Abstract
The fixation probability is determined when population size and selection change over time and differs from Kimura’s result, with long-term implications for a population. It is found that changes in population size are not equivalent to the corresponding changes in selection and can result in less drift than anticipated.
A new mutation in a finite population is subject to genetic drift and its ultimate fate is random: it may be either extinction (loss) or complete establishment (fixation). In a randomly mating population, the typical outcome for a new mutation is its loss, with fixation occurring only with small probability. Under static conditions (constant population size and constant strength of selection) a new beneficial mutation with small selective advantage, s, in a randomly mating population with discrete generations, has only a small probability of fixation: ∼2s when reproduction is treated as a branching process and the number of offspring has a Poisson distribution (Haldane 1927). A deleterious mutation has a yet smaller probability of fixation that is not calculable under a branching process. However, despite the relative rarity of fixation among the fates of all mutations, attention is largely focused on this outcome because the fixation of beneficial mutations plays a central role in the long-term adaptation of populations, and the fixation of deleterious mutations, in the absence of recombination, plays an important role in the long-term survival of populations (Muller 1964; Felsenstein 1974). Our understanding of the rate of such phenomena depends sensitively on the probability of fixation, and deviations from its static value, due to time-dependent conditions, are of particular significance. Indeed, there are a variety of reasons, both abiotic and biotic, why population size and the strength of selection do not generally remain constant over time.
Temporal changes, such as systematic trends in the composition or temperature of the atmosphere or oceans over time, although abiotic in nature, often have major implications for biological systems and may force biotic change. For example, atmospheric temperature changes may affect various biological processes within an organism, but also affect the vegetation on which an organism feeds, thereby affecting both selection and carrying capacity. Thus the general situation is complex, with selection fluctuating for multiple reasons; indeed, “… natural selection is very complicated, it is unlikely that the selection coefficient stays constant” (Ohta 1972, p. 307). Additionally, changes in, for example, resource/habitat availability or the density of parasites or predators will generally change the strength of selection as well as the size of a population. Thus generally we should expect variation in population size and the strength of selection.
The Soay sheep provide an illustration of the interplay of the various factors that affect population size and the strength of selection and the interrelation of these two quantities. The Soay sheep are an intensively studied wild mammalian population and their survival is density dependent and closely tied in with the availability of vegetation, whose quality and abundance are highly variable (Clutton-Brock and Pemberton 2003). Parasite population dynamics have been shown to regulate vertebrate populations and, in the Soay sheep, over-winter survival has been identified with a response of the host’s immune system to parasitic load (Coltman et al. 1999). Milner et al. (2003) concluded that the 20-year study of Soay sheep clearly demonstrated selection intensity fluctuating in a temporal fashion, and during years of high population density mortality was highest and hence selection most intense.
Theoretical studies of the way that population-size change affects fixation have a long history that includes an approximate diffusion analysis by Kimura and Ohta (1974) for logistic population growth and a much more recent investigation by Otto and Whitlock (1997), who considered various scenarios of population-size change, using a generalization of the branching process of Haldane (1927). Among many other things, the intuition established from the work of Otto and Whitlock (1997) is that when a beneficial mutation segregates in a population of increasing size, it has an increased probability of fixation. Subsequent work extended the calculations to include population-size changes of a stochastic nature, either within a branching-process framework (Engen et al. 2009) or for the Moran model (Parsons and Quince 2007; Parsons et al. 2010). The treatment of how changes in the strength of selection in finite populations affect fixation includes work of Kimura and Ohta (1972), while correlated fluctuations in the strength of selection, when population size is finite, have been considered by Takahata et al. (1975). Lambert (2006) combined drift and branching processes (and hence incorporates stochastic number fluctuations) and has obtained results in the regime of weak selection, which is defined by 4Ne|s| ≪ 1, where Ne denotes the effective population size.
The previous work has shown how the probability of fixation is affected by temporal variation in either population size or the strength of selection. In the present work we aim to compare and quantify the different ways that temporal changes in population size and the strength of selection affect fixation. We consider cases where selection may not be weak (i.e., 4Ne|s| may not be small compared with 1). To this end, we present a unified treatment of such temporal variations on the probability of fixation that covers beneficial, neutral/nearly neutral, and deleterious mutations. To cover this range of selective regimes we work within the framework of the diffusion approximation (Kimura 1955a), where the relative frequency of a gene is treated as a random variable that takes continuous values. This approximation derives its name from the diffusion equation that governs the distribution of the relative gene frequency, and a diffusion analysis has been used to derive many fundamental results in population genetics (Crow and Kimura 1970).
Let us begin with the standard case where a single locus determines fitness in a randomly mating diploid sexual population under static conditions (i.e., constant population size and constant strength of selection). (Asexual populations can also be studied via diffusion analysis; for a recent example, see Waxman and Loewe 2010.) The locus has two alleles, denoted A and a, and selection is on viability and is semidominant, with AA, Aa, and aa genotype individuals having relative fitnesses of 1 + 2s, 1 + s, and 1, respectively. Generations are discrete and the processes taking place in one generation are given by the life cycle
We assume that each adult contributes to a very large number zygotes, so that viability selection may be treated as being deterministic in character. Juveniles (the individuals that survive viability selection) undergo a nonselective process of ecological thinning that leads to an adult population of N individuals in each generation.
The proportion of all genes at the locus in adults that are allele A is written X(t); this is the relative frequency (henceforth termed frequency) of allele A. Because of the process of thinning in the life cycle, the frequency, X(t), generally varies randomly from generation to generation and may have different values in different replicates of a population. Statistics of X(t) can be described by a Wright–Fisher model (Fisher 1930; Wright 1931). However, to make theoretical progress, we consider an analysis based on the diffusion approximation, using methods of Kimura (1955), McKane and Waxman (2007), and Waxman (2011).
Readers not concerned with the detailed technical aspects of this work may omit the derivations/proofs contained in the main text and the material in supporting information, File S1.
A diffusion analysis is based on the diffusion equation
| (1) |
for the probability density f(x, t) of the frequency of the A allele at time t and frequency x.
Equation 1 can be solved to determine the probability of fixation of the A allele at long times, where the only possible outcomes for the locus are the A allele fixing or being lost. In terms of the frequency X(t) we have
The probability of occurrence of these different outcomes depends on the frequency, p, of the A allele at the initial time t = 0. The fixation probability can be written as where Ep[…] denotes an average over replicate populations when the A allele frequency has the value p at time t = 0. Thus Ep[…] is a shorthand for the conditional expectation E[… | X(0) = p].
The diffusion approximation of the fixation probability follows from Equation 1 and was found by Kimura (1955b) to be
| (2) |
where S = 4Nes. [The result of Equation 2 was derived assuming discrete generations. A closely related but different result is obtained if it is assumed from the outset that generations are overlapping (Moran 1958).] The numerical errors in the approximation of Equation 2 are remarkably small, even for haploid populations of size 12 (equivalent to diploid populations of size 6) as demonstrated by Ewens (1963).
The fixation probability of a single copy of an A allele in a population of census size N is obtained by setting P = 1/(2N) in Equation 2. When the population size is such that S = 4Nes is large (S >> 1) but Sp ≡ 2Nes/N is small (Sp << 1), we arrive at the approximation Pfix(p) ≃ 2Nes/N, which, when Ne = N, coincides with the leading term, 2s, of a branching process (Haldane 1927). Thus for a constant strength of selection and a constant population size, branching processes are valid for beneficial mutations in populations of suitably large size (S >> 1). Diffusion results have a broader range of applicability that includes beneficial, deleterious and neutral/nearly neutral mutations in populations of essentially arbitrary size; in such an approach, there are essentially no restrictions on the sign and size of S).
When the effective population size and the selection coefficient depend on the time t, the parameter S acquires time dependence and becomes S(t) = 4Ne(t)s(t). We initially proceed under the assumption that all changes in the composite quantity S(t) are deterministic in character and have the property that they cease after a finite time, which we denote T. That is to say for times ≥T we take S(t) to have a constant value, and schematically
| (3) |
Such an assumption on S(t) is not greatly restrictive. It could, for example, describe the situation where the strength of selection remains constant, while population size alone changes for a finite time before achieving a constant value. While logistic growth of a population does not precisely cease after a finite time, the population size can be well approximated as achieving its carrying capacity after a finite time and hence closely fits within the framework of Equation 3. Beyond such a case, Equation 3 could also describe environmental change of finite duration, which affects both population size and selection.
Generalization of Pfix(p)
The generalization of Equation 2 to the case of time-dependent population sizes, Ne(t), and time-dependent selection coefficients, s(t), can be obtained from some basic considerations. For deterministic changes in Ne(t) and s(t) that lead to the composite quantity S(t) = 4Ne(t)s(t) having the form in Equation 3, the generalization of Equation 2 is
| (4) |
(the derivation of Equation 4 is given in the following paragraph and an alternative derivation is given in Part 1 of File S1). The only statistic required in Equation 4 involves X(T), the random value of the A allele frequency at time T. The statistic in question, represents the average of over all replicate populations where the initial frequency is p at time t = 0 [i.e., having X(0) = p].
Derivation
To derive Equation 4 we note that as in the static case, the fixation probability can be written as Conditioning on the value of X(t) at time T [i.e., after changes in Ne(t) and s(t) have taken place], we have The quantity appearing in the last expression can be written as and follows from Equation 2 with the substitution p → X(T). We obtain and hence the generalization of Equation 2 is which is equivalent to Equation 4.
We note that the solution for Pfix(p) in Equation 4, which directly follows from a diffusion analysis, does not generally agree with the forms assumed by Kimura and Ohta (1972, 1974) for situations of changing selection strength or changing population size. Additionally, Equation 4 will not be compatible with a branching-process approach when S∞ is not large (S∞ ≲ 1) or indeed when S∞ is zero or negative; there is limited validity to a branching-process treatment.
Before considering the numerical estimation of the fixation probability from Equation 4, we investigate some limiting cases and properties of Equation 4. Some of the limiting cases allow us to verify that Equation 4 is in accordance with well-known/well-understood results.
Limiting cases
When the time-interval T, over which all change occurs, tends to zero there is a negligibly short period of time dependence in the parameters. It can be shown (see Part 2 of File S1) that as a consequence, the frequency X(T) is unaffected by these changes and tends to its initial value: X(T) → p. Equation 4 then collapses to Kimura’s result, Equation 2, for a population of constant size and constant selection coefficient.
When the problem is static, in the sense that neither population size nor the strength of selection changes over time [i.e., Ne(t) = Ne and s(t) = s], it should follow that Equation 4 coincides with Kimura’s result, Equation 2, which covers the static case. In this case, T can be taken to have any nonnegative value and it can be shown (see Part 3 of File S1) that under static conditions, the expectation appearing in Equation 4, namely , is independent of T and hence equals its value at T = 0, which is . As a consequence, Equation 4 collapses to Kimura’s result. The property of of being independent of T is a well-known Martingale property of the diffusion approximation of the Wright–Fisher model; it appears to have been first identified as a property of the diffusion approximation by Ewens (1964).
When S∞ becomes vanishingly small (S∞ → 0), Equation 4 reduces to Pfix(p) = Ep[X(T)]. Thus when there is selective neutrality after time T, the probability of fixation equals the mean allele frequency at time T.
When S∞ becomes large and positive (S∞ → +∞), the result in Equation 4 depends only on the probability that X(T) ≠ 0. Equation 4 leads to Pfix(p) = 1 − Ploss(T; p), where Ploss(T; p) is the probability of loss of the A allele by time T given it had a frequency of p at time t = 0 (the proof is given in the following paragraph). Thus in an environment that has strongly positive selection after time T, the probability of fixation is determined by the probability that the A allele has not been lost by time T. Any A alleles that are present in a population after time T will subsequently fix.
Proof. The result Pfix(p) = 1−Ploss(T; p) follows since corresponds to the indicator function 1{X(T)>0}, which has the value of unity when X(T) > 0 and vanishes when X(T)=0. This indicator function coincides with 1 − 1{X(T)=0} and thus Equation 4 becomes Pfix(p)=1 − Ep[1{X(T)=0}]=1 − Ploss(T; p), where Ploss(T; p) is the probability that X(T)=0, given X(0)=p.
When S∞ becomes large and negative (S∞ → −∞), the result in Equation 4 depends only on the probability that X(T) = 1. Equation 4 leads to Pfix(p) = Pfix(T; p), namely the probability of fixation of the A allele by time T, given it had a frequency of p at time t = 0 (the proof is given in the following paragraph). Thus in an environment that has strongly negative selection after time T, the probability of fixation is determined by fixations that occur up to time T; after time T any A alleles present in a population will be lost.
Proof. The result Pfix(p) = Pfix(T; p) follows since which corresponds to the indicator function 1{X(T)=1}. Equation 4 then becomes Pfix(p) = Ep[1{X(T)=1}] = Pfix(T; p), where Pfix(T; p) is the probability that X(T) = 1, given X(0) = p.
The above results may give the impression that only the time dependence of the parameter S(t) = 4Ne(t)s(t) is of significance for the fixation probability, Pfix(p), of Equation 4, and that Pfix(p) is not sensitive to the way that Ne(t) and s(t) separately change. This is not generally true. To illustrate this, let us consider two cases where either Ne(t) or s(t) linearly increases by a factor of f in time T and then remains constant after this:
- Case a:
- Case b:
Both case a and case b lead to
yet they lead to different values for the fixation probability. As a concrete example of the fixation probabilities that can arise for case a and case b, consider the parameter values N0 = 10, f = 10, T = 10, s0 = 0.04, and P = 1/(2N0) = 0.05. From simulations we find that case a leads to a fixation probability of Pfix(p) ≃ 0.363 while case b leads to Pfix(p) ≃ 0.275. [All simulations carried out in this work are made within the framework of a Wright–Fisher model (Fisher 1930; Wright 1931). In such a framework, selection is treated as a deterministic process, and only the random sampling of individuals without regard to type, i.e., the process of random genetic drift, is treated stochastically.] In this example, S(t) increases by a factor of 10 in each case but when it is just Ne(t) that varies over time, the probability of fixation is ∼30% larger than when just s(t) varies.
The results of cases a and b illustrate the more general phenomenon that, as far as fixation is concerned, the way that S(t) = 4Ne(t)s(t) varies over time is not the full story: the probability of fixation generally depends on the source of the variation of S(t).
The phenomenon that a varying Ne or a varying s has nonequivalent effects on the probability of fixation follows ultimately from the way that a changing Ne modifies the timescale of random genetic drift. Since we shall compare populations with the same initial size, Ne(0), we take Ne(0) as a fixed parameter and find it convenient to take the timescale associated with genetic drift to be
| (5) |
We call τ the drift time. We can express t as a function of the drift time, τ, and write t = t(τ). The form of t(τ) follows from solving Equation 5 for t. Let us now explain the advantage of using the drift time, τ, instead of the actual time, t.
Assuming a time-dependent population size and a time-dependent strength of selection, we transform the diffusion equation, Equation 1, so that the drift time τ (Equation 5) is used instead of the actual time t. It can be shown (see Part 4 of File S1) that under such a transformation, the quantity playing the role of the strength of selection in the resulting diffusion equation is R(τ) = 4Ne(t(τ))s(t(τ)) and we call R(τ) the overall strength of selection. The quantity Ne(t) does not appear elsewhere in the transformed diffusion equation. In a static problem, the probability of fixation is determined by just the initial frequency, p, and the overall strength of selection (see Equation 2). In a more general case, where Ne(t) and s(t) exhibit deterministic change over time, the probability of fixation is determined by p, and the entire history of R(τ), from τ = 0 onward.
We note that when population size is static (Ne(t) = Ne(0)), the drift time and the actual time coincide: t(τ) = τ, independent of any variation of s. Thus for a static population size but a changing strength of selection, the overall strength of selection is
| (6) |
By contrast, when Ne varies with time, the drift time does not generally coincide with the actual time: t(τ) ≠ τ. In such a case, the overall strength of selection, when s has the fixed value s(0), is
| (7) |
There is a key difference between Equations 6 and 7: the time that s depends on in Equation 6 is simply the time we have adopted for the diffusion equation, namely τ. By contrast, in Equation 7, the time that Ne depends upon is t(τ). It can be shown (see Part 4 of File S1) that irrespective of whether Ne(t) increases with time or whether it decreases with time, the quantity Ne(t(τ)) generally satisfies
| (8) |
This inequality stems directly from the modification of the timescale induced by an Ne that exhibits either increase or decrease. [For an Ne(t) that changes monotonically, the inequality in Equation 8 becomes replaced with Ne(t(τ)) ≥ Ne(τ).] It means the population size Ne(t(τ)) in the transformed diffusion equation is larger than the value of the population size that we might believe is relevant at time τ, namely Ne(τ).
All other things being equal, a population whose size is larger than anticipated exhibits less drift than anticipated. This is the reason case a above, for a positively selected allele, leads to a larger probability of fixation than case b. Similarly, the inequality in Equation 8 indicates that a negatively selected allele, when present in a population that increases, or one that decreases, will have a reduced probability of fixation (due to less drift) compared with the case where population size is static and all variation occurs in the strength of selection.
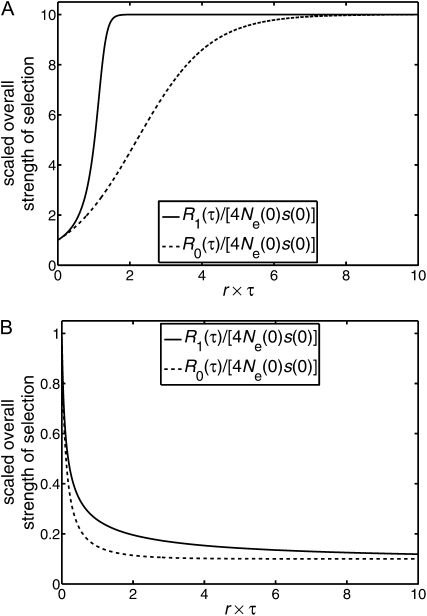
In Figure 1 we illustrate the forms of the “overall strengths of selection,” R0(τ) and R1(τ), of Equations 6 and 7 to show how different the overall strength of selection can be under “fixed Ne, varying s” and “varying Ne, fixed s.” For definiteness, Figure 1 is restricted to alleles with positive selection coefficients.
Figure 1 .
Scaled overall strength of selection. We present plots of the scaled “overall strength of selection,” namely R(τ) = 4Ne(t(τ))s(t(τ)) divided by its initial value 4Ne(0)s(0). Different scenarios of change are illustrated, where the selection coefficient s(t) is positive for all t. We used the two functions a(t) and b(t) to produce this figure. The function a(t) corresponds to logistic growth by a factor of 10; i.e., a(t) = (1/10 + 9/10e−rt)−1, where r is a positive constant, and hence a(0) = 1 while a(∞) = 10. The function b(t) corresponds to logistic decay by a factor of 10; i.e., b(t) = (10 − 9e−rt)−1 and hence b(0) = 1 while b(∞) = 1/10. (A) We compare (i) R0(τ) for a fixed effective population size and a logistically increasing selection coefficient (Ne(t) = Ne(0), s(t) = s(0)a(t)), with (ii) R1(τ) for a logistically increasing effective population size and a fixed selection coefficient (Ne(t) = Ne(0)a(t), s(t) = s(0)). (B) We compare (iii) R0(τ) for a fixed effective population size and a logistically decreasing selection coefficient (Ne(t) = Ne(0), s(t) = s(0)b(t)), with (iv) R1(τ) for a logistically decreasing effective population size and a fixed selection coefficient (Ne(t) = Ne(0)b(t), s(t) = s(0)). In A, an increasing population size leads to a larger overall strength of selection than that of an increasing selection coefficient. In B, a decreasing population size leads again to a larger overall strength of selection than that of a decreasing selection coefficient. These properties result from either an increasing population size or a decreasing population size modifying the natural timescale of the diffusion equation in such a way that there is less drift than might be anticipated (see Equation 8).
When Ne(t) exhibits periods of both increase and decrease, no inequality of the form in Equation 8 generally holds.
Estimation of the probability of fixation
Consider now how we would estimate the probability of fixation, for a case where potentially complicated deterministic changes of s(t) and Ne(t) take place up to a finite time, T. A direct approach would simply be to simulate the behavior of many replicates of a population. Each replicate population would need to be “followed” for a sufficient number of generations until either fixation or loss occurred. The fraction of such populations that fix is an estimate of the fixation probability. By contrast, using Equation 4 it is necessary to follow replicate populations for, at most, only T generations. On average it will require less than T generations, since fixation or loss will occur in some replicate populations prior to time T, and no further change will occur in such populations. An estimate of the fixation probability follows from such simulations by using the average of in Equation 4. This procedure may be significantly shorter than the direct approach, depending on the parameter values in the problem (Table 1 illustrates differences in the time required). Alternatively, the quantity that appears in Equation 4 can be estimated from numerical solution of the backward diffusion equation (see Part 5 of File S1). This thus provides an alternative route to estimation of the fixation probability.
Table 1 . Illustrative numerical results for the probability of fixation when the composite quantity S(t) = 4Ne(t)s(t) changes with time.
| Data set | Method | T | N0 | N∞ | s0 | s∞ | S0 | S∞ | Pfix | Cost/rep |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Direct simulation | 20 | 50 | 50 | 0.005 | 0.050 | 1 | 10 | 0.0687 | 12 |
| 2 | Finite T simulation | 20 | 50 | 50 | 0.005 | 0.050 | 1 | 10 | 0.0689 | 4 |
| 3 | Direct simulation | 20 | 50 | 500 | 0.005 | 0.005 | 1 | 10 | 0.0817 | 115 |
| 4 | Finite T simulation | 20 | 50 | 500 | 0.005 | 0.005 | 1 | 10 | 0.0818 | 9 |
| 5 | Direct simulation | 200 | 50 | 50 | 0.005 | 0.050 | 1 | 10 | 0.0320 | 11 |
| 6 | Finite T simulation | 200 | 50 | 50 | 0.005 | 0.050 | 1 | 10 | 0.0321 | 10 |
| 7 | Direct simulation | 200 | 50 | 500 | 0.005 | 0.005 | 1 | 10 | 0.0451 | 51 |
| 8 | Finite T simulation | 200 | 50 | 500 | 0.005 | 0.005 | 1 | 10 | 0.0447 | 18 |
| 9 | Direct simulation | 20 | 500 | 500 | 0.005 | 0.050 | 10 | 100 | 0.0672 | 18 |
| 10 | Finite T simulation | 20 | 500 | 500 | 0.005 | 0.050 | 10 | 100 | 0.0671 | 4 |
| 11 | Direct simulation | 20 | 500 | 5000 | 0.005 | 0.005 | 10 | 100 | 0.0814 | 192 |
| 12 | Finite T simulation | 20 | 500 | 5000 | 0.005 | 0.005 | 10 | 100 | 0.0812 | 8 |
| 13 | Direct simulation | 200 | 500 | 500 | 0.005 | 0.050 | 10 | 100 | 0.0298 | 14 |
| 14 | Finite T simulation | 200 | 500 | 500 | 0.005 | 0.050 | 10 | 100 | 0.0302 | 11 |
| 15 | Direct simulation | 200 | 500 | 5000 | 0.005 | 0.005 | 10 | 100 | 0.0428 | 90 |
| 16 | Finite T simulation | 200 | 500 | 5000 | 0.005 | 0.005 | 10 | 100 | 0.0425 | 17 |
We take S(t) to start at the positive value S0 at time t = 0 and then to linearly increase to the value S∞ = 10S0 by time T and then remain constant at the value S∞ for all times larger than T. Two different methods are used to estimate the probability of fixation when initially there is only a single copy of an A allele: (i) direct simulation, where we “follow” each replicate population until either fixation or loss occurs and (ii) simulations based on Equation 4, where we follow each replicate population for, at most, T generations. The column labeled “Cost/rep” gives the mean number of generations a replicate population was followed in a simulation. In the simulations, 5 × 105 replicate populations were used, and we adopted a Wright–Fisher model where the only place in the life cycle where randomness occurs is in the thinning of the number of individuals to Ne = N adults. The initial values of Ne and s are N0 and s0, while the final values are N∞ and s∞; data sets 1, 2, 5, 6, 9, 10, 13, and 14 correspond to Ne fixed and s changing with time. It is evident from the table that there are differences in the fixation probability, depending on whether Ne changed with time, at fixed s, or s changed with time at fixed Ne.
The different approaches to the calculation of the fixation probability are illustrated with an example in Table 1.
Stochastic fluctuations
So far we have presented results of the probability of fixation for cases where the time-dependent changes in the population size and the strength of selection are deterministic in character. Let us now point out a generalization of these results that includes stochastic fluctuations in population size and the strength of selection.
We note that Lambert (2006) obtained results in the regime of weak selection, when there is fluctuating population size and random genetic drift, by combining branching and Wright–Fisher processes, while Parsons et al. (2010) considered the effects of fluctuating population size, in a quasi-neutral (i.e., weak selection) situation, where different alleles have the same ratio of intrinsic birth to death rates. The work of Parsons and Quince (2007) covers the nonneutral regime and includes density dependence and fluctuations in population size that arise from uncorrelated stochastic births and deaths. By contrast, Karlin and Levikson (1974) considered the case where stochastic changes in Ne(t) and s(t) have temporal correlations that relate the values of these quantities in adjacent generations. These authors found that various statistics of Ne(t) and s(t) make contributions to the drift and diffusion coefficients of the diffusion equation. Here, we make the alternative assumption that the stochastic fluctuations of Ne(t) and s(t) have temporal autocorrelations that decay slowly, over very many generations. It is then possible to account for these fluctuations using the approach of Takahata et al. (1975); see also Huerta-Sanchez et al. (2008) where two different models of autocorrelation are incorporated into stochastic fluctuations of selection.
To generalize Equation 4, we first assume there are both deterministic changes and stochastic fluctuations in Ne(t) and s(t) for times t ≤ T, but only stochastic fluctuations for times t > T. Then the appropriate generalization of Equation 4 for this case is , where is an eigenfunction of an averaged backward diffusion operator: see Part 6 of File S1 for further details.
To summarize: in this work we have presented results, based on the diffusion approximation, which generalize Kimura’s result for the probability of fixation to cases where population size and the strength of selection are time dependent. We have provided results when the changes are deterministic and also shown their generalization when there are stochastic fluctuations with temporal autocorrelations that decay over many generations. This work has implications for the long-term adaptation of populations, demonstrating that while temporal variation in population size and the strength of selection both affect the probability of fixation, the changes are not equivalent and that generally, a population size that either increases or decreases will lead to less drift and hence less fixation of deleterious mutations and greater fixation of beneficial mutations than would otherwise be anticipated. There are many possible scenarios where population size and the strength of selection change, and the result of Equation 4 allows an efficient way to explore the implications of these for fixation.
Acknowledgments
I thank Warren Ewens, Jianfeng Feng, Guillaume Martin, Yannis Michalakis, and Andy Overall for stimulating discussions and the latter for very helpful suggestions on the manuscript. I also thank the two anonymous reviewers for comments and suggestions that substantially improved this work.
Note: See Uecker and Hermisson (pp. 915–930) in this issue, for a related work.
Literature Cited
- Coltman D. W., Pilkington J. G., Smith J. A., Pemberton J. M., 1999. Parasite-mediated selection against inbred Soay sheep in a free-living, island population. Evolution 53: 1259–1267 [DOI] [PubMed] [Google Scholar]
- Clutton-Brock T. H., Pemberton J. M., 2003. Soay Sheep: Dynamics and Selection in an Island Population. Cambridge University Press, Cambridge, UK [Google Scholar]
- Crow J. F., Kimura M., 1970. An Introduction to Population Genetics Theory. Harper & Row, New York [Google Scholar]
- Engen S., Lande R., Saether B., 2009. Fixation probability of beneficial mutations in a fluctuating population. Genet. Res. 91: 73–82 [DOI] [PubMed] [Google Scholar]
- Ewens W. J., 1963. Numerical results and diffusion approximations in a genetic process. Biometrika 50: 241–249 [Google Scholar]
- Ewens W. J., 1964. The pseudo-transient distribution and its uses in genetics. J. Appl. Probab. 1: 141–156 [Google Scholar]
- Felsenstein J., 1974. The evolutionary advantage of recombination. Genetics 78: 737–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. A., 1930. The Genetical Theory of Natural Selection. Clarendon Press, Oxford [Google Scholar]
- Haldane J. B. S., 1927. A mathematical theory of natural and artificial selection. V. Selection and mutation. Proc. Camb. Philos. Soc. 23: 838–844 [Google Scholar]
- Huerta-Sanchez M., Durrett R., Bustamante C. D.., 2008. Population genetics of polymorphism and divergence under fluctuating selection. Genetics 178: 325–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlinson S., Levikson B., 1974. Temporal fluctuations in selection intensities: Case of small population size. Theor. Pop. Biol. 6: 383–412 [Google Scholar]
- Kimura M., 1955a. Stochastic processes and distribution of gene frequencies under natural selection. Cold Spring Harbor Symp. Quant. Biol. 20: 33–53 [DOI] [PubMed] [Google Scholar]
- Kimura M., 1955b. Solution of a process of random genetic drift with a continuous model. Proc. Natl. Acad. Sci. USA 41: 141–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., Ohta T., 1972. Probability of fixation of a mutant gene in a finite population when selective advantage decreases with time. Genetics 65: 525–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., Ohta T., 1974. Probability of gene fixation in an expanding finite population. Proc. Natl. Acad. Sci. USA 71: 3377–3379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert A., 2006. Probability of fixation under weak selection: a branching process unifying approach. Theor. Popul. Biol. 69: 419–441 [DOI] [PubMed] [Google Scholar]
- Milner J. M., Albon S. D., Kruk L. E. B., Pemberton J. M., 2003. Selection on phenotype, pp. 190–216 Soay Sheep: Dynamics and Selection in an Island Population, edited by Clutton-Brock T. H., Pemberton J. M. Cambridge University Press, Cambridge, UK [Google Scholar]
- Moran P. A. P., 1958. Random processes in genetics. Math. Proc. Camb. Philos. Soc. 54: 60–71 [Google Scholar]
- McKane A. J., Waxman D., 2007. Singular solutions of the diffusion equation of population genetics. J. Theor. Biol. 247: 849–858 [DOI] [PubMed] [Google Scholar]
- Muller H. J., 1964. The relation of recombination to mutational advance. Mutat. Res. 1: 2–9 [DOI] [PubMed] [Google Scholar]
- Ohta T., 1972. Population size and the rate of evolution. J. Molec. Evol 1: 305–314 [PubMed] [Google Scholar]
- Otto S. P., Whitlock M., 1997. The probability of fixation in populations of changing size. Genetics 146: 723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons T. L., Quince C., 2007. Fixation in haploid populations exhibiting density dependence I: the non-neutral case. Theor. Popul. Biol. 72: 121–135 [DOI] [PubMed] [Google Scholar]
- Parsons T. L., Quince C., Plotkin J. B., 2010. Some consequences of demographic stochasticity in population genetics. Genetics 185: 1345–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N., Ishii K., Matsuda H., 1975. Effect of temporal fluctuation of selection coefficient on gene frequency in a population. Proc. Natl. Acad. Sci. USA 72: 4541–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman D., 2011. Comparison and content of the Wright–Fisher model of random genetic drift, the diffusion approximation, and an intermediate model. J. Theor. Biol. 269: 79–87 [DOI] [PubMed] [Google Scholar]
- Waxman D., Loewe L., 2010. A stochastic model for a single click of Muller’s ratchet. J. Theor. Biol. 264: 1120–1132 [DOI] [PubMed] [Google Scholar]
- Wright S., 1931. Evolution in Mendelian populations. Genetics 16: 97–159 [DOI] [PMC free article] [PubMed] [Google Scholar]