Abstract
Light regulates several aspects of the biology of many organisms, including the balance between asexual and sexual development in some fungi. To understand how light regulates fungal development at the molecular level we have used Aspergillus nidulans as a model. We have performed a genome-wide expression analysis that has allowed us to identify >400 genes upregulated and >100 genes downregulated by light in developmentally competent mycelium. Among the upregulated genes were genes required for the regulation of asexual development, one of the major biological responses to light in A. nidulans, which is a pathway controlled by the master regulatory gene brlA. The expression of brlA, like conidiation, is induced by light. A detailed analysis of brlA light regulation revealed increased expression after short exposures with a maximum after 60 min of light followed by photoadaptation with longer light exposures. In addition to brlA, genes flbA–C and fluG are also light regulated, and flbA–C are required for the correct light-dependent regulation of the upstream regulator fluG. We have found that light induction of brlA required the photoreceptor complex composed of a phytochrome FphA, and the white-collar homologs LreA and LreB, and the fluffy genes flbA–C. We propose that the activation of regulatory genes by light is the key event in the activation of asexual development by light in A. nidulans.
MANY organisms encounter cycles of light/darkness during their lives, and the presence of light serves as an environmental signal to regulate different aspects of their biology, even in nonphotosynthetic organisms. In fungi, light has a strong influence on development, regulates metabolic pathways, and may direct the growth of reproductive structures (Corrochano and Galland 2006; Bahn et al. 2007; Corrochano and Avalos 2010; Rodriguez-Romero et al. 2010). For example, fungi growing in dark or shaded areas use light as a signal to promote vegetative reproduction and to direct the growth of reproductive structures toward open air to facilitate spore dispersal (Corrochano and Galland 2006; Corrochano and Avalos 2010; Rodriguez-Romero et al. 2010). An excess of light can be harmful, particularly UV light. Thus, the activation of the biosynthesis of pigments, like carotenoids and the activation of genes for light-dependent DNA repair by light can be considered a protection mechanism from light (Berrocal-Tito et al. 1999; Alejandre-Duran et al. 2003; Corrochano and Avalos 2010). It appears that the capacity to receive and respond to light improves fungal adaptation and survival in nature.
Photoreceptor proteins sense light through a chromophore, a light-absorbing molecule bound to the protein that provides the sensitivity to a specific range of wavelengths. Several of these proteins have been identified in fungi (Corrochano 2007; Idnurm et al. 2010; Rodriguez-Romero et al. 2010). The molecular mechanism of fungal photoreception has been investigated in greatest detail in the ascomycete Neurospora crassa (Chen et al. 2010b). This fungus perceives light through the white-collar protein 1 (WC-1), a zinc-finger protein with a flavin-binding domain (named LOV for light–oxygen–voltage), and a PAS domain (Drosophila period, PER–vertebrate aryl hydrocarbon receptor nuclear translocator, ARNT–Drosophila single-minded, SIM) for protein–protein interactions (Ballario et al. 1996). The LOV domain binds the flavin FAD, allowing WC-1 to act as a photoreceptor for blue light (Froehlich et al. 2002; He et al. 2002). The WC-2 protein contains one zinc-finger and two PAS domains (Linden and Macino 1997). WC-2 interacts with WC-1 to form a heterodimeric complex via their PAS domains to form a WC complex (WCC) that binds to the promoters of light-inducible genes to regulate their transcription (Froehlich et al. 2002; He and Liu 2005; Belden et al. 2007b; Chen et al. 2009; Olmedo et al. 2010b; Smith et al. 2010). Another blue-light photoreceptor in N. crassa is VIVID (VVD), a small protein with a LOV domain that is required for the transient activation of gene transcription (photoadaptation) by light (Heintzen et al. 2001; Schwerdtfeger and Linden 2003). The transduction of the light signal in WC-1 and VVD requires the formation of a covalent photoadduct between a conserved cysteinyl residue of the LOV domain and the flavin cofactor (Cheng et al. 2003; Schwerdtfeger and Linden 2003). Recently, a physical interaction between VVD and the WCC has been shown to be required for the regulation of the activity of the WCC (Chen et al. 2010a; Hunt et al. 2010; Malzahn et al. 2010). Although VVD participates in the photoadaptation of gene expression in N. crassa, homologs of VVD are not widely distributed in fungi (Rodriguez-Romero et al. 2010). Phytochromes are photoreceptors that sense red and far-red light through a linear tetrapyrrole chromophore (Rockwell et al. 2006). Previous characterization of phytochrome mutants did not show any major alteration in light-dependent regulation of gene expression in N. crassa (Froehlich et al. 2005; Chen et al. 2009). Cryptochromes were initially identified as plant blue-light photoreceptors very similar to photolyases, enzymes for blue-light–dependent DNA repair (Lin and Todo 2005). The N. crassa cryptochrome binds the chromophores FAD and MTHF, and a strain with a deletion of the cryptochrome gene showed a minor change in circadian clock entrainment (Froehlich et al. 2010). N. crassa strains with deletions in the cryptochrome CRY-1, the rhodopsin NOP-1, and the phytochrome PHY-2 showed increased light-dependent accumulation of mRNA of light-regulated genes (con-6 and con-10). This observation suggests that these photoreceptors modify the activity of the WCC, presumably through a repressor (Olmedo et al. 2010b).
Several orthologs of the N. crassa photoreceptors have also been recently described in Aspergillus nidulans (Bayram et al. 2010; Rodriguez-Romero et al. 2010): a phytochrome, FphA, for red-light detection (Blumenstein et al. 2005; Purschwitz et al. 2008), a homolog of WC-1, LreA, and a homolog of WC-2, LreB, for blue-light detection (Purschwitz et al. 2008), and a cryptochrome, CryA, (Bayram et al. 2008a). Unlike N. crassa, A. nidulans can sense red light through the phytochrome to modulate development and other light-dependent processes (Blumenstein et al. 2005; Rodriguez-Romero et al. 2010). Another interesting aspect in the A. nidulans photobiology is that the phytochrome forms a large complex with the A. nidulans homologs of WC-1 and WC-2, and the velvet A (VeA) protein, a repressor of light-regulated conidiation and an activator of sexual development (Purschwitz et al. 2008). VeA interacts with additional proteins to regulate sexual development and the synthesis of secondary metabolites (Bayram et al. 2008b; Sarikaya Bayram et al. 2010). In addition, the UV/blue-light sensing cryptochrome is involved in the regulation of sexual development in A. nidulans by light (Bayram et al. 2008a).
Conidiation, the development of asexual spores, is controlled by a pathway, encompassing BrlA, AbaA, and WetA (see reviews by Adams et al. 1998; Yu et al. 2006). The first component in the regulatory cascade, BrlA, is necessary and sufficient to drive conidiation (Adams et al. 1988). Transcription of the brlA gene induces conidiation, and brlA itself is controlled by a number of genes, including the fluffy genes. Deletion of any of the fluffy genes gives a typical fluffy phenotype with cotton-like colonies and reduced levels of brlA expression (Adams et al. 1998; Yu et al. 2006). The fluffy genes are: fluG and flbA–E. fluG encodes a protein similar to glutamine synthetase and is responsible for the synthesis of an extracellular factor that induces conidiation (Lee and Adams 1994). FluG works upstream of the flbA–E genes (Yu et al. 2006). FlbA is a regulator of the protein G activity, which participates in a protein kinase A-dependent pathway promoting filamentous growth and inhibiting conidiation (Yu et al. 1996). FlbE interacts with FlbB at the fungal tip and is required for proper activation of FlbB (Garzia et al. 2009). FlbB is a bZip transcription factor that activates the transcription of flbD, a cMyb-type regulator. Then, both FlbB and FlbD jointly activate the transcription of brlA (Garzia et al. 2010). FlbC is a putative C2H2 Zn finger protein that constitutes a third path for the regulation of brlA expression (Kwon et al. 2010). These fluffy genes are expressed in vegetative mycelium and are able to respond to intracellular stimuli to induce a coordinated activation of the master regulator brlA (Etxebeste et al. 2010).
Light regulates the balance between asexual or sexual development in A. nidulans (Rodriguez-Romero et al. 2010), and it has been shown that light increases the accumulation of brlA mRNA (Mooney and Yager 1990). In N. crassa, light activates the accumulation of mRNAs for the developmental genes fluffy (fl) (Belden et al. 2007a; Olmedo et al. 2010a) and csp-1 (Chen et al. 2009). The FL protein is a transcription factor that is necessary and sufficient to induce conidiation (Bailey and Ebbole 1998; Bailey-Shrode and Ebbole 2004). fl is induced during conidiation along with many other genes in N. crassa (Greenwald et al. 2010). The light-dependent activation of fl by the WCC suggested a simple model for the activation of conidiation by light in N. crassa with light-activated WCC increasing fl transcription and the subsequent accumulation of regulatory FL protein activating the conidiation pathway (Olmedo et al. 2010a). This model does not appear to be so simple in A. nidulans.
Here we show that the expression of key upstream regulators is induced by light and traced down the path for the light-dependent induction of conidiation in A. nidulans.
Materials and Methods
Strains, media, and culture conditions
Strains used in this study are listed in Table 1. Strains were grown in complete or minimal media containing the appropriate supplements (Cove 1966). A total of 1% glucose and 10 mM NH4NO3 were used as carbon and nitrogen sources. Conidia were inoculated on the surface of 25 ml of complete liquid medium in a Petri dish. Cultures were grown for 18 hr at 37° in the dark before the light-induction experiments.
Table 1. A. nidulans strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| FGSC4 | Wild type | FGSC |
| BD205 | pyrG89; pyroA4; veA+ | Herrero-García et al. (2011) |
| SJP1 | pyrG89; ∆argB:trpC∆B; pyroA4 ∆fphA::argB; veA+ | Purschwitz et al. (2008) |
| SJP69 | yA1, pyrG89; ∆argB:trpC∆B; pyroA4; ∆lreA::argB; veA+ | This study |
| LBV+ | biA; ∆lreB::argB; pyroA4; veA+ | Purschwitz et al. (2008) |
| SJP21.3 | pyrG89; ∆argB:trpC∆B; pyroA4; ∆lreA::argB, ∆lreB::argB, ∆fphA::argB; veA+ | Purschwitz et al. (2008) |
| RNJ3.1 | biA1; ∆flbA::argB+; veA1 | Shin et al. (2009) |
| DKA91 | ∆flbA::argB+; veA+ | RNJ3.1 × BD205, this study |
| BD215 | ∆flbB::pyrG; pyrG89; pyroA4; veA+ | Herrero-García et al. (2011) |
| TNJ14.1 | biA1; methG1; ∆flbC::argB+; veA1 | Kwon et al. (2010) |
| DKA82 | ∆flbC::argB+; veA+ | TNJ14.1 × BD205, this study |
FGSC, Fungal Genetics Stock Center.
Light-induction experiments
Mycelial mats were exposed to light generated by a set of Phillips Master TL-D 36 W/865 white fluorescent bulbs for the indicated times (11 W/m2). After the exposure to light, mycelia were collected in the dark and immediately frozen in liquid nitrogen. Samples were stored at −80°. Control samples were harvested in complete darkness. All light-induction experiments were performed at 22°. Control samples were kept at the same temperature during the duration of the experiment. Light intensities were measured with a calibrated photodiode.
RNA isolation
Aspergillus mycelia (100–200 mg) were disrupted in 1 ml of TRI reagent (Sigma) with 1.5 g of zirconium beads (0.5-mm diameter) by using two 0.5-min pulses in a cell homogeneizer (FastPrep-24, MP Biomedicals). Cell debris was spinned down by centrifugation. Supernatants were extracted with chloroform and RNA was precipitated with isopropanol. The RNA samples were treated with DNase I (USB) prior to use in RT–PCR experiments. In an alternative method, Aspergillus mycelia (100–200 mg) were ground into fine powder in a mortar with a pestle. RNA was isolated from the powder by using the RNeasy plant mini kit (Qiagen) with the RLC buffer.
For the microarray experiments, total RNA was isolated with TRIZOL reagent (Invitrogen). Cells were broken up with acid-washed glass beads in a homogenizer (MM200, Retsch) working at maximum speed (frequency 25/s) for 5 min. Total RNA was further purified using RNeasy spin minicolumns (Qiagen). RNA was subjected to quality control in a 2100 Bioanalyzer (Agilent) using Eukaryote Total RNA as standard before the microarray experiments.
RNA labeling, hybridization, and microarray analysis
The A. nidulans DNA (version 2) microarray slides used in this study were obtained from the Pathogen Functional Genomics Resource Center (J. Craig Venter Institute, Rockville, MA). The slides were spotted with 70-mer oligonucleotides corresponding to 11,481 genes. The glass slide arrays have an “in-slide” replicate for each gene.
RNA was isolated from mycelia grown in the dark (control sample) or illuminated with white light for 30 min. Thirty-five micrograms of total RNA was labeled with Cy3 or Cy5 dyes in a reaction containing 5 µg oligo-dT primer, Expand RT mix, and RNAseH (Invitrogen) for 2 hr at 42°. Dye-swap experiments were done using RNA samples pooled from two different experiments. The labeled samples were purified in Microcon YM30 columns according to manufacturer instructions. Hybridizations and washings were performed according to the standard operating procedure of the Institute for Genomic Research (http://pfgrc.jcvi.org/index.php/microarray/protocols.html).
The hybridized slides were scanned with an Axon GenePix 4000B scanner (Molecular Devices, Sunnyvale, CA). The images were further processed with GenePix Pro software (Molecular Devices). Intensities were calculated from Lowess normalized M Log ratios, substracting the background using the following parameters: global, 0.4 smoothing and 10 iterations. Values of low quality were excluded according to a quality filter set to “sum of median < 200,” which corresponds to a saturated signal in both channels. Feature background intensities were calculated by using morphological opening and the following parameters: closing to three pixels, opening to two, feature separation. Data have been deposited in ArrayExpress (accesion no. E-MEXP-3218).
Quantitative RT–PCR
The primers employed for quantitative RT–PCR are detailed in supporting information, Table S1. Quantitative RT–PCR experiments were performed using one-step RT–PCR, using 25 µl 1X Power SYBR Green PCR Master mix (Applied Biosystems), 6.25 units MultiScribe Reverse Transcriptase (Applied Biosystems), 1.25 units RNase Inhibitor (Applied Biosystems), 0.2 µM of each primer, and 100 ng of RNA in a 25-µl reaction in a 7500 Real-Time PCR System (Applied Biosystems) according to the manufacturer’s directions. The reaction consisted of 30 min at 48°, 10 min at 95°, and 40 cycles of DNA amplification (15 s at 95° and 1 min at 60°). After each PCR, we performed melting curve analysis to show the specific amplification of single DNA segments and the absence of nonspecific amplified DNA.
Additional quantitative RT–PCR experiments were performed in a LightCycler 480 II (Roche) by using the One-Step SYBR PrimeScript RT–PCR kit (Takara Bio), 0.2 µM of each primer and 50 ng of RNA in a 10-µl reaction. The reaction consisted of 5 min at 42°, followed by 10 s at 95°, and then 40 cycles of DNA amplification (5 s at 95° and 20 s at 60°). After each PCR, we performed melting curve analysis to show the specific amplification of single DNA segments and the absence of nonspecific amplified DNA. Comparisions of both protocols showed consistency and reliability between both methods.
The fluorescent signal obtained for each gene was normalized to the corresponding fluorescent signal obtained with benA to correct for sampling errors. In all cases, expression data are shown relative to the wild-type mycelia grown in the dark and are the average of at least three independent biological replicates.
Results
Identification of light-regulated conidiation genes in A. nidulans
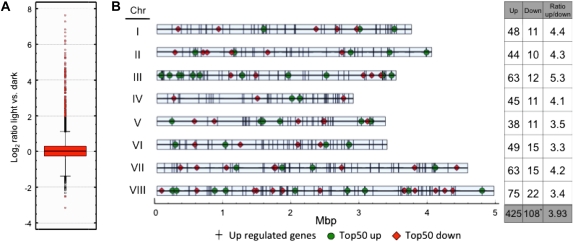
Light is a major environmental signal regulating many different biological processes. In A. nidulans, light controls asexual and sexual development as well as the production of secondary metabolites. To get a global view of genes regulated during asexual development and of genes involved in other light-regulated biological processes, a genome-wide approach was undertaken. Total RNA was isolated from surface-grown, developmentally competent mycelia of the wild-type strain FGSC4 exposed to white light (11 W/m2) for 30 min or grown in the dark, labeled, and hybridized to a spotted microarray of A. nidulans. After background correction and Lowess M normalization the threshold was set to 2-fold to identify differentially regulated genes under light vs. dark conditions (Figure 1A). Under these conditions, 533 out of 10,560 genes were differentially regulated, ∼5% of the genome. We observed large differences in the light-dependent induction and repression provoked by light: out of the 533 differentially regulated genes 425 were upregulated, but only 108 were downregulated (Figure 1 and Table S2). The highest upregulated gene, ccgB (a homolog of the clock-controlled gene 1 of N. crassa), showed an ∼240-fold increase, while the highest downregulated gene, veA (a gene required for light regulation of conidiation in A. nidulans), showed only a 8.6-fold decrease (Figure 1A and Tables 2 and 3).
Figure 1 .
Whole-genome regulation of gene expression by light. (A) Boxplot diagram of genes showing the Log2 ratio of expression in light vs. dark conditions. (B) Schematic representation of the location of the light-regulated genes in the A. nidulans chromosomes. Each vertical bar represents an upregulated gene (425 genes). The location of the highest 50 upregulated genes are shown by circles. All downregulated genes are depicted with diamonds, with the exception of (AN09511), which has not been allocated in the genome. The table on the right shows the number of up- and downregulated genes in each chromosome and the ratios of up/downregulated genes.
Table 2. Top 15 upregulated genes obtained in the microarray hybridization experiments.
| Top | Locus | Gene description | Log2 ratio light vs. dark | Fold change | SD |
|---|---|---|---|---|---|
| 1 | AN5056 | ccgB homolog to ccg-1 from N. crassa | 7.90 | 239.35 | 1.246 |
| 2 | AN9285 | ccgA homolog to ccg-1 from N. crassa | 7.78 | 219.79 | 0.263 |
| 3 | AN0045 | Solid-state culture expressed protein (Aos23) | 7.75 | 214.97 | 0.683 |
| 4 | AN0693 | Hypothetical protein | 7.26 | 152.75 | 0.519 |
| 5 | AN8339 | Hypothetical protein | 6.81 | 112.13 | 0.670 |
| 6 | AN4299 | Clock controlled and temperature regulated | 6.55 | 93.57 | 0.158 |
| 7 | AN7558 | Hypothetical protein | 6.36 | 82.03 | 0.145 |
| 8 | AN8641 | Hypothetical protein | 6.27 | 77.33 | 0.784 |
| 9 | AN8638 | Conidia enriched transcript (cetJ) | 6.19 | 73.06 | 0.494 |
| 10 | AN8018 | Auxin efflux transporter family protein | 6.07 | 67.37 | 0.076 |
| 11 | AN9310 | Hypothetical protein | 6.07 | 67.32 | 0.129 |
| 12 | AN5004 | Hypothetical protein | 6.02 | 65.03 | 0.410 |
| 13 | AN5015 | Conidiation gene (conJ) | 5.95 | 61.82 | 0.282 |
| 14 | AN3872 | Hypothetical protein | 5.93 | 60.84 | 0.423 |
| 15 | AN5764 | Hypothetical protein | 5.75 | 53.67 | 0.122 |
Average of log2 of the ratio of light vs. dark was used to calculate the fold change. SD is the standard deviations of log2 of ratio of a swap-dye experiment. The gene descriptions were determined using the Aspergillus genome database AspGD (www.aspergillusgenome.org). Hypothetical proteins have unknown function but are conserved proteins.
We analyzed the distribution of light-regulated genes in the chromosomes to identify putative light-regulated specific regions in the A. nidulans genome (Figure 1B). In addition to the visual plot of light-regulated genes, we divided the total number of upregulated genes by the total number of downregulated genes to find out whether an overrepresentation of upregulated genes was present in any of the chromosomes. Chromosome III shows a significant increase in upregulated genes, with a ratio of 5.3 in comparison to an average ratio of 4.06. Although no obvious light-regulated specific regions were identified, a high number of upregulated genes was found close to the telomeric region in chromosome III. Most of these genes belong to the top 50 most upregulated genes, among which is a previously identified cluster encoding conidia-specific mRNA of unknown function (SpoC1) (Gwynne et al. 1984; Aramayo et al. 1989). Chromosome I also shows a high number of upregulated genes. In contrast, a high proportion of downregulated genes is located on chromosomes VI and VIII.
Some of the upregulated genes were predicted to be involved in biological processes known to be regulated by light, i.e., circadian rhythm and conidiation. Some other genes were predicted to be involved in other processes such as carbon metabolism and transport, redox reactions, or stress responses. Some of these genes are transcription factors and proteins probably implicated in the activation of light-dependent signaling pathways (Table S2 and Table S3). The activation of these genes may trigger the changes in development, stress responses, and secondary metabolism that A. nidulans undergoes when living in the light. The gene with the highest level of light induction is ccgB (Table 2), a homolog of the N. crassa ccg-1 gene, a clock-controlled and glucose-repressed gene of unknown function (Arpaia et al. 1995; Bell-Pedersen et al. 1996). Interestingly, ccgB appears to be duplicated in the genome of A. nidulans but not in other related species such as A. fumigatus and A. niger. Both genes, ccgA and ccgB, are induced by light to a similar extent (219- and 239-fold, respectively). Another light-induced gene that we have identified is conJ, the homolog of the light-regulated and conidiation gene con-10 in N. crassa (Roberts et al. 1988; Olmedo et al. 2010b). The function of con-10 in N. crassa is not known (Springer and Yanofsky 1992; White and Yanofsky 1993). Other interesting upregulated genes are photoreceptors (Table S2) like cryA (induced 3.4-fold) that encodes for a UV/blue-light sensing cryptochrome involved in the regulation of sexual development in A. nidulans by light (Bayram et al. 2008a). In addition, we found that the gene nopA predicted to encode an opsin-like protein (AN3361) is induced by light (22.5-fold). This protein is related to opsins, a group of membrane-embedded proteins with seven transmembrane helices that bind to a retinal chromophore (Brown 2004). The function of NopA is still unknown but homologous proteins in other fungi show a photocycle, suggesting that they could work as sensory photoreceptors. However, an A. nidulans nopA-deletion strain did not display any phenotype (J. Rodríguez-Romero and R. Fischer, unpublished results).
Several genes involved in conidiation were found in the list of upregulated genes (e.g., AN8638, cetJ; AN5015, conJ; etc.; see Table S2). Only one gene involved in the regulation of conidiation (flbC) was found in the list of upregulated genes (3.4-fold induction). Further inspection of the data uncovered that some of the conidiation genes (brlA, fluG, and flbB) were also upregulated by light (Table S4). However, the induction of those genes was below the 2-fold threshold.
Most downregulated genes were related to transport (12%), oxidoreductase functions (10%), nuclear components (14%), and nitrogen metabolism (Table 3 and Table S2). The most downregulated gene was velvet A (veA), a component of the light regulator complex (Purschwitz et al. 2008) that has been implicated in the light response in A. nidulans (Kafer 1965; Mooney and Yager 1990; Calvo 2008). VeA represses conidiation and promotes sexual development, and therefore, the regulation of VeA affects the balance between asexual and sexual development and the coordination of morphogenesis and secondary metabolism (Calvo 2008).
Table 3. Top 15 downregulated genes obtained in the microarray hybridization experiments.
| Top | Locus | Gene description | Log2 ratio light vs. dark | Fold change | SD |
|---|---|---|---|---|---|
| 1 | AN1052 | velvetA(veA) | −3.11 | 8.60 | 0.242 |
| 2 | AN8647 | ALS family protein | −2.65 | 6.28 | 0.722 |
| 3 | AN1008 | Putative nitrate transporter (crnA) | −2.37 | 5.17 | 0.228 |
| 4 | AN5558 | Alkaline protease (prtA) | −2.31 | 4.95 | 0.255 |
| 5 | AN3304 | GABA transporter, putative | −2.20 | 4.59 | 0.611 |
| 6 | AN0231 | Conidiophore-specific phenol oxidase (ivoB) | −2.14 | 4.40 | 0.074 |
| 7 | AN8063 | Acid phosphatase activity | −1.98 | 3.94 | 0.094 |
| 8 | AN9076 | Putative adhesin function | −1.97 | 3.91 | 0.751 |
| 9 | AN2926 | 60S ribosomal protein Nsa2, putative | −1.95 | 3.86 | 0.183 |
| 10 | AN8539 | GNAT family acetyltransferase, putative | −1.92 | 3.78 | 0.309 |
| 11 | AN5353 | Hypothetical protein | −1.89 | 3.70 | 0.381 |
| 12 | AN9240 | Putative C2H2 finger domain transcription factor | −1.81 | 3.49 | 0.829 |
| 13 | AN0367 | Integral membrane protein | −1.80 | 3.49 | 0.310 |
| 14 | AN0190 | Subunit of the tRNA splicing endonuclease, | −1.77 | 3.41 | 0.117 |
| 15 | AN1131 | Cytosolic Cu/Zn superoxide dismutase, putative | −1.75 | 3.36 | 0.763 |
Average of log2 of the ratio of light vs. dark was used to calculate the fold change. SD is the standard deviations of log2 of ratio of a swap-dye experiment. The gene descriptions were determined using the Aspergillus genome database AspGD (www.aspergillusgenome.org). Hypothetical proteins have unknown function but are conserved proteins.
Light induction of the conidiation gene brlA is fast and transient
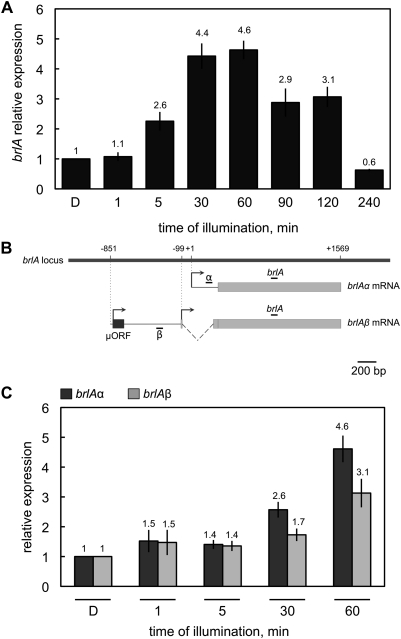
Mooney and Yager (1990) reported that brlA was activated by light (Mooney and Yager 1990) and we found brlA in the list of upregulated genes in our microarray hybridization experiments (Table S3). To characterize in detail the regulation by light of the A. nidulans conidiation genes, we assayed the response of brlA expression to light. Mycelia of A. nidulans grown in the dark for 18 hr were exposed to white light (11 W/m2) for time periods ranging from 1 min to 4 hr. After RNA extraction, the amount of brlA mRNA was determined by quantitative RT–PCR. Light activation of blrA was quick as a twofold increase of the mRNA amount was already detected after 5 min of illumination. The brlA mRNA reached a maximum of a fourfold increase after 30–60 min of light compared to the amount obtained in mycelia kept in the dark (Figure 2A). The accumulation of the brlA mRNA was transient and decreased when the exposure time was >60 min. Light-dependent brlA mRNA accumulation was not detected in mycelia exposed to light for 4 hr, suggesting that a photoadaptation event reminiscent of the one observed in N. crassa and Phycomyces blakesleeanus (Schwerdtfeger and Linden 2003; Rodriguez-Romero and Corrochano 2006) also exists in A. nidulans. Illumination for periods >4 hr and up to 12 hr did not result in photoinduction of brlA (data not shown).
Figure 2 .
The brlA gene is induced by light. Total RNA was isolated from vegetative mycelia of the wild-type strain that had been exposed to white light (11 W/m2 blue light) for various periods or kept in the dark “D” for 60, 90, 120, or 240 min prior to RNA isolation. Dark samples collected at 60, 90, 120, and 240 min of illumination gave similar values of mRNA accumulation for brlA, brlAα, and brlAβ. (A) The brlA transcript was amplified with primers that detect both transcripts. Samples were taken at time intervals ranging from 1 min to 4 hr. Maximum induction was observed after light exposures of 30–60 min. (B) Diagram depicting the organization of the brlA locus and the position of the primers used to amplify the α, the β. or both transcripts. (C) Light activates the two brlA mRNAs, brlAα and brlAβ. The plot shows the average and standard error of the mean of the relative photoactivation values with respect to the dark samples at 60 min in at least three independent experiments.
Two overlapping transcripts are produced from the brlA gene, where BrlAβ activates the transcription of brlAα and subsequently BrlAα activates the conidiation genes (Prade and Timberlake 1993). To check whether any of these transcripts was preferentially regulated upon light exposure, primer sets specific for each transcript were used in the quantitative RT–PCR experiments (Figure 2B). The induction levels of both transcripts were similar at short light-exposure times (Figure 2C). After 30 min of illumination, the α transcript accumulated to slightly higher levels than the β transcript (2.6-fold induction for the α transcript compared to 1.7-fold for the β transcript). Although the differences are not statistically significant, it was still observed after 60 min of light. This observation might be relevant for brlA regulation as the expression of brlAα is regulated by brlAβ (Barton and Prade 2008) and the β transcript predominantly accumulates over the α transcript at long light exposures (Kato et al. 2003).
The expression of the brlA upstream regulatory genes is induced by light
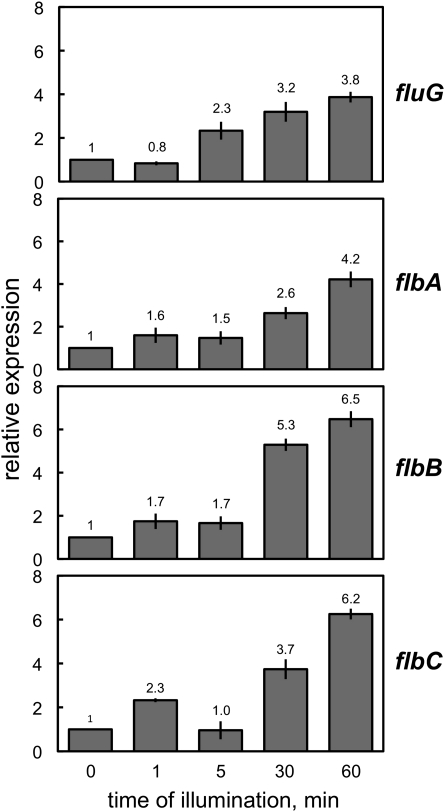
Given the fast induction by light of brlA, we asked whether the regulation of brlA expression by light is direct or whether regulatory genes upstream of brlA are additional targets for the light signal. The genes fluG and flbA–C encode developmental regulators that act upstream of brlA, and their deletion reduces the expression of brlA, resulting in aconidial, fluffy phenotypes (Adams et al. 1992, 1998; Wieser et al. 1994). Thus, our next question was whether these regulatory genes were also induced by light. Cultures of the wild-type strain were exposed to white light for different time periods and the expression of fluG, flbA, flbB, and flbC was assayed by RT–PCR. Genes fluG and flbA–C were clearly induced by light as shown by the light-dependent accumulation of the corresponding mRNAs (Figure 3). Maximum mRNA accumulation reached 4- to 6-fold after 60 min of light exposure, compared to mRNA accumulation in mycelia kept in the dark. Interestingly, the light-dependent induction of the fluffy genes displayed a similar pattern to the one observed for brlA (Figure 2), i.e., a response to short light exposures (the induction was minor but already evident after 1–5 min) and maximum accumulation (4- and 6.5-fold) after 30–60 min of illumination under our conditions (Figure 3).
Figure 3 .
Expression of the fluffy genes in response to light. Mycelia of the wild-type strain were exposed to white light for different times and the expression of the genes assayed by quantitative RT–PCR. fluG and flbA–C gene expression was induced by light. Results are shown as relative expression compared to control samples kept in the dark (0) for 60 min prior to RNA isolation. The plots represent the mean value and standard error of the mean of at least three independent experiments.
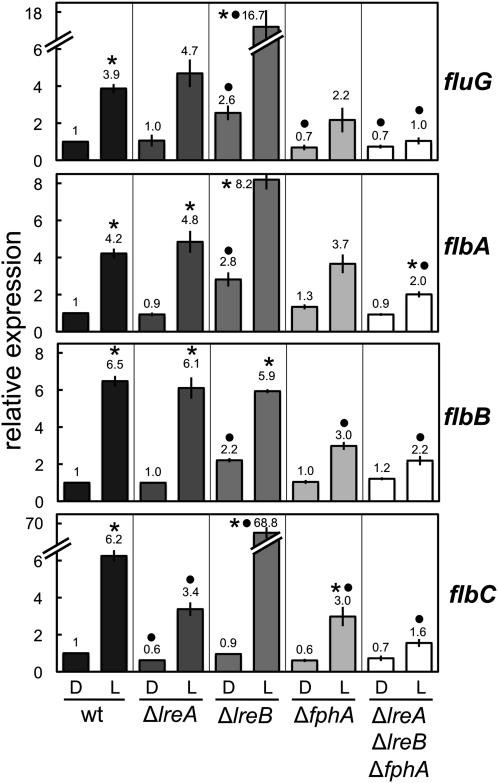
Light activation of the conidiation genes requires the photoreceptor complex
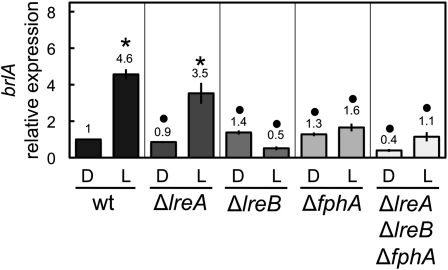
Light is perceived by a protein complex composed of the phytochrome FphA for red-light reception and the white-collar LreA/LreB proteins for blue-light reception (Purschwitz et al. 2008). To investigate whether these photoreceptors were required for the light-dependent activation of the conidiation genes, we assayed light-dependent mRNA accumulation in strains carrying single deletions of the photoreceptor genes fphA, lreA, or lreB, and in a triple deletion mutant strain (∆fphA ∆lreA ∆lreB). The light-dependent accumulation of brlA mRNA was not observed in the ΔfphA or ∆lreB strains, but was only slightly decreased in the ΔlreA mutant (Figure 4). These results suggest a role for the phytochrome and the white-collar 2 homolog LreB in brlA activation by light but not for the white-collar 1 homolog LreA. We did not detect any light-dependent brlA mRNA accumulation in the triple ∆fphA ∆lreA ∆lreB mutant (Figure 4). These results suggest a major role for FphA and LreB in the activation of brlA by light.
Figure 4 .
The activation of brlA by light requires the photoreceptor complex. Mycelia of the wild-type, ∆lreA, ∆lreB, ∆fphA, or ∆lreA∆lreB∆fphA strains were exposed to white light for 60 min or kept in the dark (D) prior to RNA extraction. The amount of brlA mRNA was assayed by quantitative RT–PCR. Results are shown as relative to control samples of the wild type kept in the dark. The plot shows the average and standard error of the mean of the relative photoactivation values in at least three independent experiments. Differences between dark and light conditions for the same strain, indicated by * and between different strains under the same conditions by •, are statistically significant according to the Wilcoxon–Mann–Whitney test and a P value <0.05.
Deletion of fphA did not prevent the light-dependent accumulation of flbA–C or fluG mRNAs but the light-dependent mRNA accumulation was reduced as compared to the wild-type levels (Figure 5). Deletion of the wc-1 gene, lreA, did not modify the expression by light of any of the genes under investigation with the exception of flbC, which showed a 50% reduction in light-dependent mRNA accumulation. Contrary to what happened with brlA expression, deletion of the wc-2 gene lreB resulted in derepression of fluG and flbC. The difference in the expression of flbA in the ∆lreB strain, although higher than in the wild-type strain, was not statistically significant (Figure 5). A triple mutant strain with deletions in fphA, lreA, and lreB still showed a limited light-dependent accumulation of flbA–C mRNAs. This suggests different roles for LreB in the regulation of brlA and the fluffy genes (flbA–C and fluG).
Figure 5 .
The activation of the upstream regulators of brlA by light requires the photoreceptor complex. Mycelia of the wild-type, ∆lreA, ∆lreB, ∆fphA, or ∆lreA∆lreB∆fphA strains were exposed to white light for 60 min or kept in the dark (D) prior to RNA extraction. The amount of fluG, flbA, flbB, or flbC mRNAs were assayed by quantitative RT–PCR. Results are shown as relative to control samples of the wild type kept in the dark. The plot shows the average and standard error of the mean of the relative photoactivation values in at least three independent experiments. Differences between dark and light conditions for the same strain, indicated by * and between different strains under the same conditions by •, are statistically significant according to the Wilcoxon–Mann–Whitney test and a P value <0.05.
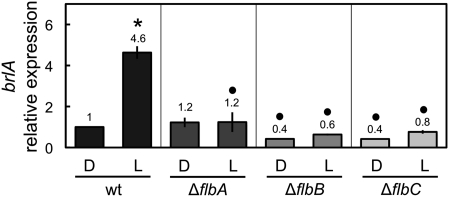
Light-dependent induction of brlA requires FlbA, FlbB, and FlbC
The induction of brlA and flbA–C gene expression required the photoreceptor complex. However, it was not known whether the light induction of brlA required the proteins FlbA–C, as these proteins are required for the developmental regulation of brlA (Adams et al. 1998). Thus, light might activate directly brlA expression to activate conidiation; in addition, light may indirectly activate brlA by directly activating the expression of brlA regulators, flbA–C. Therefore, we assayed the light-dependent induction of brlA in the wild type and the single deletion mutants of flbA, flbB or flbC (Figure 6). Under dark conditions, expression of brlA was slightly lower in the ∆flbB and ∆flbC strains. When the mycelia was illuminated with white light for 60 min, no significant increase in the expression of brlA was detected in any of the deletion mutants, showing that FlbA, FlbB, and FlbC were required for the light induction of brlA.
Figure 6 .
Light-dependent induction of the brlA gene is mediated by FlbA, FlbB, and FlbC. Mycelia of the wild-type and deletion strains were exposed to white light for 60 min and the amount of brlA mRNA was assayed by quantitative RT–PCR. Results are shown as relative to the wild-type samples kept in the dark. The plot shows the mean value and standard error of the mean of at least three independent experiments. Differences between dark and light conditions for the same strain, indicated by * and between different strains under the same conditions by •, are statistically significant according to the Wilcoxon–Mann–Whitney test and a P value <0.05.
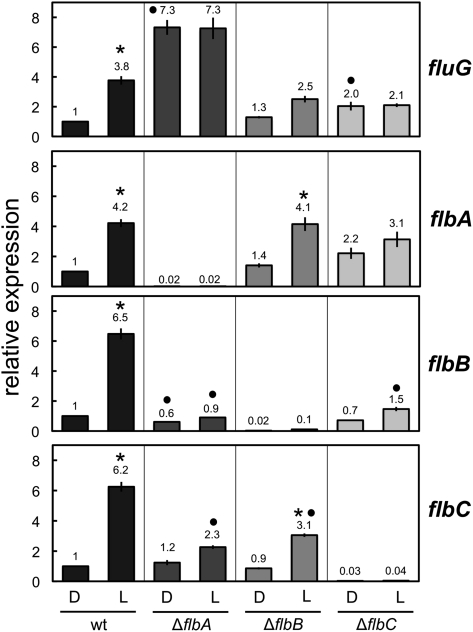
Cross-regulation of light-dependent induction in the fluffy genes
The expression of fluG was induced by light. However, the response of flbA–C to light exposure was slightly faster than the response of fluG (Figure 3). Since the regulatory role of fluG is performed upstream of flbA–C (Yu et al. 2006), we considered the possibility that FlbA–C were actually responsible for the light-dependent induction of fluG. Thus, the expression of fluG was assayed by quantitative RT–PCR in the wild type, ∆flbA, ∆flbB, or ∆flbC strains after 60 min of light exposure and compared to the accumulation of fluG mRNA in mycelia kept in the dark. The fluG gene was induced fourfold in the wild-type strain (Figure 7). In the ∆flbA strain, the expression of fluG was sevenfold higher than in the wild-type strain, regardless of light or dark conditions, showing that fluG was derepressed in the absence of flbA and unable to respond to light. A reduction of the light-dependent mRNA accumulation was observed in the ∆flbB strain, suggesting that FlbB was needed for full light-dependent induction of fluG. In the ∆flbC strain, no light-dependent induction of fluG was detected compared to the dark controls. However, the sample grown in the dark showed an expression level higher (twofold) than in the wild-type strain. The difference between the amount of fluG mRNA in light-exposed mycelia in ∆flbA or ∆flbC mutants compared to the amount observed in light-exposed mycelia of the wild type was not statistically significant. However, under dark conditions the difference between the amount of fluG mRNA in these two mutants to that observed in the wild type was statistically significant (Figure 7). These results suggest that FlbA–C were needed for the correct regulation of fluG expression, including its capacity to respond to the induction by light, and that FlbA and FlbC act as repressors of fluG.
Figure 7 .
Light-dependent cross-regulation of the fluffy genes. Mycelia of the wild-type and deletion strains were exposed to white light for 60 min and the amount of the fluG, flbA, flbB, or flbC mRNAs were assayed by quantitative RT–PCR. Results are shown as relative to the wild-type control samples kept in the dark. The plot represents the mean value and standard error of the mean of at least three independent experiments. Differences between dark and light conditions for the same strain indicated by * and between each strain and the wild type under the same conditions by •, are statistically significant according to the Wilcoxon–Mann–Whitney test and a P value <0.05.
To find out whether just one of the three fluffy genes or all (flbA, flbB, and/or flbC) were required for the reception of the signal from the photoreceptors, we also assayed the light-dependent mRNA accumulation for flbA, flbB, and flbC in each single deletion mutant (Figure 7). The expression of flbA was not affected in the ∆flbB or ∆flbC strains. However, the expression of flbB and flbC was reduced in the deletion mutants. In particular, flbB did not respond to the light stimulus in the ∆flbA and ∆flbC strains. The expression of flbC was reduced but it still retained some response to light in the ∆flbB strain. These results suggested that flbA was required for the full light-dependent response of fluG and flbB and only partially required for flbC (still showing ∼30% of induction compared to the wild-type strain). flbB was not required for the expression of the other fluffy genes, and flbC was required for the light-dependent induction of fluG, flbB, and flbA. In addition, deletion of flbC resulted in a slight derepression of fluG and flbA. The results show that FlbA and FlbC play a major role in the regulation of the fluffy genes by light.
Discussion
The response to light involves complex molecular mechanisms that regulate many different aspects of the biology of organisms. These responses and the corresponding molecular mechanisms have been extensively studied in plants and many fungi, in particular in N. crassa (Chen et al. 2004, 2010b). One of the most fascinating biological processes controlled by light is development. A. nidulans has been a model for the study of fungal development for more than three decades. However, unlike N. crassa, the study of the light-regulated processes has not been approached in detail in the Aspergilli but light sensing, including the regulation by red light of conidiation, was described in A. nidulans and other fungi many years ago (Tan 1974; Mooney and Yager 1990; Bayram et al. 2010; Rodriguez-Romero et al. 2010). Complete expression analysis has been performed by microarray hybridization experiments with different fungi (Rosales-Saavedra et al. 2006; Chen et al. 2009; Greenwald et al. 2010; Idnurm and Heitman 2010). In N. crassa changes in transcriptional levels of many different families of genes drive physiological and developmental changes (Dong et al. 2008; Greenwald et al. 2010). These changes in transcription were observed over a period of time broader than in our microarray experiment. Most of the changes in transcription levels during development and circadian cycles are not due to direct action of light but rather to a cascade of signals leading the fungus to change its physiological and/or developmental state. The number of blue-light–regulated genes was estimated to be 6% in N. crassa (Chen et al. 2009). In T. atroviride, 40 genes regulated by white light have been identified using cDNA microarrays, which represents 2.8% of the genes printed in the array. Thirty T. atroviride genes were upregulated (2%) and 10 were downregulated (0.8%) (Rosales-Saavedra et al. 2006). Here we report for the first time in A. nidulans the global effect of light exposure on transcription. In A. nidulans 5% of the genes were differentially regulated under our experimental conditions, which is similar to the response found in N. crassa. We used a short time of broad-spectrum light to discern a fast and initial transcriptional response, and to try to avoid secondary mycelial changes and adaptation to light. The transcriptomics results showed that several aspects of the biology of A. nidulans are affected by light. The response to light in A. nidulans included many upregulated genes that could be required to avoid stressing environmental changes like reactive oxygen species, osmotic stress, heat shock, etc., that may occur after light exposure or under natural conditions. In addition, we found the upregulation of genes that participate in the carbohydrate metabolism and the downregulation of genes that participate in nitrogen metabolism, suggesting that both carbohydrate and nitrogen metabolisms are regulated by light in A. nidulans.
A minimum of 15–30 min of illumination is necessary to elicit conidiation in a veA+ genetic background (Mooney and Yager 1990). This is consistent with our results showing that brlA expression reaches a plateau at ∼30 min of illumination. However, brlA is transiently activated by light as maximum light-dependent brlA mRNA accumulation was observed after 30–60 min of light, but longer exposures to light (up to 12 hr) did not increase the amount of brlA mRNA over the values obtained in the dark.
Photoadaptation has been described for light-regulated genes in N. crassa (Lauter and Yanofsky 1993; Arpaia et al. 1999; Schwerdtfeger and Linden 2003) and P. blakesleeanus (Rodriguez-Romero and Corrochano 2006). Photoadaptation in N. crassa requires the product of the vvd gene (Heintzen et al. 2001; Schwerdtfeger and Linden 2003; Zoltowski et al. 2007), such that the activity of the white-collar complex (WCC) is controlled by the VVD protein through the direct interaction of VVD with the components of the WCC (Chen et al. 2010a; Hunt et al. 2010; Malzahn et al. 2010). A homolog of vvd is not found in the genome A. nidulans. Photoadaptation of several genes also occurs in the zygomycete P. blakesleeanus despite the absence of a vvd homolog in its genome (Rodriguez-Romero and Corrochano 2006; Sanz et al. 2009). It is possible that a novel mechanism is responsible for the observed photoadaptation of brlA in the absence of a vvd homolog in A. nidulans.
The expression of the brlA gene is activated by several regulators. Recently, it has been reported that FlbB together with FlbD bind to the promoter of brlA to activate transcription (Garzia et al. 2010). FlbC is another transcriptional regulator that binds to the promoter of brlA and is involved in its activation (Kwon et al. 2010). We have found that flbB and flbC are induced by light and that deletion of flbB or flbC disrupts the activation of brlA by light. These results suggest that these two proteins are not only required for the activation of brlA but also for its regulation by light. One possibility is that the photoreceptor complex is signaling some other regulators, including FlbB and FlbC, to bind to the promoter of brlA or that the photoreceptor complex binds itself to the promoter to activate brlA transcription in a mechanism that requires FlbB and FlbC. LreA and LreB are homologs of the white-collar proteins, which are known DNA-binding proteins (Froehlich et al. 2002; He et al. 2002; Belden et al. 2007b; Olmedo et al. 2010a) and may directly regulate gene expression in A. nidulans in a similar way. It is interesting that the induction levels of all the genes that we have characterized are similar, possibly reflecting that an increase in the expression of the fluffy genes triggers an equivalent increase in brlA expression. The absence of light-dependent brlA mRNA accumulation in strains with deletions in flbA, flbB, or flbC suggests that the light-dependent activation of brlA occurs through the induction of these genes. However, deletion of fphA did not appear to have a dramatic effect in the expression of the fluffy genes, which leaves open the possibility that the photoreceptor complex is also binding to the promoter of brlA and is required for the full light-dependent activation of brlA. Thus, what remains unknown is how the photoreceptors induce the expression of the regulators of conidiation and whether it is a direct or indirect event through other components in the signaling pathway, yet to be discovered.
FluG deserves special attention. The expression of the gene is induced by illumination (Figure 3). FluG is responsible for the synthesis of an unknown diffusible factor that triggers conidiation (Lee and Adams 1994) by acting upstream of and derepressing all the fluffy genes (flbA–E). Surprisingly, our results in Figure 7 revealed that fluG is controlled by flbA, flbB, and flbC. Lee and Adams (1995) reported that fluG and flbA work interdependently to activate the transcription of brlA, i.e., each of them requires the presence of the other to promote conidiation (Lee and Adams 1995). We have found that FlbA is responsible for the correct regulation of fluG expression. In the absence of FlbA, fluG is deregulated: the gene is derepressed and also failed to be induced under illumination. FlbB and FlbC also seem to be involved in the correct light-dependent regulation of fluG expression. This shows that there is a feedback loop of regulation in the conidiation pathway. Yager et al. (1998) isolated a fluG701 mutant that failed to respond to red light, which suggests that FlbA–C could activate the transcription of another regulator that acts on FluG (Yager et al. 1998). The possibility that this feedback loop allows the amplification of the signal is possible but unlikely, since once the developmental pathways are triggered, there is a balance between them, rather than a yes/no response in A. nidulans, which would originate from signal amplification. Another possibility is that the FluG-dependent factor is sensitive to light or oxidation, and this feedback loop increases the synthesis of the unknown compound to satisfy new demands and maintain the derepressed state for conidiation.
The conidiation results obtained by Purschwitz et al. (2008) showed an interesting pattern that we could also observe with our expression data (Purschwitz et al. 2008). They observed a nonstatistically significant increase of conidial number in the ∆lreA mutant strain (both in dark and light conditions), which is consistent with the induction of some conidiation genes in the ∆lreA strain in comparison to the wild-type strain (Figure 5). Although in both cases the differences are statistically not significant, they show a clear trend that would be consistent with a dual role of LreA in induction and repression of genes. Our data on the expression of the fluffy genes in the ∆lreB mutant suggest that the role of the WC complex in A. nidulans and N. crassa differ. Deletion of lreB resulted in a strong increase of the expression of some genes compared to the wild type. However, the ∆lreB strain was blind for brlA expression, despite the fact that LreB does not have any motif for photodetection. Purschwitz et al. (2008) demonstrated the existence of a photoreceptor complex in which LreB is bridging the two photoreceptor proteins FphA and LreA (Purschwitz et al. 2008). One possibility is that LreB is required for DNA binding of the photoreceptor complex and depending on the light conditions, it will act as inducer (recruiting FphA to the promoter) or repressor through binding to LreA. WC-2 binds to hundreds of genomic regions in N. crassa. However, not all of the genes that are direct targets of the WCC are light inducible (Smith et al. 2010). This may not be surprising for genes with a complex regulation. Our data suggest that the role of WCC differs in A. nidulans and in N. crassa. LreA/LreB/FphA is a heteromeric protein with a complex role in the interplay between blue and red lights that displays a repression vs. activation activity depending on the conditions to trigger conidiation (or other biological function). In addition, the dual induction/repression role of the LreA/B could be achieved through one of the fluffy genes, acting as derepressor, while the other(s) would be inducers of the expression of brlA. This system would require the activity of all the fluffy genes. In a recent review, Etxebeste et al. (2010) discussed the possibility of a hierarchical vs. a cooperative mode of control of development in A. nidulans. Our results together with those from other laboratories (Adams et al. 1998; Yu et al. 2006; Etxebeste et al. 2010) suggest a complex combination of both modes (hierarchical plus cooperative) operating upstream of the master regulator BrlA, where some of the fluffy genes were necessary for the correct regulation of other fluffy genes, and subsequently, altogether would be required for the expression of brlA.
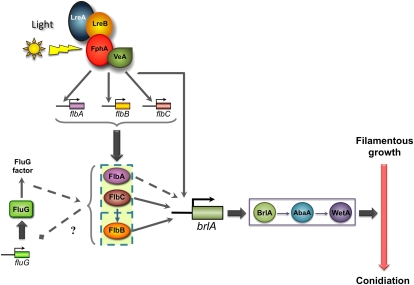
A model for the light-dependent induction of conidiation starts with light detected by the photoreceptor complex (Figure 8). Then, this complex would activate the expression of the fluffy genes flbA and flbC, which are also required for the light-dependent expression of flbB. There is cross-regulation between fluG and flbA and flbC. The fluffy genes flbA–C are essential for the light-dependent induction of brlA expression, and the activitation of these genes by light will provide the regulatory proteins for the correct activation of blrA. In addition, we propose that the photoreceptor complex interacts directly with the promoter of brlA, as deletion of lreB resulted in a complete loss of brlA expression but not fluffy genes (Figures 4 and 5). The resulting activation of brlA by light will activate a regulatory cascade that will result in the activation of the developmental program for conidiation. A. nidulans asexual development is a fascinating example of how different environmental signals are integrated and transduced into complex morphogenetic pathways. Our results provide a framework for future experimental validation that will help to understand how light acts as a signal to regulate asexual development.
Figure 8 .
A model for light-dependent induction of conidiation in A. nidulans. Light is detected by the photoreceptor complex, which in turn activates the expression of the fluffy genes flbA and flbC. They are both involved in the light-dependent expression of flbB. In addition, flbA and flbC regulate fluG expression, which creates a feedback loop. The fluffy genes flbA–C are essential for the light-dependent induction of brlA expression. The photoreceptor complex may also interact directly with brlA. The resulting activation of brlA by light activates a regulatory cascade that results in the activation of the developmental program of conidiation.
Acknowledgments
Unai Ugalde and Jae-Hyuk Yu are acknowledged for sharing strains. We thank Natalia Requena (Karlsruhe Institute of Technology) for helpful discussions. The microarray slides were obtained through National Institute of Allergy and Infectious Diseases’s Pathogen Functional Genomics Resource Center, managed and funded by Division of Microbiology and Infectious Diseases, National Institutes of Health, Department of Health and Human Services and operated by the J. Craig Venter Institute. This work was supported by European funds (European Regional Development Fund), the Spanish Ministerio de Ciencia e Innovación (BIO2009-12486) and Junta de Andalucía (P09-CVI-5027; BIO119) to L.M.C. and the Spanish Ministerio de Ciencia e Innovación (BFU2008-04306) to D.C., the German Science Foundation (DFG Fi 459), the Fonds der Chemischen Industrie, the Baden-Württemberg Stiftung, and the Centre for Functional Nanostructures to R.F. C.R.H. is a research fellow of the regional goverment (Junta de Andalucía). J.R. is a fellow of the Spanish Science Ministry (postdoctoral Spanish Ministerio de Ciencia e Innovación fellowship).
Literature Cited
- Adams T. H., Boylan M. T., Timberlake W. E., 1988. brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell 54: 353–362 [DOI] [PubMed] [Google Scholar]
- Adams T. H., Hide W. A., Yager L. N., Lee B. N., 1992. Isolation of a gene required for programmed initiation of development by Aspergillus nidulans. Mol. Cell. Biol. 12: 3827–3833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams T. H., Wieser J. K., Yu J. H., 1998. Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 62: 35–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alejandre-Duran E., Roldan-Arjona T., Ariza R. R., Ruiz-Rubio M., 2003. The photolyase gene from the plant pathogen Fusarium oxysporum f. sp. lycopersici is induced by visible light and alpha-tomatine from tomato plant. Fungal Genet. Biol. 40: 159–165 [DOI] [PubMed] [Google Scholar]
- Aramayo R., Adams T. H., Timberlake W. E., 1989. A large cluster of highly expressed genes is dispensable for growth and development in Aspergillus nidulans. Genetics 122: 65–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpaia G., Loros J. J., Dunlap J. C., Morelli G., Macino G., 1995. Light induction of the clock-controlled gene ccg-1 is not transduced through the circadian clock in Neurospora crassa. Mol. Gen. Genet. 247: 157–163 [DOI] [PubMed] [Google Scholar]
- Arpaia G., Cerri F., Baima S., Macino G., 1999. Involvement of protein kinase C in the response of Neurospora crassa to blue light. Mol. Gen. Genet. 262: 314–322 [DOI] [PubMed] [Google Scholar]
- Bahn Y. S., Xue C., Idnurm A., Rutherford J. C., Heitman J., et al. , 2007. Sensing the environment: lessons from fungi. Nat. Rev. Microbiol. 5: 57–69 [DOI] [PubMed] [Google Scholar]
- Bailey L. A., Ebbole D. J., 1998. The fluffy gene of Neurospora crassa encodes a Gal4p-type C6 zinc cluster protein required for conidial development. Genetics 148: 1813–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Shrode L., Ebbole D. J., 2004. The fluffy gene of Neurospora crassa is necessary and sufficient to induce conidiophore development. Genetics 166: 1741–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballario P., Vittorioso P., Magrelli A., Talora C., Cabibbo A., et al. , 1996. White collar-1, a central regulator of blue light responses in Neurospora, is a zinc finger protein. EMBO J. 15: 1650–1657 [PMC free article] [PubMed] [Google Scholar]
- Barton L. M., Prade R. A., 2008. Inducible RNA Interference of brlAβ in Aspergillus nidulans. Eukaryot. Cell 7: 2004–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayram O., Biesemann C., Krappmann S., Galland P., Braus G. H., 2008a More than a repair enzyme: Aspergillus nidulans photolyase-like CryA is a regulator of sexual development. Mol. Biol. Cell 19: 3254–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayram O., Krappmann S., Ni M., Bok J. W., Helmstaedt K., et al. , 2008b VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 320: 1504–1506 [DOI] [PubMed] [Google Scholar]
- Bayram O., Braus G. H., Fischer R., Rodriguez-Romero J., 2010. Spotlight on Aspergillus nidulans photosensory systems. Fungal Genet. Biol. 47: 900–908 [DOI] [PubMed] [Google Scholar]
- Belden W. J., Larrondo L. F., Froehlich A. C., Shi M., Chen C. H., et al. , 2007a The band mutation in Neurospora crassa is a dominant allele of ras-1 implicating RAS signaling in circadian output. Genes Dev. 21: 1494–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belden W. J., Loros J. J., Dunlap J. C., 2007b Execution of the circadian negative feedback loop in Neurospora requires the ATP-dependent chromatin-remodeling enzyme CLOCKSWITCH. Mol. Cell 25: 587–600 [DOI] [PubMed] [Google Scholar]
- Bell-Pedersen D., Dunlap J. C., Loros J. J., 1996. Distinct cis-acting elements mediate clock, light, and developmental regulation of the Neurospora crassa eas (ccg-2) gene. Mol. Cell. Biol. 16: 513–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrocal-Tito G., Sametz-Baron L., Eichenberg K., Horwitz B. A., Herrera-Estrella A., 1999. Rapid blue light regulation of a Trichoderma harzianum photolyase gene. J. Biol. Chem. 274: 14288–14294 [DOI] [PubMed] [Google Scholar]
- Blumenstein A., Vienken K., Tasler R., Purschwitz J., Veith D., et al. , 2005. The Aspergillus nidulans phytochrome FphA represses sexual development in red light. Curr. Biol. 15: 1833–1838 [DOI] [PubMed] [Google Scholar]
- Brown L. S., 2004. Fungal rhodopsins and opsin-related proteins: eukaryotic homologues of bacteriorhodopsin with unknown functions. Photochem. Photobiol. Sci. 3: 555–565 [DOI] [PubMed] [Google Scholar]
- Calvo A. M., 2008. The VeA regulatory system and its role in morphological and chemical development in fungi. Fungal Genet. Biol. 45: 1053–1061 [DOI] [PubMed] [Google Scholar]
- Chen M., Chory J., Fankhauser C., 2004. Light signal transduction in higher plants. Annu. Rev. Genet. 38: 87–117 [DOI] [PubMed] [Google Scholar]
- Chen C. H., Ringelberg C. S., Gross R. H., Dunlap J. C., Loros J. J., 2009. Genome-wide analysis of light-inducible responses reveals hierarchical light signalling in Neurospora. EMBO J. 28: 1029–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. H., Demay B. S., Gladfelter A. S., Dunlap J. C., Loros J. J., 2010a Physical interaction between VIVID and white collar complex regulates photoadaptation in Neurospora. Proc. Natl. Acad. Sci. USA 107: 16715–16720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. H., Dunlap J. C., Loros J. J., 2010b Neurospora illuminates fungal photoreception. Fungal Genet. Biol. 47: 922–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P., He Q., Yang Y., Wang L., Liu Y., 2003. Functional conservation of light, oxygen, or voltage domains in light sensing. Proc. Natl. Acad. Sci. USA 100: 5938–5943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrochano L. M., Galland P., 2006. Photomorphogenesis and gravitropism in fungi, pp. 233–259 The Mycota I. Growth, Differentiation and Sexuality, edited by Kües U., Fischer R. Springer-Verlag, Berlin [Google Scholar]
- Corrochano L. M., 2007. Fungal photoreceptors: sensory molecules for fungal development and behaviour. Photochem. Photobiol. Sci. 6: 725–736 [DOI] [PubMed] [Google Scholar]
- Corrochano L. M., Avalos J., 2010. Light sensing, pp. 417–441 Cellular and Molecular Biology of Filamentous Fungi, edited by Borkovich K. A., Ebbole D. J. ASM Press, Washington DC [Google Scholar]
- Cove D. J., 1966. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim. Biophys. Acta 113: 51–56 [DOI] [PubMed] [Google Scholar]
- Dong W., Tang X., Yu Y., Nilsen R., Kim R., et al. , 2008. Systems biology of the clock in Neurospora crassa. PLoS ONE 3: e3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etxebeste O., Garzia A., Espeso E. A., Ugalde U., 2010. Aspergillus nidulans asexual development: making the most of cellular modules. Trends Microbiol. 18: 569–576 [DOI] [PubMed] [Google Scholar]
- Froehlich A. C., Liu Y., Loros J. J., Dunlap J. C., 2002. White Collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science 297: 815–819 [DOI] [PubMed] [Google Scholar]
- Froehlich A. C., Noh B., Vierstra R. D., Loros J., Dunlap J. C., 2005. Genetic and molecular analysis of phytochromes from the filamentous fungus Neurospora crassa. Eukaryot. Cell 4: 2140–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froehlich A. C., Chen C. H., Belden W. J., Madeti C., Roenneberg T., et al. , 2010. Genetic and molecular characterization of a cryptochrome from the filamentous fungus Neurospora crassa. Eukaryot. Cell 9: 738–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzia A., Etxebeste O., Herrero-Garcia E., Fischer R., Espeso E. A., et al. , 2009. Aspergillus nidulans FlbE is an upstream developmental activator of conidiation functionally associated with the putative transcription factor FlbB. Mol. Microbiol. 71: 172–184 [DOI] [PubMed] [Google Scholar]
- Garzia A., Etxebeste O., Herrero-Garcia E., Ugalde U., Espeso E. A., 2010. The concerted action of bZip and cMyb transcription factors FlbB and FlbD induces brlA expression and asexual development in Aspergillus nidulans. Mol. Microbiol. 75: 1314–1324 [DOI] [PubMed] [Google Scholar]
- Greenwald C. J., Kasuga T., Glass N. L., Shaw B. D., Ebbole D. J., et al. , 2010. Temporal and spatial regulation of gene expression during asexual development of Neurospora crassa. Genetics 186: 1217–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwynne D. I., Miller B. L., Miller K. Y., Timberlake W. E., 1984. Structure and regulated expression of the SpoC1 gene cluster from Aspergillus nidulans. J. Mol. Biol. 180: 91–109 [DOI] [PubMed] [Google Scholar]
- He Q., Cheng P., Yang Y., Wang L., Gardner K. H., et al. , 2002. White collar-1, a DNA binding transcription factor and a light sensor. Science 297: 840–843 [DOI] [PubMed] [Google Scholar]
- He Q., Liu Y., 2005. Molecular mechanism of light responses in Neurospora: from light-induced transcription to photoadaptation. Genes Dev. 19: 2888–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzen C., Loros J. J., Dunlap J. C., 2001. The PAS protein VIVID defines a clock-associated feedback loop that represses light input, modulates gating, and regulates clock resetting. Cell 104: 453–464 [DOI] [PubMed] [Google Scholar]
- Herrero-García E., Garzia A., Cordobés S., Espeso E. A., Ugalde U., 2011. Carbon oxylipins inhibit germination and growth, and stimulate aerial conidiation in Aspergillus nidulans. Fungal Biol. 115: 393–400 [DOI] [PubMed] [Google Scholar]
- Hunt S. M., Thompson S., Elvin M., Heintzen C., 2010. VIVID interacts with the WHITE COLLAR complex and FREQUENCY-interacting RNA helicase to alter light and clock responses in Neurospora. Proc. Natl. Acad. Sci. USA 107: 16709–16714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A., Heitman J., 2010. Ferrochelatase is a conserved downstream target of the blue light-sensing White collar complex in fungi. Microbiology 156: 2393–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A., Verma S., Corrochano L. M., 2010. A glimpse into the basis of vision in the kingdom Mycota. Fungal Genet. Biol. 47: 881–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafer E., 1965. Origins of translocations in Aspergillus nidulans. Genetics 52: 217–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato N., Brooks W., Calvo A. M., 2003. The expression of sterigmatocystin and penicillin genes in Aspergillus nidulans is controlled by veA, a gene required for sexual development. Eukaryot. Cell 2: 1178–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon N. J., Garzia A., Espeso E. A., Ugalde U., Yu J. H., 2010. FlbC is a putative nuclear C(2)H(2) transcription factor regulating development in Aspergillus nidulans. Mol. Microbiol. 77: 1203–1219 [DOI] [PubMed] [Google Scholar]
- Lauter F. R., Yanofsky C., 1993. Day/night and circadian rhythm control of con gene expression in Neurospora. Proc. Natl. Acad. Sci. USA 90: 8249–8253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B. N., Adams T. H., 1994. The Aspergillus nidulans fluG gene is required for production of an extracellular developmental signal and is related to prokaryotic glutamine synthetase I. Genes Dev. 8: 641–651 [DOI] [PubMed] [Google Scholar]
- Lee B. N., Adams T. H., 1995. fluG and flbA function interdependently to initiate conidiophore development in Aspergillus nidulans through brlA β activation. EMBO J. 15: 299–309 [PMC free article] [PubMed] [Google Scholar]
- Lin C., Todo T., 2005. The cryptochromes. Genome Biol. 6: 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden H., Macino G., 1997. White collar 2, a partner in blue-light signal transduction, controlling expression of light-regulated genes in Neurospora crassa. EMBO J. 16: 98–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malzahn E., Ciprianidis S., Kaldi K., Schafmeier T., Brunner M., 2010. Photoadaptation in Neurospora by competitive interaction of activating and inhibitory LOV domains. Cell 142: 762–772 [DOI] [PubMed] [Google Scholar]
- Mooney J. L., Yager L. N., 1990. Light is required for conidiation in Aspergillus nidulans. Genes Dev. 4: 1473–1482 [DOI] [PubMed] [Google Scholar]
- Olmedo M., Ruger-Herreros C., Corrochano L. M., 2010a Regulation by blue light of the fluffy gene encoding a major regulator of conidiation in Neurospora crassa. Genetics 184: 651–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmedo M., Ruger-Herreros C., Luque E. M., Corrochano L. M., 2010b A complex photoreceptor system mediates the regulation by light of the conidiation genes con-10 and con-6 in Neurospora crassa. Fungal Genet. Biol. 47: 352–363 [DOI] [PubMed] [Google Scholar]
- Prade R. A., Timberlake W. E., 1993. The Aspergillus nidulans brlA regulatory locus consists of overlapping transcription units that are individually required for conidiophore development. EMBO J. 12: 2439–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purschwitz J., Muller S., Kastner C., Schoser M., Haas H., et al. , 2008. Functional and physical interaction of blue- and red-light sensors in Aspergillus nidulans. Curr. Biol. 18: 255–259 [DOI] [PubMed] [Google Scholar]
- Roberts A. N., Berlin V., Hager K. M., Yanofsky C., 1988. Molecular analysis of a Neurospora crassa gene expressed during conidiation. Mol. Cell. Biol. 8: 2411–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell N. C., Su Y. S., Lagarias J. C., 2006. Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 57: 837–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Romero J., Corrochano L. M., 2006. Regulation by blue light and heat shock of gene transcription in the fungus Phycomyces: proteins required for photoinduction and mechanism for adaptation to light. Mol. Microbiol. 61: 1049–1059 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Romero J., Hedtke M., Kastner C., Muller S., Fischer R., 2010. Fungi, hidden in soil or up in the air: light makes a difference. Annu. Rev. Microbiol. 64: 585–610 [DOI] [PubMed] [Google Scholar]
- Rosales-Saavedra T., Esquivel-Naranjo E. U., Casas-Flores S., Martinez-Hernandez P., Ibarra-Laclette E., et al. , 2006. Novel light-regulated genes in Trichoderma atroviride: a dissection by cDNA microarrays. Microbiology 152: 3305–3317 [DOI] [PubMed] [Google Scholar]
- Sanz C., Rodriguez-Romero J., Idnurm A., Christie J. M., Heitman J., et al. , 2009. Phycomyces MADB interacts with MADA to form the primary photoreceptor complex for fungal phototropism. Proc. Natl. Acad. Sci. USA 106: 7095–7100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarikaya Bayram O., Bayram O., Valerius O., Park H. S., Irniger S., et al. , 2010. LaeA control of velvet family regulatory proteins for light-dependent development and fungal cell-type specificity. PLoS Genet. 6: e1001226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwerdtfeger C., Linden H., 2003. VIVID is a flavoprotein and serves as a fungal blue light photoreceptor for photoadaptation. EMBO J. 22: 4846–4855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin K. S., Kwon N. J., Kim Y. H., Park H. S., Kwon G. S., et al. , 2009. Differential roles of the ChiB chitinase in autolysis and cell death of Aspergillus nidulans. Eukaryot. Cell 8: 738–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K. M., Sancar G., Dekhang R., Sullivan C. M., Li S., et al. , 2010. Transcription factors in light and circadian clock signaling networks revealed by genomewide mapping of direct targets for neurospora white collar complex. Eukaryot. Cell 9: 1549–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M. L., Yanofsky C., 1992. Expression of con genes along the three sporulation pathways of Neurospora crassa. Genes Dev. 6: 1052–1057 [DOI] [PubMed] [Google Scholar]
- Tan K. K., 1974. Red-far-red reversible photoreaction in the recovery from blue-light inhibition of sporulation in Botrytis cinerea. J. Gen. Microbiol. 82: 201–202 [Google Scholar]
- White B. T., Yanofsky C., 1993. Structural characterization and expression analysis of the Neurospora conidiation gene con-6. Dev. Biol. 160: 254–264 [DOI] [PubMed] [Google Scholar]
- Wieser J., Lee B. N., Fondon J., 3rd, Adams T. H., 1994. Genetic requirements for initiating asexual development in Aspergillus nidulans. Curr. Genet. 27: 62–69 [DOI] [PubMed] [Google Scholar]
- Yager L. N., Lee H. O., Nagle D. L., Zimmerman J. E., 1998. Analysis of fluG mutations that affect light-dependent conidiation in Aspergillus nidulans. Genetics 149: 1777–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. H., Mah J. H., Seo J. A., 2006. Growth and developmental control in the model and pathogenic aspergilli. Eukaryot. Cell 5: 1577–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. H., Wieser J., Adams T. H., 1996. The Aspergillus FlbA RGS domain protein antagonizes G protein signaling to block proliferation and allow development. EMBO J. 15: 5184–5190 [PMC free article] [PubMed] [Google Scholar]
- Zoltowski B. D., Schwerdtfeger C., Widom J., Loros J. J., Bilwes A. M., et al. , 2007. Conformational switching in the fungal light sensor Vivid. Science 316: 1054–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]