Abstract
The Drosophila melanogaster anterior–posterior axis is established during oogenesis by the localization of bicoid and oskar mRNAs to the anterior and posterior poles of the oocyte. Although genetic screens have identified some trans-acting factors required for the localization of these transcripts, other factors may have been missed because they also function at other stages of oogenesis. To circumvent this problem, we performed a screen for revertants and dominant suppressors of the bicaudal phenotype caused by expressing Miranda–GFP in the female germline. Miranda mislocalizes oskar mRNA/Staufen complexes to the oocyte anterior by coupling them to the bicoid localization pathway, resulting in the formation of an anterior abdomen in place of the head. In one class of revertants, Miranda still binds Staufen/oskar mRNA complexes, but does not localize to the anterior, identifying an anterior targeting domain at the N terminus of Miranda. This has an almost identical sequence to the N terminus of vertebrate RHAMM, which is also a large coiled-coil protein, suggesting that it may be a divergent Miranda ortholog. In addition, we recovered 30 dominant suppressors, including multiple alleles of the spectroplakin, short stop, a lethal complementation group that prevents oskar mRNA anchoring, and a female sterile complementation group that disrupts the anterior localization of bicoid mRNA in late oogenesis. One of the single allele suppressors proved to be a mutation in the actin nucleator, Cappuccino, revealing a previously unrecognized function of Cappuccino in pole plasm anchoring and the induction of actin filaments by Long Oskar protein.
THE subcellular localization of mRNAs is an important mechanism for restricting specific proteins to the region of a cell where they are required and plays a key role in axis formation, synaptic plasticity, and cell polarity (St. Johnston 2005; Becalska and Gavis 2009). Indeed, nearly 70% of all tested mRNAs show some pattern of localization in the early Drosophila embryo, indicating this is a widespread means for protein targeting (Lécuyer et al. 2007). mRNAs can be delivered to the correct destination in a variety of ways, such as local protection from degradation, diffusion and anchoring, or active transport along either the actin or microtubule cytoskeletons. One of the best-characterized systems for studying the latter mechanism is the formation of the anterior–posterior axis in Drosophila, which is specified by the microtubule-dependent localization of bicoid mRNA to the anterior of the oocyte and of oskar mRNA to the posterior (Bastock and St. Johnston 2008). bicoid mRNA is translated at the anterior after egg activation to produce a protein gradient that acts as a morphogen to pattern the head and thorax of the embryo (Ephrussi and Johnston 2004). oskar mRNA, on the other hand, is translated once it is localized to the posterior pole of the oocyte, where Oskar protein defines the site of pole plasm assembly, leading to the posterior recruitment of the abdominal determinant, nanos RNA (Ephrussi et al. 1991; Kim-Ha et al. 1991).
Mutants that disrupt the localization of bicoid mRNA produce embryos with defective heads, whereas oskar mRNA localization mutants result in embryos without pole cells or an abdomen, and this has allowed the identification of a number of trans-acting factors in screens for maternal-effect lethal mutations (Nusslein-Volhard et al. 1987; Schupbach and Wieschaus 1989). One limitation of these screens is that they could not identify zygotic lethal mutations in essential genes. This problem can be circumvented, however, by using the FLP/FRT system to perform screens in germ line clones, and additional factors have been identified in such screens for mutants with embryonic patterning phenotypes, as well as more targeted screens for mutations that disrupt the localization of GFP–Staufen, an RNA-binding protein that associates with oskar and bicoid mRNAs (Chou and Perrimon 1996; Martin et al. 2003; Luschnig et al. 2004).
Analyses of the mutants that affect oskar mRNA localization have revealed that its localization to the posterior of the oocyte occurs in multiple steps. The RNA is transcribed in the nurse cells of the germline cyst and first moves along microtubules through the ring canals to the anterior of the oocyte in a process that is probably mediated by dynein (Clark et al. 2007). The mRNA is then transported by kinesin along a weakly polarized microtubule cytoskeleton to the oocyte posterior (Brendza et al. 2000; Zimyanin et al. 2008). This step requires a number of factors that associate with the RNA, including HRP48, Tropomyosin II, the exon junction complex components, Mago nashi, Y14, Barentsz and eIF4AIII, and the dsRNA-binding protein, Staufen (Newmark and Boswell 1994; Hachet and Ephrussi 2001; Mohr et al. 2001; Hachet and Ephrussi 2004; Palacios et al. 2004).
Once localized, oskar mRNA is translated from two alternative in-frame start codons to produce long and short forms of Oskar that have distinct functions. Long Oskar is required for the anchoring of oskar mRNA and all other pole plasm components, whereas Short Oskar nucleates the formation of pole plasm (Vanzo and Ephrussi 2002). Oskar stimulates endocytosis at the posterior pole by recruiting a number of endocytic factors and also induces the formation of long actin filaments, and both of these activities are thought to be required for anchoring, although the exact mechanisms involved have yet to be resolved (Vanzo and Ephrussi 2002; Vanzo et al. 2007; Tanaka and Nakamura 2008).
The localization of bicoid mRNA also occurs in multiple steps during oogenesis, although genetic screens have been less successful at identifying the necessary factors (St. Johnston et al. 1989). Like oskar, bicoid mRNA is transcribed in the nurse cells and transported by dynein into the oocyte (Clark et al. 2007; Mische et al. 2007). It is then localized to the anterior cortex of the oocyte in a poorly understood process that requires Exu protein (Berleth et al. 1988; Cha et al. 2001). Several additional proteins are required to keep bicoid mRNA at the anterior of the oocyte after stage 10a of oogenesis, including Swallow, γ-tubulin 37C, the γ-tubulin ring complex components, GRIP75 and GRIP128, and the dynein light chain (St. Johnston et al. 1989; Schnorrer et al. 2002; Vogt et al. 2006; Weil et al. 2006, 2010).
Although the RNA-binding proteins that recognize bicoid mRNA to mediate early localization have not been identified, these latter stages require Staufen and the ESCRT II complex, which binds specifically to a region of the bicoid 3′ UTR (Ferrandon et al. 1994; Irion and St. Johnston 2007). The localization of bicoid mRNA changes again in mature oocytes, and the RNA becomes stably anchored to the actin cortex until the egg is activated, which releases bicoid mRNA/Staufen complexes into the egg cytoplasm and activates Bicoid translation (Weil et al. 2008).
Although many of the factors required for oskar and bicoid mRNA localization have already been identified, there are still a number of important gaps in our understanding of these processes. For example, it is not known how oskar mRNA translation is activated at the posterior of the oocyte or how increased endocytosis and actin filaments collaborate to anchor the mRNA, while only one of the specific factors required for the first phase of bicoid mRNA localization has been identified so far. There are several reasons why some important localization factors may have been missed in previous genetic screens. Some mutants might block development before the phenotype could be scored in the oocyte or embryo or might lie in regions of the genome that have not been screened in germline clone screens. As an alternative approach, we have performed a screen for dominant suppressors of a dominant bicaudal phenotype caused by expressing a Miranda–GFP fusion protein in the female germline that allows us to screen the entire genome at the same time. Because the suppressor mutations are heterozygous, this screen can also identify lethal mutations and mutants with pleiotropic phenotypes that mask an effect on mRNA localization.
Miranda is a large adaptor protein that mediates the basal targeting of the cell fate determinants, Prospero and Brat, during asymmetric neuroblast divisions (Ikeshima-Kataoka et al. 1997; Shen et al. 1997). In addition, Miranda binds directly to the C-terminal domain of Staufen to localize Staufen/prospero mRNA complexes to the basal cortex of the neuroblast (Broadus et al. 1998; Matsuzaki et al. 1998; Schuldt et al. 1998; Shen et al. 1998). Miranda is not normally expressed in the female germline, but binds to Staufen/oskar mRNA complexes when expressed ectopically and localizes with them to both the anterior and posterior of the oocyte (Irion et al. 2006). While the posterior localization reflects Miranda hitchhiking on the normal oskar mRNA localization pathway, the anterior localization occurs because Miranda couples Staufen/oskar mRNA complexes to the bicoid mRNA localization pathway, leading to the anterior formation of pole plasm and the recruitment of nanos RNA. This results in the development of embryos with pole cells at both ends and an anterior abdomen in place of the head and thorax because Nanos blocks the translation of bicoid and hunchback RNAs (Wharton and Struhl 1991; Ephrussi and Lehmann 1992).
Although the Miranda–GFP bicaudal phenotype is highly penetrant, it seems to be a good system to select for suppressors, because the normal posterior localization pathway and the Miranda-dependent anterior localization pathway compete for Staufen/oskar mRNA complexes. Any mutants that partially impair the anterior pathway will therefore result in most oskar mRNA localizing posteriorly, thereby suppressing the phenotype. The phenotype is also very sensitive to the amount of Oskar protein produced anteriorly, and mutants that reduce Oskar translation or anchoring would also be predicted to be suppressors. We therefore performed a larger-scale screen for dominant suppressors of the female sterility caused by Miranda–GFP to identify novel factors involved in bicoid mRNA localization and oskar mRNA translation and anchoring.
Materials and Methods
Drosophila stocks
As the Miranda–GFP stock, we used an insertion of P w+ matα4tub:Miranda–GFP at 18D11 on the X chromosome, which was maintained over the attached X chromosome, C(1) DX, y w f (Irion et al. 2006). The stock therefore contains only Mira–GFP/Y males and X^X/Y females, so that Miranda–GFP transgene is not expressed until the males are outcrossed to normal females. Other stocks used were dpov1; e1 (Bloomington Stock Center); UASp:oskar-bicoid 3′UTR, UASp:oskar(M1L)-bicoid 3′UTR, UASp:oskar(M139L)-bicoid 3′ UTR (Tanaka and Nakamura 2008), Df(2L)edSZ1 (Presgraves 2003); spireRP (Manseau and Schupbach 1989), spire2F (Wellington et al. 1999); capuEY13172 (Bellen et al. 2004); UASp:Capu-GFP and UASp:CapuΔN-GFP (Dahlgaard et al. 2007), nos-Gal4-VP16 (Van Doren et al. 1998); Prospero-GAL4 (Shiga et al. 1996); Df(3R)oraI9 (Shen et al. 1997); and UASp:Mira, UASp:Mira 1B2-2, UASp: Mira 1A4-1 (generated for this work). The mapping chromosomes wgSp-1 Bl1 Lrm Bc1 Pu2 PinB/SM5 and al1 dpov1 b1 pr1 c1 px1 sp1 were obtained from Bloomington Stock Center. Germline clones were generated by heat shocking hsFLP; FRT G13 Su(Mir)/ FRT G13 ovoD1 larvae for 2 hr on 3 consecutive days (Chou and Perrimon 1996).
Mutagenesis and screen for Miranda suppressors
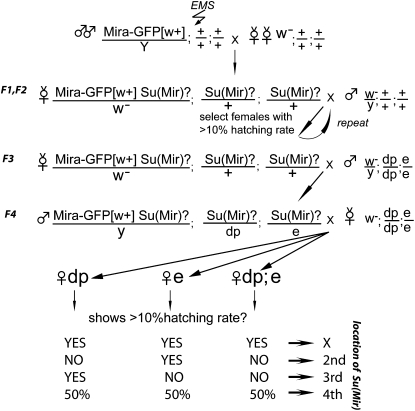
Miranda–GFP males were starved for 6 hr before being fed 30 mm EMS (Sigma) in 1% sucrose for 18 hr to induce an average of one lethal hit per chromosome. Mutagenized males were mated with w1118 virgin females. Single w− P[w+:Miranda–GFP], Su?/+; Su?/+; Su?/+ virgin females were mated with w1118 males. Females with hatching rate greater than 10% were selected for further analysis as bearing a potential suppressor mutation. The same selection procedure was repeated for an additional generation. To map suppressors to a chromosome, single w− P[w+:Miranda–GFP], Su?/+; Su?/+; Su?/+ virgin females were mated with w−; dp; e males and then single w− P[w+:Miranda–GFP], Su?/+; Su?/dp; Su?/e males were mated to w-; dp; e virgin females. The hatching rate of several females of genotypes dp, e or dp;e were tested. If suppression was observed, e or dp males from the same cross were mated to virgin females with a corresponding balancer (second or third chromosomes) (see Figure 1) and balanced stocks were established. For X chromosome linked suppressors, virgin females from F3 (see Figure 1) were used to establish balanced stocks. Identified suppressors were rescreened to confirm the presence of suppression and chromosomal location.
Figure 1 .
Crossing scheme for isolating and mapping Suppressors of Miranda.
Staining and microscopy
Antibody stainings and in situ hybridizations were performed according to standard protocols. Actin was visualized by fixing ovaries in 4% formaldehyde for 20 min and staining with rhodamine-phalloidin (1:500; Invitrogen). For better visualization of Long Oskar-induced actin filaments, the actin mesh fixation protocol was used (Dahlgaard et al. 2007). Ovaries were fixed in 10% formaldehyde for 30 min and stained with rhodamine-phalloidin overnight. Guinea pig anti-Oskar antibody was raised against amino acid residues 292–606 and used at 1:200; mouse anti-pH 3 was used at 1:200 (Cell Signaling), rabbit anti-GFP was used at 1:200 (AMS Biotechnology). Antisense oskar RNA probes for in situ hybridizations were synthesized using DIG RNA Labeling mix (Roche). Cy3-conjugated IgG mouse anti-DIG antibody (Jackson ImmunoResearch Laboratories) was used at 1:200.
Imaging was performed using a Zeiss LSM510 confocal microscope (Carl Zeiss MicroImaging, Inc.) and LSM510 AIM software. Images were processed using ImageJ and Adobe Photoshop.
Molecular biology
NotI–GFP–Strep–ProtA–SacII was cloned into pUASp-PL to make UAS-GFP-Strep-ProtA construct. “Wild-type” miraRR127 (coding region corresponding to amino acids 1–727), and the mira1B2-2 and mira1A4-1 revertants were cloned via KpnI/NotI to pUASp–GFP–Strep–ProtA vector to make UAS–mira–GFP–Strep–ProtA constructs.
Results
We initially set out to map the domains of Miranda that mediate its localization to the anterior of the oocyte by screening for revertants of the dominant bicaudal phenotype caused by Miranda–GFP expression. In addition to the expected revertants, we identified a number of dominant suppressors (data not shown). These suppressors must reduce the amount of anterior Nanos activity, and this could occur either by reducing the amount of anterior Miranda–GFP and oskar mRNA by impairing the bicoid mRNA localization pathway or by reducing the amount of anterior Oskar protein by impairing its translation or anchoring. Indeed, we found that exu and sww mutants functioned as moderate suppressors of Miranda–GFP, while heterozygosity for an RNA null allele of oskar (osk87; Jenny et al. 2006)) almost completely suppressed the formation of an anterior abdomen (99.2%). Although the embryos from Mira–GFP/+; osk87/+ females formed normal heads, only very few of them hatched (0.6%). This appeared to be due posterior defects caused by a reduction of Oskar activity at the posterior pole and can be explained by the fact that Mira–GFP already leads to the mislocalization of about half of oskar mRNA to the anterior, while the osk87 allele reduces oskar mRNA levels by a further 50%. Nevertheless, these results indicated that one could identify novel factors involved in bicoid mRNA localization as dominant suppressors of the dominant maternal-effect lethal phenotype of Mira–GFP and that one might also identify factors involved in Oskar translation or anchoring, provided that they are not so strong that they produce posterior defects. We therefore performed a large-scale screen for dominant suppressors of the Miranda–GFP maternal effect lethal phenotype (Su (Mir) mutations).
Screen for revertants and dominant suppressors of Mira–GFP
Males of the genotype w1118 P(Mira–GFP w+)/Y were mutagenized with the chemical mutagen, EMS, and were crossed to w1118 stock that had been isogenized for the autosomes. The F1 female progeny were then placed in individual egg-laying tubes, and the hatching rate of their progeny was determined. In the genetic background of the screen, the progeny of Mira–GFP/+ females have a hatching rate of <1%, and we selected any females whose progeny had a hatching rate of >10% as potential suppressors or revertants. After one generation of backcrossing to remove extraneous mutations, the mutations were mapped to a chromosome using recessive markers (Figure 1). We identified six revertants from the pilot and large-scale screens, and 144 putative suppressors (from a screen of 7062 females), 82 of which gave fertile progeny. Unfortunately, homozygosity for the dp chromosome strongly enhanced the bicaudal phenotype, leading to the loss of most suppressors on the third chromosome. Nevertheless, 30 suppressor lines survived the mapping procedure and still showed robust suppression after they had been mapped to a chromosome and retested.
Analysis of Mira–GFP revertants
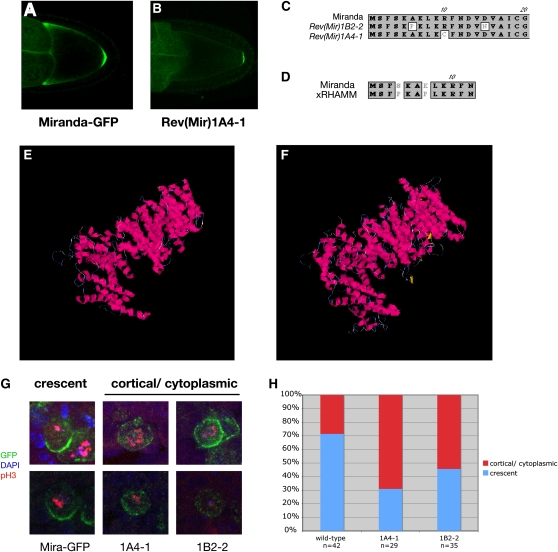
We were primarily interested in revertants that specifically disrupt the anterior localization of Mira–GFP rather than mutations that abolish Mira–GFP expression or its ability to bind to Staufen, and we therefore examined the localization of the GFP fusion protein at mid-oogenesis. Since the GFP is fused to the C terminus of Miranda, nonsense mutations will produce no detectable GFP signal, while mutations that disrupt Staufen-binding cannot enter the oocyte by hitchhiking with Staufen/oskar mRNA complexes. By contrast, Miranda–GFP revertants that specifically disrupt the anterior localization domain still localize normally to the posterior pole at stage 9, but fail to accumulate at the anterior (Figure 2, A and B). Two of the six revertants fell into this category and we therefore sequenced the coding regions of each Miranda transgene to identify the responsible mutations. In each case, the mutation fell in the first 13 amino acids of Miranda (Figure 2C). Although Miranda is a large coiled-coil domain protein with no significant homologies to other proteins over its full length, BLAST searches revealed that the very N terminus of the protein show significant similarity with the same N-terminal region of vertebrate RHAMM, and both revertants disrupt amino acids that are identical in Miranda and RHAMM (Figure 2D). Like Miranda, RHAMM is a large coiled-coil domain protein, and the NCBI conserved domain database predicts that both proteins contain SMC prok B domains that form an extended helical structure (Marchler-Bauer et al. 2009). Furthermore, structural modeling predicts that Miranda and RHAMM form very similar donut-shaped structures (Figure 2, E and F). Although there are no well-conserved orthologs of Miranda in vertebrates, these striking similarities raise the possibility that RHAMM represents such an ortholog that has diverged significantly at the sequence level while retaining the same overall structure.
Figure 2 .
Characterization of Miranda revertants. (A and B) Miranda–GFP localization in stage 9–10 oocytes. (A) Wild type. (B) 1A4–1 Miranda–GFP revertant. (C) Scheme of the N terminus of Miranda. White boxes indicate point mutations identified in the two Miranda revertans. (D) Alignment of the N terminus of Miranda and Xenopus RHAMM. (E and F) Predicted 3D structures for Miranda (E) and Xenopus RHAMM (F). The predictions were generated using the I-TASSER Internet service (Zhang 2008, 2009). (G) Localization of Miranda–GFP and Miranda–GFP revertants in Df(3R)oraI9 embryonic neuroblasts. (H) Quantification of Miranda–GFP localization in embryonic neuroblasts.
The N-terminal region of Miranda is required for its localization to the basal cortex of mitotic neuroblasts (Fuerstenberg et al. 1998). Furthermore, although the localization of Miranda depends on actin, mutants that disrupt the astral microtubules of the neuroblast cause a variable defect in Miranda accumulation at the basal cortex, suggesting that microtubules also play a role (Giansanti et al. 2001; Basto et al. 2006; Giansanti et al. 2008). To test whether the revertants also affect the basal localization of Miranda in the neuroblast, we introduced the mutations found in two of the revertants (1A4-1 and 1B2-2) into a UAS–Miranda1-727–GFP construct, as the original Miranda transgene is not expressed in neuroblasts. Miranda forms homodimers, and we therefore examined the ability of each transgene to localize in mira mutant background to preclude the formation of heterodimers between the revertants and wild-type Miranda that might rescue any defects (Yousef et al. 2008). Wild-type Miranda–GFP driven by Pros-Gal4 rescued Df(3R)oraI9 (miranda null) homozygotes to pupal stages, whereas Df(3R)oraI9 animals expressing the revertant proteins were embryonic or larval lethal. Furthermore, although both revertant Miranda proteins formed basal crescents in metaphase neuroblasts, they both showed a significantly higher frequency of cells with a diffuse cytoplasmic signal (Figure 2, G and H). Thus, the very N-terminal domain of Miranda is not essential for anchoring to the basal cortex, but may contribute to the efficient delivery of Miranda to this region.
Analysis of Su(Mir) mutations
The 30 surviving suppressor mutations were crossed to the other suppressors on the same chromosome and tested for lethality or sterility. This identified three complementation groups on the second chromosome, which we named Su(Mir)1–3, two of which are lethal with 3 and 11 alleles respectively and a third that is female sterile with 4 alleles (Table 1).
Table 1. Description of the complementation groups.
| Suppressor/gene name | Alleles | Chromosome location/map position | Homozygous viability/fertility |
|---|---|---|---|
| Mira–GFP Revertants | Rev(Mir)1A4-1, Rev(Mir)1B2-2 | First | Fertile; Mira–GFP localization to the anterior is abolished but posterior localization remains. |
| shot | 1E2-5, 1J3-2, 1J3-3, 2C2-3, 2A2-4, 2C3-1, 3C2-2, 3F1-1, x3I2-1, 2F2-2, 2B2-8 | 50C | Lethal |
| Su(Mir)2 | 1C2-2, 2H3-1, 1E3-5, 1J3-4b | Between b and c | 1C2-2, 2H3-1 viable, female sterile. 1E3-5, 1J3-4b lethal. All transheterozgous combinations viable, female sterile. |
| Su(Mir)3 | 1H2-1a, 1A2-1, 1G3-1a | Between L and Pin | Lethal |
| Single alleles | |||
| capu | 3G3-1 | 24C | Viable, female sterile |
| 1A2-2 | First | Lethal | |
| 1E2-4 | First | ||
| 1A4-1 | First | ||
| 3D1-1 | Second | Lethal | |
| 2A2-1 | Second | Lethal | |
| 3C2-1 | Second | Lethal | |
| 2E2-3 | Second | Lethal | |
| 1E3-2 | Third | Viable, female sterile | |
| 2C2-1 | Third | ||
| 1F2-3 | Third | ||
| 1A2-2 | Third |
All alleles of Su(Mir)1 are lethal over Df(2R) CX1, which removes 49C1-4 to 50C23-D2, and the position of the locus was further refined by crossing alleles to smaller deficiencies in this region, which revealed that Su(Mir)1 falls in the 50C5-C9 region uncovered by Df(2R) Exel71299 (Parks et al. 2004). Su(Mir)1 alleles failed to complement mutations in short stop (shot), a very large gene that spans most of this interval. Furthermore, a null allele of the locus, shot3, dominantly suppresses Miranda–GFP as well as the new alleles, giving a hatching rate of 11.2%. The large size of Shot (4100–8800 amino acids, depending on the isoform) and the fact that loss-of-function alleles suppress the bicaudal phenotype probably explains why we recovered so many more alleles of this complementation group than the others.
short stop encodes a large spectroplakin protein that binds to both actin and microtubules, making it a good candidate for a factor involved in either bicoid or oskar mRNA localization or anchoring(Kolodziej et al. 1995; Gregory and Brown 1998; Roper et al. 2002). Shot also performs an essential function in stabilizing microtubules on the fusome earlier in oogenesis, however, and shot mutant germline clones fail to make an oocyte and arrest development (Roper and Brown 2004). Indeed, homozygous germline clones of the shot alleles that we have tested from the screen block oocyte determination in the germarium, making it impossible to assess their effects on mRNA localization later in oogenesis. Thus, further investigation of the role of Shot in mRNA localization will require methods to specifically knock down its activity at later stages of oogenesis.
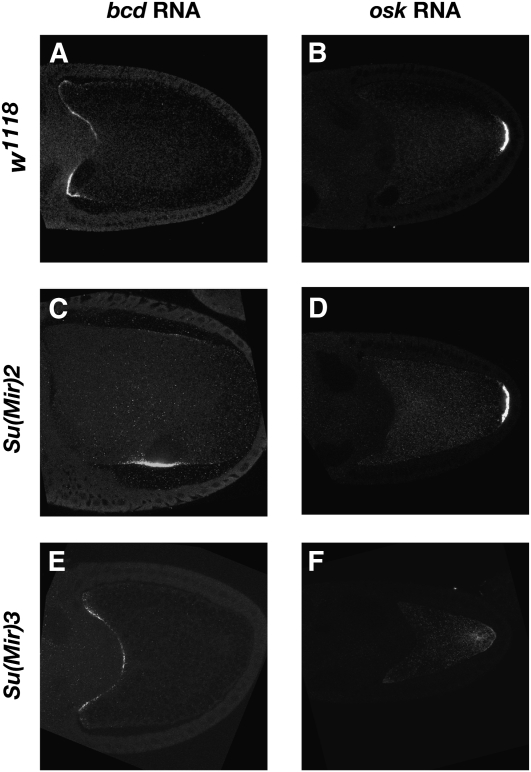
We have not yet identified the genes affected in the two other complementation groups, but both have been mapped to an approximate region. This has allowed us to cross alleles of the lethal complementation group, Su(Mir)3, onto a FRT G13 chromosome and examine their phenotype in germline clones. While one allele blocks oogenesis at an early stage, the two other alleles show a defect in the anchoring of oskar mRNA and protein at the posterior of the oocyte (Figure 3, E and F). This suggests that Su(Mir)3 mutants suppress the Miranda–GFP phenotype by reducing the amount of ectopic Oskar anchored at the oocyte anterior. By contrast, females trans-heterozygous for viable mutant combinations of Su(Mir)2 alleles show normal oskar mRNA localization, but bicoid mRNA is mislocalized in a ring in the center of the oocyte next to the misplaced oocyte nucleus at stage 10b (Figure 3, C and D). This phenotype is similar to that seen in cap’n collar (cnc) and skittles germline clones (Guichet et al. 2001; Gervais et al. 2008). Su(Mir)2 does not correspond to either of these loci, however, indicating that it represents a new gene required for the anterior anchoring of bicoid mRNA during the later stages of oogenesis.
Figure 3 .
Su(Mir)2 disrupts bcd mRNA localization and Su(Mir)3 disrupts oskar RNA localization. In situ hybridization to bicoid (A, C, E) and oskar (B, D, F) mRNAs in wild type (A and B), Su(Mir)2 (C and D) and Su(Mir)3 (E and F) oocytes.
Cappuccino and Spire play a role in Oskar anchoring
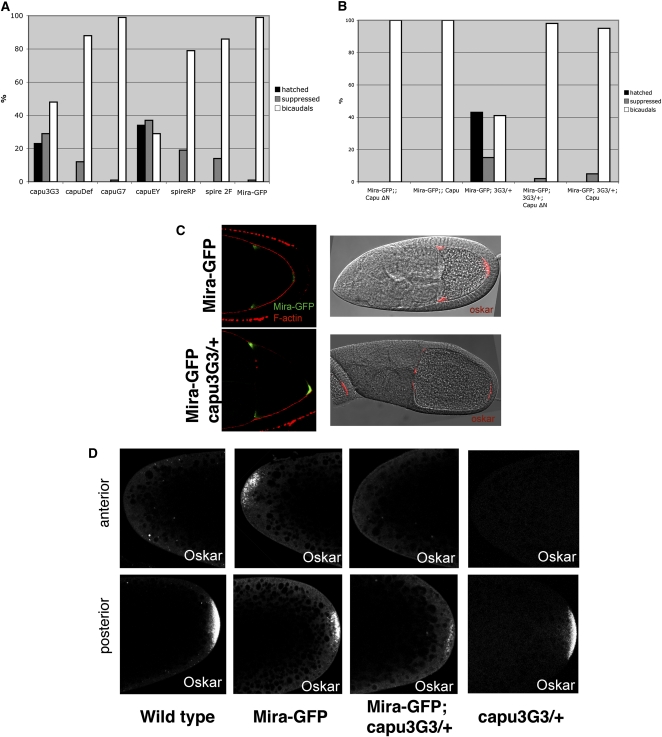
We also examined the phenotypes produced by the two single alleles that are homozygous viable. 1E3-2 homozygotes are maternal-effect lethal, but show no obvious defects in bicoid or oskar mRNA localization. By contrast, 3G3-1 homozygous females are sterile, and their ovaries contain ventralized oocytes that show premature streaming of the oocyte cytoplasm at mid-oogenesis. This phenotype is very similar to that produced by cappuccino and spire mutants (Manseau and Schupbach 1989; Manseau et al. 1996). Complementation tests and sequence analysis indicated that 3G3-1 is a new allele of capu caused by the insertion of the juan element into the first common exon of the Capu coding region. This should result in the expression of a truncated protein with only the first 103 amino acids of Capu-PA followed by 20 amino acids encoded by juan and removes all of the conserved formin homology domains of the protein. To confirm that this mutation is responsible for the suppression, we first tested whether other capu alleles suppress the Miranda–GFP phenotype. Although heterozygosity for a deletion of capu reduced the frequency of bicaudal embryos laid by Mira–GFP/+ females, it did not give any hatching larvae and capuG7 did not suppress at all (Figure 4A). However, a P-element insertion in the same exon as capu3G3-1, capuEY12344 also acted as a strong suppressor, suggesting that expression of just the N-terminal 100 amino acids of Capu might exert a dominant negative effect. Capu functions with Spire to assemble a cytoplasmic network of actin in the oocyte and binds directly to Spire protein (Rosales-Nieves et al. 2006; Dahlgaard et al. 2007; Quinlan et al. 2007). We therefore also tested two alleles of spire for suppression, and both gave similar levels of suppression to the deficiency for capu, suggesting that Spire and Capu act together in this context. To confirm that the suppression was due to a reduction in Capu activity, we examined whether expression of full-length Capu or a constitutively active form of Capu lacking the N-terminal inhibitory domain (CapuΔN) could block the suppression of Miranda–GFP by capu3G3-1 (Dahlgaard et al. 2007). Although overexpression of either form of Capu had no effect on the phenotype of Miranda–GFP alone, both reversed the suppression by capu3G3-1, confirming that the suppression is caused by a reduction in Capu activity (Figure 4B).
Figure 4 .
A new allele of cappuccino suppresses the Miranda–GFP bicaudal phenotype. (A) Bar diagram showing the level of suppression of the Miranda–GFP bicaudal phenotype by various capu and spire alleles (Miranda–GFP/+;capu3G3/+, Miranda–GFP/+;Df(2L)edSZ1/+, Miranda–GFP/+;capuG7/+, Miranda–GFP/+;capuEY13172/+, Miranda–GFP/+;spireRP/+, Miranda–GFP/+;spire2F/+, and Miranda–GFP/+). Suppressed embryos were scored as embryos that did not form abdominal structures or posterior spiracles at the anterior, but still failed to hatch. (B) Bar diagram showing the effect of Capu overexpression using a maternal Gal4 driver on the suppression of the Miranda–GFP bicaudal phenotype by capu3G3. Capu represents a full-length Capu protein, and Capu ∆N is a truncated form that is constitutively active. (C) Localization of Miranda–GFP (left) and oskar mRNA (right) in Miranda–GFP/+ (top) or Miranda–GFP/+; capu3G3/+ egg chambers (bottom). (D) Localization of Oskar protein in wild-type, Miranda–GFP/+, Miranda–GFP/+; capu3G3/+, or capu3G3/+ embryos.
Capu and Spire are both actin nucleators that work together to assemble an actin mesh in the oocyte cytoplasm from stage 5–10b of oogenesis that limits kinesin-dependent cytoplasmic flows (Emmons et al. 1995; Pruyne et al. 2002; Quinlan et al. 2005; Dahlgaard et al. 2007). In the absence of the mesh, premature cytoplasmic streaming washes the microtubules to the cortex and the prevents the kinesin-dependent transport of oskar mRNA to the oocyte posterior, while bicoid mRNA localization is unaffected (Serbus et al. 2005; Dahlgaard et al. 2007; Zimyanin et al. 2008). To investigate the basis for Miranda suppression by capu3G3-1, we examined the localization of Miranda–GFP, oskar mRNA and Oskar protein in Mira–GFP/+; capu3G3-1/+ oocytes and eggs. Miranda–GFP and oskar mRNA localize to the anterior and posterior poles of the oocyte, as they do in the absence of capu3G3-1 (Figure 4C). Oskar protein is not translated at the anterior of Miranda–GFP/+ oocytes until the end of oogenesis, and we therefore examined its distribution in freshly aid eggs (Irion et al. 2006). Oskar is localized only to the posterior pole of wild type and capu3G3-1/+ eggs, but is localized symmetrically at the anterior and posterior poles in Miranda–GFP/+ eggs (Figure 4D). By contrast, no Oskar protein is detectable at the anterior of Mira–GFP/+; capu3G3-1/+ eggs, and Oskar levels at the posterior are also significantly reduced (Figure 4D). Thus, heterozygosity for capu3G3-1 appears to disrupt the anchoring of Oskar protein at the anterior and reduces the anchoring of Oskar at its normal position at the posterior of the oocyte.
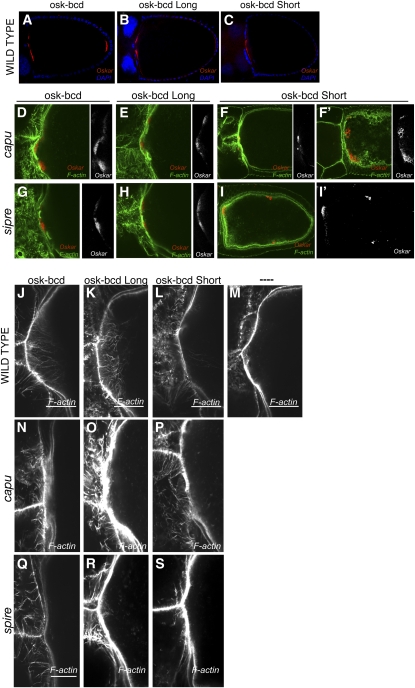
These results suggest that Capu and Spire play a role in the anchoring of Oskar protein and the pole plasm that has been obscured by their earlier requirement in the localization of oskar mRNA to the posterior of the oocyte. To circumvent this problem, we took advantage of a series of oskar-bicoid 3′−UTR constructs that express Long Oskar (required for anchoring), Short Oskar (nucleator of the pole plasm), or both Oskar isoforms at the anterior of the oocyte at mid-oogenesis (Figure 5, A–C)(Tanaka and Nakamura 2008). Long Oskar remains stably anchored at the anterior cortex of the oocyte in capu or spire null mutants, and most Short Oskar also seems to be anchored at the anterior in the presence of Long Osk (Figure 5, D, E, G, and H). However, Short Oskar is not stably anchored at the anterior of the oocyte in capu or spire mutants when it is expressed in the absence of the long isoform (Figure 5, F, F′, I, and I′). Thus, the anchoring of Short Oskar at the anterior seems to require either Capu and Spire or Long Oskar protein.
Figure 5 .
Cappuccino and Spire are required for the anchoring of Short Oskar and the formation of Long Oskar-dependent F-actin filaments. (A–I′) Localization of Oskar protein in oocytes expressing of oskar-bcd 3′ UTR (A, D, G), oskar (M139L)-bcd 3′ UTR (B, E, H), or oskar (M1L)-bcd 3′ UTR (C, F, F′, I, I′) in wild type (A–C), capu3G3/ Df(2L)edSZ1 (D–F′), or spireRP (G–I′) mutants. (J–S) High magnification views of F-actin staining at the anterior of oocytes expressing oskar-bcd 3′ UTR (J, N, Q), oskar (M139L)-bcd 3′ UTR (K, O, R), or oskar (M1L)-bcd 3′ UTR (L, P, S), in wild ype (J–M), capu3G3/ Df(2L)edSZ1 (N–P), or spireRP (Q–S). (M) High-magnification view of F-actin staining at the anterior of a wild-type oocyte.
Since Capu and Spire are actin nucleators and Oskar has been shown to trigger the formation of long actin filaments, we examined F-actin organization in oocytes expressing the osk-bcd constructs with or without Capu and Spire activity (Figure 5, J–S) (Vanzo et al. 2007; Tanaka and Nakamura 2008). Long Oskar induces the formation of long arrays of actin filaments extending from the anterior cortex, whereas Short Oskar alone does not (Figure 5, J–M). These filaments do not form in capu and spire mutants, however, indicating that Capu and Spire act downstream of Long Oskar to nucleate these F-actin structures (Figure 5, N–S).
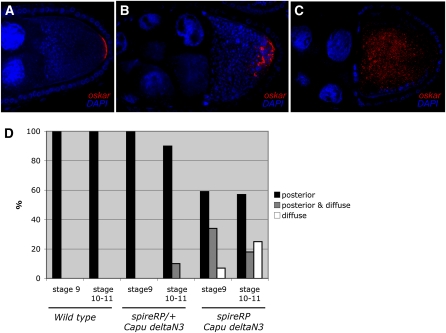
To test directly whether Spire plays a role in the normal anchoring of oskar mRNA at the posterior of the oocyte, we took advantage of the fact that expression of constitutively active CapuΔN3 suppresses the premature streaming phenotype of spire mutants, allowing the posterior localization of oskar mRNA in the absence of Spire activity (Dahlgaard et al. 2007). A third of the stage 9 spireRP; CapuΔN3/+ egg chambers show a typical oskar mRNA anchoring defect, in which the RNA is localized to the posterior region of the oocyte, but is not tightly apposed to the posterior cortex (Figure 6, A–D). Once cytoplasmic streaming starts, this mRNA is often washed away from the posterior and shows a diffuse distribution throughout the oocyte cytoplasm (Figure 6C). These observations support the view that Spire plays a similar role in the normal process of Oskar anchoring at the posterior of the oocyte as it does at the anterior in Miranda–GFP and osk–bcd females.
Figure 6 .
Effect of spire and cappuccino mutants on oskar mRNA anchoring. (A–C) oskar mRNA localization in wild type (A) and spireRP; nos-Gal4 UAS-Capu∆N3/+ egg chambers (B and C). (D) Bar diagram showing a quantification of oskar mRNA localization in stage 9 and stage 10–11 oocytes in wild type, spireRP/+; nos-Gal4 UAS-Capu∆N3/+, and spireRP; nos-Gal4 UAS-Capu∆N3/+. Posterior refers to wild-type posterior localization (A), posterior and diffuse refers to some posterior localization with the mRNA extending away from the posterior pole (B), and diffuse refers to no posterior enrichment (C).
Discussion
The goal of the screen reported here was to identify novel factors involved in bicoid and oskar mRNA localization and to isolate revertants of Miranda that define its anterior localization domain. The screen itself was very simple to perform, as it was actually a selection, with only revertants and suppressors producing viable progeny. On the other hand, the downstream analysis of the new mutants proved challenging, since one cannot follow the mutations in males and must therefore establish and retest multiple lines for each mutant. Furthermore, the extent of the suppression varied with genetic background, and the suppression became too weak to maintain a number of mutants during the crosses to map them to a chromosome. Nevertheless, the screen succeeded in identifying revertants at the expected frequency (∼1/1000), as well as three Su (Mir) complementation groups and a number of single alleles.
The revertants of the Miranda–GFP transgene identified the very N terminus of the protein as the region responsible for its anterior localization. This region showed a short but significant homology to the very N terminus of vertebrate RHAMM, which is also a large coiled-coiled domain protein, which is predicted to fold into a remarkably similar structure to Miranda. Thus, RHAMM may represent a very divergent ortholog of Miranda that has conserved its structure, but retained only a very small region of primary sequence similarity. This idea is supported by the similar properties of both proteins. Although RHAMM was originally identified as a cell-surface hyaluronic acid receptor, the protein lacks a signal peptide and localizes to the mitotic spindle and centrosomes, where it plays an essential role in the chromatin-mediated spindle assembly pathway (Hofmann et al. 1998; Maxwell et al. 2003; Groen et al. 2004). Miranda also localizes to centrosomes in the Drosophila embryo and decorates the mitotic spindle in neuroblasts that are mutant for lgl, dlg, or scribble (Ohshiro et al. 2000; Peng et al. 2000; Albertson and Doe 2003). Furthermore, N terminus of Miranda is essential for its microtubule-dependent localization to the anterior of the oocyte, as it is disrupted by single amino acid mutations in the short N-terminal region with homology to RHAMM, and the region of RHAMM that associates with microtubules has also been mapped to its N-terminal domain (Maxwell et al. 2003; Irion et al. 2006). It would therefore be interesting to test whether Miranda also plays a role in the chromatin-mediated spindle assembly pathway, which is redundant with the centrosomal pathway under normal conditions and has not been well characterized in Drosophila.
The domain that targets Miranda to the basal cortex of the neuroblast has also been mapped to the N-terminal 290 amino acids of the protein (Fuerstenberg et al. 1998; Matsuzaki et al. 1998; Shen et al. 1998). The mutations that block anterior Miranda localization in the oocyte do not disrupt its basal localization in the neuroblast, however, consistent with the data that the latter occurs by a distinct actin-dependent mechanism (Shen et al. 1998; Erben et al. 2008). Nevertheless, these single amino acid changes appear to delay the formation of the Miranda basal crescent, since the protein is more frequently observed to be cytoplasmic. This suggests that the mutations either partially impair the ability of Miranda to associate with the actin-rich cortex or that they inhibit a redundant localization pathway that increases the efficiency of the targeting of Miranda to the basal cortex. In support of the second possibility, it has recently been found that redundant pathways localize the key polarity factor, Pins, to the apical cortex: it is directly recruited by Inscuteable, but is still delivered apically in the absence of Inscuteable by a microtubule-dependent pathway that involves the microtubule motor protein, Khc-73 and Dlg (Schaefer et al. 2000; Yu et al. 2000; Siegrist and Doe 2005). Since mutations that disrupt the astral microtubules impair Miranda localization, it is possible that a similar microtubule-dependent mechanism plays a redundant role in delivering Miranda to the basal cortex (Giansanti et al. 2001; Basto et al. 2006; Giansanti et al. 2008).
Modifier screens are based on the idea that recessive loss-of-function mutations can become dominant in a sensitized genetic background in which the process that they affect is limiting (Simon 1994; St. Johnston 2002). Because Miranda–GFP links some oskar mRNA to the bicoid mRNA localization pathway, we expected the suppressors to be dosage-sensitive factors involved in the transport of bicoid mRNA to the anterior or regulators of oskar mRNA anchoring and translation that reduce the amount of anterior pole plasm produced. Although we have only partially analyzed the three complementation groups, Su(Mir)2 and 3 appear to fall into each expected class, with the former disrupting bicoid mRNA localization, and the latter giving an oskar mRNA anchoring phenotype. By far the most frequent class of suppressor, however, was alleles of the giant actin and microtubule-binding protein, Shot. Shot is therefore an excellent candidate to play a role in one or both of these processes, although we have been unable to test this directly because all of the alleles we have tested block oogenesis at an early stage.
In addition to the complementation groups, we also recovered a number of single mutations, including the capu3G3-1 allele. Interestingly, capu3G3-1 and the very similar allele, capuEY12344 suppress the Miranda–GFP bicaudal phenotype much more strongly than a deficiency for the locus, indicating that they have an antimorphic effect, and this may explain why we recovered only a single allele at this locus. This revealed an unexpected role for Capu and its binding partner Spire in Oskar anchoring that was not detected previously, because oskar mRNA is not localized to the posterior in capu and spire mutants. capu3G3-1 dominantly disrupts the anchoring of Oskar at the anterior of Miranda–GFP eggs and also reduces the normal anchoring of Oskar at the posterior.
The anchoring of Short Oskar and associated pole plasm depends on F-actin and Long Oskar (Jankovics et al. 2002; Polesello et al. 2002; Vanzo and Ephrussi 2002; Babu et al. 2004). In addition, the localization of Oskar to either the anterior or posterior of the oocyte induces the formation of actin filaments from the adjacent cortex (Vanzo et al. 2007; Tanaka and Nakamura 2008). Our results demonstrate that it is Long Oskar that induces actin filaments and that their formation requires both Capu and Spire. Thus, these two actin nucleators act downstream of Long Oskar to nucleate actin filaments. Since, Long Oskar, Capu, Spire, and F-actin are all required for the anchoring of Short Oskar, it seems likely that Short Oskar is tethered in some way to this actin structures.
Long Oskar recruits a number of endocytic factors to its site of localization to locally stimulate endocytosis, and endocytic mutants also disrupt Short Oskar anchoring at the oocyte posterior (Tanaka and Nakamura 2008). This raises the question of the relationship between Capu- and Spire-dependent actin filament formation and Oskar-dependent endocytosis. One possibility is that the Capu and Spire are activated as a consequence of endocytosis. However, F-actin is still enriched at the anterior of osk-bcd 3′−UTR oocytes when endocytosis is disrupted, although the actin forms aggregates rather than filaments. Thus, Long Oskar induces actin polymerization in the absence of increased endocytosis (Tanaka and Nakamura 2008). The interpretation of the relationship between Long Oskar, endocytosis, Capu and Spire, and anchoring is further complicated by the fact that the requirements for Short Oskar anchoring vary according to stage and position within the oocyte. Capu and Spire are required for the anterior anchoring of Short Oskar at stage 10b in the absence of Long Oskar in oskM1L-bcd 3′ UTR oocytes, indicating that they must be able to nucleate actin filaments that anchor Short Oskar in the absence of Long Oskar. On the other hand, they are dispensable for the anterior anchoring of Short Oskar at this stage when Long Oskar is present. There must therefore be another parallel mechanism to hold Short Oskar at the anterior under these conditions. Similarly, although the induction of increased endocytosis by Long Oskar is required for anchoring of Short Oskar at the posterior, this is not the case at the anterior. Thus, Long Oskar-dependent endocytosis and actin filament formation may act redundantly to anchor Short Oskar at the anterior in mid-oogenesis. This redundancy does not appear to exist at the posterior of the oocyte, however, as Long Oskar, Capu and Spire, and endocytosis are all necessary for efficient anchoring, and the same is true at later stages for the anterior of Miranda–GFP-expressing oocytes. This difference may reflect the presence or absence of cytoplasmic flows at each stage and position, since Short Oskar anchoring at the posterior is important only after stage 10b when cytoplasmic streaming begins. As this movement is weaker very close to the anterior cortex, there may be less force causing Short Oskar to spread away from this position.
Our results show that Capu and Spire play a key role in the normal anchoring of Short Oskar at the posterior, and hence in pole plasm formation, but also indicate the existence of partially redundant anchoring pathways downstream of Long Oskar. These results are consistent with the observations of Babu et al. who found that there are two overlapping anchoring pathways for Oskar, an actin-dependent pathway that involves the Bifocal protein, and an actin-independent pathway that requires the Homer protein (Babu et al. 2004). The anchoring of Short Oskar probably, therefore, depends on a complex network of interactions involving Long Oskar, endocytic vesicles, and actin filaments, all of which contribute some anchoring activity. A complete understanding of this process will therefore require deciphering the molecular links between these components.
Acknowledgments
We thank Akira Nakamura, Nick Brown, and the Bloomington Stock Center for Drosophila stocks and Rebecca Bastock for the anti-Oskar antibody. This work was supported by a Wellcome Trust Principal Research Fellowship to D.S.J. and core funding from the Wellcome Trust and Cancer Research UK. D.N. was supported by a postdoctoral fellowship from the Swedish Research Council.
Literature Cited
- Albertson R., Doe C. Q., 2003. Dlg, Scrib and Lgl regulate neuroblast cell size and mitotic spindle asymmetry. Nat. Cell Biol. 5: 166–170 [DOI] [PubMed] [Google Scholar]
- Babu K., Cai Y., Bahri S., Yang X., Chia W., 2004. Roles of Bifocal, Homer, and F-actin in anchoring Oskar to the posterior cortex of Drosophila oocytes. Genes Dev. 18: 138–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R., Lau J., Vinogradova T., Gardiol A., Woods C. G., et al. , 2006. Flies without centrioles. Cell 125: 1375–1386 [DOI] [PubMed] [Google Scholar]
- Bastock R., St. Johnston D., 2008. Drosophila oogenesis. Curr. Biol. 18: R1082–R1087 [DOI] [PubMed] [Google Scholar]
- Becalska A. N., Gavis E. R., 2009. Lighting up mRNA localization in Drosophila oogenesis. Development 136: 2493–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen H. J., Levis R. W., Liao G., He Y., Carlson J. W., et al. , 2004. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167: 761–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berleth T., Burri M., Thoma G., Bopp D., Richstein S., et al. , 1988. The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. EMBO J. 7: 1749–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendza R., Serbus L., Duffy J., Saxton W., 2000. A function for kinesin I in the posterior transport of oskar mRNA and Staufen protein. Science 289: 2120–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadus J., Fuerstenberg S., Doe C. Q., 1998. Staufen-dependent localization of prospero mRNA contributes to neuroblast daughter-cell fate. Nature 391: 792–795 [DOI] [PubMed] [Google Scholar]
- Cha B., Koppetsch B., Theurkauf W., 2001. In vivo analysis of Drosophila bicoid mRNA localization reveals a novel microtubule-dependent axis specification pathway. Cell 106: 35–46 [DOI] [PubMed] [Google Scholar]
- Chou T., Perrimon N., 1996. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics 144: 1673–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A., Meignin C., Davis I., 2007. A Dynein-dependent shortcut rapidly delivers axis determination transcripts into the Drosophila oocyte. Development 134: 1955–1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlgaard K., Raposo A. A., Niccoli T., St. Johnston D., 2007. Capu and Spire assemble a cytoplasmic actin mesh that maintains microtubule organization in the Drosophila oocyte. Dev. Cell 13: 539–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmons S., Phan H., Calley J., Chen W., James B., et al. , 1995. Cappuccino, a Drosophila maternal effect gene required for polarity of the egg and embryo, is related to the vertebrate limb deformity locus. Genes Dev. 9: 2482–2494 [DOI] [PubMed] [Google Scholar]
- Ephrussi A., Johnston D., 2004. Seeing is believing the bicoid morphogen gradient matures. Cell. 116: 143–152 [DOI] [PubMed] [Google Scholar]
- Ephrussi A., Lehmann R., 1992. Induction of germ cell formation by oskar. Nature 358: 387–392 [DOI] [PubMed] [Google Scholar]
- Ephrussi A., Dickinson L. K., Lehmann R., 1991. Oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell 66: 37–50 [DOI] [PubMed] [Google Scholar]
- Erben V., Waldhuber M., Langer D., Fetka I., Jansen R. P., et al. , 2008. Asymmetric localization of the adaptor protein Miranda in neuroblasts is achieved by diffusion and sequential interaction of Myosin II and VI. J. Cell Sci. 121: 1403–1414 [DOI] [PubMed] [Google Scholar]
- Ferrandon D., Elphick L., Nusslein-Volhard C., St. Johnston D., 1994. Staufen protein associates with the 3′UTR of bicoid mRNA to form particles that move in a microtubule-dependent manner. Cell 79: 1221–1232 [DOI] [PubMed] [Google Scholar]
- Fuerstenberg S., Peng C. Y., Alvarez-Ortiz P., Hor T., Doe C. Q., 1998. Identification of Miranda protein domains regulating asymmetric cortical localization, cargo binding, and cortical release. Mol. Cell. Neurosci. 12: 325–339 [DOI] [PubMed] [Google Scholar]
- Gervais L., Sandra C., Januschke J., Roth S., Guichet A., 2008. PIP5K-dependent production of PIP2 sustains microtubule organization to establish polarized transport in the Drosophila oocyte. Development. 135: 3829–3838 [DOI] [PubMed] [Google Scholar]
- Giansanti M. G., Gatti M., Bonaccorsi S., 2001. The role of centrosomes and astral microtubules during asymmetric division of Drosophila neuroblasts. Development 128: 1137–1145 [DOI] [PubMed] [Google Scholar]
- Giansanti M. G., Bucciarelli E., Bonaccorsi S., Gatti M., 2008. Drosophila SPD-2 is an essential centriole component required for PCM recruitment and astral-microtubule nucleation. Curr. Biol. 18: 303–309 [DOI] [PubMed] [Google Scholar]
- Gregory S. L., Brown N. H., 1998. kakapo, a gene required for adhesion between and within cell layers in Drosophila, encodes a large cytoskeletal linker protein related to plectin and dystrophin. J. Cell Biol. 143: 1271–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groen A. C., Cameron L. A., Coughlin M., Miyamoto D. T., Mitchison T. J., et al. , 2004. XRHAMM functions in ran-dependent microtubule nucleation and pole formation during anastral spindle assembly. Curr. Biol. 14: 1801–1811 [DOI] [PubMed] [Google Scholar]
- Guichet A., Peri F., Roth S., 2001. Stable anterior anchoring of the oocyte nucleus is required to establish dorsoventral polarity of the Drosophila egg. Dev. Biol. 237: 93–106 [DOI] [PubMed] [Google Scholar]
- Hachet O., Ephrussi A., 2001. Drosophila Y14 shuttles to the posterior of the oocyte and is required for oskar mRNA transport. Curr. Biol. 11: 1666–1674 [DOI] [PubMed] [Google Scholar]
- Hachet O., Ephrussi A., 2004. Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature 428: 959–963 [DOI] [PubMed] [Google Scholar]
- Hofmann M., Fieber C., Assmann V., Gottlicher M., Sleeman J., et al. , 1998. Identification of IHABP, a 95 kDa intracellular hyaluronate binding protein. J. Cell Sci. 111(12): 1673–1684 [DOI] [PubMed] [Google Scholar]
- Ikeshima-Kataoka H., Skeath J. B., Nabeshima Y., Doe C. Q., Matsuzaki F., 1997. Miranda directs Prospero to a daughter cell during Drosophila asymmetric divisions. Nature 390: 625–629 [DOI] [PubMed] [Google Scholar]
- Irion U., St. Johnston D., 2007. bicoid RNA localization requires specific binding of an endosomal sorting complex. Nature 445: 554–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irion U., Adams J., Chang C., St. Johnston D., 2006. Miranda couples oskar mRNA/Staufen complexes to the bicoid mRNA localization pathway. Dev. Biol. 297: 522–533 [DOI] [PubMed] [Google Scholar]
- Jankovics F., Sinka R., Lukacsovich T., Erdelyi M., 2002. MOESIN crosslinks actin and cell membrane in Drosophila oocytes and is required for OSKAR anchoring. Curr. Biol. 12: 2060–2065 [DOI] [PubMed] [Google Scholar]
- Jenny A., Hachet O., Závorszky P., Cyrklaff A., Weston M. D. J., et al. , 2006. A translation-independent role of oskar RNA in early Drosophila oogenesis. Development 133: 2827–2833 [DOI] [PubMed] [Google Scholar]
- Kim-Ha J., Smith J. L., Macdonald P. M., 1991. oskar mRNA is localized to the posterior pole of the Drosophila oocyte. Cell 66: 23–35 [DOI] [PubMed] [Google Scholar]
- Kolodziej P. A., Jan L. Y., Jan Y. N., 1995. Mutations that affect the length, fasciculation, or ventral orientation of specific sensory axons in the Drosophila embryo. Neuron 15: 273–286 [DOI] [PubMed] [Google Scholar]
- Lécuyer E., Yoshida H., Parthasarathy N., Alm C., Babak T., et al. , 2007. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131: 174–187 [DOI] [PubMed] [Google Scholar]
- Luschnig S., Moussian B., Krauss J., Desjeux I., Perkovic J., et al. , 2004. An F1 genetic screen for maternal-effect mutations affecting embryonic pattern formation in Drosophila melanogaster. Genetics 167: 325–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manseau L. J., Schupbach T., 1989. cappuccino and spire: two unique maternal-effect loci required for both the anteroposterior and dorsoventral patterns of the Drosophila embryo. Genes Dev. 3: 1437–1452 [DOI] [PubMed] [Google Scholar]
- Manseau L., Calley J., Phan H., 1996. Profilin is required for posterior patterning of the Drosophila oocyte. Development 122: 2109–2116 [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A., Anderson J. B., Chitsaz F., Derbyshire M. K., DeWeese-Scott C., et al. , 2009. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37: D205–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S., Leclerc V., Smith-Litiere K., St. Johnston D., 2003. The identification of novel genes required for Drosophila anteroposterior axis formation in a germline clone screen using GFP-Staufen. Development 130: 4201–4215 [DOI] [PubMed] [Google Scholar]
- Matsuzaki F., Ohshiro T., Ikeshima-Kataoka H., Izumi H., 1998. Miranda localizes staufen and prospero asymmetrically in mitotic neuroblasts and epithelial cells in early Drosophila embryogenesis. Development 125: 4089–4098 [DOI] [PubMed] [Google Scholar]
- Maxwell C. A., Keats J. J., Crainie M., Sun X., Yen T., et al. , 2003. RHAMM is a centrosomal protein that interacts with dynein and maintains spindle pole stability. Mol. Biol. Cell 14: 2262–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mische S., Li M., Serr M., Hays T. S., 2007. Direct observation of regulated ribonucleoprotein transport across the nurse cell/oocyte boundary Mol Biol Cell; 18: 2254–2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr S., Dillon S., Boswell R., 2001. The RNA-binding protein Tsunagi interacts with Mago Nashi to establish polarity and localize oskar mRNA during Drosophila oogenesis. Genes Dev. 15: 2886–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmark P., Boswell R., 1994. The mago nashi locus encodes an essential product required for germ plasm assembly in Drosophila. Development 120: 1303–1313 [DOI] [PubMed] [Google Scholar]
- Nusslein-Volhard C., Frohnhofer H. G., Lehmann R., 1987. Determination of anteroposterior polarity in Drosophila. Science 238: 1675–1681 [DOI] [PubMed] [Google Scholar]
- Ohshiro T., Yagami T., Zhang C., Matsuzaki F., 2000. Role of cortical tumour-suppressor proteins in asymmetric division of Drosophila neuroblast. Nature 408: 593–596 [DOI] [PubMed] [Google Scholar]
- Palacios I., Gatfield D., St. Johnston D., Izaurralde E., 2004. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature 427: 753–757 [DOI] [PubMed] [Google Scholar]
- Parks A. L., Cook K. R., Belvin M., Dompe N. A., Fawcett R., et al. , 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36: 288–292 [DOI] [PubMed] [Google Scholar]
- Peng C. Y., Manning L., Albertson R., Doe C. Q., 2000. The tumour-suppressor genes lgl and dlg regulate basal protein targeting in Drosophila neuroblasts. Nature 408: 596–600 [DOI] [PubMed] [Google Scholar]
- Polesello C., Delon I., Valenti P., Ferrer P., Payre F., 2002. Dmoesin controls actin-based cell shape and polarity during Drosophila melanogaster oogenesis. Nat. Cell Biol. 4: 782–789 [DOI] [PubMed] [Google Scholar]
- Presgraves D. C., 2003. A fine-scale genetic analysis of hybrid incompatibilities in Drosophila. Genetics 163: 955–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne D., Evangelista M., Yang C., Bi E., Zigmond S., et al. , 2002. Role of formins in actin assembly: nucleation and barbed-end association. Science 297: 612–615 [DOI] [PubMed] [Google Scholar]
- Quinlan M. E., Heuser J. E., Kerkhoff E., Mullins R. D., 2005. Drosophila Spire is an actin nucleation factor. Nature 433: 382–388 [DOI] [PubMed] [Google Scholar]
- Quinlan M. E., Hilgert S., Bedrossian A., Mullins R. D., Kerkhoff E., 2007. Regulatory interactions between two actin nucleators, Spire and Cappuccino. J. Cell Biol. 179: 117–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper K., Brown N. H., 2004. A spectraplakin is enriched on the fusome and organizes microtubules during oocyte specification in Drosophila. Curr. Biol. 14: 99–110 [PubMed] [Google Scholar]
- Roper K., Gregory S. L., Brown N. H., 2002. The 'spectraplakins': cytoskeletal giants with characteristics of both spectrin and plakin families. J. Cell Sci. 115: 4215–4225 [DOI] [PubMed] [Google Scholar]
- Rosales-Nieves A. E., Johndrow J. E., Keller L. C., Magie C. R., Pinto-Santini D. M., et al. , 2006. Coordination of microtubule and microfilament dynamics by Drosophila Rho1, Spire and Cappuccino. Nat. Cell Biol. 8: 367–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M., Shevchenko A., Shevchenko A., Knoblich J. A., 2000. A protein complex containing Inscuteable and the Galpha-binding protein Pins orients asymmetric cell divisions in Drosophila. Curr. Biol. 10: 353–362 [DOI] [PubMed] [Google Scholar]
- Schnorrer F., Luschnig S., Koch I., Nusslein-Volhard C., 2002. γ-Tubulin 37C and γ-tubulin ring complex protein 75 are essential for bicoid RNA localization during Drosophila oogenesis. Dev. Cell 3: 685–696 [DOI] [PubMed] [Google Scholar]
- Schuldt A., Adams J., Davidson C., Micklem D., Haseloff J., et al. , 1998. Miranda mediates asymmetric protein and RNA localization in the developing nervous system. Genes Dev. 12: 1847–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupbach T., Wieschaus E., 1989. Female sterile mutations on the second chromosome of Drosophila melanogaster. I. Maternal effect mutations. Genetics 121: 101–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serbus L., Cha B., Theurkauf W., Saxton W., 2005. Dynein and the actin cytoskeleton control kinesin-driven cytoplasmic streaming in Drosophila oocytes. Development 132: 3743–3752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C. P., Jan L. Y., Jan Y. N., 1997. Miranda is required for the asymmetric localization of Prospero during mitosis in Drosophila. Cell 90: 449–458 [DOI] [PubMed] [Google Scholar]
- Shen C. P., Knoblich J. A., Chan Y. M., Jiang M. M., Jan L. Y., et al. , 1998. Miranda as a multidomain adapter linking apically localized Inscuteable and basally localized Staufen and Prospero during asymmetric cell division in Drosophila. Genes Dev. 12: 1837–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiga Y., Tanaka-Matakatsu M., Hayashi S., 1996. A nuclear GFP/ beta- galactosidase fusion protein as a marker for morphogenesis in living Drosophila. Dev. Growth Differ. 38: 99–106 [Google Scholar]
- Siegrist S. E., Doe C. Q., 2005. Microtubule-induced Pins/Galphai cortical polarity in Drosophila neuroblasts. Cell 123: 1323–1335 [DOI] [PubMed] [Google Scholar]
- Simon M. A., 1994. Signal transduction during the development of the Drosophila R7 photoreceptor. Dev. Biol. 166: 431–442 [DOI] [PubMed] [Google Scholar]
- St. Johnston D., 2002. The art and design of genetic screens: Drosophila melanogaster. Nat. Rev. Genet. 3: 176–188 [DOI] [PubMed] [Google Scholar]
- St. Johnston D., 2005. Moving messages: the intracellular localization of mRNAs. Nat. Rev. Mol. Cell Biol. 6: 363–375 [DOI] [PubMed] [Google Scholar]
- St. Johnston D., Driever W., Berleth T., Richstein S., Nusslein-Volhard C., 1989. Multiple steps in the localization of bicoid RNA to the anterior pole of the Drosophila oocyte. Development 107(Suppl): 13–19 [DOI] [PubMed] [Google Scholar]
- Tanaka T., Nakamura A., 2008. The endocytic pathway acts downstream of Oskar in Drosophila germ plasm assembly. Development 135: 1107–1117 [DOI] [PubMed] [Google Scholar]
- Van Doren M., Williamson A. L., Lehmann R., 1998. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr. Biol. 8: 243–246 [DOI] [PubMed] [Google Scholar]
- Vanzo N., Ephrussi A., 2002. Oskar anchoring restricts pole plasm formation to the posterior of the Drosophila oocyte. Development 129: 3705–3714 [DOI] [PubMed] [Google Scholar]
- Vanzo N., Oprins A., Xanthakis D., Ephrussi A., Rabouille C., 2007. Stimulation of endocytosis and actin dynamics by Oskar polarizes the Drosophila oocyte. Dev. Cell 12: 543–555 [DOI] [PubMed] [Google Scholar]
- Vogt N., Koch I., Schwarz H., Schnorrer F., Nusslein-Volhard C., 2006. The gammaTuRC components Grip75 and Grip128 have an essential microtubule-anchoring function in the Drosophila germline. Development 133: 3963–3972 [DOI] [PubMed] [Google Scholar]
- Weil T. T., Forrest K. M., Gavis E. R., 2006. Localization of bicoid mRNA in late oocytes is maintained by continual active transport. Dev. Cell 11: 251–262 [DOI] [PubMed] [Google Scholar]
- Weil T. T., Parton R., Davis I., Gavis E. R., 2008. Changes in bicoid mRNA anchoring highlight conserved mechanisms during the oocyte-to-embryo transition. Curr. Biol. 18: 1055–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil T. T., Xanthakis D., Parton R., Dobbie I., Rabouille C., et al. , 2010. Distinguishing direct from indirect roles for bicoid mRNA localization factors. Development 137: 169–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellington A., Emmons S., James B., Calley J., Grover M., et al. , 1999. Spire contains actin binding domains and is related to ascidian posterior end mark-5. Development 126: 5267–5274 [DOI] [PubMed] [Google Scholar]
- Wharton R. P., Struhl G., 1991. RNA regulatory elements mediate control of Drosophila body pattern by the posterior morphogen nanos. Cell 67: 955–967 [DOI] [PubMed] [Google Scholar]
- Yousef M. S., Kamikubo H., Kataoka M., Kato R., Wakatsuki S., 2008. Miranda cargo-binding domain forms an elongated coiled-coil homodimer in solution: implications for asymmetric cell division in Drosophila. Protein Sci. 17: 908–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Morin X., Cai Y., Yang X., Chia W., 2000. Analysis of partner of inscuteable, a novel player of Drosophila asymmetric divisions, reveals two distinct steps in inscuteable apical localization. Cell 100: 399–409 [DOI] [PubMed] [Google Scholar]
- Zhang Y., 2008. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., 2009. I-TASSER: fully automated protein structure prediction in CASP8. Proteins 77(Suppl. 9): 100–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimyanin V. L., Belaya K., Pecreaux J., Gilchrist M. J., Clark A., et al. , 2008. In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell 134: 843–853 [DOI] [PMC free article] [PubMed] [Google Scholar]