Abstract
lag-2 encodes a ligand for LIN-12/Notch and is a component of the lateral signal that activates LIN-12/Notch during Caenorhabditis elegans vulval precursor cell (VPC) fate patterning. lag-2 is specifically transcribed in one VPC, named P6.p, in response to activation of EGFR/Ras/MAPK by the inductive signal that initiates vulval development. Here, we show that a critical molecular event linking inductive and lateral signaling is the relief of VPC-wide lag-2 repression in P6.p. We find that the lag-2 promoter contains an element, VPCrep, which mediates repression in all VPCs when the inductive signal is absent, and another promoter element, VPCact, which is required for activation when repression is relieved by the inductive signal. We show that repression through VPCrep is mediated by the Elk1 ortholog LIN-1, and that the level and subcellular accumulation of a functional LIN-1::GFP protein is similar in all six VPCs before and after vulval induction, suggesting that relief of LIN-1–mediated repression in P6.p is likely due to the known MAPK-dependent phosphorylation of LIN-1. We also provide evidence that the factor(s) acting through VPCact is present in all VPCs but is not modulated by the inductive signal, and that transcription of lag-2 requires the Hth/Meis ortholog UNC-62 and the Mediator complex component SUR-2. Relief of repression of lag-2 in P6.p offers a plausible mechanistic basis for spatial restriction of lag-2 in generating the precise spatial pattern of VPC fates.
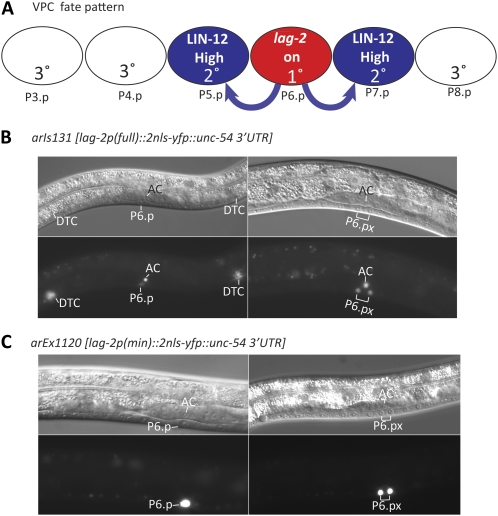
PATTERNING of vulval precursor cell (VPC) fates has provided an important paradigm for elucidating the components, roles, and regulation of conserved signal transduction pathways in cell fate specification. The VPC fates are patterned during the L3 larval stage (Figure 1A). Each of six VPCs, numbered P3.p–P8.p, has the potential to generate cells that contribute to the vulva, but in wild-type hermaphrodites, only P5.p–P7.p do so. These three VPCs have a symmetrical spatial fate pattern, such that P6.p, the central most VPC, adopts a fate called “1°,” and the flanking VPCs adopt a fate called “2°.” The 2°-1°-2° pattern of fates reflects the net outcome of LET-23/EGF Receptor and LIN-12/Notch signaling (reviewed in Sternberg 2005; Sundaram 2005). P6.p adopts the 1° fate because it is closest to the source of an EGF-like ligand produced by the anchor cell of the gonad, which activates a canonical EGFR/Ras/MAPK cascade. P6.p also produces a “lateral signal” that activates LIN-12/Notch in the neighboring VPCs, P5.p and P7.p, promoting the 2° fate, and completing the 2°-1°-2° spatial pattern.
Figure 1 .
VPC fate pattern and lag-2 expression. (A) Schematic view of VPC patterning. The inductive signal from anchor cell (AC) of the gonad activates the EGFR/Ras/MAPK cascade in P6.p in the early L3 stage to promote the 1° fate and lateral signal gene transcription. The lateral signal activates LIN-12/Notch to promote the 2° fate in P5.p and P7.p. Descendants of 1° and 2° cells form the vulva. VPCs that adopt the 3° fate generate two daughters that fuse with the major hypodermal syncytium, hyp7, but initially have the potential to adopt vulval fates if the EGFR or LIN-12/Notch is ectopically activated. (B) arIs131[lag-2p::2nls-yfp::unc-54 3′UTR], generated from the full-length lag-2p::2nls-yfp reporter used for deletion analysis (see Figure 2A), is expressed in P6.p, the AC, and the distal tip cells (DTCs), consistent with known functional roles for lag-2. Expression in the VPCs is restricted to P6.p and its descendants. We note that we did not observe the weak initial expression reported in all six VPCs by Chen and Greenwald (2004); we have not systematically assessed the numerous differences between the way transgenes were marked, generated, maintained, or visualized. All of the reporters presented in this study were made using similar conditions and therefore are directly comparable to each other. Furthermore, reporters in both studies behave identically for observations relating to vulval induction: both are strongly expressed in P6.p and depend on sur-2 for expression upon vulval induction. (C) arEx1120 [lag-2p(min)::2nls-yfp::unc-54] minimal reporter (Figure 2G) confers expression in P6.p and its descendants, but lacks expression in the AC and DTCs.
Genetic analysis implicated three genes, lag-2, apx-1, and dsl-1, as functionally redundant components of the lateral signal. All three genes encode “DSL” (Delta-Serrate-LAG-2) proteins, distinguished by an amino-terminal DSL domain followed by EGF-like motifs—the hallmarks of ligands that activate LIN-12/Notch. All three genes are transcribed in P6.p in response to the inductive signal (Chen and Greenwald 2004), suggesting that the key patterning event underlying the spatial pattern is the regulated transcription of the lateral signal genes in P6.p (Figure 1A).
Here, we have studied transcriptional regulation of the lateral signal gene lag-2 during VPC specification. Our results suggest that LIN-1, the Caenorhabditis elegans Elk1 ortholog, links inductive and lateral signaling by directly regulating lag-2 transcription. lin-1 has been extensively studied for its role in vulval induction. In lin-1 null mutants, all six VPCs adopt vulval fates even in the absence of the inductive signal, suggesting that lin-1 is a negative regulator of vulval fate (Ferguson et al. 1987; Beitel et al. 1995), although, lin-1 also appears to have a positive role in promoting the 1° fate (Howard and Sundaram 2002; Tiensuu et al. 2005). LIN-1 binds to the core Ets binding consensus sequence and is a direct target of phosphorylation by MPK-1/MAPK (Jacobs et al. 1998, 1999; Miley et al. 2004). In addition, if the MAPK phosphorylation site is disrupted, LIN-1 prevents P6.p from adopting the 1° fate (Jacobs et al. 1998). These observations indicate that LIN-1 is a direct target of the inductive signaling pathway, and that MAPK-dependent phosphorylation of LIN-1 relieves its ability to negatively regulate vulval fates. The Hox gene lin-39, which promotes vulval competence and may also promote the 1° fate, may be a direct target of LIN-1 for vulval induction (Maloof and Kenyon 1998; Wagmaister et al. 2006; Guerry et al. 2007).
Several studies have addressed the molecular mechanism by which LIN-1 is regulated as a target of the inductive signaling pathway. During vulval induction, MPK-1–mediated phosphorylation of the Forkhead protein LIN-31 (Miller et al. 1993) disrupts a complex with LIN-1 and promotes the 1° fate in P6.p (Tan et al. 1998). Phosphorylation of human Elk1 by MAPK promotes its function as a transcriptional activator (Yang et al. 2003), so by homology, phosphorylated LIN-1 may in some contexts, or on some targets, act as a transcriptional activator. However, LIN-1, like Elk1, has also been demonstrated to function as a transcriptional repressor. Importantly, Leight et al. (2005) showed that sumoylation of LIN-1 mediates its role as a transcriptional repressor and promotes inhibition of vulval fates, leading to the proposal that MPK-1/MAPK phosphorylation of LIN-1 relieves repression of lin-1 targets that promote the 1° vulval cell fate. Both modes may be relevant for the regulation of some targets, with LIN-1 potentially having a repressor role prior to induction and an activator role afterward.
Here, we implicate lag-2 as a direct target of LIN-1 transcriptional repression in the VPCs. In combination with available biochemical and functional information, our analysis leads to a model in which MAP kinase-mediated phosphorylation of LIN-1/Elk1 relieves repression to allow transcriptional activation of lag-2 in P6.p. This role for LIN-1 in lag-2 regulation does not require LIN-31/Forkhead, and thus differs in this molecular detail from the mechanism by which LIN-1 regulates induction per se. In addition, while relief of LIN-1–mediated repression is an essential event promoted by the inductive signal, we find that inductive signal-independent transcriptional activation by another factor, or factors, through an adjacent cis-acting site is also required. The activity of sur-2, a component of the Mediator complex, and unc-62, the ortholog of the Hox cofactor Hth/Meis, are required for transcriptional activation.
Materials and Methods
Reporters and transgenic lines
The wild-type parent of all strains used in this study, C. elegans var. Bristol strain N2, was described by Brenner (1974). Information about mutations used can be found via WormBase (www.wormbase.org). Reporters were generated by germline injection (Mello et al. 1991) of either fusion PCR products or PCR products from plasmids into GE24 pha-1(e2123), together with plasmid pCW2.1 [ceh-22::gfp] (20 μg/ml) (Okkema et al. 1997) and pBX [pha-1(+)] (50 μg/ml) (Granato et al. 1994). The expression reporter constructs were injected at 20 μg/ml, unless otherwise noted. Transgenic worms were maintained at 25° for selection of pha-1(+).
Strains carrying extrachromosomal arrays are listed in supporting information, Table S1 and Table S3. GS4892, containing arIs131 [lag-2p::2nls-yfp::unc-54 3′UTR], is an integrated line formed from an extrachromosomal array created using a fusion PCR product corresponding to Figure 2A.
Figure 2 .
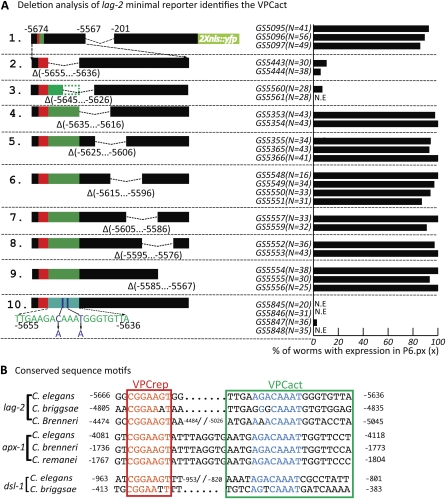
Deletion analysis of the lag-2 promoter identified a distal element conferring P6.p-specific expression. Strains carrying independent arrays formed from the different constructs were scored for expression as indicated. (A) Schematic representation of the full-length lag-2p::2nls-yfp::unc-54 3′UTR reporter. (B–H) Subsets of the lag-2 5′ flanking region fused to the lag-2 promoter-proximal sequence. The minimal promoter reporter used for further analysis of regulatory elements is shown in G. (I and J) The gcy-5 promoter-proximal region was used as a heterologous promoter. The red box represents VPCrep and the green box, VPCact. The translational start (ATG) is +1, and the fusion point is at −1. To insure the VPC induction has been completed, L3 hermaphrodites were scored at the Pn.px or Pn.pxx stage for YFP expression in VPCs, anchor cell (AC), and distal tip cells (DTCs). N.E., no expression.We note that there appears to be an element in the region from −1029 to −201 that is necessary for AC and DTC expression. We did not investigate this further, except for ascertaining that mutation of the E box at −365 in construct 2E abolishes DTC but not P6.p or AC expression, suggesting that this region may be involved in the known bHLH-mediated regulation of lag-2 expression in the DTC (Karp and Greenwald 2004; Chesney et al. 2009).
Constructs generated by fusion PCR
All DNA fragments from the 5′ flanking region of lag-2 were PCR amplified from N2 genomic DNA. Fusion PCR was performed to generate the transcription reporter constructs as described previously (Hobert 2002). Briefly, the template “2Xnls-yfp::unc-54 3′UTR” was PCR amplified from a 2Xnls-yfp plasmid (Yoo et al. 2004) using primer C (AGCTTGCATGCCTGCAGGTCGACT) and primer D (AAGGGCCCGTACGGCCGACTAGTAGG). The template for the promoter region was amplified using forward primer A and reverse primer B. Using the two templates above, the final reporter construct was generated by fusion PCR amplification using primer A*, nested to primer A and primer D* (GGAAACAGTTATGTTTGGTATATTGGG), nested to primer D. To generate reporter constructs with deletion, primer pairs “5F” (forward)/“5R” (reverse) and “3F” (forward)/“3R” (reverse) were used to PCR amplify the 5′ fragment and 3′ fragment of the targeted deletion, respectively. Primer A, nested to 5F and primer B, nested to 3R were used to amplify the first fusion product, the promoter template with deletion. A second fusion PCR was performed using primer A* and reverse primer D* to generate the final reporter construct as described above. Table S1 lists the primer sets used to generate each of the constructs in this study by fusion PCR. arEx763 and arEx764 were generated by injecting a concentration of 100 μg/ml of construct, and arEx791, arEx792, arEx774, arEx776, arEx777, arEx869, and arEx870 were generated at 50 μg/ml.
Plasmids
To generate plasmids containing promoter of interest driving 2nls-yfp, the promoter was generated by fusion PCR from N2 genomic DNA (Table S2: primer pairs 5F-PstI/5R for distal element, primer pairs 3F/3R-KpnI for proximal element, primer pairs 5F-PstI/ and 3R-KpnI for fusion PCR). The fusion PCR product was then cloned into the 2nls-yfp::unc-54 3′UTR plasmid at the sites of PstI and KpnI. The following plasmids were made using this strategy:
p847, encompassing 5′ flanking regions of lag-2 from −5674 to −5567 and from −201 to −1; and
P849, encompassing 5′ flanking regions of lag-2 from −5674 to −5567 and 5′ flanking regions of gcy-5 from −200 to −1.
Site-directed mutagenesis was performed on p847 to generate plasmids in this study and PCR products from those plasmids were used at 20 μg/ml for germline injection to generated arrays (Table S3).
The PCR product of 7.1 kb 5′ flanking region of lag-2 was cloned into 2nls-yfp::unc-54 3′UTR plasmid at the sites of HindIII and KpnI to make p859, using the following primers:
F-HindIII (AAAAAGCTTTGTCAGAATGTCCCATGTAGG)
R-KpnI (GAGCTCGGTACCCGGGTTTCTGAAAAAAGGCAAATTTGAAAAGTG).
P862, in which the lag-2 5′ flanking region from −5674 to −5567 was deleted, was generated using p859 as template. Strains GS5840(arEx1352), GS5841(arEx1353), GS5842(arEx1354), GS5843(arEx1355), and GS5844(arEx1356) were made by using the 9-kb HindIII/SpeI fragment from p862 for injection at 50 μg/ml.
Sequence analysis
The 5′ flanking region sequences of lag-2, apx-1, and dsl-1 orthologs in other species were retrieved from WormBase using WormMart, and motifs were identified using Vector NTI.
In addition to the sequence analysis described below, we also analyzed VPCact using MatInspector (Genomatrix) but did not find any compelling candidates for transcriptional activators acting through this site. We used default settings, 10 bp of additional flanking sequence on either side of the 20-bp VPCact region boxed in Figure 4, and weight matrixes derived from vertebrates. This analysis identified two candidates. One is Brachyury, a T-box protein; there is no C. elegans ortholog of Brachyury, and none of the 21 T-box genes (Woollard 2005) have been implicated in vulval development to date. The other is NeuroD; in addition to the arguments against the involvement of bHLH proteins advanced in Results, the C. elegans ortholog of NeuroD is expressed only in the nervous system (Hallam et al. 2000).
Figure 4 .
Identification of VPCact. (A) Schematic representation of the reporter series, each with an internal 20-bp deletion, derived from the minimal construct identified in Figure 2G and here as construct 1. The lack of expression in reporters 2 and 3 define VPCact. VPCrep (red box) is intact; the green box shows the inferred position of VPCact based on these deletions, corresponding to the deleted region in construct 2 and the boxed region in B. (B) Alignments of VPCrep and VPCact sequences in lag-2, apx-1, and dsl-1 from C. elegans C. briggsae, C. brenneri, and C. remanei. Red box, VPCrep (Elk1 site). Green box, sequence deleted in reporter 2. Sequences similar to VPCact were not identified in the 5′ flanking regions of other C. elegans dsl genes (X. Zhang, unpublished observations).
LIN-1::GFP fosmid reporter and rescue
Recombineering was used to generate p845, which encodes C-terminal tagged LIN-1::GFP, as described in Tursun et al. (2009). Fosmid WRM0629cG08 was used as the template. Fusion PCR was performed to generate GFPint-FgF C-terminal fusion cassette from pBALU1 using the following primers:
5primer, AGTACCCATAAAAATGCCAACTTTGATGAGTAAAGGAGAAGAACTTTTCAC;
5PrimerNest, CAACGGATTCTTTAAAAACACCTACAGTACCCATAAAAATGCCAACTTTG;
3primer, TTTTCTCGCTCCAAAGTTCAAACTATTTGTATAGTTCATCCATGCCATG; and
3PrimerNest, AAAATTTGCTTTTCGAAAATTTTTATTTTTTTCTCGCTCCAAAGTTCAA.
The correct insertion of gfp was confirmed by sequencing analysis using the following primers:
5seq, CCAATTCCCGCCGGTCTCCGCATTC and
3seq, TCATCCGGTGGGGGAGATCATGGAG.
P845 was injected into GE24 pha-1(e2123) at 10 μg/ml, together with N2 genomic DNA (50 μg/ml), plasmid pGC204[ceh-22::tdimer2::let-858 3′UTR] (1 μg/ml) (Voutev and Hubbard 2008) and pBX [pha-1(+)] (1 μg/ml). To assess lin-1(0) rescue, the same injection mix was injected into GS6015 pha-1(e2123); lin-1(n304), and all viable worms were non-Muv.
RNAi
Strain GS5891 arIs131 [lag-2p::2nls-yfp::unc-54 3′UTR]; nre-1(hd20) lin-15b(hd126), sensitized for RNAi (Schmitz et al. 2007), was used for feeding RNAi. Briefly, gravid adults were bleached and eggs were placed on RNAi plates, prepared as described in Choi et al. (2010), at 20°. To score the lin-1(RNAi) positive control, hermaphrodites were scored 3 days after the eggs were placed on the plates for a Multivulva phenotype. Worms were scored at L3 Pn.px stage for expression of arIs131.
Laser ablation
The four cells of the gonad primordium were ablated using a laser microbeam in newly hatched L1 larvae as described (Bargmann and Avery 1995). Success of ablation was confirmed by the lack of gonad development.
Results
Identification of a regulatory module that confers correct VPC expression
The coding region of the gene located 5′ of lag-2 begins ∼7.2 kb upstream of the coding region of lag-2. Most previous studies of lag-2 regulation utilized transcriptional reporters containing only 3.4 kb of 5′ flanking region. These reporters were based on the amount of 5′ sequence sufficient to achieve rescue of the lag-2(0) lethal phenotype in a genomic context (e.g., Henderson et al. 1994; Wilkinson et al. 1994; Karp and Greenwald 2003), and most such reporters are not expressed in VPCs. Another previous reporter, consisting of 6.2 kb of 5′ flanking region driving nuclearly localized β-galactosidase, displayed strong expression in P6.p after vulval induction (Chen and Greenwald 2004), suggesting that sequences located between 3.4 kb and 6.2 kb upstream of the protein coding region would mediate transcription of lag-2 in response to the inductive signal.
To visualize lag-2 expression in live worms, we created a reporter containing 7.1 kb 5′ flanking region of lag-2 driving a nuclearly localized form of YFP (Figure 2A). Extrachromosomal arrays and an integrant, arIs131[lag-2p::2Xnls::yfp], revealed expression in P6.p upon its EGF-mediated induction in the L3 stage as well as expression in the anchor cell (AC) and distal tip cells (DTCs) of the somatic gonad (Figure 1B). Subsequent deletion analysis to identify the sequence elements conferring P6.p-specific expression upon induction used comparable ways of marking, generating, maintaining, and visualizing transgenic arrays to minimize variations in expression pattern due to factors other than the engineered change.
Deletion of 1679 bp from the distal end of the fragment abolishes P6.p expression, indicating the distal region contains sequences that are necessary for P6.p expression (Figure 2B), consistent with the previous inference of a necessary sequence located between −6.4 kb and −3.4 kb. This deletion also abolishes AC expression, but does not alter DTC expression (Figure 2B). Further deletion analysis (Figure 2, C–G) identified a “distal element,” comprising the lag-2 5′ flanking region from −5674 to −5567, which confers P6.p-specific expression, but not AC expression, when combined with a 201-bp minimal promoter region from lag-2 (Figures 1C and 2G). We refer to the combination of the distal element fused to the lag-2 minimal promoter region as lag-2p(min) and performed further deletion analysis as described below to identify specific sites that mediate repression and activation.
When the distal element is deleted from an otherwise full-length 5′ flanking region construct, P6.p expression is completely abolished, but the expression in the AC and DTCs remains unchanged, indicating that the distal element is necessary and specific for P6.p expression (Figure 2H). Although the distal element alone does not drive expression in P6.p when placed directly adjacent to the 2Xnls::yfp coding region (data not shown), the distal element conferred P6.p-specific expression when combined with a 200-bp minimal promoter region from gcy-5 (Figure 2J). gcy-5 is normally expressed in neurons but not in VPCs or any other hypodermal cells (Yu et al. 1997), and we verified that this promoter region alone does not drive expression in VPCs (Figure 2I). The results from our promoter analysis indicate that the distal −5674 to −5567 element contains the minimal sequences that are necessary and sufficient, when supplied with any basal promoter, to promote transcription in P6.p.
VPCrep
We looked for potential transcription factor binding sites in lag-2p(min) computationally and by sequence conservation with other Caenorhabditis species and identified a conserved module that includes the core Elk1/Ets binding site consensus sequence, CGGAAGT (Wei et al. 2010) (Figures 3A and 4B). We note that additional sequences conforming to this consensus are also found elsewhere in the full-length lag-2 5′ flanking region (data not shown) as well as in the 5′ flanking region of apx-1 and dsl-1 (Figure 4B), the two other lateral signal genes.
Figure 3 .
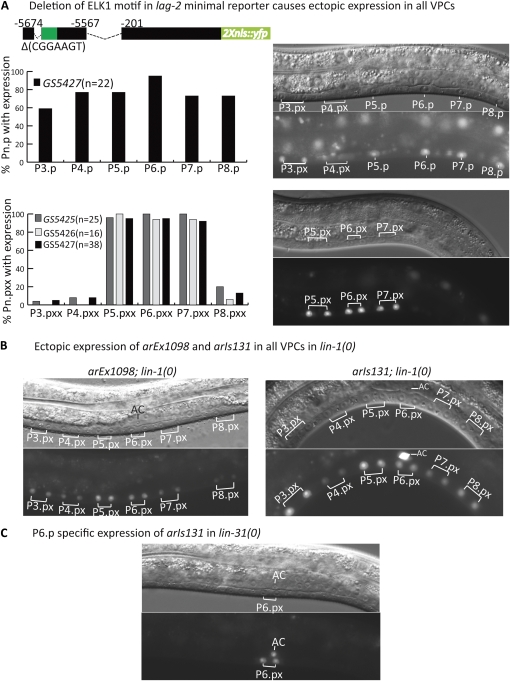
Identification of VPCrep. (A) Deletion of the Elk1 motif from the distal element causes ectopic, uniform expression in all VPCs, indicating it serves as a site of repression (VPCrep). The Elk1 motif, (CGGAAGT), is located in position −5664 to −5658 and corresponds to the red box in Figure 2. The green box represents the activator site, VPCact, identified as described in Figure 4. (Left) Quantitation of strains carrying ∆VPCrep reporters. (Right) Photomicrographs of L3 hermaphrodites showing VPC or Pn.px expression. At the Pn.px stage, expression is not seen in the daughters of P3.p, P4.p and P8.p because they have fused with hyp7. (B) In the absence of LIN-1, achieved using the lin-1(n304) null allele (Beitel et al. 1995), arEx1098[lag-2p(min)::2nls-yfp::unc-54 3′UTR] and arIs131[lag-2p::2nls-yfp::unc-54 3′UTR] are uniformly expressed in all VPCs. (C) In the absence of LIN-31, achieved using the lin-31(n301) null allele (Miller et al. 2000), arIs131[lag-2p::2nls-yfp::unc-54 3′UTR] expression remains P6.p specific.
Deletion of the Elk1 site from lag-2p(min) results in strong ectopic expression in P5.p and P7.p and their descendants, as well as in P3.p, P4.p, and P8.p and their daughters prior to their fusion with hyp7 (Figure 3A). Deletion of the Elk1 site from the distal element in the context of the gcy-5 promoter also results in derepression in all six VPCs (Figure S1). These observations indicate that this site, “VPCrep,” mediates repression of lag-2 in VPCs. In addition, the observation that the ∆VPCrep reporters become derepressed even in P3.p, P4.p, and P8.p, which in an otherwise wild-type background do not receive the inductive signal, suggests that VPCrep-mediated repression occurs in all VPCs and is independent of the inductive signal. The strong and uniform expression seen when VPCrep is deleted has important implications for modeling the mechanism by which lag-2 is activated, as considered further in Discussion.
VPCact
We performed further deletion analysis on lag-2p(min) and found that the conserved sequence immediately adjacent to VPCrep is required for expression in P6.p (Figure 4A, 2), suggesting that it encompasses a sequence required for transcriptional activation. This sequence, VPCact, comprises the lag-2 5′ flanking region from −5655 to −5636. Deletion of VPCact along with VPCrep in the context of the full-length reporter (Figure 2H) or lag-2p(min) (our unpublished observations) abolishes expression in P6.p as well as the other VPCs, suggesting that positive input through VPCact is required for transcription even in the absence of repression through VPCrep. Sequences similar to VPCact are also found in the apx-1 and dsl-1 5′ flanking sequences (Figure 4B). Mutation of 2 bp within this region, chosen because they are part of an E-box binding site consensus (but see below), abolished P6.p-specific expression (Figure 4A, 10), establishing it as required for VPCact activity.
VPCrep, but not VPCact, is regulated by the inductive signal
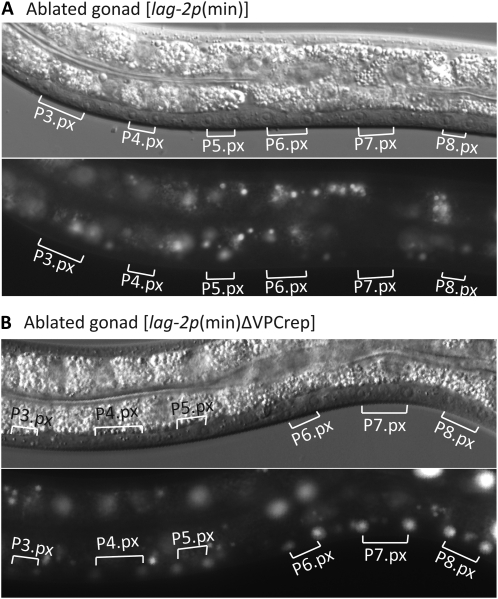
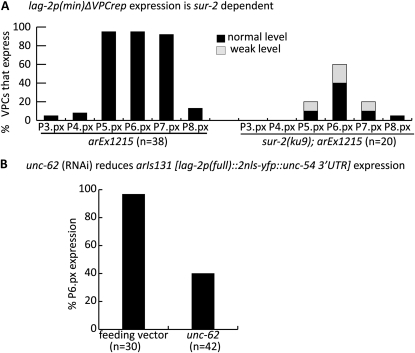
To assess the response of these sites to the inductive signal and their contribution to the final pattern of lag-2 expression, we ablated the gonad in the early L1 stage using a laser microbeam (see Materials and Methods) and examined the effect on the expression of trangenes carrying either an intact lag-2p(min) or a mutant form lacking VPCrep. As is also true for a full-length reporter (Chen and Greenwald 2004), the intact lag-2p(min) reporter requires the inductive signal for expression in P6.p: 0/11 operated lag-2p(min) worms showed P6.p expression (Figure 5A). In contrast, when VPCrep was deleted from this reporter [lag-2p(min)∆VPCrep], all operated worms (9/9) showed uniform, strong derepression in all VPCs (Figure 5B), with levels comparable to what was observed for the lag-2p(min) reporter in P6.p in unoperated worms.
Figure 5 .
∆VPCrep causes gonad-independent lag-2 expression in all VPCs. The gonad was ablated in the L1 stage and hermaphrodites were examined in the L3 Pn.px stage for lag-2 expression in VPCs. (A) arEx1120 [lag-2p(min)::2nls-yfp::unc-54 3′UTR]. (B) arEx1215 [lag-2p(min∆VPCrep)::2nls-yfp::unc-54 3′UTR].
The gonad independence of lag-2p(min)∆VPCrep expression leads to two important conclusions about the trans-acting factors that pattern lag-2 expression. First, it demonstrates that VPCrep is not required for transcriptional activation, thereby ruling out a model in which the repressor that binds to VPCrep is turned into an activator by the inductive signal, promoting transcription in conjunction with a VPCact-binding activator. Second, the observation that lag-2p(min)∆VPCrep transcription is seen in all VPCs in the absence of the gonad suggests that the activator that works through VPCact is always present in all VPCs and does not require activation per se. Together, these conclusions imply that the main spatial-patterning role for the EGFR/Ras/MAPK inductive pathway in P6.p is mediated by the relief of VPCrep-mediated repression specifically in P6.p, rather than by cell-specific expression of an activator.
LIN-1/Elk1 and repression through VPCrep
VPCrep conforms to the consensus Elk1 site; LIN-1, the C. elegans Elk1 ortholog (Beitel et al. 1995), has been demonstrated to bind to this consensus sequence (Miley et al. 2004), suggesting that LIN-1 may mediate negative regulation through this site. When we examined the effect of removing lin-1 activity on the expression of the full-length reporter arIs131[lag-2p::2Xnls::yfp] or the lag-2p(min) reporter arEx1098[lag-2p(min):: 2nls-yfp], we observed strong and uniform derepression of lag-2 in all six VPCs in lin-1(0) mutants (Figure 3B), similar to when VPCrep was deleted in the context of lag-2p(min) (Figure 3A). These observations, combined with the known binding properties of Elk1 and LIN-1, suggest that LIN-1 is the repressor that acts through VPCrep. Furthermore, when combined with the gonad-ablation results discussed above, our results indicate that lin-1 does not play a positive role in promoting lag-2 transcription, as might have been the case if phosphorylation of LIN-1 turned it into a transcriptional activator of lag-2.
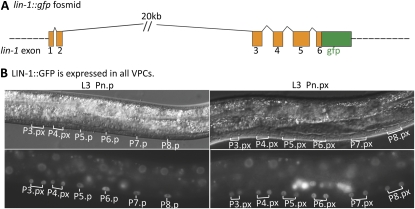
To explore the mechanism by which the inductive signal relieves repression of lag-2 by LIN-1, we generated a fosmid-based reporter in which GFP was fused to the C terminus of LIN-1 in a full genomic context (Figure 6A and Materials and Methods). Transgenes carrying this reporter fully rescue the Multivulva phenotype of lin-1(0) (see Materials and Methods), indicating that LIN-1::GFP is functional and regulated normally. LIN-1::GFP accumulated to a similar level in the nuclei of all six VPCs both before and after vulval induction (Figure 6B), suggesting that modification of LIN-1 activity by MAPK phosphorylation (Tan et al. 1998), rather than differential stability or subcellular localization, is likely to underlie the relief of LIN-1–mediated repression of lag-2 in P6.p.
Figure 6 .
LIN-1::GFP is uniformly expressed in all VPCs. (A) The lin-1 genomic region, with the position of gfp insertion. (B) GS5958, carrying the fosmid-based lin-1::gfp reporter, shows LIN-1::GFP expression in all VPCs before (L3 Pn.p) and after induction (Pn.px).
LIN-1 works together with LIN-31/Forkhead in negatively regulating vulval induction (Miller et al. 1993) and also appears to play a positive role in promoting the 1° fate (Howard and Sundaram 2002; Tiensuu et al. 2005). However, we found that arIs131[lag-2p::2Xnls::yfp] is expressed specifically in P6.p in a lin-31(0) null mutant, as in wild type (Figure 3C). The observation that there is no ectopic expression in other VPCs suggests that LIN-1 does not require LIN-31 to repress lag-2 through VPCrep. Furthermore, if phosphorylated LIN-31 is able to promote lag-2 transcription through VPCact, then it must be functionally redundant with other activators, as loss of lin-31 does not cause loss of expression in P6.p.
Assessment of factors that may activate through VPCact
As described above, the inductive signal does not appear to regulate transcription through VPCact, indicating that activation is constitutive when repression is relieved. Thus, studies of the transcriptional activator are not likely to yield further understanding into the nature of the link between inductive and lateral signaling. Nevertheless, we explored a potential link between VPCact and selected trans factors.
We began with SUR-2, a component of the Mediator coactivator complex. sur-2 is expressed in all VPCs, and a null allele results in a lateral signaling defect without a major effect on the 1° fate (Singh and Han 1995); loss of expression of lag-2 and the other lateral signal genes can account for the sur-2(0) vulval defect (Chen and Greenwald 2004). When we examined the effect of sur-2(0) on the expression of lag-2p(min)∆VPCrep, we found that expression in all six VPCs and unfused Pn.px cells was greatly reduced (Figure 7A). This observation both suggests that the transcription factor(s) functioning through VPCact requires the Mediator complex for transcriptional activation and supports the inference that the transcription factor mediating activation through this site is present and functional in all of the VPCs.
Figure 7 .
Expression of lag-2p(min) depends on sur-2 and unc-62. (A) arEx1215[lag-2p(min∆VPCrep)::2nls-yfp::unc-54 3′UTR], which is uniformly expressed in all VPCs (Figure 3A), shows reduced expression in all VPCs in the sur-2(ku9) null background. The difference in expression in P5.p, P6.p, and P7.p between sur-2(+) and sur-2(0) is significant (Fisher's exact test, P < 0.002). (B) arIs131[lag-2p::2nls-yfp::unc-54 3′UTR] expression is reduced by unc-62(RNAi) (see Materials and Methods for complete genotype). The difference in expression between the feeding vector and unc-62(RNAi) is significant (Fisher's exact test, P < 0.0001).
The Mediator complex works in conjunction with sequence-specific DNA binding proteins to promote transcription. We therefore examined the VPCact sequence for a clue to the factor that might recruit the Mediator complex. A general sequence analysis program did not identify any compelling candidates (see Materials and Methods), so we considered specific factors. First, we considered the possibility that the C. elegans E ortholog HLH-2, which regulates lag-2 in the anchor cell and distal tip cells (Krause et al. 1997; Karp and Greenwald 2003, 2004), is involved. As described above, VPCact contains an E-box consensus sequence, and mutating this sequence abrogates VPCact function (Figure 4A). However, it seems unlikely that HLH-2 is a general transcriptional activator of the lateral signal genes because the E-box consensus sequence is not present in the VPCact-homologous region of apx-1 (Figure 4B) and a fosmid-based translational reporter for the C. elegans E ortholog HLH-2, the obligatory dimerization partner for class II bHLH proteins, is not detectable in the VPCs in the L3 stage (J. Ohlmeyer and I. Greenwald, unpublished observations).
We also considered the potential involvement of members of Hox genes because lin-39 (Maloof and Kenyon 1998) and ceh-13 (Tihanyi et al. 2010) are known to function in the VPCs. In particular, LIN-39 is present in all VPCs and prevents premature fusion of VPCs with the major hypodermal syncytium hyp7, so that the VPCs are competent to be induced by patterning signals (Clark et al. 1993). lin-39 may be required for basal expression of lin-12 and lag-2 in VPCs (Takacs-Vellai et al. 2007) [through a putative binding site, that is not present, however, in our lag-2p(min) constructs]. In addition, LIN-39 is upregulated in P6.p during vulval induction (Maloof and Kenyon 1998). When we examined VPCact with Target Explorer (Sosinsky et al. 2003), using position weight matrixes based on data from Noyes et al. (2008), we found that it does contain a potential Hox binding site. However, since P6.p fuses with hyp7 in lin-39(0) mutant larvae, we could not simply assess the effect on lag-2 expression.
As an approach to circumventing premature fusion, we examined the effect of depleting the Hox cofactor UNC-62, a member of the Homothorax (Hth)/Meis family of proteins (reviewed in Mann et al. 2009). unc-62 is expressed in all VPCs (Jiang et al. 2009) and since VPCs in unc-62(RNAi) do not fuse with hyp7 and instead adopt vulval fates (Yang et al. 2005), we could score the effect of unc-62(RNAi) on expression of arIs131 [lag-2p::2Xnls::yfp]. We found that only 40% of hermaphrodites displayed expression in P6.px (n = 42) as compared to feeding vector control (100%, n = 30). In some animals, we observed ectopic induction along with loss of lag-2 reporter expression (data not shown). Our results are consistent with the possibility that Meis/Hth/UNC-62–dependent Hox activity may be required directly or indirectly for VPC-wide activation via the VPCact element. However, we cannot rule out a more general effect on 1° cell fate or a Hox-independent activity of Hth/Meis (Mann et al. 2009).
Finally, we considered the possibility that EOR-1, a BTB and C2H2 zinc finger domain transcription factor (Howard and Sundaram 2002), might promote lag-2 transcription. EOR-1, together with its binding partner EOR-2 (Howell et al. 2009), functions redundantly with LIN-1 and the Mediator complex to promote certain Ras pathway outputs. Genetic evidence suggests LIN-1 acts as a positive regulator in conjunction with the Mediator complex but independently of, and in parallel to, EOR-1 (Howard and Sundaram 2002).
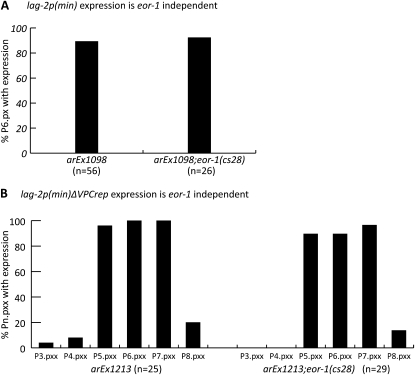
eor-1(0) does not have a lateral signaling defect, suggesting that EOR-1 is not the sole activator of lateral signal gene transcription. However, it is possible that there are redundant activators, obscuring a role for EOR-1 through VPCact. We crossed the lag-2p(min) and lag-2p(min∆VPCrep) reporters into an eor-1(0) background and found that their transcription was not affected by loss of eor-1 (Figure 8). Therefore, EOR-1 is not the activator acting through VPCact. We note that using lag-2p(min∆VPCrep) to eliminate the potential contribution of LIN-1 to transcriptional activation bypasses potential problems of cell fate transformation and/or synthetic lethality associated with concomitant removal of eor-1 and lin-1 function (Howard and Sundaram 2002). The observation that even in the absence of the LIN-1 binding site in VPCrep eor-1(0) does not affect lag-2 transcription suggests that a redundant function for EOR-1 is not masked by LIN-1 and supports the conclusion that neither of these transcription factors activates lag-2 expression.
Figure 8 .
Expression of lag-2p(min) and lag-2p(min∆VPCrep) is not eor-1 dependent. (A) In the absence of eor-1, arEx1098[lag-2p(min)::2nls-yfp::unc-54 3′UTR] expression remains P6.p specific. The difference in expression between eor-1(+) and eor-1(−) is not significant (Fisher's exact test, P = 1.0). (B) In the absence of eor-1, the ectopic expression of arEx1213[lag-2p(min∆VPCrep)::2nls-yfp::unc-54 3′UTR] remains uniform in all VPCs. The difference in expression between eor-1(+) and eor-1(−) is not significant (Fisher's exact test, P > 0.24).
Discussion
We have investigated the mechanism by which the lateral signal genes are expressed specifically in a single VPC in response to EGFR/Ras/MAPK activation by studying the regulation of the lateral signal gene lag-2. Our results suggest a model for how lag-2 is transcribed specifically in P6.p in response to inductive signaling (Figure 9). There are three key elements of this model. First and foremost, the critical patterning event is loss of repression by LIN-1/Elk1 in P6.p. Second, relief of repression allows a transcriptional activator (or activators) present in all VPCs, and which is not itself directly regulated by the inductive signal, to promote transcription through VPCact. Third, LIN-1 does not appear to function as a transcriptional activator of lag-2, either alone or redundantly with other factors. We consider here the rationale for each of these elements and additional implications of our findings.
Figure 9 .
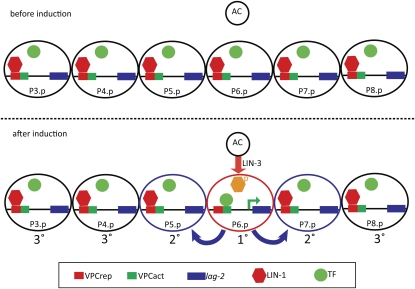
Model for how relief of LIN-1 repression through VPCrep leads to P6.p-specific expression of lag-2. (Top) Before induction, lag-2 is repressed in all VPCs through the action of LIN-1 via VPCrep. The inferred activator is drawn as not associated with VPCact, but it may instead be bound without promoting transcription. (Bottom) The inductive signal from the anchor cell (AC) activates the EGFR/Ras/MAPK cascade in P6.p. Phosphorylation of LIN-1/Elk1 in P6.p relieves repression through VPCrep, allowing a transcriptional activator that is present in all six VPCs to function. If VPCrep is deleted or LIN-1 removed mutationally, transcriptional activation can occur uniformly in all six VPCs.
The assertion that the critical patterning event is loss of repression by LIN-1/Elk1 in P6.p is based on the observations that loss of LIN-1 or deletion of VPCrep, which conforms to the Elk1 binding site consensus, results in strong, uniform expression of lag-2 in all six VPCs. This expression does not depend on the inductive signal, so the activator itself is not directly regulated by the inductive signal; loss of repression is sufficient to allow expression in all VPCs, and deletion of VPCact abrogates expression even in the absence of repression, so the activator (or activators) must be present in all VPCs. Together, our data indicate that the role of the inductive signal in promoting lag-2 expression in a normal developmental context reflects relief of LIN-1–mediated repression in P6.p.
Since a functional LIN-1::GFP protein is present at a uniform level in the nuclei of all VPCs and does not change in P6.p upon induction, LIN-1 activity appears to be regulated post-translationally. LIN-1 phosphorylation has been proven to occur downstream of EGFR/Ras/MAPK activation in P6.p (Jacobs et al. 1998, 1999), suggesting that phosphorylation may lead to abrogation of LIN-1–mediated repression of lag-2. This proposal is supported by the similar mechanisms of transcriptional repression of LIN-1 and its mammalian ortholog Elk-1: in both cases, sumoylation promotes repressor activity (Yang et al. 2003; Leight et al. 2005). For Elk1, sumoylation and repressor activity is lost upon activation of the MAP kinase pathway (Yang et al. 2003), supporting the inference that repressor activity is also lost upon MAPK phosphorylation of LIN-1.
Loss of repression by phosphorylation of LIN-1 may be common to both vulval induction and lag-2 regulation. However, a significant difference is that LIN-31 is an obligate partner of LIN-1 in preventing VPCs from adopting vulval fates (Tan et al. 1998), but the absence of lin-31 does not lead to derepression of lag-2 in other VPCs. Furthermore, phosphorylated LIN-31 has been proposed to promote transcriptional activation of some targets that promote vulval fate (Tan et al. 1998), but the absence of lin-31 does not cause a loss of lag-2 transcription in P6.p, which instead requires an activator that does not depend on inductive signaling for activity. Thus, our results suggest a novel, LIN-31–independent function of LIN-1 in modulating lag-2 transcription.
There is evidence that LIN-1 plays a positive role in vulval induction in conjunction with another transcription factor, EOR-1, and it has been proposed that LIN-1 is converted from a transcriptional repressor to a transcriptional activator by MPK-1 phosphorylation (Howard and Sundaram 2002). It remains possible that phosphorylated LIN-1 is a transcriptional activator of other targets that promote vulval fate. However, our results indicate that phosphorylated LIN-1 is unlikely to be a direct transcriptional activator of lag-2 either alone or redundantly with another transcription factor: loss of LIN-1 or deletion of VPCrep in the context of the minimal promoter does not block transcription, indicating that LIN-1 binding to that site is not necessary for transcription, and deletion of VPCact is sufficient to abrogate expression, indicating that activation through this site is necessary for transcription.
In sum, our data indicate that lag-2 is a direct target of LIN-1 and that lag-2 transcription in P6.p results from loss of repression via post-translational regulation of LIN-1 in response to the inductive signal. In addition to establishing a plausible mechanistic basis for how inductive signaling leads to transcription of lag-2 in P6.p, the differences in the requirements for other factors that have been established as contributing to vulval induction suggest that there are different genetic circuits and formal logic underlying different aspects of the 1° fate.
Acknowledgments
We gratefully acknowledge Dan Shaye for many valuable suggestions and much discussion. We are also indebted to Xinlan Zhou and Richard Ruiz for expert technical assistance, and we thank Luisa Cochella and Baris Tursun for advice about fosmid recombineering; Katie Lelli and Vincent Bertrand for help with sequence analysis; Meera Sundaram for eor-1(cs28) and discussion; and Xantha Karp, Maria Sallee, and Dan Shaye for helpful comments that improved this manuscript. X.Z. was a postdoctoral associate and I.G. is an investigator of the Howard Hughes Medical Institute.
Literature Cited
- Bargmann C. I., Avery L., 1995. Laser killing of cells in Caenorhabditis elegans. Methods Cell Biol. 48: 225–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beitel G. J., Tuck S., Greenwald I., Horvitz H. R., 1995. The Caenorhabditis elegans gene lin-1 encodes an ETS-domain protein and defines a branch of the vulval induction pathway. Genes Dev. 9: 3149–3162 [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Greenwald I., 2004. The lateral signal for LIN-12/Notch in C. elegans vulval development comprises redundant secreted and transmembrane DSL proteins. Dev. Cell 6: 183–192 [DOI] [PubMed] [Google Scholar]
- Chesney M. A., Lam N., Morgan D. E., Phillips B. T., Kimble J., 2009. C. elegans HLH-2/E/Daughterless controls key regulatory cells during gonadogenesis. Dev. Biol. 331: 14–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi M. S., Yoo A. S., Greenwald I., 2010. sel-11 and cdc-42, two negative modulators of LIN-12/Notch activity in C. elegans. PLoS ONE 5: e11885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S. G., Chisholm A. D., Horvitz H. R., 1993. Control of cell fates in the central body region of C. elegans by the homeobox gene lin-39. Cell 74: 43–55 [DOI] [PubMed] [Google Scholar]
- Ferguson E. L., Sternberg P. W., Horvitz H. R., 1987. A genetic pathway for the specification of the vulval cell lineages of Caenorhabditis elegans. Nature 326: 259–267 [DOI] [PubMed] [Google Scholar]
- Granato M., Schnabel H., Schnabel R., 1994. pha-1, a selectable marker for gene transfer in C. elegans. Nucleic Acids Res. 22: 1762–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerry F., Marti C. O., Zhang Y., Moroni P. S., Jaquiery E., et al. , 2007. The Mi-2 nucleosome-remodeling protein LET-418 is targeted via LIN-1/ETS to the promoter of lin-39/Hox during vulval development in C. elegans. Dev. Biol. 306: 469–479 [DOI] [PubMed] [Google Scholar]
- Hallam S., Singer E., Waring D., Jin Y., 2000. The C. elegans NeuroD homolog cnd-1 functions in multiple aspects of motor neuron fate specification. Development 127: 4239–4252 [DOI] [PubMed] [Google Scholar]
- Henderson S. T., Gao D., Lambie E. J., Kimble J., 1994. lag-2 may encode a signaling ligand for the GLP-1 and LIN-12 receptors of C. elegans. Development 120: 2913–2924 [DOI] [PubMed] [Google Scholar]
- Hobert O., 2002. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques 32: 728–730 [DOI] [PubMed] [Google Scholar]
- Howard R. M., Sundaram M. V., 2002. C. elegans EOR-1/PLZF and EOR-2 positively regulate Ras and Wnt signaling and function redundantly with LIN-25 and the SUR-2 Mediator component. Genes Dev. 16: 1815–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell K., Arur S., Schedl T., Sundaram M. V., 2010. EOR-2 is an obligate binding partner of the BTB-zinc finger protein EOR-1 in Caenorhabditis elegans. Genetics 184: 899–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs D., Beitel G. J., Clark S. G., Horvitz H. R., Kornfeld K., 1998. Gain-of-function mutations in the Caenorhabditis elegans lin-1 ETS gene identify a C-terminal regulatory domain phosphorylated by ERK MAP kinase. Genetics 149: 1809–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs D., Glossip D., Xing H., Muslin A. J., Kornfeld K., 1999. Multiple docking sites on substrate proteins form a modular system that mediates recognition by ERK MAP kinase. Genes Dev. 13: 163–175 [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Shi H., Liu J., 2009. Two Hox cofactors, the Meis/Hth homolog UNC-62 and the Pbx/Exd homolog CEH-20, function together during C. elegans postembryonic mesodermal development. Dev. Biol. 334: 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp X., Greenwald I., 2003. Post-transcriptional regulation of the E/Daughterless ortholog HLH-2, negative feedback, and birth order bias during the AC/VU decision in C. elegans. Genes Dev. 17: 3100–3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp X., Greenwald I., 2004. Multiple roles for the E/Daughterless ortholog HLH-2 during C. elegans gonadogenesis. Dev. Biol. 272: 460–469 [DOI] [PubMed] [Google Scholar]
- Krause M., Park M., Zhang J. M., Yuan J., Harfe B., et al. , 1997. A C. elegans E/Daughterless bHLH protein marks neuronal but not striated muscle development. Development 124: 2179–2189 [DOI] [PubMed] [Google Scholar]
- Leight E. R., Glossip D., Kornfeld K., 2005. Sumoylation of LIN-1 promotes transcriptional repression and inhibition of vulval cell fates. Development 132: 1047–1056 [DOI] [PubMed] [Google Scholar]
- Maloof J. N., Kenyon C., 1998. The Hox gene lin-39 is required during C. elegans vulval induction to select the outcome of Ras signaling. Development 125: 181–190 [DOI] [PubMed] [Google Scholar]
- Mann R. S., Lelli K. M., Joshi R., 2009. Hox specificity unique roles for cofactors and collaborators. Curr. Top. Dev. Biol. 88: 63–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D., Ambros V., 1991. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10: 3959–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miley G. R., Fantz D., Glossip D., Lu X., Saito R. M., et al. , 2004. Identification of residues of the Caenorhabditis elegans LIN-1 ETS domain that are necessary for DNA binding and regulation of vulval cell fates. Genetics 167: 1697–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L. M., Gallegos M. E., Morisseau B. A., Kim S. K., 1993. lin-31, a Caenorhabditis elegans HNF-3/fork head transcription factor homolog, specifies three alternative cell fates in vulval development. Genes Dev. 7: 933–947 [DOI] [PubMed] [Google Scholar]
- Miller L. M., Hess H. A., Doroquez D. B., Andrews N. M., 2000. Null mutations in the lin-31 gene indicate two functions during Caenorhabditis elegans vulval development. Genetics 156: 1595–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes M. B., Christensen R. G., Wakabayashi A., Stormo G. D., Brodsky M. H., et al. , 2008. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell 133: 1277–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkema P. G., Ha E., Haun C., Chen W., Fire A., 1997. The Caenorhabditis elegans NK-2 homeobox gene ceh-22 activates pharyngeal muscle gene expression in combination with pha-1 and is required for normal pharyngeal development. Development 124: 3965–3973 [DOI] [PubMed] [Google Scholar]
- Schmitz C., Kinge P., Hutter H., 2007. Axon guidance genes identified in a large-scale RNAi screen using the RNAi-hypersensitive Caenorhabditis elegans strain nre-1(hd20) lin-15b(hd126). Proc. Natl. Acad. Sci. USA 104: 834–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N., Han M., 1995. sur-2, a novel gene, functions late in the let-60 ras-mediated signaling pathway during Caenorhabditis elegans vulval induction. Genes Dev. 9: 2251–2265 [DOI] [PubMed] [Google Scholar]
- Sosinsky A., Bonin C. P., Mann R. S., Honig B., 2003. Target Explorer: An automated tool for the identification of new target genes for a specified set of transcription factors. Nucleic Acids Res. 31: 3589–3592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg P. W., 2005. Vulval development (June, 25, 2005), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.6.1, http://www.wormbook.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram M. V., 2005. The love-hate relationship between Ras and Notch. Genes Dev. 19: 1825–1839 [DOI] [PubMed] [Google Scholar]
- Takacs-Vellai K., Vellai T., Chen E. B., Zhang Y., Guerry F., et al. , 2007. Transcriptional control of Notch signaling by a HOX and a PBX/EXD protein during vulval development in C. elegans. Dev. Biol. 302: 661–669 [DOI] [PubMed] [Google Scholar]
- Tan P. B., Lackner M. R., Kim S. K., 1998. MAP kinase signaling specificity mediated by the LIN-1 Ets/LIN-31 WH transcription factor complex during C. elegans vulval induction. Cell 93: 569–580 [DOI] [PubMed] [Google Scholar]
- Tiensuu T., Larsen M. K., Vernersson E., Tuck S., 2005. lin-1 has both positive and negative functions in specifying multiple cell fates induced by Ras/MAP kinase signaling in C. elegans. Dev. Biol. 286: 338–351 [DOI] [PubMed] [Google Scholar]
- Tihanyi B., Vellai T., Regos A., Ari E., Muller F., et al. , 2010. The C. elegans Hox gene ceh-13 regulates cell migration and fusion in a non-colinear way. Implications for the early evolution of Hox clusters. BMC Dev. Biol. 10: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tursun B., Cochella L., Carrera I., Hobert O., 2009. A toolkit and robust pipeline for the generation of fosmid-based reporter genes in C. elegans. PLoS ONE 4: e4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voutev R., Hubbard E. J., 2008. A “FLP-Out” system for controlled gene expression in Caenorhabditis elegans. Genetics 180: 103–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagmaister J. A., Miley G. R., Morris C. A., Gleason J. E., Miller L. M., et al. , 2006. Identification of cis-regulatory elements from the C. elegans Hox gene lin-39 required for embryonic expression and for regulation by the transcription factors LIN-1, LIN-31 and LIN-39. Dev. Biol. 297: 550–565 [DOI] [PubMed] [Google Scholar]
- Wei G. H., Badis G., Berger M. F., Kivioja T., Palin K., et al. , 2010. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 29: 2147–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson H. A., Fitzgerald K., Greenwald I., 1994. Reciprocal changes in expression of the receptor lin-12 and its ligand lag-2 prior to commitment in a C. elegans cell fate decision. Cell 79: 1187–1198 [DOI] [PubMed] [Google Scholar]
- Woollard A., 2005. Gene duplications and genetic redundancy in C. elegans. WormBook, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Sym M., Kenyon C., 2005. The roles of two C. elegans HOX co-factor orthologs in cell migration and vulva development. Development 132: 1413–1428 [DOI] [PubMed] [Google Scholar]
- Yang S. H., Jaffray E., Hay R. T., Sharrocks A. D., 2003. Dynamic interplay of the SUMO and ERK pathways in regulating Elk-1 transcriptional activity. Mol. Cell 12: 63–74 [DOI] [PubMed] [Google Scholar]
- Yoo A. S., Bais C., Greenwald I., 2004. Crosstalk between the EGFR and LIN-12/Notch pathways in C. elegans vulval development. Science 303: 663–666 [DOI] [PubMed] [Google Scholar]
- Yu S., Avery L., Baude E., Garbers D. L., 1997. Guanylyl cyclase expression in specific sensory neurons: a new family of chemosensory receptors. Proc. Natl. Acad. Sci. USA 94: 3384–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]