Abstract
Zinc-finger nucleases (ZFNs) are targetable DNA cleavage reagents that have been adopted as gene-targeting tools. ZFN-induced double-strand breaks are subject to cellular DNA repair processes that lead to both targeted mutagenesis and targeted gene replacement at remarkably high frequencies. This article briefly reviews the history of ZFN development and summarizes applications that have been made to genome editing in many different organisms and situations. Considerable progress has been made in methods for deriving zinc-finger sets for new genomic targets, but approaches to design and selection are still being perfected. An issue that needs more attention is the extent to which available mechanisms of double-strand break repair limit the scope and utility of ZFN-initiated events. The bright prospects for future applications of ZFNs, including human gene therapy, are discussed.
Genetics is driven by the ability to connect genotype with phenotype. The classical approach is to identify a novel phenotype, whether occurring spontaneously or derived by mutagenesis, to identify the responsible gene(s) and to discover why mutations at that locus have the observed effect. A more modern approach, sometimes called reverse genetics, is to identify a gene from a genomic sequence to make mutations specifically in that gene and to characterize the resulting phenotype.
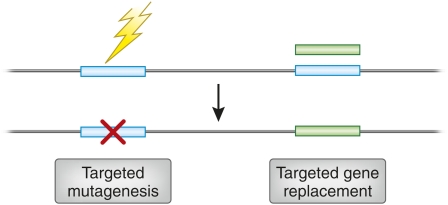
Two types of gene-specific manipulations can be envisioned (Figure 1). In one, which we can call “targeted gene replacement,” the goal is to make localized sequence changes, often ones that will create a null mutation. In targeted gene replacement, the goal is to replace an existing sequence with one designed in the laboratory. The latter allows the introduction of both more subtle and more extensive alterations.
Figure 1 .
Illustration of two types of genome engineering. In the top portion, the horizontal line represents a genome segment, and the open rectangles, two individual genes. The jagged arrow on the left indicates an unspecified mutagenic agent targeted to one gene. The shaded rectangle on the right is a manipulated version of the second gene that has been supplied by the experimenter. The outcomes below are targeted mutagenesis, resulting in a localized sequence alteration (“x”), and targeted gene replacement, produced by homologous recombination between the original and exogenous gene copies.
Making directed genetic changes is often called “gene targeting.” It sounds simple enough, but targeting a single gene within a large genome presents a substantial challenge. Procedures for gene replacement in baker’s yeast, Saccharomyces cerevisiae, have been available for several decades (Scherer and Davis 1979; Rothstein 1983). Success in this case depends on several features: the ability to manipulate segments of yeast DNA in the laboratory, the ability to introduce DNA into yeast cells, interaction between donor and target DNA by homologous recombination, the near absence of competing reactions that would integrate the donor into alternative sites in the genome, and the ability to apply strong selection for the desired product. These properties are shared by some other fungi and many bacteria, but not by the majority of eukaryotic organisms.
Making targeted gene replacements has also become standard practice in mice, thanks to the availability of embryonic stem (ES) cells that can be manipulated in culture and the development of powerful selection procedures (Capecchi 2005). Like targeting in yeast, the process in mice depends on homologous recombination between the donor and the target. In addition, selection must be applied against the more common products of random integration. This is accomplished by placing a positive selectable marker inside the donor homology and a negative selectable marker outside the homology (Mansour et al. 1988). Double selection yields the desired replacements, and the pluripotency of the ES cells allows them to populate all cell lineages after injection into early embryos.
In both yeast and mouse cells, the absolute frequency of homologous recombination between donor and target sequences is quite low—on the order of one in every 104 to 107 cells. Selection in culture allows the recovery of the rare cells that have enjoyed the desired event. With other experimental organisms, ES cells are not available, screening or selection procedures are not adequate, and development of useful gene-targeting approaches is impeded by the low frequency of recombination.
Stimulating Gene Targeting With Double-Strand Breaks
The challenge in extending gene targeting to other organisms and situations could be viewed largely as one of increasing the frequency of recombination. This could be done a priori by manipulating the donor DNA, the genomic target, or the genetic background. Both in yeast and in murine ES cells, a linear donor DNA is more efficient than a circular donor. This makes sense, as DNA ends are typically recombinagenic, but the effect is rather modest. Increasing the amount of donor DNA has little effect, and in mammalian cells seems largely to increase the frequency of nonhomologous integration (Vasquez et al. 2001). The use of oligonucleotide donors to introduce very localized changes has been somewhat successful, but the high frequencies claimed in early reports have not proved robust or reproducible. Some attempts have been made to increase the levels of proteins involved in recombination reactions, again with limited success.
The greatest impediment to efficient targeting is the fact that an intact target is essentially inert. This has been demonstrated by damaging the target and observing increased levels of recombination. Early experiments showed that DNA-damaging agents stimulated homologous exchanges between sister chromatids (Latt 1981). Most compelling, however, were studies showing that a single double-strand break (DSB) dramatically increased the frequency of local recombination.
These experiments were inspired by the discovery that natural recombination events, including meiotic crossing over and mating-type switching in yeast, are initiated by DSBs. In this approach, pioneered by Haber in yeast (Rudin et al. 1989; Plessis et al. 1992) and by Jasin and others in mammalian cells (Rouet et al. 1994; Choulika et al. 1995), a recognition site for a very specific DNA endonuclease was inserted at a unique site in the genome and then cut by introduction of the corresponding enzyme. Recombination with a homologous donor DNA was stimulated by several orders of magnitude. Other means of damaging the target have also shown some utility, but nothing as effective as making a DSB.
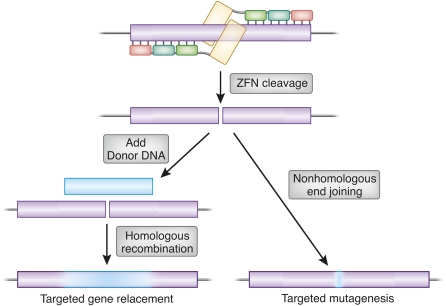
Chromosomal breaks are detected in cells as potentially lethal damage, and one natural pathway of DSB repair is copying from a homologous template. From this perspective, DSB-stimulated gene targeting simply provides an exogenous template for a natural repair process. An alternative repair pathway for DSBs, nonhomologous end joining, often joins the broken ends inaccurately, creating deletions, insertions, and substitutions at the break site. Thus, both mutagenesis and gene replacement are stimulated locally by DSBs (Figure 2).
Figure 2 .
Repair outcomes of a genomic double-strand break, illustrated for the case of ZFN cleavage. A pair of three-finger ZFNs is shown at the top in association with a target gene (open box). If a homologous donor DNA is provided (solid box, left), repair can proceed by homologous recombination using the donor as template. The amount of donor sequence ultimately incorporated will typically decline with distance from the original break, as illustrated by the shading. Alternatively, the break can be repaired by nonhomologous end joining, leading to mutations at the cleavage site. These may be deletions, insertions, and base substitutions, usually quite localized, but sometimes extending away from the break.
Addressable Gene-Targeting Reagents
The prototype enzymes for demonstrating DSB stimulation of gene targeting were I-SceI and HO, both of which have long recognition sites (18 bp for I-SceI, 24 bp for HO). While they provided very useful information on the efficiency and mechanisms of DSB repair, they were limited in their utility because their recognition sites had to be inserted in the genome by a low-efficiency process before they could be used to effect high-efficiency recombination. Reagents were needed that could be designed to attack arbitrarily chosen, preexisting genomic sequences.
A number of research groups focused on small compounds that would find their targets essentially by base recognition. These included oligonucleotides that could form DNA triplexes by adding a synthetic strand to a duplex target (Chin and Glazer 2009). Variations on the theme included peptide nucleic acids that substitute a peptide backbone for the usual sugar–phosphate linkage (Kim et al. 2006) and synthetic compounds designed to recognize base pairs with novel functional groups (Doss et al. 2006). These recognition moieties were linked to reactive groups that would cut or locally damage the DNA, thereby stimulating repair by homologous recombination. These efforts have yielded some success, but they are limited in the number of target sequences that they can access, and the frequencies of site-specific damage have not been consistently high.
Another approach has been to modify the recognition specificity of enzymes such as I-SceI (homing endonucleases, also called meganucleases) (Ashworth et al. 2006; Pâques and Duchateau 2007). This has proved very successful in some cases, but the intimate connection between the recognition and cleavage elements in the protein structures makes it challenging to alter one without affecting the other.
Zinc-Finger Nucleases
The class of targeting reagents that has proved the most versatile and effective in recent years is that of the zinc-finger nucleases (ZFNs), which have separate DNA-binding and DNA-cleavage domains (Figures 3 and 4). These synthetic proteins originated in the observation by Chandrasegaran that the natural type IIS restriction enzyme, FokI, has physically separable binding and cleavage activities (Li et al. 1992). The cleavage domain has no apparent sequence specificity, and Chandrasegaran showed that cutting could be redirected by substituting alternative recognition domains for the natural one (Kim and Chandrasegaran 1994; Kim et al. 1996, 1998). The most useful of these was a set of Cys2His2 zinc fingers (ZFs) in which each unit of ∼30 amino acids bound a single atom of zinc. The crystal structure of a set of three fingers bound to DNA showed that each finger contacts primarily 3 bp of DNA in remarkably modular fashion (Pavletich and Pabo 1991). This suggested that many different sequences could be attacked by making novel assemblies of ZFs.
Figure 3 .
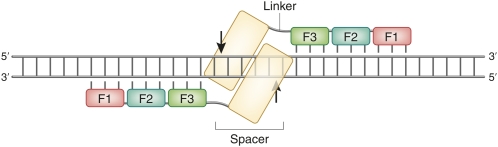
Illustration of a pair of ZFNs bound to DNA. Zinc fingers are shown as open boxes, with short vertical lines indicating the main contacts with the DNA base pairs. FokI cleavage domains are shown as shaded boxes, with common cleavage sites, spaced by 4 bp, and indicated by vertical arrows. Zinc fingers are numbered from the N terminus. The linker between the binding and cleavage domains of one protein is labeled. The spacer between the zinc-finger binding sites, 6 bp in this case, is also indicated.
Figure 4 .
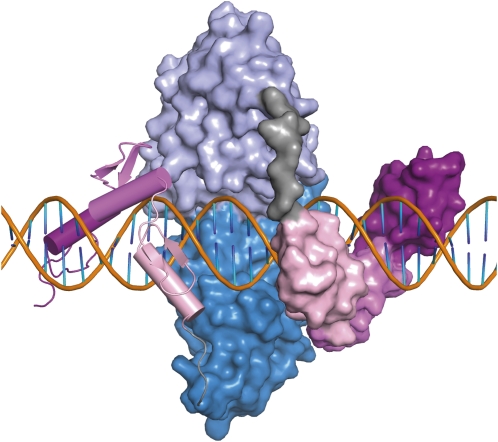
Model of a pair of ZFNs bound to DNA. Each zinc finger is shown in a shade of pink, in ribbon representation on the left and space-filling representation on the right. The FokI cleavage domains are shown in shades of blue. The four-amino-acid linker between the binding and cleavage domains is gray. DNA is shown with the sugar–phosphate backbone in orange and the bases in orange and blue. The separation between ZF binding sites is 6 bp. This model (Smith et al. 2000) was compiled from crystal structures of zinc fingers bound to DNA (Protein Database 1MEY) and the FokI restriction endonuclease in the absence of DNA (2FOK). I am grateful to Dr. Frank Whitby for help with the modeling.
Although it was not recognized initially (Kim et al. 1996), the FokI cleavage domain must dimerize to cut DNA (Bitinaite et al. 1998; Smith et al. 2000). The dimer interface is weak, and the best way to achieve cleavage is to construct two sets of fingers directed to neighboring sequences and join each to a monomeric cleavage domain (Figure 3). When both sets of fingers bind to their recognition sequences, high local concentration facilitates dimerization and cleavage. Several studies have shown that the optimum configuration uses a short linker between the domains of the protein and a spacer of 5 or 6 bp (7 can also work) between binding sites that lie in inverted orientation (Bibikova et al. 2001; Handel et al. 2009; Shimizu et al. 2009).
The requirement for dimerization is a great advantage for this reason: because a monomer is not active, cleavage does not occur at single binding sites. The cleavage reagent is assembled only at the target if the fingers have adequate specificity, and the combined requirement for binding two proteins brings the overall specificity into a very useful range; e.g., two three-finger proteins specify the location of 18 bp, which is sufficient, in principle, to pick out a single target, even in a complex genome.
Gene Targeting With ZFNs
The first ZFNs were created as chimeric restriction endonucleases and were shown to have in vitro activity (Kim et al. 1996). It was not clear that the prokaryotic cleavage domain would be able to act on DNA assembled into chromatin, but experiments in Xenopus oocytes with a synthetic, extrachromosomal substrate and ZFNs of known specificity showed very high efficiency of cleavage and recombination (Bibikova et al. 2001). The first success with a ZFN pair designed de novo for a genomic target occurred in Drosophila. Both targeted mutagenesis (Bibikova et al. 2002) and targeted gene replacement (Bibikova et al. 2003) were demonstrated at the yellow locus in the soma and, most importantly, in the germline. Since then, ZFN pairs have been designed, constructed, and used successfully for individual genes in quite a variety of organisms and cell types (Table 1). While the frequencies of target modification vary, yields in the vicinity of 10% of all targets are quite common.
Table 1 . Reported instances of successful ZFN-induced gene targeting.
| Organism | Latin name | Method | TM | TGR | References |
|---|---|---|---|---|---|
| Animals | |||||
| Fruit fly | Drosophila melanogaster | Heat-shock induction | + | + | Bibikova et al. (2002, 2003), Beumer et al. (2006) |
| Embryo injection | + | + | Beumer et al. (2008) | ||
| Nematode | C. elegans | Gonad injection | + | Morton et al. (2006) | |
| Silkworm | Bombyx mori | Embryo injection | + | Takasu et al. (2010) | |
| Zebrafish | Danio rerio | Zygote injection | + | Meng et al. (2008), Doyon et al. (2008), Foley et al. (2009) | |
| Sea urchin | Hemicentrotus pulcherrimus | Embryo injection | + | Ochiai et al. (2010) | |
| Frog | Xenopus tropicalis | Embryo injection | + | Young et al. (2011) | |
| Rat | Rattus norvegicus | Zygote injection | + | + | Geurts et al. (2009), Mashimo et al. (2010) |
| Mouse | Mus musculus | Zygote injection | + | + | Meyer et al. (2010), Carbery et al. (2010), Cui et al. (2011) |
| Plants | |||||
| Cress | A. thaliana | Agrobacterium | + | Carbery et al. (2010), Cui et al. (2011), Lloyd et al. (2005), Zhang et al. (2010), Osakabe et al. (2010), De Pater et al. (2009) | |
| Tobacco | Nicotiana sp. | Protoplasts | + | + | Wright et al. (2005), Townsend et al. (2009) |
| Agrobacterium | + | + | Cai et al. (2009) | ||
| Viral delivery | + | Marton et al. (2010) | |||
| Maize | Zea mays | Cell culture | + | + | Shukla et al. (2009) |
| Petunia | Petunia sp. | Viral delivery | + | Marton et al. (2010) | |
| Mammalian cells in culture | |||||
| Human | Homo sapiens | DNA transformation | + | + | Porteus and Baltimore (2003), Urnov et al. (2005), Alwin et al. (2005), Perez et al. (2008), Hockemeyer et al. (2009), Kim et al. (2009), Zou et al. (2009), Dekelver et al. (2010) |
| Viral delivery | + | + | Lombardo et al. (2007) | ||
| Mouse | M. musculus | DNA transformation | + | + | Goldberg et al. (2010), Connelly et al. (2010) |
| Hamster | Cricetulus griseus | DNA transformation | + | + | Santiago et al. (2008), Liu et al. (2010), Cost et al. (2010) |
| Pig | Sus domestica | DNA transformation | + | Watanabe et al. (2010) | |
TM refers to targeted mutagenesis by nonhomologous end joining TGR is targeted gene replacement by homologous recombination. In addition to the examples shown here, I have heard reliable, but unpublished, reports of successful ZFN-induced targeting in several other organisms. The list of references is not exhaustive, but provides guidance to key publications.
Key to these successes have been methods for the delivery of the ZFNs and, when desired, a donor DNA. In cultured cells, expression constructs for ZFNs use promoters appropriate to the cell type and vectors that can be introduced by transfection of DNA or infection by viruses. The same methods also serve to introduce the donor. In Drosophila, early experiments relied on genomic integration of ZFN-coding sequences and donor DNA via P-element-mediated transformation (Bibikova et al. 2002, 2003; Beumer et al. 2006). This required rather elaborate strain construction, and a welcome breakthrough occurred when it was demonstrated that excellent efficiencies of both homologous and nonhomologous events could be obtained by injecting ZFN mRNAs and donor DNA into embryos (Beumer et al. 2008).
Embryo injection of mRNAs for ZFN expression has proved practical in several other organisms. This is a well-established method in zebrafish, and very usable frequencies of ZFN-induced mutagenesis have been achieved in quite a number of genes (Doyon et al. 2008; Meng et al. 2008; Foley et al. 2009). Recent experiments with embryos of rat (Geurts et al. 2009; Mashimo et al. 2010), mouse (Carbery et al. 2010; Meyer et al. 2010), sea urchin (Ochiai et al. 2010), and frog (Young et al. 2011) have resulted in similar success. Injection into silkworm embryos, very much in parallel with the Drosophila method, also works (Takasu et al. 2010). Homologous recombination with donor DNA has been achieved in rats and mice (Meyer et al. 2010; Cui et al. 2011). In all these cases, viable adults carrying germline mutations were grown from the treated embryos.
In other organisms, more specialized approaches to delivery have been taken. In plants—both the favored experimental cress, Arabidopsis thaliana, and some crop species—ZFN expression was achieved by delivering coding sequences under the control of a viral promoter by agrobacterial transformation (Lloyd et al. 2005; Cai et al. 2009; De Pater et al. 2009; Osakabe et al. 2010; Zhang et al. 2010). Direct DNA transformation (Wright et al. 2005; Cai et al. 2009; Shukla et al. 2009; Townsend et al. 2009) and viral delivery (Marton et al. 2010) have also succeeded in plants.
Various studies have also revealed some of the challenges of delivering the targeting materials. Initial experiments with Caenorhabditis elegans achieved high levels of somatic mutagenesis in targets both in the genome and on extrachromosomal arrays by using a heat-shock promoter to drive ZFN expression from a DNA template (Morton et al. 2006). Parallel expression in the germline was undetectable, presumably due to suppression by well-known RNA interference mechanisms. It must be possible to escape this limitation, but it has certainly proved challenging.
As noted in Table 1, ZFN-targeted mutagenesis has been achieved in many cases, but gene replacement has not occurred in all of them. In at least some situations, this is not for lack of trying. Despite obviously high efficiencies of cleavage and mutagenesis in zebrafish, no homologous gene replacement has yet been reported. There is no problem with co-injecting a plausible donor DNA, yet recombination with the cut target does not ensue. It appears that DSB repair is different in different cell types and developmental stages, and novel strategies, based on an understanding of the biology of each system, will be necessary to overcome the limitations encountered.
Genetics of Gene Targeting
In most targeting systems, little effort has been made to understand in any detail nor to manipulate the molecular processes of DNA repair. It seems very likely that the standard processes of homologous recombination and nonhomologous end joining operate in most situations, but there could be important variations and specialized components that could be adjusted. One study in Drosophila Bozas et al. (2009) showed that most of the homologous replacement was dependent on the usual suspects—Rad51 (spnA in Drosophila) and Rad54 (okr)—but that a significant minority apparently proceeded by a Rad51-independent process, presumably single-strand annealing. Much of the nonhomologous end joining depended on the specialized DNA ligase, Lig4, and in its absence, repair shifted strongly toward homologous events. This feature also characterized the mRNA injection protocol, and larger yields of gene replacement products were obtained from injection of lig4− embryos (Beumer et al. 2008). In both situations, however, it was clear that some mutant end-joined products were recovered in the absence of Lig4, indicating the presence of an alternative pathway. These observations should help inform experiments in other systems, although the roles of the various components may differ.
It may also be possible to influence the balance between homologous and nonhomologous events by providing activities that encourage the former. Zebrafish embryos may lack Rad51 or other essential components. Perhaps recombination proteins from simpler systems could be introduced along with the ZFNs and donor DNA.
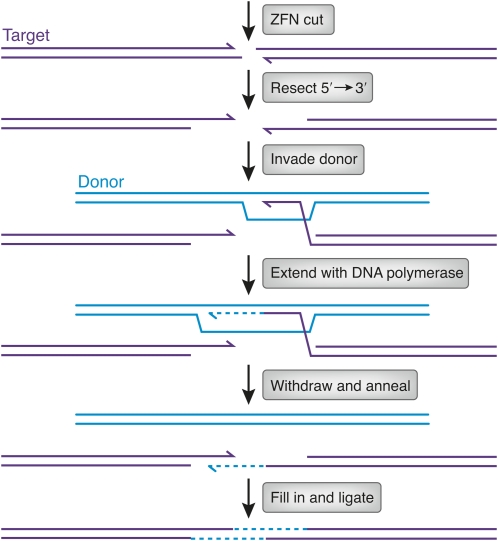
Another feature of gene replacement that needs analysis is the extent of conversion from the donor DNA during recombination. In many organisms, including Drosophila, homologous recombination proceeds by a mechanism known as synthesis-dependent strand annealing (Kurkulos et al. 1994; Nassif et al. 1994) (Figure 5). The ends at the target break are resected in the 5′ → 3′ direction, leaving a 3′-ending single strand that invades the donor. This 3′ end is extended by DNA polymerase for some distance and then withdraws and anneals with the other end from the break. The extent of donor sequence ultimately incorporated depends on the extent of synthesis, the degree of degradation of target sequence, and the direction of mismatch repair in the final heteroduplex. While each of these contributions is unknown, the lengths of ultimate conversion tracts have been measured in relevant experiments. They are quite long in Drosophila; several kilobases of donor are incorporated, albeit at decreasing frequency at greater distances from the break (Nassif et al. 1994). In mammalian cells, similar experiments revealed very short tracts, so that beyond ∼100–200 bp from the break, very little donor sequence appears after repair (Elliott et al. 1998). A thorough understanding of the homologous recombination process could reveal approaches to enlarging these tracts.
Figure 5 .
Illustration of the synthesis-dependent strand annealing mechanism of homologous recombination. After ZFN cleavage, the ends of the target DNA are resected by 5′ → 3′ exonuclease action (3′ ends are shown with half arrowheads). One of the resulting single-stranded 3′ ends invades homologous sequences in the donor (thick lines). The invading 3′ end is extended by DNA polymerase (dashed line). After some synthesis, the extended end withdraws and anneals to the other end at the original break. The gaps are filled in (dashed lines; thick lines denote donor sequence, thin lines target sequence), and continuity of the strands is restored by ligation. The extent of donor sequence incorporated at the target depends on (1) the extent of synthesis after invasion, (2) whether the invading 3′ end had been chewed back, and (3) the direction of mismatch repair in the heteroduplex formed by annealing.
ZFN Specificity
Up to this point I have made it seem that there is a smooth path from ZFN design to targeted genetic modifications. In fact, a substantial proportion of ZFN pairs fail, whether they are produced by design or selection (Ramirez et al. 2008; Joung et al. 2010; Kim et al. 2010). Even scientists at Sangamo Biosciences and Sigma-Aldrich, who have access to the largest and best-characterized archive of ZFs, make multiple pairs for sequences within a single target gene and test them extensively.
It can be effective in some cases to treat fingers as independent modules and assemble them in new combinations for new targets (Carroll et al. 2006). Subtle effects of context can, however, defeat this approach. Methods for selecting new three-finger sets from partially randomized libraries have been developed (Meng et al. 2007; Maeder et al. 2008), but can be quite time-consuming. Sangamo designs ZFNs using libraries of two-finger modules (Moore et al. 2001), which addresses the context issue. Members of the Zinc Finger Consortium have recently derived fingers for some DNA triplets that work well in neighbor combination (Sander et al. 2011), and a group at ToolGen describes the individual fingers in their collection that are best behaved in modular assembly (Kim et al. 2011). Continued experience should provide deeper insight into critical features of ZF recognition.
Another issue is the affinity of a particular ZF set. At least three fingers in each ZFN are required to provide adequate affinity, but not all fingers make equal contributions. More fingers can be added, and examples up to six fingers have been used. It is also possible that some genomic regions, even particular sequences within a single gene, are inaccessible due to compact chromatin structure, DNA modification, or other factors. Chromatin structure is responsible, for example, for preventing cleavage of intact recognition sites by the HO endonuclease during mating-type switching in S. cerevisiae (Rusche et al. 2003). This would be difficult to assess in many situations, and it has not been addressed experimentally for any ZFN target. It is possible that ZFN cleavage occurs largely during S phase of the cell cycle, when all genomic sequences are exposed for replication. Experiments with ZFNs in definitively nondividing cells would be very informative in this regard. In many ways, we are fortunate that the ZF framework comes from natural transcription factors that must find their targets within a chromatin context.
Specificity of ZF binding is another challenge. Some fingers bind equally well to triplets other than their supposed preference, and even the best ones have some affinity for related sequences. Adding fingers can improve specificity, as well as affinity, but there is also the possibility that subsets of fingers in a polydactyl domain will mediate binding to off-target sites. Separating two-finger modules with a very short linker has been shown to improve specificity (Moore et al. 2001), and this is the approach used routinely by Sangamo and Sigma-Aldrich.
When off-target cleavage is extensive, the number of breaks outstrips the DNA repair capacity and leads to death of the treated cells or organisms (Bibikova et al. 2002; Porteus and Baltimore 2003; Alwin et al. 2005). Typically, a single member of a ZFN pair is responsible for most of this toxicity (Beumer et al. 2006). The effect has been greatly ameliorated by the introduction of substitutions in the dimer interface of the cleavage domain that prevent homodimerization, but allow heterodimers to form (Miller et al. 2007; Szczepek et al. 2007; Sollu et al. 2010). In some situations, the efficiency of cleavage is reduced by these modifications, but they seem quite effective in other contexts. New designs that retain activity while suppressing homodimerization have been reported very recently (Doyon et al. 2011).
For use in genetic analysis of model organisms, a low frequency of off-target cleavage and mutagenesis is tolerable, since the desired allele can usually be isolated by repeated out-crossing. When ZFNs are contemplated for use in human gene therapy, much greater care must be taken to avoid potentially harmful unintended genome alterations. The ZFNs employed in a current clinical trial have been selected, refined, and tested extensively (Perez et al. 2008; Urnov et al. 2010). Still, it would be useful to have a direct analysis of where off-target cuts occur, even at very low levels.
Prospects for ZFN-Based Gene Targeting
In principle, any gene in any organism can be targeted with a properly designed pair of ZFNs. Zinc-finger recognition depends only on a match to DNA sequence, and mechanisms of DNA repair, both homologous recombination and nonhomologous end joining, are shared by essentially all species. As noted above, methods for effective delivery of ZFNs and donor DNA will differ among applications, and biological variations in the availability of particular DNA repair pathways may affect the outcome. Nonetheless, it seems very likely that ZFN-based targeting will be applied to additional organisms in the future, including ones of economic and medical importance.
Among experimental organisms, ZFN technology is having its greatest impact on species that previously had no effective gene-targeting procedure. The community of zebrafish investigators has adopted this approach for creating gene knockouts, and it is hoped that continuing research will uncover methods that encourage homologous gene replacement. The rat was a favored mammalian model for physiological research, but lost ground to the mouse when powerful genetic methods, including gene targeting, were developed for the latter. Now embryo injection of ZFN mRNAs offers the prospect of creating targeted mutations in the rat (Geurts et al. 2009; Mashimo et al. 2010). ZFNs could also be used in conjunction with the recently described rat ES cell procedures (Tong et al. 2010). The very recent demonstration of mutagenesis and gene replacement in mouse by embryo injection of ZFNs (Carbery et al. 2010; Meyer et al. 2010) indicates that the time-consuming ES cell procedures could be avoided in some instances.
Applications of ZFNs to crop plants create alterations in normal genomic loci, which may prove more acceptable to consumers than strains genetically modified by gene addition. Delivery will be a key issue. Both tobacco (Townsend et al. 2009) and maize (Shukla et al. 2009), the species favored to date, can be regrown from cells or callus that have been modified in culture by ZFNs. Pursuit of targeting studies in Arabidopsis should help define the best methods to use in other species.
The current clinical trial, involving ZFN knockout of the CCR5 gene, represents the first of many possible therapeutic applications to humans (Urnov et al. 2010). The CCR5 protein is a coreceptor for HIV-1. Natural human variants lacking the protein are healthy and resist progression to AIDS after HIV infection. The clinical protocol involves isolation of T-cell precursors, high-efficiency mutagenesis of the CCR5 gene with a ZFN pair, expansion of the cells, and reimplantation back to the donor. This provides an HIV-resistant population to reconstitute the patient’s immune system.
Future treatments based on culture and transplantation are easy to envision. Hematopoietic stem cells are well suited to this approach. Other precursor cells—human ES and induced pluripotent stem (iPS) cells, for example—are also excellent candidates, once they have been thoroughly characterized and their requirements for proper differentiation are revealed. The prospects for therapies that require delivery of ZFNs, and perhaps donor DNA, to intact tissues seem more distant, particularly if efficacy would require modification of a substantial fraction of affected cells. In such cases, one can imagine germline modifications performed in conjunction with in vitro fertilization, much as has been achieved in rats and mice. At present, such an approach is likely to cause more harm than good, with the prospect of off-target cleavage and possibly other unforeseen consequences.
Closing Comments
Like other breakthroughs, both technical and conceptual, the development of ZFNs as gene targeting tools has depended on prior discoveries from unrelated sources. It has been important to know that DNA double-strand breaks are both recombinagenic and mutagenic. Study of the FokI restriction enzyme revealed that it consisted of two functionally separable domains, which opened the door to manipulating its specificity. The discovery of zinc fingers and their modular association with DNA identified them as prime candidates for targeting moieties with a broad range of specificities.
I will note in passing that another DNA-recognition module with promising characteristics has recently been identified. The TAL effector domain, found in some Xanthomonas proteins involved in manipulating host gene expression, is composed of modules of ∼34 amino acids, each of which contacts a single base pair (Boch et al. 2009; Moscou and Bogdanove 2009). Fusions of TAL domains to the FokI nuclease domain (TALNs or TALENs) direct cleavage to specific sites both in vitro and in vivo (Christian et al. 2010; Li et al. 2011; Miller et al. 2011). Whether these modules will prove to have similar or greater utility than zinc fingers remains to be explored.
The extension of ZFN technology to other organisms and situations will depend on both practical and mechanistic studies. Delivery may be largely a trial-and-error issue in many cases. Finding optimal conditions for mutagenesis and gene replacement likely will require more molecular and genetic analysis. Remarkably, the first description of genomic modification with ZFNs appeared <10 years ago, and progress has accelerated dramatically in the last few years. The prospects for continuing developments seem bright.
Acknowledgments
I am grateful to the people who have worked with me over the years on ZFNs, including those who, through collaboration, have opened the door to organisms previously unfamiliar to me. I also acknowledge the outstanding scientists working on ZFN development and applications, in both the public and private sectors, who have pushed the technology forward and freely shared their progress with me. Research in my lab has been supported by National Institutes of Health awards GM58504 and GM078571, by a contract from Dow AgroSciences, and by core facilities funded in part by the University of Utah Cancer Center Support Grant.
Literature Cited
- Alwin S., Gere M. B., Gulh E., Effertz K., Barbas C. F., III, et al. , 2005. Custom zinc-finger nucleases for use in human cells. Mol. Ther. 12: 610–617 [DOI] [PubMed] [Google Scholar]
- Ashworth J., Havranek J. J., Duarte C. M., Sussman D., Monnat R. J., Jr, et al. , 2006. Computational redesign of endonuclease DNA binding and cleavage specificity. Nature 441: 656–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer K., Bhattacharyya G., Bibikova M., Trautman J. K., Carroll D., 2006. Efficient gene targeting in Drosophila with zinc finger nucleases. Genetics 172: 2391–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer K. J., Trautman J. K., Bozas A., Liu J.-L., Rutter J., et al. , 2008. Efficient gene targeting in Drosophila by direct embryo injection with zinc-finger nucleases. Proc. Natl. Acad. Sci. USA 105: 19821–19826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M., Carroll D., Segal D. J., Trautman J. K., Smith J., et al. , 2001. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol. Cell. Biol. 21: 289–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M., Golic M., Golic K. G., Carroll D., 2002. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics 161: 1169–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M., Beumer K., Trautman J. K., Carroll D., 2003. Enhancing gene targeting with designed zinc finger nucleases. Science 300: 764. [DOI] [PubMed] [Google Scholar]
- Bitinaite J., Wah D. A., Aggarwal A. K., Schildkraut I., 1998. FokI dimerization is required for DNA cleavage. Proc. Natl. Acad. Sci. USA 95: 10570–10575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch J., Scholze H., Schornack S., Landgraf A., Hahn S., et al. , 2009. Breaking the code of DNA binding specificity of TAL-Type III effectors. Science 326: 1509–1512 [DOI] [PubMed] [Google Scholar]
- Bozas A., Beumer K. J., Trautman J. K., Carroll D., 2009. Genetic analysis of zinc-finger nuclease-induced gene targeting in Drosophila. Genetics 182: 641–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C. Q., Doyon Y., Ainley W. M., Miller J. C., DeKelver R. C., et al. , 2009. Targeted transgene integration in plant cells using designed zinc finger nucleases. Plant Mol. Biol. 69: 699–709 [DOI] [PubMed] [Google Scholar]
- Capecchi M. R., 2005. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat. Rev. Genet. 6: 507–512 [DOI] [PubMed] [Google Scholar]
- Carbery I. D., Ji D., Harrington A., Brown V., Weinstein E. J., et al. , 2010. Targeted genome modification in mice using zinc-finger nucleases. Genetics 186: 451–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D., Morton J. J., Beumer K. J., Segal D. J., 2006. Design, construction and in vitro testing of zinc finger nucleases. Nat. Protoc. 1: 1329–1341 [DOI] [PubMed] [Google Scholar]
- Chin J. Y., Glazer P. M., 2009. Repair of DNA lesions associated with triplex-forming oligonucleotides. Mol. Carcinog. 48: 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choulika A., Perrin A., Dujon B., Nicolas J.-F., 1995. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol. Cell. Biol. 15: 1968–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M., Cermak T., Doyle E. L., Schmidt C., Zhang F., et al. , 2010. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186: 757–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly J. P., Barker J. C., Pruett-Miller S., Porteus M. H., 2010. Gene correction by homologous recombination with zinc finger nucleases in primary cells from a mouse model of a generic recessive genetic disease. Mol. Ther. 18: 1103–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cost G. J., Freyvert Y., Vafiadis A., Santiago Y., Miller J. C., et al. , 2010. BAK and BAX deletion using zinc-finger nucleases yields apoptosis-resistant CHO cells. Biotechnol. Bioeng. 105: 330–340 [DOI] [PubMed] [Google Scholar]
- Cui X., Ji D., Fisher D. A., Wu Y., Briner D. M., et al. , 2011. Targeted integration in rat and mouse embryos with zinc-finger nucleases. Nat. Biotechnol. 29: 64–67 [DOI] [PubMed] [Google Scholar]
- DeKelver R. C., Choi V. M., Moehle E. A., Paschon D. E., Hockemeyer D., et al. , 2010. Functional genomics, proteomics, and regulatory DNA analysis in isogenic settings using zinc finger nuclease-driven transgenesis into a safe harbor locus in the human genome. Genome Res. 20: 1133–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pater S., Neuteboom L. W., Pinas J. E., Hooykaas P. J., van der Zaal B. J., 2009. ZFN-induced mutagenesis and gene-targeting in Arabidopsis through Agrobacterium-mediated floral dip transformation. Plant Biotechnol. J. 7: 821–835 [DOI] [PubMed] [Google Scholar]
- Doss R. M., Marques M. A., Foister S., Chenoweth D. M., Dervan P. B., 2006. Programmable oligomers for minor groove DNA recognition. J. Am. Chem. Soc. 128: 9074–9079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon Y., McCammon J. M., Miller J. C., Faraji F., Ngo C., et al. , 2008. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat. Biotechnol. 26: 702–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon Y., Vo T. D., Mendel M. C., Greenberg S. G., Wang J., et al. , 2011. Enhancing zinc-finger-nuclease activity with improved obligate heterodimer architectures. Nat. Methods 8: 74–79 [DOI] [PubMed] [Google Scholar]
- Elliott B., Richardson C., Winderbaum J., Nickoloff J. A., Jasin M., 1998. Gene conversion tracts from double-strand break repair in mammalian cells. Mol. Cell. Biol. 18: 93–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley J. E., Yeh J. R., Maeder M. L., Reyon D., Sander J. D., et al. , 2009. Rapid mutation of endogenous zebrafish genes using zinc-finger nucleases made by Oligomerized Pool ENgineering (OPEN). PLoS ONE 4: e4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts A. M., Cost G. J., Freyvert Y., Zeitler B., Miller J. C., et al. , 2009. Knockout rats via embryo microinjection of zinc-finger nucleases. Science 325: 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg A. D., Banaszynski L. A., Noh K.-M., Lewis P. W., Elsaesser S. J., et al. , 2010. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell Stem Cell 140: 678–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Händel E.-M., Alwin S., Cathomen T., 2009. Expanding or restricting the target site repertoire of zinc-finger nucleases: the inter-domain linker as a major determinant of target site selectivity. Mol. Ther. 17: 104–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D., Soldner F., Beard C., Gao Q., Mitalipova M., et al. , 2009. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol. 27: 851–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J. K., Voytas D. F., Cathomen T., 2010. Reply to “Genome editing with modularly assembled zinc-finger nucleases.” Nat. Methods 7: 91–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. J., Lee H. J., Kim H., Cho S. W., Kim J.-S., 2009. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 19: 1279–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.-S., Lee H. J., Carroll D., 2010. Genome editing with modularly assembled zinc-finger nucleases. Nat. Methods 7: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. H., Nielsen P. E., Glazer P. M., 2006. Site-specific gene modification by PNAs conjugated to psoralen. Biochemistry 45: 314–323 [DOI] [PubMed] [Google Scholar]
- Kim S., Lee M. J., Kim H., Kang M., Kim J.-S., 2011. Preassembled zinc-finger arrays for rapid construction of ZFNs. Nat. Methods 8: 7. [DOI] [PubMed] [Google Scholar]
- Kim Y.-G., Chandrasegaran S., 1994. Chimeric restriction endonuclease. Proc. Natl. Acad. Sci. USA 91: 883–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.-G., Cha J., Chandrasegaran S., 1996. Hybrid restriction enzymes: zinc finger fusions to FokI cleavage domain. Proc. Natl. Acad. Sci. USA 93: 1156–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.-G., Smith J., Durgesha M., Chandrasegaran S., 1998. Chimeric restriction enzyme: Gal4 fusion to FokI cleavage domain. Biol. Chem. 379: 489–495 [DOI] [PubMed] [Google Scholar]
- Kurkulos M., Weinberg J. M., Roy D., Mount S. M., 1994. P element-mediated in vivo deletion analysis of white-apricot: deletions between direct repeats are strongly favored. Genetics 136: 1001–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latt S. A., 1981. Sister chromatic exchange formation. Annu. Rev. Genet. 15: 11–55 [DOI] [PubMed] [Google Scholar]
- Li L., Wu L. P., Chandrasegaran S., 1992. Functional domains in FokI restriction endonuclease. Proc. Natl. Acad. Sci. USA 89: 4275–4279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Huang S., Jiang W. Z., Wright D., Spalding M. H., et al. , 2011. TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res. 39: 359–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P.-Q., Chan E., Cost G. J., Zhang L., Wang J., et al. , 2010. Generation of a triple-gene knockout mammalian cell line using engineered zinc-finger nucleases. Biotechnol. Bioeng. 106: 97–105 [DOI] [PubMed] [Google Scholar]
- Lloyd A., Plaisier C. L., Carroll D., Drews G. N., 2005. Targeted mutagenesis using zinc-finger nucleases in Arabidopsis. Proc. Natl. Acad. Sci. USA 102: 2232–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo A., Genovese P., Beausejour C. M., Colleoni S., Lee Y.-L., et al. , 2007. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat. Biotechnol. 25: 1298–1306 [DOI] [PubMed] [Google Scholar]
- Maeder M. L., Thibodeau-Beganny S., Osiak A., Wright D. A., Anthony R. M., et al. , 2008. Rapid “Open-Source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol. Cell 31: 294–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour S. L., Thomas K. R., Capecchi M. R., 1988. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature 336: 348–352 [DOI] [PubMed] [Google Scholar]
- Marton I., Zuker A., Shklarman E., Zeevi V., Tovkach A., et al. , 2010. Non-transgenic genome modification in plant cells. Plant Physiol. 154: 1079–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashimo T., Takizawa A., Voigt B., Yoshimi K., Hiai H., et al. , 2010. Generation of knockout rats with X-linked severe combined immunodeficiency (X-SCID) using zinc-finger nucleases. PLoS ONE 5: e8870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Thibodeau-Beganny S., Jiang T., Joung J. K., Wolfe S. A., 2007. Profiling the DNA-binding specificities of engineered Cys2His2 zinc finger domains using a rapid cell-based method. Nucleic Acids Res. 35: e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Noyes M. B., Zhu L. J., Lawson N. D., Wolfe S. A., 2008. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat. Biotechnol. 26: 695–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M., Hrabé de Angelis M., Wurst W., Kühn R., 2010. Gene targeting by homologous recombination in mouse zygotes mediated by zinc-finger nucleases. Proc. Natl. Acad. Sci. USA 107: 15022–15026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. C., Holmes M. C., Wang J., Guschin D. Y., Lee Y.-L., et al. , 2007. An improved zinc-finger nuclease architecture for highly specific genome cleavage. Nat. Biotechnol. 25: 778–785 [DOI] [PubMed] [Google Scholar]
- Miller J. C., Tan S., Qiao G., Barlow K. A., Wang J., et al. , 2011. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol . 29: 143–148 [DOI] [PubMed] [Google Scholar]
- Moore M., Klug A., Choo Y., 2001. Improved DNA binding specificity from polyzinc finger peptides by using strings of two-finger units. Proc. Natl. Acad. Sci. USA 98: 1437–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton J., Davis M. W., Jorgensen E. M., Carroll D., 2006. Induction and repair of zinc-finger nuclease-targeted double-strand breaks in Caenorhabditis elegans somatic cells. Proc. Natl. Acad. Sci. USA 103: 16370–16375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscou M. J., Bogdanove A. J., 2009. A simple cipher governs DNA recognition by TAL effectors. Science 326: 1501. [DOI] [PubMed] [Google Scholar]
- Nassif N. A., Penney J., Pal S., Engels W. R., Gloor G. B., 1994. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol. Cell. Biol. 14: 1613–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiai H., Fujita K., Suzuki K., Nishikawa M., Shibata T., et al. , 2010. Targeted mutagenesis in the sea urchin embryo using zinc-finger nucleases. Genes Cells 15: 875–885 [DOI] [PubMed] [Google Scholar]
- Osakabe K., Osakabe Y., Toki S., 2010. Site-directed mutagenesis in Arabidopsis using custom-designed zinc finger nucleases. Proc. Natl. Acad. Sci. USA 107: 12034–12039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pâques F., Duchateau P., 2007. Meganucleases and DNA double-strand break-induced recombination: perspectives for gene therapy. Curr. Gene Ther. 7: 49–66 [DOI] [PubMed] [Google Scholar]
- Pavletich N. P., Pabo C. O., 1991. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A resolution. Science 252: 809–817 [DOI] [PubMed] [Google Scholar]
- Perez E. E., Wang J., Miller J. C., Jouvenot Y., Kim K. A., et al. , 2008. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat. Biotechnol. 26: 808–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plessis A., Perrin A., Haber J. E., Dujon B., 1992. Site-specific recombination determined by I-SceI, a mitochondrial group I intron-encoded endonuclease expressed in the yeast nucleus. Genetics 130: 451–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porteus M. H., Baltimore D., 2003. Chimeric nucleases stimulate gene targeting in human cells. Science 300: 763. [DOI] [PubMed] [Google Scholar]
- Ramirez C. L., Foley J. E., Wright D. A., Muller-Lerch F., Rahman S. H., et al. , 2008. Unexpected failure rates for modular assembly of engineered zinc fingers. Nat. Methods 5: 374–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J., 1983. One-step gene disruption in yeast. Methods Enzymol. 101: 202–211 [DOI] [PubMed] [Google Scholar]
- Rouet P., Smih F., Jasin M., 1994. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol. Cell. Biol. 14: 8096–8106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin N., Sugarman E., Haber J. E., 1989. Genetic and physical analysis of double-strand break repair and recombination in Saccharomyces cerevisiae. Genetics 122: 519–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche L. N., Kirchmaier A. L., Rine J., 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72: 461–516 [DOI] [PubMed] [Google Scholar]
- Sander J. D., Dahlborg E. J., Goodwin M. J., Cade L., Zhang F., et al. , 2011. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA). Nat. Methods 8: 67–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago Y., Chan E., Liu P.-Q., Orlando S., Zhang L., et al. , 2008. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc. Natl. Acad. Sci. USA 105: 5809–5814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer S., Davis R. W., 1979. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc. Natl. Acad. Sci. USA 76: 4951–4955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y., Bhakta M. S., Segal D. J., 2009. Restricted spacer tolerance of a zinc finger nuclease with a six amino acid linker. Bioorg. Med. Chem. Lett. 19: 3970–3972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla V. K., Doyon Y., Miller J. C., DeKelver R. C., Moehle E. A., et al. , 2009. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature 459: 437–441 [DOI] [PubMed] [Google Scholar]
- Smith J., Bibikova M., Whitby F. G., Reddy A. R., Chandrasegaran S., et al. , 2000. Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains. Nucleic Acids Res. 28: 3361–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söllü C., Pars K., Cornu T. I., Thibodeau-Beganny S., Maeder M. L., et al. , 2010. Autonomous zinc-finger nuclease pairs for targeted chromosomal deletion. Nucleic Acids Res. 38: 8269–8276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczepek M., Brondani V., Buchel J., Serrano L., Segal D. J., et al. , 2007. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat. Biotechnol. 25: 786–793 [DOI] [PubMed] [Google Scholar]
- Takasu Y., Kobayashi I., Beumer K., Uchino K., Sezutsu H., et al. , 2010. Targeted mutagenesis in the silkworm Bombyx mori using zinc-finger nuclease mRNA injections. Insect Biochem. Mol. Biol. 40: 759–765 [DOI] [PubMed] [Google Scholar]
- Tong C., Li P., Wu N. L., Yan Y., Ying Q.-L., 2010. Production of p53 knockout rats by homologous recombination in embryonic stem cells. Nature 467: 211–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend J. A., Wright D. A., Winfrey R. J., Fu F., Maeder M. L., et al. , 2009. High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature 459: 442–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov F. D., Miller J. C., Lee Y.-L., Beausejour C. M., Rock J. M., et al. , 2005. Highly efficient endogenous gene correction using designed zinc-finger nucleases. Nature 435: 646–651 [DOI] [PubMed] [Google Scholar]
- Urnov F. D., Rebar E. J., Holmes M. C., Zhang H. S., Gregory P. D., 2010. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 11: 636–646 [DOI] [PubMed] [Google Scholar]
- Vasquez K. M., Marburger K., Intody Z., Wilson J. H., 2001. Manipulating the mammalian genome by homologous recombination. Proc. Natl. Acad. Sci. USA 98: 8403–8410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M., Umeyama K., Matsunari H., Takayanagi S., Haruyama E., et al. , 2010. Knockout of exogenous EGFP gene in porcine somatic cells using zinc-finger nucleases. Biochem. Biophys. Res. Commun. 402: 14–18 [DOI] [PubMed] [Google Scholar]
- Wright D. A., Townsend J. A., Winfrey R. J., Jr, Irwin P. A., Rajagopal J., et al. , 2005. High-frequency homologous recombination in plants mediated by zinc-finger nucleases. Plant J. 44: 693–705 [DOI] [PubMed] [Google Scholar]
- Young J. J., Cherone J. M., Doyon Y., Ankoudinova I., Faraji F. M., et al. , 2011. Efficient targeted gene disruption in the soma and germ line of the frog Xenopus tropicalis using engineered zinc-finger nucleases. Proc. Natl. Acad. Sci. USA 108: 7052–7057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Maeder M. L., Unger-Wallace E., Hoshaw J. P., Reyon D., et al. , 2010. High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proc. Natl. Acad. Sci. USA 107: 12028–12033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J., Maeder M. L., Mali P., Pruett-Miller S. M., Thibodeau-Beganny S., et al. , 2009. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell 5: 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]