B; compare rows 5 and 6). H2A-V101I therefore significantly corrects the spt16-11 defect most closely associated with DNA replication, but it has less effect on the transcription defect. Integrated H2A-V101I had no effect in a wild-type strain (Table 2, Figure S1B), but partially suppressed the Ts− and HU sensitivity caused by pob3-Q308K (Table 2, Figure S1A, Figure 3C), generally recapitulating the results from the plasmid-based assays.
In contrast, the locus expressing H2B-A84D expression significantly influenced the effect of this mutant. In a wild-type strain, expressing H2B-A84D from HTB1 caused a very mild Spt− phenotype (Figure 4A; compare the medium lacking lysine (−lys) growth at 2 and 6 days with the 2-day incubation of an spt16-11 mutant in Figure 4B). Expressing H2B-A84D from HTB2 did not cause this phenotype and did not enhance the effect of htb1-A84D (Figure 4A; compare rows 2 and 4). In an spt16-11 strain, the htb1-A84D allele suppressed the Ts− and hydroxyurea sensitivity phenotypes but did not affect the Spt− phenotype; this was true in both an HTA1htb1-A84D HTA2HTB2 strain (normal H2B available from the second copy of the gene) and an HTA1htb1-A84D hta2-htb2-∆ strain (only H2B-A84D available; Figure 4B; compare row 1 with 2 or 6). However, the htb2-A84D allele had no effect when paired with HTB1 (Figure 4B, row 3) and caused variable effects when paired with htb1-A84D (Figure 4B, rows 3 and 4). (HTB1 is essential for viability so the effect of htb1-∆ htb2-A84D could not be assessed.) Relative to htb1-A84D HTB2spt16-11, the htb1-A84D htb2-A84D spt16-11 strain displayed markedly reduced suppression of the Ts− phenotype, increased suppression of the Spt− phenotype, and enhanced sensitivity to low concentrations of HU (Figure 4B, rows 2 and 4).
Figure 4 .
The source of expression of H2B-A84D affects the resulting phenotypes. Isogenic strains from the A364a genetic background with the spt16-11 mutation (Table 1) were tested as in Figure 1, with the concentration of HU indicated. Panel A shows strains with the WT SPT16 allele and B shows strains with the spt16-11 mutation. Incubation times are listed in days to allow comparison of the level of growth at comparable times. In particular, htb1-A84D causes a weak Spt− phenotype, as very little growth on medium lacking lysine is observed after 2 days but substantial growth is visible after 6 days. This is weak relative to the effect of spt16-11, which causes substantial growth on medium lacking lysine after 2 days. Both HTB1 and HTB2 are tagged in all cases, whether wild-type (+) or mutant (A84D). “∆” indicates deletion of both HTA2 and HTB2.
The effects of the source of the H2B-A84D mutation are therefore quite complex and vary with the phenotype being observed. When challenged with HU or elevated temperature, expressing H2B-A84D only from the HTB1 locus was strongly advantageous to an spt16-11 strain (Figure 4B) but had no apparent effect in a wild-type strain (Figure 4A). However, supplying H2B-A84D only from the HTB2 locus under these stress conditions had no effect on either spt16-11 or wild-type strains. htb1-A84D caused the same effects whether normal HTB2 was available or not, suggesting that HTB2 expression is irrelevant in these tests. However, cells with both htb1-A84D and htb2-A84D mutations displayed distinct phenotypes compared to the single mutants, so HTB2 expression is important in this context. While both HTB1 and HTB2 are transcribed at similar levels under standard growth conditions (David et al. 2006), these results show that the two loci have distinct functions under some circumstances, as observed previously for the heat-shock response (Norris and Osley 1987) and the ability to suppress the effects of different Ty1 delta-element insertions (Clark-Adams et al. 1988). These differences may result from the slightly different proteins produced by the two genes (Figure S3), or they may indicate that the two genes are differentially activated by stress conditions. In any case, this shows that testing variants only from a plasmid-borne copy of HTB1 can produce an incomplete picture.
Histone mutations that suppress spt16-11 form unstable nucleosomes in vitro
The clustering of spt16-11 suppressors in the nucleosome structure (Figure 2) suggests that the Spt16-11 protein is defective in a process that includes disruption of the H2A-H2B interface with (H3-H4)2 tetramers. To investigate this possibility as well as other potential mechanisms, we reconstituted nucleosomes containing H2A-V101I or H2B-A84D and tested their stability in vitro. A 181-bp DNA fragment including the 146-bp sea urchin 5S rDNA nucleosome positioning sequence was labeled with Cy5 and assembled into nucleosomes using recombinant yeast histones expressed in bacteria as described previously (Xin et al. 2009). The H2A-Q114C mutation was introduced into the HTA1 gene to provide a unique cysteine residue for labeling this subunit with a maleimide derivative of the fluorescent dye Oregon Green 488 prior to assembly of octamers. The resulting nucleosomes therefore contained two fluorescent dyes that could be detected independently, allowing us to follow the DNA and H2A-H2B dimer components of the nucleosome separately.
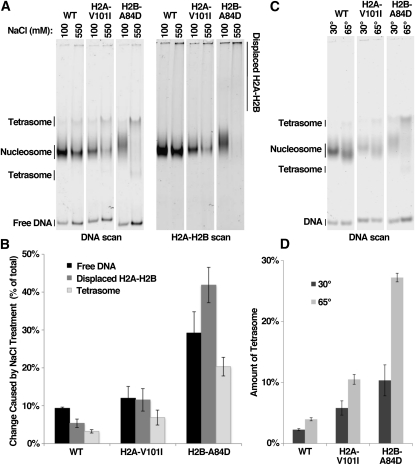
To test whether suppressor mutations affect the physical stability of nucleosomes, we used native polyacrylamide gel electrophoresis to measure the amount of H2A-H2B displaced from the nucleosomes, the amount of tetrasome formed, and the amount of free DNA released under various conditions. Over 90% of a sample of wild-type nucleosomes remained intact by all three measurements after a 1-hr incubation at 30° in 550 mm NaCl (Figure 5, A and B). In contrast, nucleosomes constructed with H2A-V101I lost dimers and formed tetrasomes more readily than wild type, and these effects were even more pronounced with H2B-A84D. In the latter case, even nucleosomes not exposed to high salt migrated aberrantly in native polyacrylamide gels, consistent with the observed spontaneous loss of dimers during preparation and storage (Figure 5A, lane 5 and not shown). Incubation of nucleosomes for 1 hr at 65° also caused low levels of tetrasome formation with wild-type nucleosomes, but substantially elevated levels for each mutant nucleosome (Figure 5, C and D). Thus, both H2A-V101I and H2B-A84D cause nucleosome instability in vitro, including an increase in dimer displacement.
Figure 5 .
Histone gene mutations destabilize nucleosomes in vitro. (A) Nucleosomes were constructed using recombinant yeast histones (normal or the mutant indicated) and a 181-bp 5S rDNA fragment with Cy5 at the 5′ end and Oregon Green 488 (Molecular Probes) attached to H2A residue C114 (originally Q114) (Xin et al. 2009). Samples were prepared in triplicate and incubated for 1 hr at 30° in 100 or 550 mm NaCl and then separated by electrophoresis through a native 4% polyacrylamide gel in 0.25× TBE as described (Xin et al. 2009). The single gel was then scanned to detect the fluorescent dyes on the DNA and the H2A independently using a Typhoon scanner (GE). One of the three repeats for each condition is shown. (B) Each lane was scanned and the amount of signal corresponding to free DNA, displaced H2A-H2B, or tetrasome forms was determined as a percentage of the total signal in the lane. The regions assigned as free DNA and tetrasomes are shown in the DNA scan in A, as determined using pure reference samples (tetrasomes migrate as two main bands using this combination of DNA and histones). The amount of displaced H2A-H2B was determined by calculating the signal in the top 40% of the H2A-H2B scan in A, as described previously (Xin et al. 2009). The change in the level of each form caused by treatment with high salt is plotted, with error bars indicating the standard deviation among the three samples. (C and D) As in A and B, except samples were incubated at 30° or 65° for 1 hr and only the total amount of tetrasomes detected is displayed.
FACT might either induce nucleosomes to reorganize or selectively bind to and stabilize spontaneously reorganized nucleosomes. In the first case, a weakened dimer:tetramer interface would make it easier to overcome resistance to reorganization, and in the second case, it would provide a larger subpopulation of complexes for binding. In either case, these results show that the stability of the dimer:tetramer interface is an important element of the core reaction promoted by FACT in vivo.
FACT(Spt16-11) has a nucleosome-binding defect in vitro
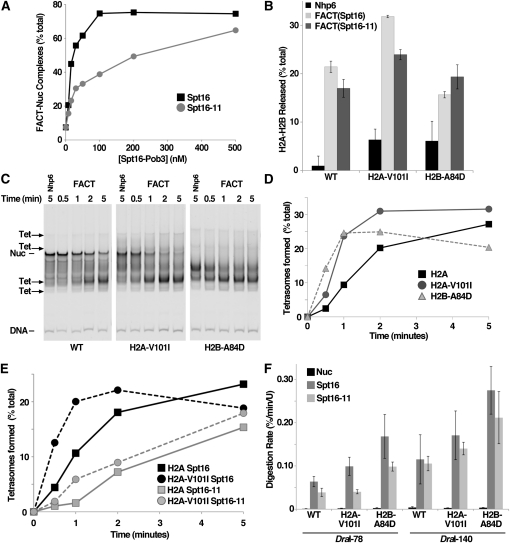
We used purified nucleosomes mixed with normal and mutant FACT complexes to examine the nature of the suppression observed in vivo. We first measured the apparent affinity of FACT for nucleosomes using an electrophoretic mobility shift assay (EMSA) (Rhoades et al. 2004). The DNA-binding protein Nhp6 is required for complex formation in this assay (Formosa et al. 2001; Xin et al. 2009), with 50% saturation occurring at ∼460 nM Nhp6 (Ruone et al. 2003). This same value was observed for both Spt16-Pob3 and (Spt16-11)-Pob3 with wild-type and mutant nucleosomes (not shown), so Spt16-11 and Spt16 proteins each require about the same amount of Nhp6 to support stable complex formation. In contrast, Spt16-11 was three- to fivefold less effective than Spt16 in this assay (Figure 6A). This could mean that Spt16-11 has a lower affinity for nucleosomes, that it is less effective in performing reorganization, or that it is unable to maintain the reorganized state stably enough for detection by EMSA. Nucleosomes with H2A-V101I or H2B-A84D were the same as wild-type H2A in this assay (Figure S5), so while this reveals a defect in Spt16-11 activity, it does not reveal the mechanism of genetic suppression.
Figure 6 .
Spt16-11 protein and its suppressors alter the rates of reactions on the basis of reorganization in vitro. (A) Nucleosomes constructed as in Figure 5 were mixed with different concentrations of Spt16-Pob3 heterodimers containing normal Spt16 or Spt16-11 protein, along with 10 µM Nhp6. Samples were incubated for 10 min at 30° and then separated by native PAGE, and the fraction of the total DNA signal in FACT–nucleosome complexes was determined as described previously (Xin et al. 2009). The nucleosomes shown contained the H2B-A84D mutation and are typical of results obtained with wild type and H2A-V101I. Multiple preparations of wild-type and mutant FACT were tested in independent experiments; values for half saturation varied somewhat between experiments, but the defect for Spt16-11 protein was reproducible. (B) Nucleosomes constructed as in Figure 5 were treated for 10 min at 30° with 5 µM Nhp6 alone, 5 µM Nhp6 and 200 nM Spt16-Pob3 [“FACT(Spt16)”], or 5 µM Nhp6 and (Spt16-11)-Pob3 [“FACT(Spt16-11)”]. Three independent samples for each condition were analyzed as in Figure 5, A and B, with the average and standard deviation of the three measurements presented. (C and D) Nucleosomes were constructed using recombinant histones from X. laevis (with normal histones or the mutation indicated) and a 255-bp MMTV DNA fragment (Flaus et al. 2004) labeled with Cy5. Samples were treated with FACT for the amount of time indicated, excess unlabeled genomic DNA was added to disrupt the FACT–nucleosome complexes, and then products were separated by native PAGE (Rhoades et al. 2004). Conversion from initial forms (“Nuc”) to the tetrasomal products (indicated by arrows) was quantitated and plotted in D. (E) As in D, except FACT(Spt16) or FACT(Spt16-11) were mixed with nucleosomes containing H2A or H2A-V101 as indicated. (F) Nucleosomes were constructed using wild-type or mutant yeast histones and 181-bp 5S rDNA fragments with DraI recognition sites 78 or 140 bp from the left edge of the nucleosome (Xin et al. 2009). The initial rate of digestion by DraI in the absence of other factors or with wild-type or mutant FACT was determined by examining samples taken at 8-min intervals with denaturing PAGE (Xin et al. 2009). Each condition was tested in three independent experiments with the average and standard deviation shown.
spt16-11 and histone mutants have opposing effects on reorganization rates
Reorganized nucleosomes are more prone to losing one or both H2A-H2B dimers in vitro (Belotserkovskaya et al. 2003; Xin et al. 2009). Nuclease sensitivity can be detected in complete octameric nucleosomes, so dimer loss is not a necessary feature of reorganization, but reorganization can lead to dimer loss so it serves as an assay for the ability of FACT to promote this structural change.
We measured the total dimer displacement after a 10-min incubation. As expected, FACT caused more dimer displacement than Nhp6 alone, but the levels for Spt16 and Spt16-11 were comparable (Figure 6B). Histone mutants displayed increased dimer loss in the presence of Nhp6 alone, consistent with the results described above showing inherent instability of these nucleosomes. Nucleosomes with wild-type histones or with the H2A-V101I protein displayed slightly lower dimer displacement with Spt16-11 than with Spt16, but nucleosomes with H2B-A84D had a similarly small but opposite effect. Spt16-11 therefore appears to have a small effect on dimer loss after a 10-min incubation.
We next examined the rate of dimer displacement. Instead of the 181-bp 5S rDNA nucleosomes containing recombinant yeast histones used above, we used a 255-bp MMTV (Mouse Mammary Tumor Virus) sequence (Flaus et al. 2004), recombinant Xenopus laevis histones, and low Mg2+ ion concentrations. These conditions increase the total dimer loss caused by FACT (Xin et al. 2009) and increase the electrophoretic separation of octameric and tetrameric nucleosomes. These nucleosomes migrate as one major band in native gels (Flaus et al. 2004) (Figure 6C, “Nuc”) along with several minor translational variants, while reconstructions show that tetrasomes migrate to four main bands (Figure 6C, “Tet”). FACT does not appear to promote translocation of nucleosomes (Rhoades et al. 2004), so we infer that the multiple forms observed with this larger MMTV fragment are corresponding pairs of octasomal and tetrasomal species occupying the same preferred translational positions.
Samples taken during a 5-min incubation show that FACT promotes tetrasome formation more rapidly with mutant histones than with wild-type histones (Figure 6, C–E). Conversely, FACT with Spt16-11 caused displacement more slowly than FACT with wild-type Spt16 protein (Figure 6E, Figure S6). Combining histone mutants with the FACT mutant resulted in a slight suppression of the Spt16-11 defect (Figure 6E, Figure S6), but the amount of compensation was minimal.
Together, these results show that FACT containing Spt16-11 protein has significant defects in vitro in forming stable complexes with nucleosomes and in promoting the normal rapid rate of dimer loss. Mutant histones identified as suppressors of the spt16-11 allele are lost from nucleosomes at an abnormally high rate, but do not strongly reverse the Spt16-11 defects in these assays.
Nuclease sensitivity reveals a potential mechanism of suppression
We have proposed that FACT promotes equilibration of nucleosomes between canonical and reorganized forms, with the rate of digestion by nucleases being proportional to the fraction of time that the nucleosomes spend in the reorganized state (Xin et al. 2009). We therefore used restriction endonuclease sensitivity to examine whether Spt16-11 and histone mutants alter the persistence of the reorganized state.
We measured the rates of DraI digestion of 181-bp 5S rDNA nucleosomes with yeast histones to probe two distinct physical contexts [DraI-78, near the dyad of symmetry, and DraI-140, near an entry/exit point, as described in Xin et al. (2009)]. Both FACT and FACT(Spt16-11) enhanced the rate of digestion near the dyad significantly, although FACT(Spt16-11) consistently produced less of an effect (Figure 6F). The histone mutants had the opposite effect of increasing the rate of DraI digestion. The same overall pattern was observed near the entry/exit points, although the rates of digestion were generally higher (Xin et al. 2009) and the defect of FACT(Spt16-11) was less significant. These results show that FACT(Spt16-11) can reorganize nucleosomes, but it either produces a structure with less accessibility to the DNA or fails to achieve or maintain the reorganized state for the normal length of time.
Notably, the combination of FACT(Spt16-11) with H2B-A84D nucleosomes produced a rate of digestion that is as high or higher than the rate with wild-type FACT combined with wild-type nucleosomes (Figure 6F). This also appears to be true for FACT(Spt16-11) with H2A-V101I nucleosomes at the entry/exit point, but not near the dyad. These two histones behave differently in vivo and may use overlapping but distinct mechanisms of suppression. These results suggest that FACT with Spt16-11 protein fails to achieve or maintain the open reorganized nucleosome form long enough to allow the normal rate of restriction endonuclease digestion, but this defect is counterbalanced by histone mutants that achieve this state more rapidly or maintain it for a longer amount of time than usual.
Discussion
We have shown that histone gene mutations can compensate for defects in FACT activity, with the spt16-11 mutation being suppressed by H2A-H2B mutants that destabilize the interface between these dimers and the (H3-H4)2 tetramer. In tests with purified components, these histone mutants caused increased dimer displacement in the absence of FACT and more rapid or more persistent nucleosome reorganization in the presence of FACT. Genetic suppression therefore appears to result from combining a FACT complex that is inefficient at nucleosome reorganization with nucleosomes that are more rapidly or more easily reorganized. By comparing the properties of the same FACT and histone gene mutations in vivo and in vitro, these results provide insight into the mechanism of FACT activity in its physiological settings. Initial reports indicated that FACT removes one H2A-H2B dimer from a nucleosome (Belotserkovskaya et al. 2003), and our experiments showed that dimer displacement can be an outcome of FACT action but is not a necessary feature of the mechanism of reorganization (Xin et al. 2009). Both sets of in vitro investigations indicate that FACT activity involves disruption of the H2A-H2B:(H3-H4)2 interface, and the results presented here confirm that this is an important feature of FACT function in vivo.
FACT activity has been implicated in both DNA replication and transcription (Formosa 2008). FACT’s ability to promote equilibration between stable and dynamic forms of histone:DNA complexes could reduce the barrier to polymerase progression posed by existing nucleosomes, but it could also promote establishment of chromatin through the reverse reaction by converting loosely associated components into mature nucleosomes. Consistent with a nucleosome assembly function, FACT has been implicated in the deposition of new nucleosomes after DNA replication (Belotserkovskaya et al. 2003; Vandemark et al. 2006) as well as in the reestablishment of repressive chromatin after transcription (Jamai et al. 2009). Defects in this nucleosome deposition activity are likely to be the cause of cryptic promoter activation (Kaplan et al. 2003), failed repression of SER3 expression (Hainer et al. 2010), and the Spt− phenotype (Malone et al. 1991; Rowley et al. 1991). Most FACT gene mutations (including spt16-11) cause phenotypes associated with chromatin quality defects, indicating that maintaining appropriately repressive chromatin throughout the genome requires optimal levels of FACT activity. The histone mutants described in this report did not suppress the Spt− phenotype caused by spt16-11, so by this assay they do not enhance the ability of the Spt16-11 protein to assemble normal chromatin.
Essentially all known mutations in FACT genes cause the Spt− phenotype, but only a subset of these mutations causes the HU sensitivity that is likely to reflect a defect in replication fork progression or stability. The results described here suggest that spt16-11 causes HU sensitivity because Spt16-11 protein is unable to promote nucleosome reorganization rapidly enough to allow a normal rate of fork progression through a normal chromatin template. The additional delay that results from decreased availability of dNTPs when RNR is inhibited by HU might be intolerable when FACT is defective. Histone mutants that promote faster nucleosome reorganization by destabilizing the histone dimer:tetramer interface then diminish the impaired progression and restore viability under HU stress, but do not resolve the defect in chromatin quality, so the Spt− phenotype persists.
Our tests of an H2B mutation integrated into the genome reveal a clear functional difference between HTB1 and HTB2. In normal cells, each gene is transcribed at about the same level on about the same cell cycle schedule. However, the A84D mutant is effective only at suppressing the HU sensitivity (likely to be a replication defect) caused by spt16-11 if it is expressed from HTB1 alone. Curiously, the effects of htb1-A84D are similar when it is paired with normal HTB2 or a deletion of HTB2. In contrast, expressing H2B-A84D from both loci begins to suppress the Spt− phenotype (transcription defect) caused by spt16-11, but enhances the HU sensitivity (probable replication defect). These differences could be due to the different primary sequences of Htb1 and Htb2 proteins, or they could indicate that the two genes have different expression profiles under some circumstances such as nucleosome replacement outside of S phase. In either case, Htb1 and Htb2 proteins have different functional roles, and this becomes particularly important in an spt16-11 strain. It is therefore necessary to consider the expression context when examining the effects of histone gene mutations, especially when they affect H2B.
The results presented here provide support for the model that FACT activity promotes or requires destabilization of histone dimer:tetramer interaction. The analysis suggests that promoting the reversible, rapid oscillation of nucleosomes between a stable canonical form and a more open reorganized form is a key function of FACT in vivo, highlighting the dynamic nature of nucleosomes under physiological conditions.
Acknowledgments
We thank Mitch Smith for providing the initial strain used in our screen, Liz Kendall and Peter Winters for technical assistance, and members of the Formosa and Stillman labs for valuable input. This project was supported by a grant from the National Institutes of Health to T.F. and D.J.S.
Literature Cited
- Ahn S. H., Cheung W. L., Hsu J. Y., Diaz R. L., Smith M. M., et al. , 2005. Sterile 20 kinase phosphorylates histone H2B at serine 10 during hydrogen peroxide-induced apoptosis in S. cerevisiae. Cell 120: 25–36 [DOI] [PubMed] [Google Scholar]
- Belotserkovskaya R., Oh S., Bondarenko V. A., Orphanides G., Studitsky V. M., et al. , 2003. FACT facilitates transcription-dependent nucleosome alteration. Science 301: 1090–1093 [DOI] [PubMed] [Google Scholar]
- Biswas D., Yu Y., Prall M., Formosa T., Stillman D. J., 2005. The yeast FACT complex has a role in transcriptional initiation. Mol. Cell. Biol. 25: 5812–5822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas D., Dutta-Biswas R., Mitra D., Shibata Y., Strahl B. D., et al. , 2006. Opposing roles for Set2 and yFACT in regulating TBP binding at promoters. EMBO J. 25: 4479–4489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas D., Takahata S., Xin H., Dutta-Biswas R., Yu Y., et al. , 2008. A role for Chd1 and Set2 in negatively regulating DNA replication in Saccharomyces cerevisiae. Genetics 178: 649–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., Trueheart J., Natsoulis G., Fink G. R., 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154: 164–175 [DOI] [PubMed] [Google Scholar]
- Brewster N. K., Johnston G. C., Singer R. A., 2001. A bipartite yeast SSRP1 analog comprised of Pob3 and Nhp6 proteins modulates transcription. Mol. Cell. Biol. 21: 3491–3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson T. W., Sikorski R. S., Dante M., Shero J. H., Hieter P., 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110: 119–122 [DOI] [PubMed] [Google Scholar]
- Clapier C. R., Cairns B. R., 2009. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78: 273–304 [DOI] [PubMed] [Google Scholar]
- Clark-Adams C. D., Norris D., Osley M. A., Fassler J. S., Winston F., 1988. Changes in histone gene dosage alter transcription in yeast. Genes Dev. 2: 150–159 [DOI] [PubMed] [Google Scholar]
- David L., Huber W., Granovskaia M., Toedling J., Palm C. J., et al. , 2006. A high-resolution map of transcription in the yeast genome. Proc. Natl. Acad. Sci. USA 103: 5320–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollard C., Ricupero-Hovasse S. L., Natsoulis G., Boeke J. D., Winston F., 1994. SPT10 and SPT21 are required for transcription of particular histone genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 14: 5223–5228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duina A. A., Rufiange A., Bracey J., Hall J., Nourani A., et al. , 2007. Evidence that the localization of the elongation factor Spt16 across transcribed genes is dependent upon histone H3 integrity in Saccharomyces cerevisiae. Genetics 177: 101–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaus A., Rencurel C., Ferreira H., Wiechens N., Owen-Hughes T., 2004. Sin mutations alter inherent nucleosome mobility. EMBO J. 23: 343–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa T., 2008. FACT and the reorganized nucleosome. Mol. Biosyst. 4: 1085–1093 [DOI] [PubMed] [Google Scholar]
- Formosa T., Eriksson P., Wittmeyer J., Ginn J., Yu Y., et al. , 2001. Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J. 20: 3506–3517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa T., Ruone S., Adams M. D., Olsen A. E., Eriksson P., et al. , 2002. Defects in SPT16 or POB3 (yFACT) in Saccharomyces cerevisiae cause dependence on the Hir/Hpc pathway: polymerase passage may degrade chromatin structure. Genetics 162: 1557–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainer S. J., Pruneski J. A., Mitchell R. D., Monteverde R. M., Martens J. A., 2010. Intergenic transcription causes repression by directing nucleosome assembly. Genes Dev. 25: 29–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess D., Winston F., 2005. Evidence that Spt10 and Spt21 of Saccharomyces cerevisiae play distinct roles in vivo and functionally interact with MCB-binding factor, SCB-binding factor and Snf1. Genetics 170: 87–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamai A., Puglisi A., Strubin M., 2009. Histone chaperone spt16 promotes redeposition of the original h3-h4 histones evicted by elongating RNA polymerase. Mol. Cell 35: 377–383 [DOI] [PubMed] [Google Scholar]
- Kaplan C. D., Laprade L., Winston F., 2003. Transcription elongation factors repress transcription initiation from cryptic sites. Science 301: 1096–1099 [DOI] [PubMed] [Google Scholar]
- Libuda D. E., Winston F., 2006. Amplification of histone genes by circular chromosome formation in Saccharomyces cerevisiae. Nature 443: 1003–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lolas I. B., Himanen K., Gronlund J. T., Lynggaard C., Houben A., et al. , 2010. The transcript elongation factor FACT affects Arabidopsis vegetative and reproductive development and genetically interacts with HUB1/2. Plant J. 61: 686–697 [DOI] [PubMed] [Google Scholar]
- Lycan D., Mikesell G., Bunger M., Breeden L., 1994. Differential effects of Cdc68 on cell cycle-regulated promoters in Saccharomyces cerevisiae. Mol. Cell. Biol. 14: 7455–7465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone E. A., Clark C. D., Chiang A., Winston F., 1991. Mutation in SPT16/CDC68 suppress cis- and trans-acting mutations that affect promoter function in Saccharomyces cerevisiae. Mol. Cell. Biol. 11: 5710–5717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris D., Osley M. A., 1987. The two gene pairs encoding H2A and H2B play different roles in the Saccharomyces cerevisiae life cycle. Mol. Cell. Biol. 7: 3473–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell A. F., Brewster N. K., Kurniawan J., Minard L. V., Johnston G. C., et al. , 2004. Domain organization of the yeast histone chaperone FACT: the conserved N-terminal domain of FACT subunit Spt16 mediates recovery from replication stress. Nucleic Acids Res. 32: 5894–5906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell A. F., Stevens J. R., Kepkay R., Barnes C. A., Johnston G. C., et al. , 2009. New mutant versions of yeast FACT subunit Spt16 affect cell integrity. Mol. Genet. Genomics 282: 487–502 [DOI] [PubMed] [Google Scholar]
- Orphanides G., LeRoy G., Chang C.-H., Luse D. S., Reinberg D., 1998. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 92: 105–116 [DOI] [PubMed] [Google Scholar]
- Osley M. A., 1991. The regulation of histone synthesis in the cell cycle. Annu. Rev. Biochem. 60: 827–861 [DOI] [PubMed] [Google Scholar]
- Osley M. A., Lycan D., 1987. Trans-acting regulatory mutations that alter transcription of Saccharomyces cerevisiae histone genes. Mol. Cell. Biol. 7: 4204–4210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinberg D., Sims R. J., III, 2006. de FACTo nucleosome dynamics. J. Biol. Chem. 281: 23297–23301 [DOI] [PubMed] [Google Scholar]
- Rhoades A. R., Ruone S., Formosa T., 2004. Structural features of nucleosomes reorganized by yeast FACT and its HMG box component, Nhp6. Mol. Cell. Biol. 24: 3907–3917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley A., Singer R. A., Johnston G., 1991. CDC68, a yeast gene that affects regulation of cell proliferation and transcription, encodes a protein with a highly acidic carboxyl terminus. Mol. Cell. Biol. 11: 5718–5726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruone S., Rhoades A. R., Formosa T., 2003. Multiple Nhp6 molecules are required to recruit Spt16-Pob3 to form yFACT complexes and reorganize nucleosomes. J. Biol. Chem. 278: 45288–45295 [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P., 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simchen G., Winston F., Styles C. A., Fink G. R., 1984. Ty-mediated gene expression of the LYS2 and HIS4 genes of Saccharomyces cerevisiae is controlled by the same SPT genes. Proc. Natl. Acad. Sci. USA 81: 2431–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulmay A., Schneiter R., 2006. A two-step method for the introduction of single or multiple defined point mutations into the genome of Saccharomyces cerevisiae. Yeast 23: 825–831 [DOI] [PubMed] [Google Scholar]
- VanDemark A. P., Blanksma M., Ferris E., Heroux A., Hill C. P., et al. , 2006. The structure of the yFACT Pob3-M domain, its interaction with the DNA replication factor RPA, and a potential role in nucleosome deposition. Mol. Cell 22: 363–374 [DOI] [PubMed] [Google Scholar]
- VanDemark A. P., Xin H., McCullough L., Rawlins R., Bentley S., et al. , 2008. Structural and functional analysis of the Spt16p N-terminal domain reveals overlapping roles of yFACT subunits. J. Biol. Chem. 283: 5058–5068 [DOI] [PubMed] [Google Scholar]
- White C. L., Suto R. K., Luger K., 2001. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 20: 5207–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler D. D., Luger K., 2011. The histone chaperone FACT: structural insights and mechanisms for nucleosome reorganization. J. Biol. Chem. 286: 18369–18374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin H., Takahata S., Blanksma M., McCullough L., Stillman D. J., et al. , 2009. yFACT induces global accessibility of nucleosomal DNA without H2A–H2B displacement. Mol. Cell 35: 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Kim U. J., Schuster T., Grunstein M., 1992. Identification of a new set of cell cycle-regulatory genes that regulate S-phase transcription of histone genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 12: 5249–5259 [DOI] [PMC free article] [PubMed] [Google Scholar]