Abstract
Mutator alleles, which elevate an individual’s mutation rate from 10 to 10,000-fold, have been found at high frequencies in many natural and experimental populations. Mutators are continually produced from nonmutators, often due to mutations in mismatch-repair genes. These mutators gradually accumulate deleterious mutations, limiting their spread. However, they can occasionally hitchhike to high frequencies with beneficial mutations. We study the interplay between these effects. We first analyze the dynamics of the balance between the production of mutator alleles and their elimination due to deleterious mutations. We find that when deleterious mutation rates are high in mutators, there will often be many “young,” recently produced mutators in the population, and the fact that deleterious mutations only gradually eliminate individuals from a population is important. We then consider how this mutator–nonmutator balance can be disrupted by beneficial mutations and analyze the circumstances in which fixation of mutator alleles is likely. We find that dynamics is crucial: even in situations where selection on average acts against mutators, so they cannot stably invade, the mutators can still occasionally generate beneficial mutations and hence be important to the evolution of the population.
BIOLOGY has evolved sophisticated machinery to avoid errors in replication. Clearly, it is worth much effort to make sure that mutations are rare. Yet at the same time, higher mutation rates are selected for in many natural and experimental populations (Denamur and Matic 2006; Baer et al. 2007). Since mutations can change mutation rates, and this heritable variation in mutation rates is acted on by selection, the mutation rates we observe are a result of a balance between different evolutionary forces. Our understanding of these forces remains incomplete.
In this article we consider large-effect mutator alleles that increase the mutation rate by a factor of 10 to 10,000. These are often mutations in the mismatch-repair system (Denamur and Matic 2006; Baer et al. 2007). Recent evidence suggests that in some circumstances these mutator alleles can be selected for, in both natural (Gross and Siegel 1981; Leclerc et al. 1996, 1998; Matic et al. 1997; Oliver et al. 2000; Bjorkholm et al. 2001; Denamur et al. 2002; Giraud et al. 2002; Richardson et al. 2002; Prunier et al. 2003; Watson et al. 2004; Del Campo et al. 2005; Labat et al. 2005) and experimental populations (Cox and Gibson 1974; Trobner and Piechocki 1981; Chao and Cox 1983; Chao et al. 1983; Mao et al. 1997; Sniegowski et al. 1997; Giraud et al. 2001; Notley-McRobb et al. 2002; Shaver et al. 2002; Thompson et al. 2006; Pal et al. 2007). Since mismatch-repair systems are maintained in many populations, there must also be selection operating against mutator alleles.
Mutator alleles provide a laboratory for understanding the forces acting on the evolution of mutation rates. Mutators can be created and monitored in laboratory environments, allowing for experimental tests of theoretical predictions. A variety of theoretical (Tanaka et al. 2003; Lynch 2008; Soderberg and Berg 2011), simulation-based (Taddei et al. 1997; Tenaillon et al. 1999, 2000; Travis and Travis 2002), and experimental (Cox and Gibson 1974; Trobner and Piechocki 1981; Chao and Cox 1983; Chao et al. 1983; Mao et al. 1997; Sniegowski et al. 1997; Boe et al. 2000; Giraud et al. 2001; Notley-McRobb et al. 2002; Shaver et al. 2002; Thompson et al. 2006) studies have begun to explore mutator dynamics.
Several previous studies have analyzed the evolution of mutation rates from a game theory perspective (Kimura and Maruyama 1966; Kimura 1967; Leigh 1970, 1973; Painter 1975; Gillespie 1981; Ishii et al. 1989; Johnson 1999b; Dawson 1998, 1999; Andre and Godelle 2006). These analyses assume that the population is fixed for a particular mutation rate and ask if an allele that modifies the mutation rate can invade. They assume that a modifier invades if the net effect of selection against deleterious mutations and for beneficial mutations, averaged over time, increases the frequency of the modifier. They calculate the “evolutionary stable” mutation rate at which no modifier can invade the population.
This work does not investigate modifier dynamics in detail. It merely assumes that if the time-averaged selection pressure is against a modifier, the modifier cannot invade. Yet at an evolutionary stable mutation rate, mutators will continually arise. Even if they cannot take over, they are maintained in the population at some frequency. When beneficial mutations are available, the modifiers will occasionally sweep through the population and can have a significant influence on the overall evolution. Thus even when mutator alleles cannot stably invade a population, their population dynamics may affect its evolution.
In this article, we focus on the dynamics of mutator alleles, regardless of whether they are stable. We assume the population is fixed for a relatively low (nonmutator) mutation rate and examine the dynamics of mutator alleles arising within this population. In the absence of beneficial mutations, we calculate how the constant creation of mutators is balanced by selection against them due to deleterious mutations. We then outline the effects of beneficial mutations in increasing mutator frequencies.
We show that mutation rates can be controlled by dynamic effects. In this picture, mutation rates can fluctuate with time. When no beneficial mutation has recently been linked to a mutator, mutation rates will be low because deleterious mutations have selected against higher mutation rates. But at some point, mutators may get a beneficial mutation and sweep through a population before once more being selected against. Populations may typically be in this sort of transient regime. When they are, their mutation rates will typically not be at the “stable” rate, but rather much higher or lower depending on the recent history. Our analysis provides a framework for understanding these dynamics.
We must consider four evolutionary forces in understanding the dynamics of mutator alleles. First, mutators are continually produced by mutations from nonmutators. When they are created, they inherit the genetic background of the nonmutator that they arose from. That is, they initially have the same fitness (apart from direct effects of the mutator allele) as the nonmutator individual in which they arose.
Second, mutators have a disadvantage because they produce more deleterious mutations than nonmutators. This tends to purge them from a population. Yet this disadvantage is felt only slowly as the mutator accumulates deleterious mutations, declines in fitness, and is selected against. The speed of this decline depends on the cost of the deleterious mutations.
Third, mutators can have an advantage over nonmutators because they acquire beneficial mutations faster. When beneficial mutations are rare, this advantage may only occasionally be felt, but can have dramatic effects when it is. When deleterious mutations are common, this effect is mitigated because beneficial mutations in mutators will often carry a larger deleterious load than their counterparts in nonmutators.
Finally, mutators may have a direct fitness advantage, because the mismatch repair machinery presumably carries a physiological cost. The balance between the physiological cost of lowering the mutation rate and the cost of deleterious mutations could be what sets most “normal” mutation rates in the wild. However, the direct physiological advantage to a mutator is unlikely to play a major role in mutator dynamics, because if it was comparable in magnitude to the cost of an increased deleterious mutation rate, the two effects would balance and mutators would remain fixed in many natural populations. Since they do not, we focus on the other three effects, and neglect any direct benefit to being a mutator.
These four evolutionary forces on mutation rates exist in both sexual and asexual populations. In asexuals, the alleles that modify mutation rates remain perfectly linked to the beneficial and deleterious mutations that they cause. In a sexual population, this linkage is imperfect and hence these forces are tempered. In this article, we focus exclusively on asexual populations. For simplicity, we focus on haploids; in an asexual population the analysis proceeds identically for diploids provided that there is no dominance (although ploidy will affect the parameters, particularly the rate and effects of deleterious mutations; for a more general discussion of mutators in sexual and asexual populations including the effects of dominance see Lynch 2008). We also assume that there is no epistasis between deleterious mutations; if epistasis is indeed pervasive our results could change dramatically.
We begin with a full treatment of the balance between the constant production of mutators and the accumulation of a deleterious load that selects against them. We describe the growth of the mutator population, the evolution of its fitness distribution, and the steady-state mutation–selection balance. Our analysis is similar to that of Johnson (1999a), but uses a different model that allows us to find simpler and more intuitive results. We then connect our results to the dynamics of a single mutator individual. We next consider the dynamics starting from a population composed entirely of mutators and describe how the nonmutator population can become reestablished.
Finally, we consider the effects of beneficial mutations. We outline the considerations that determine whether beneficial mutations help the mutators increase in frequency. In certain simple parameter regimes, we calculate the probability that a beneficial mutation occurs and spreads in a mutator.
Deleterious Mutations and the Mutator–Nonmutator Balance
We begin by considering the first two forces outlined above: the constant production of mutators from the nonmutator population and the accumulation of deleterious mutations. Since in this section we consider only deleterious mutations, and hence random fluctuations have no significant impact except on the long timescales associated with Muller’s ratchet, we use a deterministic infinite population approximation. We note, however, that if the population is small enough or we are interested in sufficiently long timescales, the finite-population effects of Muller’s ratchet can be relevant. Soderberg and Berg (2011) recently analyzed these interactions between mutator dynamics and the ratchet in detail; in this article we neglect these effects and focus instead instead on the dynamics of mutator alleles in large asexual populations.
We denote the log fitness of an individual as −x, where by convention the larger the x, the less fit the individual. We use a continuous-time model, so that the proportion of the population with log fitness x grows or shrinks exponentially at rate , where is the population-averaged log fitness. We assume that deleterious mutations occur at rate Ud in the nonmutator population and λUd in the mutator population and that their effect is drawn from a probability distribution ρ(x) (ρ(x) is normalized to 1). We assume that ρ(x) does not depend on how many mutations an individual has; this will be roughly true provided that individuals do not typically acquire a significant fraction of all the possible deleterious mutations of some particular effect. We also neglect backmutations. We assume that mutators are created from nonmutators at a rate γ and neglect mutations that create nonmutators from mutators (we discuss this approximation in more detail below). These and other parameters relevant for our analysis are summarized in Table 1.
Table 1 . Summary of commonly used variables and parameters.
| Descriptions | |
|---|---|
| Ud | Deleterious mutation rate in nonmutators |
| λUd | Deleterious mutation rate in mutators |
| x | Negative log fitness of an individual (larger x corresponds to lower fitness) |
| ρ(x) | Distribution of fitness effects of deleterious mutations |
| R(k) | Laplace transform of ρ(x) |
| M | Difference between mutator and nonmutator deleterious mutation rates |
| γ | Mutation rate from nonmutators to mutators |
| fn(x,t) | Distribution of fitnesses of nonmutators at time t |
| fm(x,t) | Distribution of fitnesses of mutators at time t |
| F{n,m}(k,t) | Laplace transform of fn(x,t) or fm(x,t) |
| σ | Effect of a typical deleterious mutation |
| α | Defined as M/σ |
| δn,δm | Average deleterious load of nonmutators or mutators respectively |
| pm | Fraction of mutators in the total population |
| τ | Average lifetime of a single mutator lineage |
| η | Backmutation rate from mutators to nonmutators |
| Ub,λUb | Beneficial mutation rate in nonmutators or mutators respectively |
| N | Population size |
| sb | Fitness advantage provided by a beneficial mutation |
| s | Fitness advantage of a beneficial mutant relative to population mean fitness |
Our analysis assumes that the mutator population is always rare compared to the nonmutator population. We expect that the rate at which mutators are created is small and that the deleterious mutations they incur select against them, so unless we start from a situation where mutators are common this assumption will typically be valid. Naturally, our assumption can fail whenever beneficial mutations arise, so we will treat these beneficial mutations separately. We define M ≡ (λ − 1)Ud to be the difference in deleterious mutation rate between the mutator and nonmutator population.
We define the distribution of fitnesses of the nonmutator population, fn(x, t)dx, to be the fraction of the overall population that are nonmutators with fitness between x and x + dx at time t. We define fm(x, t)dx analogously as the fraction of the overall population that are mutators with fitness between x and x + dx at time t. The dynamics of the mutator and nonmutator populations are given by
| (1) |
| (2) |
The terms involving reflect the fact that only relative fitnesses matter (they keep the population size constant). Provided that the mutators are rare, we have
| (3) |
Because we neglect mutations that create nonmutators from mutators and assume that the mutator population is always rare compared to the nonmutator population, the dynamics of the nonmutator population is independent of the mutators. We can thus solve for the behavior of fn(x, t) neglecting the mutators and then plug this solution into the equation for and solve for the mutator dynamics.
Before doing this, it is useful to introduce unnormalized fitness distributions gn(x, t) and gm(x, t) for the nonmutators and mutators, respectively. We define these to be functions that satisfy Equation 1 and Equation 2 without the terms:
| (4) |
| (5) |
It is straightforward to show that if gn and gm satisfy Equation 4 and Equation 5, then
| (6) |
satisfy Equation 1 and Equation 2.
We can solve these equations by passing to Laplace transforms. We define Gn(k, t) and Gm(k, t) to be the Laplace transforms of gn(x,t) and gm(x,t); we have , with an analogous expression for Gm. Similarly we define Fn(k,t) and Fm(k,t) as the Laplace transforms of fn(x, t) and fm(x, t). Note that
| (7) |
With this notation, we have
| (8) |
| (9) |
where R(k) is the Laplace transform of ρ(x).
We can solve Equation 8 using the method of characteristics. Assuming we start with a situation where the population is composed entirely of nonmutators with no deleterious mutations, the solution is
| (10) |
We can now substitute this expression into Equation 9 and solve this with the method of characteristics. We find
| (11) |
To solve for Fn and Fm, we simply divide these expressions by Gn(k = 0, t).
Note that because of our assumption that the mutators are always rare compared to the nonmutators, we will always have Fn(k = 0, t) = 1. The fraction of mutators in the population at time t is given by Fm(k = 0, t).
Equation 10 and Equation 11 provide a complete solution for the dynamics of both the mutator and nonmutator population, starting from a clonal nonmutator population. However, this formal solution offers very little understanding of the dynamics.
We now turn to several specific examples to get an intuitive sense of the behavior. We solve for the fraction of the population that are mutators, and their average fitness, for three cases: when all deleterious mutations have the same effect, when there is an exponential distribution of the fitness costs of deleterious mutations, and when there are two types of deleterious mutations (a common weak-effect one and a rarer strong-effect one). We then present an approximation for general ρ(x) that is valid for λ ≫ 1 and finally describe an alternative approach to the problem that treats each mutator clone independently.
All deleterious mutations have the same effect
We begin by considering the case when all deleterious mutations have the same effect. This corresponds to ρ(x) = δ(x − σ), where δ is the Dirac delta function and σ is the cost of each deleterious mutation.
Solving Equation 10 for this case and calculating the resulting Fn(k, t), we find that the Laplace transform of the fitness distribution of nonmutators is
| (12) |
We can invert Fn(k, t) and find that the number of deleterious mutations is Poisson distributed with mean , consistent with the classical result of Haigh (1978).
Using this solution, we can solve for the Laplace transform of the fitness distribution of the mutator population. We find
| (13) |
where we have defined
| (14) |
This solution is rather opaque, but we can calculate from it two quantities of particular interest: the fraction of individuals in the total population that are mutators, which we call pm, and the average deleterious load carried by each mutator, δm. From the properties of Laplace transforms, we have that
| ,. |
We find that the fraction of individuals that are mutators is given by
| (15) |
The behavior of pm depends on how large α is relative to 1. For α ≪ 1 we can get an approximate solution for the full time-dependent behavior, . In this case, the number of mutators increases linearly for , pm ≈ γt, eventually saturating at a steady-state value . For α ≫ 1 the result is more complex. However, asymptotic analysis of the integral in Equation 15 using the method of steepest descent shows that for , pm increases linearly with time, pm ≈ γt. For longer times, pm saturates at the steady-state value . The dynamics are thus relatively simple: pm increases linearly with time at rate γ until saturating at the steady-state value
| (16) |
This approximation for the steady-state pm as well as the time dependence are compared with the exact result Equation 15 in Figure 1. The result Equation 16 is worth examining in detail. As we would expect, the number of mutators is proportional to the rate at which they are produced from nonmutators. However, we can also see that is a key parameter. When the mutator deleterious mutation rate is small compared to the effect of the mutations (i.e., α ≪ 1), then our result has a simple intuitive explanation: individuals that acquire deleterious mutations are effectually dead. Thus mutators produce “dead” offspring at a rate M higher than the nonmutator population and therefore have an effective selective disadvantage M compared to the nonmutators. Thus this intuition predicts a steady-state frequency of mutators equal to the rate at which they are produced divided by their effective selective disadvantage, or .
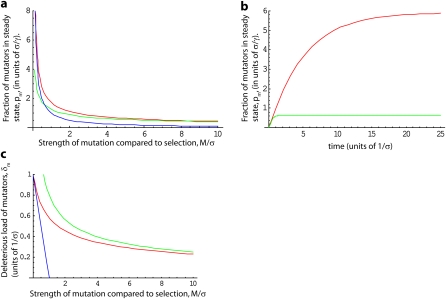
Figure 1 .
Expected behavior when all deleterious mutations have the same effect σ. (a) Steady-state fraction of mutators in the population, pm, in units of . The exact result, Equation 15 is shown in red. The approximations for weak and strong selection relative to mutation, M/σ ≪ 1 and M/σ ≫ 1, in Equation 16, are shown in blue and green, respectively. (b) Dynamics of pm, from Equation 15, in units of . In red, we show α = 0.2 (strong selection relative to mutation); in green we show the opposite case of α = 5 (weak selection relative to mutation). Note that δm increases linearly at rate γ for (red line) or (green line), before saturating at its steady-state value. (c) Average deleterious load δm of a mutator. The exact result, Equation 17 is shown in red. The approximations for M/σ ≪ 1 and M/σ ≫ 1 in Equation 18 are shown in blue and green, respectively. Note the results in b and c make use of the approximation λ ≫ 1.
However, as we can see from our result, this intuition is wrong whenever the mutator deleterious mutation rate is large compared to the effect of these mutations (i.e., α ≫ 1). In this case, mutators are much more common than ; the mutator frequency instead depends on the square root of the mutation rate times the selective disadvantage. This is consistent with the calculations of Johnson (1999a). We return to the intuition behind this result below.
It is also interesting to calculate from Equation 13 the average deleterious load, δm, carried by the mutators. We find
| (17) |
This does not depend on γ, as expected. Asymptotic expansions of the integrals in these expressions show that for small t, δm increases linearly from 0 with time at a rate proportional to the deleterious mutation rate. For large t, δm saturates at the steady-state value
| (18) |
We thus see that for M ≪ σ, the mutators come to their steady-state mutation–selection balance, with roughly equal to their deleterious mutation rate, as predicted from classical population genetics. However, when M ≫ σ, the mutator population is dominated by individuals that have recently been created from nonmutators. Thus the average deleterious load δm is approximately equal to that of nonmutators, Ud (set by their mutation–selection balance). These results are illustrated in Figure 1.
An exponential distribution of deleterious mutations
We now turn to the case where deleterious mutations have an effect given by an exponential distribution with mean σ. That is, .
Solving Equation 10, we find that the Laplace transform of the fitness distribution of nonmutators is
| (19) |
This gives a nonmutator average fitness
| , |
as expected. Substituting this solution into Equation 11, we find that the Laplace transform of the mutator fitness distribution is given by
| (20) |
As before, we can calculate the expected fraction of the population that are mutators and the average mutator deleterious load. We find
| (21) |
This result is very similar to the case of a δ-function distribution of deleterious mutations. Again the number of mutators increases linearly with time at rate γ for small t. And again for long times the number of mutators is given by
| (22) |
We can also calculate the average deleterious load in mutators and find
| (23) |
Again this result is very similar to the previous case. The average fitness of the mutators increases linearly from 0 at small times and eventually reaches the steady-state value
| (24) |
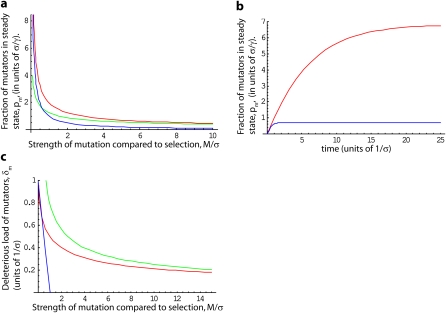
These results are illustrated in Figure 2. Note that both the exact results and the analytical approximations are very similar to those shown in Figure 1, indicating that an exponential distribution of fitness effects with average effect σ leads to almost identical behavior as if all deleterious mutations had the same fitness effect σ.
Figure 2 .
Expected behavior when the effects of deleterious mutations are exponentially distributed with mean σ. (a) Steady-state fraction of mutators in the population, pm, in units of . The exact result, Equation 21, is shown in red. The approximations for weak and strong selection relative to mutation, M/σ ≪ 1 and M/σ ≫ 1 in Equation 22, are shown in blue and green, respectively. (b) Dynamics of pm, from Equation 21, in units of . In red, α = 0.2, and in blue α = 5. Note that δm increases linearly at rate γ for (red line) or (blue line), before saturating at its steady-state value. (c) Average deleterious load δm of a mutator. The exact result, Equation 23, is shown in red. The approximations for M/σ ≪ 1 and M/σ ≫ 1 in Equation 24 are shown in blue and green, respectively. Note the results in b and c make use of the approximation λ ≫ 1.
Two types of deleterious mutations
We have analyzed the mutator–nonmutator mutation–selection balance for two particularly important distributions of deleterious mutations. To gain more intuition for the general case, it is useful to consider the situation in which two different types of deleterious mutations are possible: a large-effect mutation that is relatively rare, and a small effect one that is relatively common. We denote the cost of these two mutations as s1 and s2, respectively, and their mutation rates as U1 and U2, where s2 > s1 and U1 > U2. We define M1 and M2 in the obvious way, M1 ≡ (λ − 1)U1 and M2 ≡ (λ − 1)U2.
We can analyze this situation with the same methods that we have used above. There are four possible distinct regimes of the relative sizes of the various parameters. For the number of mutators in steady state, we find
| (25) |
These results initially seem opaque, but they are actually straightforward: the number of mutators at steady-state pm is roughly equal to the lesser of the two results for pm that would be obtained from considering the cases in which only one or the other mutation was possible. That is, we can calculate pm using the result for when all deleterious mutations have effect s2 and occur at rate U2 and using the result for when all deleterious mutations have effect s1 and occur at rate U1. The smaller of these two values of pm is the number of mutators at steady state in the case when both mutations are possible. We obtain analogous results when we consider the mean deleterious load in mutators, δm.
These results suggest that when multiple deleterious mutations are possible, those for which produce a pm of , while those for which produce a pm of . The actual value of pm is then dominated by whichever deleterious mutation would produce the smallest pm.
General ρ(x)
Since we are focusing on strong-effect mutators (λ ≫ 1), we can to a good approximation assume that the deleterious mutation rate in nonmutators is 0. This greatly simplifies our results and allows us to give relatively simple expressions for , pm, and δm for general distributions of deleterious mutations ρ(x). This will enable us to see directly how the deleterious mutations of various different effects contribute to the mutator dynamics.
Making the approximation that there are no deleterious mutations in nonmutators, we have
| (26) |
We can use this result in solving for Fm; we find
| (27) |
Note that because we are assuming that Ud in nonmutators is 0, we have M = λUd rather than (λ − 1)Ud. From this expression we calculate
| (28) |
We can gain insight into the mutator dynamics by studying Equation 28. By approximating for and for , we can approximate pm by
| (29) |
We denote the size of a “typical” deleterious mutation as σ. More precisely, σ is the value of for which the first term in the above expression becomes becomes equal to the second two. Assuming that ρ(x) falls off rapidly with x (i.e., that large-effect deleterious mutations are rare compared to small-effect ones), σ will be roughly the effect of the average deleterious mutation. For , the first integral in the integrand is large compared to other two. For , the first is small compared to the others. So for the steady-state we have
| (30) |
In the first term in this expression, z is always less than , so . In the second term, on the other hand, z is always greater than so and . Thus we have
| (31) |
We can evaluate both of these integrals and find that the first term is dominant if M ≫ σ, while the second dominates if M ≪ σ. We thus have
| (32) |
This general result is valid for any distribution ρ(x) that falls off rapidly with x; it confirms the intution developed from the specific cases we considered above.
We can use the same analysis to understand the time dependence of pm. For , pm is given by the first term in Equation 31, with the upper limit of integration replaced by t. For , pm is given by Equation 31 with the upper limit of integration in the second term replaced by t. Analyzing this equation with the same methods described above, we find that for , we have pm ∼ γt for . For , we find pm ∼ γt for .
This means that the full dynamics of the number of mutators is quite simple: mutators accumulate approximately linearly with time at a rate γ until they reach the steady-state value of either
| or . |
We also note that in some contexts it may be useful to understand how the dynamics of pm depends on the moments of ρ(x). We can calculate this by using the full Taylor expansion of in Equation 28. We find
| (33) |
We can also analyze the dynamics of the mean fitness of the mutator population in this approximation that there are no deleterious mutations in the nonmutators. From Equation 27, we find
| (34) |
We can analyze this with similar techniques to those described above. In steady state, we find that for , δm ≈ M. For , in steady state δm is small compared to M (on the order of ).
The fate of each mutator lineage
Thus far we have understood mutator population dynamics by focusing on the entire mutator population at once and calculating the time evolution of its fitness distribution. An alternative approach is to focus on the fate of each individual mutator lineage. The overall mutator population is composed of many such mutator lineages, created by mutations in nonmutators at a variety of different times. If we can understand the dynamics of each individual mutator lineage, we can simply add up these results to understand the overall mutator population dynamics.
This approach was recently used by Andre and Godelle (2006), who studied the case in which a number of mutators are created all at once, and no further mutators ever arise. These mutators eventually reach a steady-state fitness distribution, which Andre and Godelle (2006) calculate for the case in which all deleterious mutations have the same effect. However, we consider a situation in which mutators are constantly being produced from nonmutators, so there will always be some mutators that were recently produced and that therefore have not yet reached their individual steady-state fitness distribution. Even when the overall mutator population is in steady state, this overall steady-state fitness distribution is a sum of transient distributions for individual mutators. We must therefore also understand the probability that a mutator lineage is eliminated before its fitness distribution reaches steady state, and its transient fitness distribution, before we can understand the overall mutator dynamics.
Imagine that we know the expected number of descendants of a single mutator a time t after it was produced. Call this h(t). Since there are γ mutators produced per nonmutator per unit time, starting from 0 at t = 0, at some later time t the fraction of mutators in the population is
| (35) |
The part of this integral near x = 0 corresponds to the mutators that have been produced recently, while the part near x = t corresponds to the mutators that originated near t = 0. In steady state, this becomes Note that this steady-state result depends on the form of h(t) near t = 0, which corresponds to recently produced mutators that are not yet in their individual steady state; this is why we must understand the transient dynamics of h(t).
However, from Equation 28 we can immediately see in the approximation that deleterious mutations in nonmutators can be neglected,
| (36) |
This leads to a new interpretation of Equation 29. Following the same logic leading to Equation 29, we find
| (37) |
The first term here dominates for small t, , where σ is defined as before as a typical effect of a deleterious mutation. This corresponds to the situation in which the mutator has not yet come to its individual mutation–selection balance (which occurs only after the order of generations). The second and third terms dominate for and correspond to mutators that were created long enough ago to be in their individual mutation–selection balance. We have
| (38) |
A simple argument shows that this solution makes intuitive sense. For , deleterious mutations are accumulating roughly linearly with time, since selection has not had time to eliminate them. Thus the deleterious load of the mutator individual is Mtσ. The probability the mutator has been eliminated due to selection, p, is thus given by the equation , which gives . This result is consistent with our small-t result for h(t). On the other hand, for , the expected deleterious load carried by this mutator has reached its mutation–selection balance M. Thus , and P = exp[−Mt]. This is consistent with our large-t expression for h(t).
As we would expect, the part of our result for h(t) is identical to the result of Andre and Godelle (2006), since for these large times the mutator lineage has reached its steady state. Our expressions differ for younger mutators that have not yet reached this point. We can connect these results to our previous analysis by plugging Equation 38 into the steady-state version of Equation 35 to find pm. We find identical results for pm, both in steady state and for the time dependence, as we did with the other methods described above.
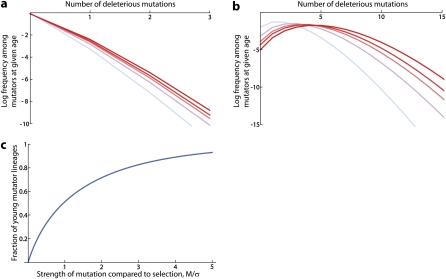
This analysis provides intuitive insight into our results for pm and δm and helps us understand why is a key parameter. Each individual mutator lineage takes on the order of generations to come to its individual steady-state fitness distribution, and then generations to be eliminated from the population by selection (because its deleterious load in this steady state is M). We illustrate this in Figure 3, which shows how each mutator lineage approaches its individual steady states and the fraction of old vs. young mutators in the population as a whole. When M ≪ σ, we have , so mutators reach their individual steady-state fitness distribution quickly compared to the time that it takes to then eliminate them from the population. Hence most mutators in the population have reached their individual steady-state fitness distribution (see Figure 3, a and c). This means that the mean fitness of the mutators is roughly −M, and since each mutator takes on the order of generations to eliminate from the population, . On the other hand, when M ≫ σ, most mutators in the population are eliminated before reaching their individual steady-state fitness distribution, and hence mutators which have not yet come to equilibrium dominate the overall mutator population (Figure 3, b and c). Since the mutators are accumulating deleterious mutations roughly linearly in time in this regime, they take a time on the order of to eliminate from the population. Hence we get . The average fitness of the mutators will be the wild-type mutation rate plus the fitness decline accumulated in generations, which is . In other words, as M increases past , the fact that a mutator lineage has a larger deleterious load in its individual steady state is partially offset by the fact that on average the mutators present in the population are younger and have not fully reached this steady state. These results are all consistent with our earlier analysis.
Figure 3 .
(a) The distribution of the number of deleterious mutations in an individual mutator lineage of a given age τ, for M/σ = 0.1, for the case in which all deleterious mutations have the same effect σ. Each line represents a different age; shown from lighter to darker are , , , , and the steady-state τ = ∞. Blue lines represent “young” mutators where , while red lines represent “old” mutators; the marginal case is shown in purple. (b) The analogous distribution of the number of deleterious mutations in an individual mutator lineage of a given age τ, for M/σ = 10. (c) The fraction of the total mutator population that is young as a function of M/σ, from Equation 36. For M/σ = 0.1 the mutator population is mostly old, while for M/σ = 10 the mutator population is almost all young. Note that the fact that each mutator lineage has a larger steady-state deleterious load as M/σ increases is partially offset by the fact that a larger fraction of the mutators are young.
We can get a final perspective on how different effect deleterious mutations contribute to the mutator dynamics from a different estimate of pm. We have thus far described the dynamics as depending on whether M is large or small compared to the typical effect of a deleterious mutation, σ. However, when deleterious mutations have a variety of effects, an individual mutator lineage can be in steady state with respect to larger-effect mutations, but not with respect to smaller-effect ones. We can understand this by noting that there is some average time τ that a single mutator survives after being created. In steady state, we have pm = γτ. We can thus find the steady-state pm by calculating τ. Large- and small-effect mutations contribute to τ in different ways. Mutations with reach their steady state quickly, so the selection pressure they exert on the mutator lineage is equal to the mutation rate to these mutations, . Smaller mutations with , however, do not reach their steady state during the lifetime of an individual mutator. An individual lineage has a deleterious load of from these mutations (increasing with time as they accumulate), so averaged over the the lifetime of the lineage these provide a selective pressure . The lifetime of the mutant lineage τ is determined by the sum of the selection pressure from the small and large effect mutations. This gives a self-consistent solution for τ,
| (39) |
For any particular ρ(x), we can solve this for τ and hence calculate the steady-state pm. The result is the same as the other calculations above. However, this perspective highlights the fact that there is a certain cutoff size of deleterious mutations. Above this size, the mutations can be treated as being effectively in mutation–selection balance, but below it they cannot. The M ≪ σ and M ≫ σ regimes correspond to the cases in which most deleterious mutations are or are not in balance, respectively.
Reestablishment of the nonmutators
Thus far our analysis has assumed that mutators are rare. This is appropriate for studying the deleterious mutation–selection balance. However, mutators may occasionally sweep to fixation or near-fixation due to linkage with beneficial mutations. If no further beneficial mutations are available, the nonmutators will then reestablish themselves as the majority of the population. In this section, we describe this process. We imagine starting from a population in which the mutator has swept to fixation and ask the timescale on which the nonmutator reestablishes itself.
We assume that mutations within the mutator population create nonmutators at rate η. These mutations could be reversions of the mismatch-repair mutations that originally created the mutators, or they could be other compensatory mutations. We expect η < γ, because there are likely more targets for disabling mismatch-repair genes than there are for repairing them. However, η may not be much smaller than γ because reversions occur at the elevated mutation rate in mutators.
When the mutator population has swept, the nonmutators are rare. Thus we can use the exact same analysis described above to understand the dynamics. We simply relabel the mutator and nonmutator populations, redefine Ud to be the deleterious mutation rate in mutators, redefine λ < 1 to be the reduction in mutation rate in nonmutators, and take γ → η.
This redefined system has no steady state, since the nonmutator population will take over and the approximation that it is rare will cease to be true. However, we can still use this analysis to get the timescale on which the nonmutator population reestablishes. We find that initially it increases linearly with time, pn ≈ ηt. Once , we find pn ∼ eMt. This makes intuitive sense: the mutators will on average be M fitter than the mutators, so they reestablish exponentially at rate M.
For smaller populations, this will be an overestimate of the reestablishment rate. A mutation that creates a nonmutator is a beneficial mutation, so the deterministic description above may not be valid in a smaller population. Rather, a mutation creating a nonmutator must first occur and survive random drift before the nonmutator can begin to expand exponentially. We can calculate the time it takes for this to occur in the special case in which all deleterious mutations have the same effect σ. In this case, after a mutator sweep, once the mutators are in their steady-state fitness distribution, the number of mutators with i deleterious mutations is , where N is the population size. This means that nonmutators with i deleterious mutations are created and survive drift at a rate . Thus it takes on the order of
| (40) |
generations for a nonmutator with no deleterious mutations to arise. It is only after this time that the exponential growth of the nonmutators at rate M can begin. For small N or large , when , nonmutators with some deleterious mutations will reestablish instead of the most-fit possible nonmutator. In this case, the reestablishment of the nonmutator population will still occur exponentially, but at a slower rate M − iσ, where i is the smallest number of deleterious mutations in a nonmutator.
Note that these results imply that generally speaking the stronger the effect of the mutator, the more quickly selection will act to reestablish the nonmutators after a mutator sweep. Thus, other things being equal, larger-effect mutators will tend to be shorter lived once they have swept, consistent with the observations of Denamur et al. (2005).
Although we have assumed that mutations within the mutator population create nonmutators at some fixed rate η, this may not always be the case. Many possible mutations can lead to mutator phenotypes. These mutations happen in nonmutators at rate γ, so they presumably happen in nonmutator individuals at rate λγ in mutators. This means that mutator individuals can acquire additional mutations in mismatch-repair genes, and if these additional mutations reach high frequency along with the mutators (e.g., by hitchhiking with a beneficial mutation), it may require more than one mutation to revert to a nonmutator phenotype. This means that over time a population dominated by mutators can become “trapped” in a mutator state. On the other hand, it may be possible for a population to escape such traps through horizontal gene transfer from nonmutators, particularly since mutator alleles also often increase recombination rates (Denamur et al. 2000). Our analysis in this section does not account for these interesting possibilities.
When are mutators rare?
Throughout most of this analysis, we have assumed that the mutator population is rare. For this assumption to be valid, the contribution of the mutators to the mean fitness of the population must be negligible. This means that we require δmpm ≪ δn, which reduces to
| (41) |
Whenever this is true, initially rare mutators will always remain rare, and our analysis will be valid. Since we expect that the overall deleterious mutation rate will typically be large compared to the mutation rate that produces mutators, this is likely a good approximation in natural situations. We also require that mutations that generate nonmutators from mutators can be neglected compared to the selection on nonmutators, but this will always be true for reasonable parameters when mutators are rare.
Beneficial Mutations
When beneficial mutations are possible, mutators may have an advantage because they can acquire these beneficial mutations more quickly than nonmutators. The size of this advantage is complex. Because beneficial mutations are likely to be rare, but have a dramatic effect on the population dynamics when they occur, stochastic effects are crucial and the behavior will often depend strongly on population size. This is particularly true in asexual populations, where the fixation of a beneficial mutation causes the genetic background on which that mutation occurred to hitchhike to fixation.
There are a variety of different scenarios involving beneficial mutations that we could study. Leigh (1970) considers a model in which beneficial mutations are possible at any time and cause a selective sweep with a probability K per generation. Johnson (1999b) considers a similar model. Andre and Godelle (2006) consider this model as well as the situation in which beneficial mutations are always available but their rate is proportional to the population size and mutation rate. They also consider a model involving a changing environment, as do Gillespie (1981) and Ishii et al. (1989); Painter (1975) also considers a related situation.
Much of this work aims to calculate the average advantage or disadvantage felt by an allele modifying the mutation rate, and hence an evolutionary stable mutation rate (Kimura and Maruyama 1966; Kimura 1967; Leigh 1970, 1973; Gillespie 1981; Ishii et al. 1989; Dawson 1998, 1999; Johnson 1999b; Andre and Godelle 2006). As we have pointed out, this is not the only relevant result for an asexual population. The mutator frequency may fluctuate widely, increasing when a beneficial mutation happens to occur in a mutator and then gradually decreasing to the mutator–nonmutator balance before increasing again when another beneficial mutation occurs. The population parameters and the particular model for beneficial mutations all affect the dynamics.
In this article, we make no attempt to explore the wide variety of possible models for beneficial mutations. Instead, we consider the simplest possible situation and use this to highlight some of the important factors involved in mutator dynamics in the presence of beneficial mutations. Our analysis is much less detailed than recent work by Tanaka et al. (2003) and Wylie et al. (2009). However, while these studies provide a more complete theoretical description of mutator dynamics in the presence of beneficial mutations, they are valid only when deleterious mutations have very strong effects and can be treated as effectively lethal. Our work, by contrast, is intended to provide a more general overview of how the interplay between beneficial and deleterious mutations influences mutator dynamics. We imagine a population that lacks beneficial mutations and has come to its mutator–nonmutator balance. We then imagine that a beneficial mutation of effect sb becomes available, and occurs at rate Ub in the nonmutators (λUb in the mutators). We want to understand the subsequent dynamics. For simplicity, we assume that deleterious mutations in the nonmutators can be neglected.
At first, it may seem that this situation is very simple: the beneficial mutation will occur either first in a mutator or in a nonmutator and cause a selective sweep that eliminates the other population. The rate at which the beneficial mutation occurs in the mutator subpopulation is NpmλUb, while in the nonmutator subpopulation it occurs at rate N(1 − pm) Ub ≈ NUb. Thus the probability that the beneficial mutation occurs first in a mutator is
| (42) |
According to this simple intuition, the beneficial mutation should cause the mutators to sweep a fraction P of the time and the nonmutators to sweep the remaining 1 − P of the time. This result is sometimes used in studies of mutator dynamics and mutation rate evolution.
Unfortunately, this simple intuition is incorrect. A beneficial mutation that occurs in a mutator will tend to be saddled with a larger deleterious load than one occurring in a nonmutator and will be further hampered by the additional deleterious mutations it continues to accumulate. This means that beneficial mutations occurring in mutators are less likely to survive random genetic drift while they are rare than those occurring in nonmutators.
Further, even if we knew the rates at which beneficial mutations occur and survive drift in the mutator and nonmutator subpopulations, the probability the first occurs in one or the other population is not the only important quantity to consider. Even if a beneficial mutation occurs in a mutator first, it will tend to carry a deleterious load, so if a later mutation occurs in the nonmutator population it can outcompete the earlier mutator mutation. Conversely, even if the first beneficial mutation occurs in a nonmutator, a later beneficial mutation in a mutator can increase the mutator frequency transiently before the nonmutator sweeps (because the mutators are at a lower overall frequency in the population). While this mutator population is transiently more frequent, it is much more likely to get additional beneficial mutations, which in turn increase the mutator frequency even more. This process naturally depends on the model of beneficial mutations we are considering. If only one beneficial mutation is possible, it cannot occur. However, if multiple beneficial mutations are possible, this effect means that the mutators may sweep even though they would only rarely do so if only one mutation was available at a time.
Much of the earlier work described above has explored aspects of these dynamics in great detail in specific parameter regimes. Here we do not aim to give a full analysis of all possible results above across the wide range of plausible situations. Instead, we provide an outline of the relevant effects, with the aim of developing a general understanding of how various evolutionary forces interact in different ways in different parameter regimes to determine the fate of mutator alleles.
The establishment probability of a beneficial mutant
When it first occurs, a beneficial mutation is present in only one individual. Its lineage is very likely to go extinct due to random genetic drift, but there is some probability that it will survive and grow to a large enough population size that selection dominates drift. Thereafter its behavior is mostly deterministic. We refer to this process by which a lucky mutation occurs and survives genetic drift the establishment of the mutation. We begin by considering the probability that a beneficial mutation survives genetic drift given that it occurs in either the mutator or the nonmutator population.
In the absence of deleterious mutations, the probability that a beneficial mutation establishes, given that it occurs, is 2s, where s is the fitness advantage of the mutant relative to the mean fitness in the population (Ewens 2004). Since we neglect deleterious mutations in the nonmutators, this establishment probability in nonmutators is .
The establishment probability in mutators is more complex, for two reasons. First, the beneficial mutation may occur in an individual that carries some deleterious load. Thus its fitness advantage relative to the mean fitness in the population is 2s, where s ≡ sb − x and x is the deleterious load in the mutator individual in which the beneficial mutation occurred. Second, while the mutant lineage is rare it can accumulate more deleterious mutations, further reducing its establishment probability. Johnson and Barton (2002) studied this second effect and developed an algorithmic way of calculating how it reduces the establishment probability. Unfortunately, there is no closed-form expression for this probability in terms of the population parameters, and their result is further restricted to the case where all deleterious mutations have the same effect. This makes it impossible to find a general result, but fortunately in certain parameter regimes we can still calculate .
The lineage with the beneficial mutation starts as a single individual and will either establish or die out due to these stochastic effects on the order of generations. On the other hand, it takes on order of generations for deleterious mutations to reach their steady-state distribution in this lineage. Thus if σ ≫ s, the deleterious mutations reach their steady state long before the mutant lineage establishes or dies out. On the other hand, if σ ≪ s the mutant lineage establishes or dies out long before the deleterious mutations reach their steady-state distribution in this lineage.
We begin with the first case, σ ≫ s. This corresponds roughly to the “ruby in the rubbish” case considered by Peck (1994), where any deleterious mutation dooms a lineage. In this case, when M > s, the mean fitness of the mutant lineage is reduced to s − M < 0 before the lineage has a chance to establish. Thus it can never establish, and On the other hand, if s ≫ M, the deleterious mutations quickly reduce the fitness of the mutant lineage to s − M ≈ s, and hence it establishes with probability .
In the opposite case, σ ≪ s, on average the deleterious load in the mutant lineage increases linearly with time at rate Mσ. Thus after a time on the order of , the deleterious load in the mutant lineage is on the order of . Thus if (i.e., ), the beneficial mutant can never establish, and If , it establishes with probability In this case, however, the mutant lineage can establish and yet be deterministically eliminated later if M > s, because once the full deleterious load is felt in this lineage it will be less fit than a nonmutator without the beneficial mutation.
We must now ask what s is. When a beneficial mutation occurs in a mutator, it carries a deleterious load x with probability fm(x, t), and s = sb − x. We found above that whenever s is small compared to M or , then Since s ≤ sb, this is also true whenever sb is small compared to M or . On the other hand, we found that when s is large, . In these large-s regimes, s ≫ δm, so we also have sb ≫ δm. In other words, the deleterious load of a typical mutator is small compared to sb. Thus s ≈ sb, and hence in these regimes. We can estimate the corrections to this result by using our results for the Laplace transform of fm(x, t). For sb ≫ σ, the average s, ≈ sb − δm, and hence . For sb ≪ σ, , because the mutation must occur in a deleterious mutation-free background to survive. These results are consistent with earlier calculation in these parameter regimes (Peck 1994; Johnson 1999b; Johnson and Barton 2002; Andre and Godelle 2006).
These parameter regimes represent the possible simple cases. To summarize, we have found that when σ ≫ sb we have
| (43) |
On the other hand, when σ ≪ sb we have
| (45) |
It may sometimes be the case that none of these situations apply and that sb is on the order of σ, M, or . These intermediate regimes must be analyzed using the algorithmic methods of Johnson and Barton (2002), and no simple formulas exist.
The probability a mutator sweeps to fixation
The next step in our analysis is to calculate the rates at which the beneficial mutation establishes in both the mutator and nonmutator population. These rates are proportional to the probability of a beneficial mutation arising and then establishing in each of these populations. In the nonmutators this is In the mutators it is In the simple parameter regimes we considered above, is particularly simple. For the small-sb cases, , and for the large-sb cases . Given these rates, we can understand the effect of beneficial mutations on the dynamics. This depends on the size of the establishment times relative to the fixation time of the beneficial mutation once it is established,
| . |
If the establishment times in both mutators and nonmutators are large compared to the fixation time, then the first beneficial mutation to occur dominates the dynamics. The mutators sweep to fixation if the beneficial mutation happens first in a mutator, or the nonmutators sweep if the beneficial mutation happens first in a nonmutator. The probability that the first mutation happens in a mutator is
| (47) |
Thus the beneficial mutation leads to a mutator sweep after a time on the order of with probability P. If this happens, the nonmutators are eliminated from the population until they eventually take over again or another beneficial mutation occurs.
In the small-sb regimes we have considered, P = 0. In the large-sb regimes, we have
| . | (48) |
(The only exception is when but sb ≪ M, in which case the beneficial mutation can establish in the mutator but will never fix, so P = 0.) This result for P is identical to the naive expectation described above. Thus we see that the naive result is an approximation valid only when beneficial mutations are rare (so that the establishment time is long compared to the fixation time, and multiple beneficial mutations do not arise), and sb is large compared to the deleterious mutation rate.
On the other hand, if the establishment times in both mutators and nonmutators are small compared to the fixation time, the nonmutator is never eliminated by the first beneficial mutation. A beneficial mutation will always arise in the nonmutator, and because it suffers less from deleterious mutations, this mutant will always outcompete any beneficial mutations occurring in the mutator population. However, the mutator population can become transiently common in the interim and, if additional beneficial mutations are possible, may be able to eliminate the nonmutators.
In the large-sb case, we can calculate how beneficial mutations increase the mutator frequency in this interim. When the establishment times of the beneficial mutations are small compared to their fixation times, we can analyze their dynamics deterministically. We find that the number of individuals with the beneficial mutation in the nonmutator population increases with time as , while in the mutators the beneficial mutations increase as . This implies that the mutators reach a maximum frequency of . When , this means that the mutator frequency increases transiently
| (49) |
After this transient increase in the mutator frequency, the nonmutators take over again if no additional beneficial mutations are available. The mutator population is reduced until the steady-state mutator–nonmutator balance is reestablished. However, this transient success of the mutator population becomes particularly important if multiple beneficial mutations are possible. While it is transiently more common due to a first beneficial mutation, the mutator population is much more likely to get an additional beneficial mutation. This second mutation may make it more fit than any nonmutator, even accounting for the cost of deleterious mutations. This then makes mutators even more common, and even more likely to get future beneficial mutations. This process naturally depends strongly on assumptions about the availability of beneficial mutations. Tanaka et al. (2003) analyzed this effect in one situation, showing that it can substantially increase the probability of mutator sweeps, although this analysis did not fully account for the effects of deleterious mutations. This qualitative result was also found by earlier simulation studies (Taddei et al. 1997; Tenaillon et al. 1999; Travis and Travis 2002). To fully address these multiple-mutation possibilities, we can use our earlier analysis of Desai and Fisher (2007) to understand the random distribution of times at which multiple mutations occur, from which we can calculate the probabilities of mutator and nonmutator sweeps. This is a broad topic, which we leave for future work.
A final possibility is that the nonmutator establishment time is short compared to the fixation time but the opposite is true in the mutators, or vice versa. When only one beneficial mutation is available, this is a simple situation: the subpopulation with the shorter establishment time will acquire the beneficial mutation first and sweep to fixation. When multiple mutations are possible, this case becomes more interesting. If the establishment rate is faster in the mutator population, the mutators will still always win. However, if the establishment rate is faster in the nonmutators, occasionally by chance the mutator population will get a beneficial mutation and increase transiently in frequency. During this period, its establishment time is reduced substantially and it is more likely to get additional mutations, which reduce the establishment time of triple mutations further. This can create a runaway process that causes a mutator sweep.
Discussion
In this article, we analyzed the evolutionary forces controlling the dynamics of mutator alleles. We began by calculating how the constant production of mutators is balanced by the selection against deleterious mutants. We analyzed a continuous-time model in which mutators are produced from an initially clonal nonmutator population at rate γ.
Using this continuous-time model, we derived differential equations describing the effects of deleterious mutations and selection on the dynamics of the fitness distribution of both the mutator and nonmutator populations. We solved these differential equations using Laplace transform methods, assuming that the mutator population is rare. This yielded expressions for the Laplace transforms of the fitness distributions of the mutator and nonmutator populations. From these expressions we calculated the dynamics of the frequency of the mutator allele as well as its mean fitness. This approach is similar in spirit to that of Johnson (1999a) but is less general, since we must assume that the mutator population is rare. However, this restriction allows us to calculate explicit results for more quantities of interest and to find simpler and more intuitive formulas.
From these solutions, we calculated the full time-dependent fate of each individual mutator lineage. This extends earlier work by Andre and Godelle (2006), who calculated the fate of individual mutator lineages once they reach their mutation–selection equilibrium. Since these mutator lineages are continually being produced, there are always young lineages in the population that are not in this equilibrium. Thus our extension of Andre and Godelle (2006)’s solution connects their results with the full mutator dynamics.
We found that for any distribution of deleterious mutations ρ(x), there is a typical effect of a deleterious mutation σ, which is on the order of the mean fitness effect of a deleterious mutation. Starting from a clonal nonmutator population, the mutators initially increase linearly with time,
| (50) |
This persists until the mutator frequency reaches a steady state, which takes the order of generations if M ≪ σ and the order of generations if M ≫ σ. At steady state, the mutator frequency is
| (51) |
The mean fitness of a mutator individual in this steady state (in the approximation that deleterious mutations in the nonmutators can be neglected) is
| (52) |
Note here that the comparison between M and σ is between the deleterious mutation rate and the typical fitness effect of a deleterious mutation (as defined in the analysis). Naturally in any real situation there will be some deleterious mutations with fitness effect that is large compared to M and others with fitness effects that are small compared to M; the key question is whether most deleterious mutations have fitness cost greater or less than M.
When M ≪ σ, these results are consistent with many earlier analyses of the evolution of mutation rates, which treated all individuals with deleterious mutations as effectively dead (Leigh 1970; Dawson 1998, 1999; Johnson 1999b) (i.e., that all deleterious mutations can be considered lethal). Since mutators produce such dead offspring at a rate M greater than nonmutators, this work assumed that the mutator effective fitness is lower by M, and hence that they are maintained in the population at a frequency . This assumption is implicit in work on mutators that approximates the effect of deleterious mutations as being simply a constant reduction in the fitness of mutators (e.g., Tanaka et al. (2003)).
As we can see from our result above, this is indeed reasonable when M ≪ σ. This makes intuitive sense: in this case, a deleterious mutation takes much longer to arise than it does to kill an individual, so treating the mutation as instantaneously lethal is not far wrong. Another perspective on this is to note that each individual mutator lineage reaches its own mutation–selection balance in a time on the order of . It is then eliminated from the population after a time . When M ≪ σ, the former time is short compared to the latter time. Thus most mutators in the population have reached their steady-state mutation–selection balance, where they are M less fit than the nonmutators (see Figure 3, a and c). Hence the mutator population as a whole is M less fit than the nonmutators, so it is maintained at a frequency . Thus we have δm = M and .
However, as our analysis shows, the assumption that all deleterious mutations are effectively dead is not justified when M ≫ σ. In this case, deleterious mutations arise quickly compared to the rate at which they are selected against. Although it may be true that individuals that have deleterious mutations do not leave any offspring in the long term, the fact that it takes time for them to be selected against is important, because it means that mutators persist at higher frequencies for longer. We note that this general effect is described in a somewhat different context by Gerrish et al. (2007) as the potential basis for runaway evolution of increasing mutation rates in certain situations.
In this M ≫ σ regime, the time it takes for each individual mutator to reach its own mutation–selection balance is long compared to the time over which it will be eliminated from the population. Thus most individual mutator lineages in the population will not have reached their steady-state fitness distribution. Most of them are still much more fit than −M; they are closer to the mean fitness of the nonmutator population from which they arose (see Figure 3, b and c). This is why δm is much smaller than M when M ≫ σ. The dependence on arises because well before each individual mutator lineage reaches its steady-state mutation–selection balance, it simply accumulates deleterious mutations linearly with time. Thus its average fitness at time t after being created is −Mσt, which means that the cumulative strength of selection against this mutation over its lifetime τ is just the integral of −Mσt, or . Hence the lineage lasts generations, and during this time has average fitness . Thus the average fitness of the mutator individuals is , and the mutator frequency is . Note that these results show that when M ≫ σ, there are many more mutators, with much lower deleterious load, than the assumption that all deleterious mutations are effectively dead would predict.
Our result for the dynamics of the frequency of the mutators, starting from a clonal nonmutator population, is remarkably simple. The mutator frequency increases linearly with time, at rate γ, for small t, or , whichever is larger. There is a more complex crossover regime when t becomes on the order of or , but by then the mutator frequency has already become on the order of its steady-state value. Note that in contrast to the work of Johnson (1999a), our results show that the mutators do not come to equilibrium on a timescale on the order of generations. Rather, they take or generations. This makes intuitive sense: although each individual mutator lineage comes to its mutation–selection equilibrium on a timescale on the order of generations, the overall mutator population is not in steady state until these lineages begin to be eliminated from the population and new lineages arise to replace them. Since the lineages take either or generations to be selected against, this is the timescale for the whole mutator population to come to steady state.
The balance between the generation of mutator alleles and the selection against them due to deleterious mutations can be disrupted when beneficial mutations are available. A variety of experimental work has found that mutator alleles can be selected for when beneficial mutations are possible (Cox and Gibson 1974; Trobner and Piechocki 1981; Chao and Cox 1983; Chao et al. 1983; Mao et al. 1997; Sniegowski et al. 1997; Giraud et al. 2001; Notley-McRobb et al. 2002; Shaver et al. 2002; Thompson et al. 2006). However, we do not yet have a good understanding about under what conditions we expect beneficial mutations to occur in mutators.
This problem has been addressed by several simulation studies (Taddei et al. 1997; Tenaillon et al. 1999; Travis and Travis 2002). This work suggests that larger populations, stronger effect mutator alleles, and a large number of available beneficial mutations all tend to increase the probability that mutator alleles will increase in frequency due to beneficial mutations. However, it is not entirely clear how these various parameters interact. For example, when only a few beneficial mutations are available, the effect of population size may be weaker than when there are many. Tanaka et al. (2003) combined simulations with theoretical analysis to analyze a situation in which periodic environmental shifts allow for new beneficial mutations. Their work gives a deeper understanding of how multiple mutations affect the possibilities of mutator success at different population sizes and mutation rates. More recently, Wylie et al. (2009) developed a detailed and fully stochastic analysis of the fixation probabilities of mutator alleles in asexual populations. Both of these analyses explored the effects of beneficial mutations on mutator dynamics in much more detail than our work here, but under a more restricted set of assumptions. Most importantly, they were both limited primarily to the regime in which deleterious mutations are strongly selected, and treating them as instantly lethal is a good approximation.
When deleterious mutations are less strongly selected, the interaction between deleterious and beneficial mutations is more complex. In intermediate regimes where the effect of a beneficial mutation sb is on the order of the mean fitness of the mutators, we have no analytical expression for the probability that a beneficial mutation can survive drift, or for its fitness if it does so. To answer this question, we have to turn to the algorithmic approach of Johnson and Barton (2002), which unfortunately does not allow for an analytical expression that we can use in further analysis.
However, in certain extreme cases we can explore how beneficial mutations affect mutator success. We have used this to develop a general outline of the different evolutionary forces affecting mutator success. We have seen that for beneficial mutations of small effect sb, sb ≪ M, or (depending on the size of σ relative to M), beneficial mutations can never establish in mutators. In this case, beneficial mutations do not contribute to mutator success, and mutators cannot help a population adapt. On the other hand, in the opposite regime of large sb, deleterious mutations do not hinder the establishment of beneficial mutations in mutators, nor are they hampered by a significant deleterious load in the mutator individual they occurred in.
In this large-sb case, we calculated the probability of mutator sweeps or episodes of transiently common mutators. The way in which these situations affects the overall role and dynamics of mutators depends on the specific model of beneficial mutations, including details such as how many beneficial mutations are possible and the distribution of their fitness effects, among other factors.
In the situation where establishment times of beneficial mutations are long compared to the fixation times (e.g., small population size or rare beneficial mutations), each time a beneficial mutation occurs it creates a sweep by either the mutator or nonmutator population. This is then followed by a period of either increase or reduction in the mutator frequency toward the mutator–nonmutator balance, until another beneficial mutation occurs. If we imagine that beneficial mutations sweep through the population on a timescale τ generations, then the dynamics depends on the relative size of τ and the timescales for reestablishment of the mutator–nonmutator deleterious steady state. If τ is long compared to (if M ≪ σ) or (if M ≫ σ), then after each nonmutator sweep there is time for the mutators to increase to their steady-state frequency before the next beneficial mutation occurs. Thus the probability of a mutator sweep the next time a beneficial mutation occurs will remain
| (53) |
However, if τ is instead short compared to or , then the mutators will only have reached a frequency pm = γτ by the next time a beneficial mutation occurs, and P will be correspondingly reduced. Thus when τ is short, initial nonmutator success makes future nonmutator success more likely. For analogous reasons, initial mutator success makes future mutator success more likely as well.
The value of τ could be set by forces external to the population, such as the rate of changes in the environment that make new beneficial mutations possible. Alternatively, a number of beneficial mutations could always be available and τ could simply be the establishment time of a beneficial mutation in either the mutator or nonmutator population, . In the latter case, a mutator sweep would then lead to a drastic reduction in τ (and could make the establishment time short compared to the fixation time of beneficial mutations, leading to a period of rapid accumulation of beneficial mutations). On the other hand, a nonmutator sweep would lead to a slight increase in τ until the mutator population reestablished itself.
In the opposite case in which establishment times are short compared to fixation times (e.g., large N or high beneficial mutation rates), then the particular model for the availability of beneficial mutations becomes even more important. An initial beneficial mutation causes the mutators to become transiently more common by a factor of roughly λ for a period on the order of generations. If additional beneficial mutations become available only when the environment shifts, then this has no further effects unless the environment shifts quickly compared to the reduction in the mutator frequency. However, if the environment changes more quickly, then at subsequent times the mutators will again become transiently more common by another factor of λ, and so on. On the other hand, if multiple beneficial mutations are possible all at once, limited only by their establishment rates, the transient increase in the mutator frequency from the first such mutation will make further beneficial mutations in the mutators likely, and so on. This can lead to a runaway process that leads to mutator sweeps. After the first few such multiple mutations, however, stochastic effects become important and the analysis becomes difficult. Paradoxically, this situation in which beneficial mutations are common and establishment times are short is very favorable for the mutators, despite this being precisely the case in which the population does not really “need” mutators in order to adapt. That is, mutators are particularly likely to succeed in situations where clonal interference and multiple mutation effects mean that raising the mutation rate only slightly increases the rate of adaptation. This has been noted previously in numerical simulations (Tenaillon et al. 1999), and contrasts with the results of Wylie et al. (2009), presumably because the latter authors studied a model in which the effects of multiple beneficial mutations are neglected.
A variety of experimental work has shown that in natural and laboratory situations, the conditions are in fact right for mutators to at least sometimes sweep. This includes cases in which mutators are found at high frequencies in natural populations (Gross and Siegel 1981; Leclerc et al. 1996, 1998; Matic et al. 1997; Oliver et al. 2000; Bjorkholm et al. 2001; Denamur et al. 2002; Giraud et al. 2002; Richardson et al. 2002; Prunier et al. 2003; Watson et al. 2004; Del Campo et al. 2005; Labat et al. 2005). It also includes experimental situations in which mutators spontaneously arose (Mao et al. 1997; Sniegowski et al. 1997; Notley-McRobb et al. 2002; Shaver et al. 2002; Pal et al. 2007), and experimental work in which labeled mutator strains were placed in competition with nonmutator strains to directly investigate mutator success (Cox and Gibson 1974; Trobner and Piechocki 1981; Chao and Cox 1983; Chao et al. 1983; Giraud et al. 2001; Thompson et al. 2006). Both types of study relate to our analysis, but each has important limitations.
Experiments in which mutators appear spontaneously correspond well to nature and the analysis of this article. Here mutators are presumably present at the low frequencies given by the mutator–nonmutator balance and occasionally spontaneously sweep due to beneficial mutations. But since the mutator strains arise naturally and are unlabeled, it is difficult to follow the dynamics of the mutator population, particularly while it is rare.
Experiments involving deliberate mixtures of mutator and nonmutator strains that carry neutral genetic markers may be more useful in studying the the quantitative aspects of mutator behavior needed to test the results of this article. These studies are limited, however, because mutations that generate additional mutators from the nonmutators are not genetically marked. Thus the steady-state mutator–nonmutator balance cannot be observed; rather, the initial lineage of marked mutators steadily declines in frequency until they are eliminated, unless they acquire one or more beneficial mutations. However, we can use this decline of the marked mutators to test our expression for the fate of an individual mutator lineage h(t). This analysis might be complicated by beneficial mutations and would need to be done at small enough population sizes that they would not occur.
These marked-mutator studies can also be used to test the dynamics of mutators in the presence of beneficial mutations, looking for transient increases in mutator frequencies (and possible additional beneficial mutations leading to mutator sweeps) for large N where the establishment times are short, but either mutator or nonmutator sweeps at smaller N where establishment times are longer.
We note that throughout our analysis we have considered purely asexual populations, and like many earlier studies we have neglected the effects of recombination. This assumption reflects our focus on microbial populations. However, microbial populations are not purely asexual, and horizontal gene transfer could potentially play a role in mutator dynamics. In general, recombination acts to reduce the length of time that mutator alleles remain linked to the beneficial and deleterious mutations that they produce, and hence it reduces the strength of the hitchhiking effects our analysis describes. However, provided that recombination rates are sufficiently small compared to selection pressures, mutators remain linked to the mutations they produce over the timescale in which these mutations change substantially in frequency. Thus we expect our asexual analysis to accurately describe the dynamics in this regime. For weaker selection pressures, however, further work is needed to analyze the interplay between recombination and mutator dynamics.
Finally, we note that throughout this article we have assumed that there is no epistasis among either beneficial or deleterious mutations. That is, the fitness cost of two deleterious mutations is the sum of the individual fitness costs of each one. If epistasis among mutations is pervasive, our results could change dramatically. Broadly speaking, if deleterious mutations interact synergistically (the fitness of the double mutant is less than the product of the fitnesses of the two single mutants), then mutator frequencies will tend to be lower than we have predicted. Alternatively, if deleterious mutations interact antagonistically (the fitness of the double mutant is higher than the product of the fitnesses of the two single mutants), then mutator frequencies will tend to be higher. The details of this analysis, however, are complex and difficult to calculate. The main approach of this article to the mutator–nonmutator deleterious mutation–selection balance cannot be easily extended to the case when epistasis is common, so alternative approaches are necessary to study this situation.
The dynamics of mutator alleles offer a way to test experimentally for overall patterns of epistasis among deleterious mutations. We could look at the dynamics by which marked mutator lineages are eliminated from a population. Differences in the time dependence of this process (and inconsistencies between the mean fitness of the mutator lineage and the time dependence of its decline) from the predictions described in this article would suggest that epistasis among deleterious mutations is common. Given solutions for the expected dynamics with different types of epistasis, it might be possible to tell what types of epistasis are prevalent in the experimental population.
Acknowledgments
We thank Andrew Murray, John Wakeley, and Paul Sniegowski for many useful discussions.
Literature Cited
- Andre J.-B., Godelle B., 2006. The evolution of mutation rate in finite asexual populations. Genetics 172: 611–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer C. F., Miyamoto M. M., Denver D. R., 2007. Mutation rate variation in multicellular eukaryotes: causes and consequences. Nat. Rev. Genet. 8: 619–631 [DOI] [PubMed] [Google Scholar]
- Bjorkholm B., Sjolund M., Falk P. G., Berg O. G., Engstrand L., et al. , 2001. Mutation frequency and biological cost of antibiotic resistance in helicobacter pylori. Proc. Natl. Acad. Sci. USA 98: 14607–14612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boe L., Danielsen M., Knudsen S., Petersen J. B., Maymann J., et al. , 2000. The frequency of mutators in populations of Escherichia coli. Mutat. Res. 448: 47–55 [DOI] [PubMed] [Google Scholar]
- Chao L., Cox E. C., 1983. Competition between high and low mutating strains of Escherichia coli. Evolution 37: 125–134 [DOI] [PubMed] [Google Scholar]
- Chao L., Vargas C., Spear B. B., Cox E. C., 1983. Transposable elements as mutator genes in evolution. Nature 303: 633–635 [DOI] [PubMed] [Google Scholar]
- Cox E. C., Gibson T. C., 1974. Selection for high mutation rates in chemostats. Genetics 77: 169–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson K. J., 1998. Evolutionarily stable mutation rates. J. Theor. Biol. 194: 143–157 [DOI] [PubMed] [Google Scholar]
- Dawson K. J., 1999. The dynamics of infinitesimally rare alleles, applied to the evolution of mutation rates and the expression of deleterious mutations. Theor. Popul. Biol. 55: 1–22 [DOI] [PubMed] [Google Scholar]
- del Campo R., Morosini M. I., de la Pedrosa E. G. G., Fenoll A., Munoz-Almagro C., et al. , 2005. Population structure, antimicrobial resistance, and mutation frequencies of Streptococcus pneumoniae isolates from cystic fibrosis patients. J. Clin. Microbiol. 43: 2207–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denamur E., Matic I., 2006. Evolution of mutation rates in bacteria. Mol. Microbiol. 60: 820–827 [DOI] [PubMed] [Google Scholar]
- Denamur E., Lecointre G., Darlu P., Tenaillon O., Acquaviva C., et al. , 2000. Evolutionary implications of the frequent horizontal transfer of mismatch repair genes. Cell 103: 711–721 [DOI] [PubMed] [Google Scholar]
- Denamur E., Bonacorsi S., Giraud A., Duriez P., Hilali F., et al. , 2002. High frequency of mutator strains among human uropathogenic Escherichia coli isolates. J. Bacteriol. 184: 605–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denamur E., Tenaillon O., Deschamps C., Skurnik D., Ronco E., et al. , 2005. Intermediate mutation frequencies favor evolution of multidrug resistance in escherichia coli. Genetics 171: 825–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai M. M., Fisher D. S., 2007. Beneficial mutation-selection balance and the effect of linkage on positive selection. Genetics 176: 1759–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewens W. J., 2004. Mathematical Population Genetics. I. Theoretical Introduction. Springer, New York, NY [Google Scholar]
- Gerrish P. J., Colato A., Perelson A. S., Sniegowski P. D., 2007. Complete genetic linkage can subvert natural selection. Proc. Natl. Acad. Sci. USA 104: 6266–6271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie J. H., 1981. Mutation modification in a random environment. Evolution 35: 468–476 [DOI] [PubMed] [Google Scholar]
- Giraud A., Matic I., Tenaillon O., Clara A., Radman M., et al. , 2001. Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science 291: 2606–2608 [DOI] [PubMed] [Google Scholar]
- Giraud A., Matic I., Radman M., Fons M., Taddei F., 2002. Mutator bacteria as a risk factor in treatment of infectious diseases. Antimicrob. Agents Chemother. 46: 863–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross M. D., Siegel E. C., 1981. Incidence of mutator strains in Escherichia coli and coliforms in nature. Mutat. Res. 91: 107–110 [DOI] [PubMed] [Google Scholar]
- Haigh J., 1978. The accumulation of deleterious genes in a population: Muller's ratchet. Theor. Popul. Biol. 14: 251–267 [DOI] [PubMed] [Google Scholar]
- Ishii K., Matsuda H., Iwasa Y., Sasaki A., 1989. Evolutionarily stable mutation rate in a periodically changing environment. Genetics 121: 163–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T., 1999a. The approach to mutation-selection balance in an infinite asexual population, and the evolution of mutation rates. Proc. Biol. Sci. 266: 2389–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T., 1999b. Beneficial mutations, hitchhiking and the evolution of mutation rates in sexual populations. Genetics 151: 1621–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T., Barton N. H., 2002. The effect of deleterious alleles on adaptation in asexual populations. Genetics 162: 395–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., 1967. On the evolutionary adjustment of spontaneous mutation rates. Genet. Res. 9: 23–34 [Google Scholar]
- Kimura M., Maruyama T., 1966. The mutational load with epistatic gene interactions in fitness. Genetics 54: 1337–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labat F., Pradillon O., Garry L., Peuchmaur M., Fantin B., et al. , 2005. Mutator phenotype confers advantage in escherichia coli chronic urinary tract infection pathogenesis. FEMS Immunol. Med. Microbiol. 44: 317–321 [DOI] [PubMed] [Google Scholar]
- LeClerc J. E., Li B. G., Payne W. L., Cebula T. A., 1996. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science 274: 1208–1211 [DOI] [PubMed] [Google Scholar]
- LeClerc J. E., Payne W. L., Kupchella E., Cebula T. A., 1998. Detection of mutator subpopulations in salmonella typhimurium lt2 by reversion of his alleles. Mutat. Res. 400: 89–97 [DOI] [PubMed] [Google Scholar]
- Leigh E. G., 1970. Natural selection and mutability. Am. Nat. 104: 301 [Google Scholar]
- Leigh E. G., 1973. Evolution of mutation rates. Genetics 73: 1–18 [PubMed] [Google Scholar]
- Lynch M., 2008. The cellular, developmental and population-genetic determinants of mutation-rate evolution. Genetics 180: 933–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao E. F., Lane L., Lee J., Miller J. H., 1997. Proliferation of mutators in a cell population. J. Bacteriol. 179: 417–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matic I., Radman M., Taddei F., Picard B., Doit C., et al. , 1997. Highly variable mutation rates in commensal and pathogenic Escherichia coli. Science 277: 1833–1834 [DOI] [PubMed] [Google Scholar]
- Notley-McRobb L., Seeto S., Ferenci T., 2002. Enrichment and elimination of muty mutators in Escherichia coli populations. Genetics 162: 1055–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver A., Canton R., Campo P., Baquero F., Blazquez J., 2000. High frequency of hypermutable Pseudomonas aerunginosa in cystic fibrosis lung infection. Science 288: 1251–1253 [DOI] [PubMed] [Google Scholar]
- Painter P. R., 1975. Mutator genes and selection for the mutation rate in bacteria. Genetics 79: 649–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal C., Macia M. D., Oliver A., Schachar I., Buckling A., 2007. Coevolution with viruses drives the evolution of bacterial mutation rates. Nature 450: 1079–1081 [DOI] [PubMed] [Google Scholar]
- Peck J. R., 1994. A ruby in the rubbish: beneficial mutations, deleterious mutations and the evolution of sex. Genetics 137: 597–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunier A. L., Malbruny B., Laurans M., Brouard J., Duhamel J. F., et al. , 2003. High rate of macrolide resistance in Staphylococcus aureus strains from patients with cystic fibrosis reveals high proportions of hypermutable strains. J. Infect. Dis. 187: 1709–1716 [DOI] [PubMed] [Google Scholar]
- Richardson A. R., Yu Z., Popovic T., Stojiljkovic I., 2002. Mutator clones of neisseria meningitidis in epidemic serogroup a disease. Proc. Natl. Acad. Sci. USA 99: 6103–6107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaver A. C., Dombrowski P. G., Sweeney J. Y., Treis T., Zappala R. M., et al. , 2002. Fitness evolution and the rise of mutator alleles in experimental Escherichia coli populations. Genetics 162: 557–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sniegowski P. D., Gerrish P. J., Lenski R. E., 1997. Evolution of high mutation rates in experimental populations of E. coli. Nature 387: 703–705 [DOI] [PubMed] [Google Scholar]
- Soderberg R. J., Berg O. G., 2011. Kick-starting the ratchet: the fate of mutators in asexual populations. Genetics 187: 1129–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei F., Radman M., Maynard-Smith J., Toupance B., Gouyon P. H., et al. , 1997. Role of mutator alleles in adaptive evolution. Nature 387: 700–702 [DOI] [PubMed] [Google Scholar]
- Tanaka M. M., Bergstrom C. T., Levin B. R., 2003. The evolution of mutator genes in bacterial populations: the roles of environmental change and timing. Genetics 164: 843–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon O., Toupance B., Le Nagard H., Taddei F., Godelle B., 1999. Mutators, population size, adaptive landscape and the adaptation of asexual populations of bacteria. Genetics 152: 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon O., Le Nagard H., Godelle B., Taddei F., 2000. Mutators and sex in bacteria: conflict between adaptive strategies. Proc. Natl. Acad. Sci. USA 97: 10465–10470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson D. A., Desai M. M., Murray A. W., 2006. Ploidy controls the success of mutators and nature of mutations during budding yeast evolution. Curr. Biol. 16: 1581–1590 [DOI] [PubMed] [Google Scholar]
- Travis J., Travis E., 2002. Mutator dynamics in fluctuating environment. Proc. Biol. Sci. 269: 591–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trobner W., Piechocki R., 1981. Competition growth between Escherichia coli mutl and mut+ in continuously growing cultures. Z. Allg. Mikrobiol. 21: 347–349 [PubMed] [Google Scholar]
- Watson M. E., Burns J. L., Smith A. L., 2004. Hypermutable haemophilus influenzae with mutations in muts are found in cystic fibrosis sputum. Microbiol. Sgm. 150: 2947–2958 [DOI] [PubMed] [Google Scholar]
- Wylie C. S., Ghim C.-M., Kessler D., Levine H., 2009. The fixation probability of rare mutators in finite asexual populations. Genetics 181: 1595–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]