Abstract
What determines the genetic contribution that an individual makes to future generations? With biparental reproduction, each individual leaves a “pedigree” of descendants, determined by the biparental relationships in the population. The pedigree of an individual constrains the lines of descent of each of its genes. An individual’s reproductive value is the expected number of copies of each of its genes that is passed on to distant generations conditional on its pedigree. For the simplest model of biparental reproduction (analogous to the Wright–Fisher model), an individual’s reproductive value is determined within ∼10 generations, independent of population size. Partial selfing and subdivision do not greatly slow this convergence. Our central result is that the probability that a gene will survive is proportional to the reproductive value of the individual that carries it and that, conditional on survival, after a few tens of generations, the distribution of the number of surviving copies is the same for all individuals, whatever their reproductive value. These results can be generalized to the joint distribution of surviving blocks of the ancestral genome. Selection on unlinked loci in the genetic background may greatly increase the variance in reproductive value, but the above results nevertheless still hold. The almost linear relationship between survival probability and reproductive value also holds for weakly favored alleles. Thus, the influence of the complex pedigree of descendants on an individual’s genetic contribution to the population can be summarized through a single number: its reproductive value.
THE most obvious feature of sexual reproduction is that each individual has two parents. Yet, the pedigrees that describe biparental relationships have received surprisingly little attention, compared with the genealogies that describe the uniparental relationships of genes. (Throughout, we refer to relationships between genes as their “genealogy”, in contrast to the “pedigree” of biparental relationships; genealogy should be understood as a shorthand for “gene genealogy”.) Following the rediscovery of Mendelian genetics, attention focused on the random genetic drift of discrete alleles and on the converse process of inbreeding, by which genes become identical by descent. There has of course been substantial work on the fate of genes within a given pedigree (e.g., Smith 1976, Cannings et al. 1978; Thompson et al. 1978), but relatively little on the pedigrees themselves.
Pedigrees are of interest in their own right: it is natural to ask who our ancestors were (Chang 1999; Rohde et al. 2004) and, conversely, how many descendants we will each leave. But, from a genetic point of view, the pedigree constrains what genes can be passed on: with Mendelian inheritance, selection acts solely through the different contributions made by individuals to the pedigree. The recent availability of genomic sequences may focus more attention on pedigrees: given sufficient sequence, we can infer the pedigree many generations back; and given this pedigree, we can ask what contribution is likely to be made to future generations by each ancestral genome. These questions are long standing (Thompson et al. 1978; Thompson 1979a, b), but it has become feasible to answer them only in the past few years (Huff et al. 2011).
The notion of reproductive value was introduced by Fisher (1930) to study populations structured by age. The reproductive value of an individual of a given age is its expected future contribution to the population (conditional on having survived to that age). Caswell (1982) generalized this to populations with an arbitrary structure (for example, where individuals vary in size or microhabitat). Grafen (2006) emphasizes that reproductive value can be ascribed to individuals as well as classes and shows rigorously that reproductive value is the target of selection. In the long term, alleles that increase the reproductive value will be the ones that increase, and traits will evolve that tend to maximize an individual’s reproductive value. In this setting, an individual’s reproductive value is defined to be its expected genetic contribution, that is, the expected number of copies of one of its alleles that it leaves in distant future generations, conditional on its pedigree of descendants. Once a pedigree is specified, one can superpose the passage of neutral alleles: offspring, independently, sample one allele from each parent. In this way an individual’s reproductive value is defined to be a function of its pedigree. Thus, we structure the population by the pedigree that connects every individual, rather than with a coarser structure by age or class.
An individual’s reproductive value is determined within ∼10 generations, whereas its ultimate genetic contribution is determined over very long timescales. Here, we examine the relationship between pedigrees and genealogies over intermediate timescales of a few tens of generations.
It is crucial to realize that overall genetic contribution to future generations is much more complex than simply the reproductive value, which gives the expected contribution at any one locus. The key result of this article is that the reproductive value of an individual determines the survival probability of its genes, but conditional on survival, the distribution of the number of copies of an allele in future generations is the same for all individuals, independent of their reproductive value. This result applies to a single genetic locus. Most of an individual ancestor’s genome is lost, but some small blocks survive in large numbers (Baird et al. 2003). By investigating simple summary statistics of the distribution of surviving blocks, we illustrate that the influence of the pedigree on the whole complex distribution of genetic contribution of an individual is also determined by its reproductive value. Thus, over these intermediate timescales, from the point of view of allele frequencies, the tangled web of relationships that forms an individual’s pedigree can be completely captured in a single number: the reproductive value.
Previous work modeling the evolution of pedigrees
The spread of single genes is often represented by the Wright–Fisher model, in which the single parent of a gene is chosen at random from the gene pool in the previous generation. The obvious analog for pedigrees is to choose two parents at random; if this is done with replacement, then selfing is possible. Surprisingly, this biparental model has only relatively recently been analyzed; it behaves quite differently from the uniparental Wright–Fisher model. Chang (1999) shows that ∼log2 N generations back into the past, an individual will have existed who was ancestral to every present-day individual; going back ∼1.77 log2 N generations, all those individuals who are ancestors will be ancestors of every present-day individual. [See Appendix C for an explanation of the mathematical notations f(N) ∼ g(N), f(N) = (g(N)), and f(N) ≍ g(N).] Moreover, for large N, the time when a common ancestor first appears and the time when every individual shares the same set of ancestors cluster very closely around their expected values. Rohde et al. (2004) show that the rapid mixing of the pedigree is not greatly slowed by the degree of population subdivision thought likely for humans: a single migrant is enough to link the ancestry of an isolated deme to that of the whole species.
This rapid mixing of biparental ancestry contrasts with the very slow process through which single genetic loci come to share common ancestry. The time since the most recent common ancestor of the whole population at a single genetic locus has mean ≍ 4N generations under the Wright–Fisher model (Möhle 2004), but the standard deviation is of the same order (with a contribution of ∼2N generations from the approximately exponentially distributed time during which there are exactly two lineages ancestral to the population). Similarly, looking forward in time, an individual’s contribution to the pedigree is decided within a few generations, whereas its ultimate genetic contribution is decided by drift over a much longer timescale, of (N) generations. Our work is concerned with intermediate timescales. We shall see (both mathematically and through simulations) that the reproductive value of an individual is determined within a few tens of generations. Our analytic results for the number of copies of surviving alleles require somewhat longer timescales, long enough that the alleles at a given locus represented in the current population are inherited from a small fraction of ancestors, but not so long that the fate of the population has been determined by genetic drift.
Derrida et al. (1999, 2000) investigate the distribution of reproductive value (which they term the “weight”) numerically. They show that it rapidly settles to a stationary distribution, which is close to that obtained by considering, at large times t (t > 50, say), 2−t times the number of offspring in a Galton–Watson branching process with Poisson offspring distribution with mean 2 (which can be investigated analytically). To understand this result, first observe that the probability that a particular gene is passed down a particular line of descent spanning t successive generations is 2−t and so the expected number of copies of that gene after t generations is just 2−t times the number of distinct lines of descent through the pedigree. In a large population, the pedigree of descendants of an individual will initially grow like a branching process in which each individual has a Poisson number of offspring with mean 2 and under this branching process approximation, the number of distinct lines of descent through a pedigree spanning t generations is just the number of pedigree descendants after t generations. In other words, in a population of moderate size (N > 100, say), the distribution of reproductive value can be calculated simply by assuming that the descendants of a single individual never meet.
Matsen and Evans (2008) study quantitatively the relationship between the genetic contribution and pedigree. Although for most of their article they relate the number of pedigree descendants to the number of genetic descendants, in their section 4 they consider a multigraph that corresponds precisely to the pedigree that we consider here and show that, for large populations and for t ≪ log2N generations, one can couple the pedigree to a Galton–Watson branching process with a Poisson offspring distribution with mean 2. Combining this with the extremely rapid convergence of the quantity corresponding to Derrida et al.’s weight for a branching process (see section 3 in Matsen and Evans 2008) is a mathematically rigorous route to the results of Derrida et al. (1999).
Following the usual convention, we refer to an individual’s reproductive value as v, rather than using Derrida et al.’s w. We shall also talk about the “relative contribution” of an individual a few generations later. For example, over one generation, the relative contribution of an individual is just the number of its offspring divided by the expected number of offspring for individuals in the population. For populations like the diploid Wright–Fisher model of Chang (1999) and Derrida et al. (1999, 2000), in which the expected number of offspring of each individual is two, the relative contribution converges as the number of generations grows to the reproductive value, but even in this case, for shorter times the relative contribution is not, in general, the same as the reproductive value.
An individual’s genetic contribution consists of a series of blocks of genome that are passed down to its descendants via a tangled web of relationships. Conditional on the pedigree of an individual in the ancestral population, the reproductive value, v, of that ancestor is the expected number of copies, many generations later, of one of its genes. Our aim is to investigate the relationship between this single number and the total genetic contribution of the ancestor.
Cannings et al. (1978) consider the probability that a set of genes carried by one or more individuals will survive to some later time; Thompson (1979a,b) shows how this survival probability is correlated between related individuals. Both deal with contributions of individuals in specific pedigrees, but they do not discuss reproductive value specifically. Derrida et al. (2000) study an extension of reproductive value in a model that attempts to capture the genetic contribution made by each individual across multiple loci. They suppose that offspring inherit a fraction f of their genes from one parent and 1 − f from the other, with f following some distribution ρ(f) on [0, 1], but they ignore the linear structure of the genome. Chapman and Thompson (2003) analyze the distribution of blocks that are identical by descent between contemporary genomes. Baird et al. (2003) consider a linear genetic map and follow the descent of a single ancestral genome, forward in time; they obtain the generating function for the complete distribution of sizes and the location of blocks of genome that are passed on. Like Derrida et al. (1999, 2000), their analysis is based on a branching process approximation, which in effect assumes an infinite population, but in fact is accurate for populations of moderate size.
Summary
The timescales for evolution of the pedigree [(log2N) generations] and of the genes [(N) generations] are very different. Individual contributions to the pedigree, quantified through their reproductive values, are decided early on, over such a short timescale that unless selection on individual genes is very strong its effect on the pedigree can be ignored. We analyze the relation between an individual’s reproductive value, v, and its genetic contribution. We find that even under weak selection, the probability that an ancestral allele contributes to future generations is determined by the reproductive value. Moreover, with neutral evolution, conditional on survival, the distribution of the number of copies of an ancestral allele seen in generation t, where is independent of the reproductive value. We extend the analysis to a linear genome and consider the distribution of blocks of genetic material passed on to future generations by an individual. For simple summary statistics of this distribution we show how the influence of the pedigree is encoded in the reproductive value. These results remain true when there is inherited variation in fitness on unlinked loci, represented by the infinitesimal model (Fisher 1918; Bulmer 1971). Thus the influence of pedigree on genetic contribution is entirely summarized in the reproductive value. This makes it much simpler to understand the origins of the genetic material that we see in present-day populations. All the calculations in this paper are contained in a Mathematica notebook, available as supporting information, File S1.
Model and Methods
Definitions
We assume a population of N diploid individuals. The pedigree spanning t generations is represented by a sequence of N × N matrices M0, M1, … , Mt. Here we count time back into the past. The ith row of Mt specifies the parents (alive in generation t + 1) of the individual labeled i in generation t, so that the matrix Mt connects generation t before the present with generation t + 1 before the present. If an individual has two different parents, then the row has two nonzero elements, each set at ; if it is produced by self-fertilization, then there is a single nonzero entry, with value 1. We represent the matrix in Mathematica as a sparse array (Wolfram 1991); this allows large populations (N ∼ 1000, say) to be handled efficiently, since only 2N elements are stored, rather than N2.
In the simplest neutral model, each parent is chosen at random, with replacement, so that a fraction averaging 1/N are produced by selfing and each parent has approximately a Poisson number of offspring with mean 2. In Appendix D we see how our results can be extended to more general offspring distributions. Once the pedigree is determined, genotypes are then chosen randomly. Genes may be labeled 0 or 1 to indicate their allelic state, or they may be given a unique integer in the first generation, so that identity by descent can be followed [“gene dropping” (Edwards 1968; MacCluer et al. 1986)].
Distribution of reproductive value, v
Rather than simulating genes on the pedigree, we can calculate quantities of interest directly for any given pedigree (Cannings et al. 1978; Derrida et al. 1999; Vindenes et al. 2009). First, consider the reproductive values, which are represented as a vector, v, with N elements. By definition this is the large time limit of the vector of relative contributions (scaled to sum to N). Write 1 for the vector in all of whose entries are 1. We set v0 = 1 for the vector of relative contributions of the current individuals to the present generation, at time t = 0. We can define the relative contributions vt by working backward in time. As we add an additional ancestral generation, each individual ancestor’s relative contribution is just half the sum of its offspring’s contributions (Derrida et al. 1999). In matrix notation,
| (1) |
where taking the product with 1 corresponds to taking a sum over descendants. The (i, j)th entry in the product of random matrices M0M1M2, … , Mt−1 gives the expected contribution to descendant i from the ancestor labeled j, t generations before. This matrix M0M1, … , Mt−1 rapidly settles to a constant form in which the contribution from ancestor j is the same for each descendant i, corresponding to the entries in each column being constant (Appendix D). Each ancestor is thus characterized by a single number, which is the same for every distant descendant and is independent of time t. The reproductive value of the jth ancestor is the sum of the elements in the jth column of the matrix. Note that the vector of reproductive values is given exactly by Equation 1: there is no need to assume that the population is large. Moreover, it is not necessary that the matrices Mt correspond to the diploid Wright–Fisher model. In Appendix D we consider pedigrees corresponding to more general offspring distributions and in Appendix F we introduce simple forms of structure into our model. The form of Equation 1 does not change and the rapid convergence of the relative contributions to the reproductive value is robust to these extensions.
The probabilities of loss or survival of each allele
Unless the pedigree is small, it is not feasible to find an analytic expression for the probability that an allele will survive. In Appendix E we obtain a simple approximation for the survival probability of an allele carried in single copy in an ancestor generations in the past, in terms of the reproductive value of that individual and the corresponding survival probability for the descendants of a single individual in a haploid population of size 2N.
The analytic results in Appendix E are asymptotic in population size N. To compute the survival probability numerically, we consider times that are short relative to the population size (t ≪ N), so that descendants of a single gene are unlikely to become common. In an unstructured population, there are a very large number of paths through the pedigree and so over this timescale different descendants of a given gene do not meet each other, so that they will be lost independently. (That is, the chance that every descendant is lost is the product of the chance that each one of them is lost separately.) Thus, the descent of genes through a large unstructured population can be approximated as a branching random walk through the pedigree. Given a pedigree generated by matrices Mt we can then write down recursions for loss probabilities and this enables us to plot the relationship between reproductive value and survival probability. Two copies of a gene within a diploid individual are not passed on independently to the next generation, since they both depend on the reproduction of the same individual. Therefore, to find the probability that an allele present in an ancestor alive t generations in the past will be lost by the current generation, we must follow two vectors: the chance Qt,j that an allele present in one copy in ancestor j (alive at time t before the present) will be lost and the chance that an allele present in two copies in ancestor j will be lost. Under the assumption that genes in different descendants are lost independently, these follow the recursions
| (2) |
and
| (3) |
with initial conditions
In Appendix A we show how these probabilities can be calculated.
The distribution of the number of copies
Under the assumption t ≪ N, one can use the same approach to approximate the number of copies of a gene passed down through a pedigree. The generating function for the number of copies is given by the same recursion (Equations 2 and 3) as the loss probabilities, but with different initial conditions (see Vindenes et al. 2009, Appendix 2). Define the generating function for the number of copies nt in generation 0 descended from a single copy in ancestor j at time t as and similarly define as the generating function for the number of copies at time 0 descended from two copies within the ancestor j alive t generations in the past. These quantities follow the recursions in Equations 2 and 3, with initial conditions Q0,i(y) = y and The probability of loss is the special case where y = 0.
The variance in the number of copies left by ancestor j after t generations, Vt,j, can be found by differentiating the generating function. It is simplest to rewrite y = ew to recover Q = E[ewn], the moment generating function of the copy number, from Qt,j. Then, the variance of n is
The corresponding recursion is presented in Appendix A.
In Appendix E we work directly with the diploid Wright–Fisher model to find, analytically, an approximation for the distribution of the number of copies of a neutral allele, conditional on survival, and as a function of reproductive value, for In particular, we establish that this is in agreement with the numerical predictions obtained through the “branching random walk” approximation described above.
Inherited variation in fitness at unlinked loci
More generally, parents are chosen independently, with probability proportional to their individual fitness. We use the infinitesimal model to describe the aggregate effect of selection acting on large numbers of unlinked loci (Fisher 1918; Bulmer 1971). The probability that an individual is chosen as parent is proportional to ez where z, the log fitness, is an additive trait. Offspring have a normally distributed trait value with mean equal to the mean of their two parents and variance fixed at half the additive genetic variance, V/2. If we consider discrete unlinked loci, then we can always superpose the passage of neutral genes onto the pedigree (no matter how it is generated). Each diploid parent passes on one or the other of its genes with equal probability, independently across loci. In Appendix B we extend our branching random walk approximation to find the generating function for the distribution of reproductive values under the infinitesimal model.
The probability of fixation of a favored allele
When we follow an allele that itself affects fitness, we can no longer separate the growth of the pedigree from that of the allele: we cannot follow the flow of genes through a given pedigree, when those genes themselves influence the structure of that pedigree. Therefore, to find the relation between individual reproductive value and the fate of a selectively favored allele, we must simulate jointly the genotype and the pedigree. However, we need simulate only for a few tens of generations, over which time the relative contribution of an individual approaches a fixed reproductive value.
After that time, we find the probability of ultimate fixation as a function of the number of copies, n, of the allele that survive. In moderately large populations, and with weak selection, this number is small (n ≪ N, s ≪ 1), and so the ultimate fixation probability is given accurately by the branching process approximation 1 − (1 − P)n, where P is the probability of survival of the descendants of a single allele. If there is no selection at other loci, then under our simple model P ∼ 2s for small s. For a large population, we can further improve efficiency by starting with several copies of the favorable allele, each labeled by a distinct integer, on the assumption that these will be lost independently of each other.
Surviving blocks of ancestral genome
Baird et al. (2003) consider the fate of a single block of ancestral genome as it is passed through a pedigree generated by a branching process. Although most of the block is lost, typically some small subblocks survive and are represented in large numbers in the population. Starting from a single block of length y embedded in an interval of length 1 that experiences one crossover per genome per generation, they investigate the distribution of the numbers of surviving blocks of different sizes after t generations. Simple statistics of this distribution are expressed through its “moment densities”, which can be integrated to establish moments and mixed moments of numbers of blocks of different sizes.
In Appendix G we show how an arbitrary process of recombination can be superposed on a pedigree through our matrix recursions. We then specialize to the single crossover model and show that the first two moment densities depend linearly on reproductive value.
Results
The distribution of reproductive value
Each ancestor leaves a random number of descendants. If any survive the first few generations, their number will become so large as to grow almost deterministically. The relative contribution of each individual to future generations therefore quickly settles to a constant—its reproductive value—which is determined by fluctuations in the first few generations. In Appendix D we prove that under the simple neutral model, the change in the relative contribution of an ancestor over a single generation is (10−3) after ≈10 generations, independent of population size (Equation D12). Moreover, we show that replacing the multinomial sampling of our Wright–Fisher model by a more general offspring distribution does not significantly change this result except that the number of generations required scales linearly with the variance of the number of offspring of each individual.
Until the number of pedigree descendants of an individual form an appreciable portion of the population, the pedigree can be approximated by a branching process. In a finite population, the number of descendants is bounded, and so there must ultimately be inbreeding: descendants will be connected with each ancestor via many overlapping routes through the pedigree. However, when the population is large, the reproductive value v is determined before finite population size has any significant effect and so we may approximate its distribution by that of the corresponding quantity for a branching process.
For a branching process with growth rate λ, the relative contribution of an ancestor alive t generations in the past is just Zt/λt, where Zt is its total number of descendants in the present population. From the classical theory of branching processes (see, e.g., Harris 1963, Chap. I, section 8) we know that this converges extremely quickly to a nonnegative random variable v. If the number of offspring of an individual has probability generating function f(s), then v can be characterized through its moment generating function, ϕ(s) = E[exp(−sv)], which satisfies
| (4) |
With a Poisson offspring distribution, it is convenient to consider the probability generating function of the reproductive value. Writing for the generating function of the distribution of the relative contribution of an ancestor after t generations, we obtain the recursion
| (5) |
which quickly converges to the fixed point where gt+1(y) = gt(y); λ = 2 for a stable population. When λ = 2, Equation 5 corresponds to Equation 7 of Derrida et al. (1999). Their equation takes a slightly different form since they consider (recall that they use the notation wt for our vt and we have substituted θ for their λ to avoid confusion with the growth rate above). Moreover, since they compute the reproductive value by working backward through the pedigree, as opposed to forward from a fixed ancestor, their initial condition differs from ours.
Equation 5 does not have an explicit solution. However, simple summary statistics of the limiting random variable v are readily calculated. For example E[v] = 1 and var(v) = 1. [For Equation 4 these become E[v] = 1, var(v) = var(Z1)/(λ2 − λ).] If we condition a critical branching process on nonextinction, after normalizing by t, we have convergence to an exponential distribution. For the supercritical branching processes considered here, instead of t, we normalize by λt to obtain the nontrivial limit, v. The distribution of v, conditional on being nonzero, is “narrower” than exponential. For the Poisson offspring distribution with λ = 2, conditional on there being some descendants [probability P = 1 − g(0), which is ≈0.80 for λ = 2], the distribution has mean 1/P ≈ 1.26 and a variance 2/P − 1/P2 ≈ 0.935. (An exponential with the same mean would have variance 1/P2 ≈ 1.58.)
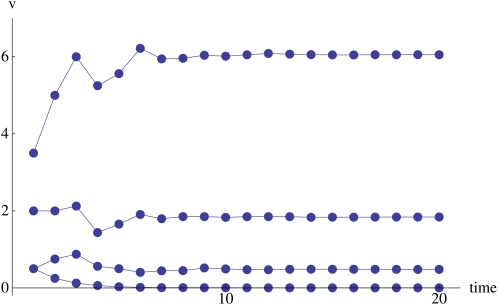
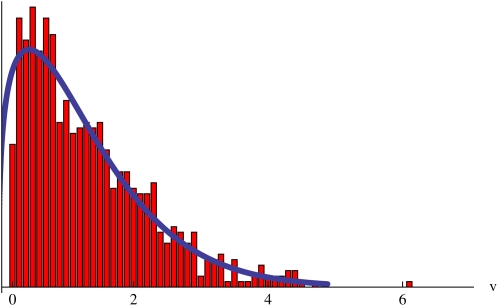
The rapid convergence to a constant reproductive value is illustrated in Figure 1, which shows (for the neutral model) the numbers of descendants over time, scaled relative to the expected number, 2t, for four ancestors. The upper set of points shows the ancestor with highest reproductive value. In the first generation, it has seven offspring and thus makes a relative contribution of 7/2. These offspring themselves have more offspring than average, and so the relative contribution continues to increase, converging to v = 6.056. With a Poisson number of offspring, with mean 2, the variance in offspring number is 2, and so the variance in relative contribution to the first generation is 1/2. This is half of the total variance in reproductive value, var(v) = 1. The distribution of reproductive value across ancestors is close to the theoretical expectation, even in a single realization (Figure 2).
Figure 1 .
The relative contribution of four ancestors from a population of N = 1000, plotted against time. This is defined as the number of pedigree descendants, divided by 2t. The top and bottom sets of points show the ancestors with highest and lowest reproductive value, respectively; the two intermediate sets are randomly chosen individuals. Note that the distribution of v has mean and variance equal to one.
Figure 2 .
The bars show the distribution of reproductive values for a single population of N = 1000, compared with the theoretical expectation (curve). The latter is calculated by expanding the generating function for the distribution of numbers, E[yn] as a Taylor series in y and then rescaling using n = v2t. This calculation was done at t = 10 generations; however, the correlation with the ultimate reproductive value is extremely close (0.99957).
The ancestry of each individual quickly becomes homogeneous: every individual receives almost exactly the same contribution from each ancestor, which is defined by those ancestors’ reproductive values. To quantify this, for each ancestor we scale the contribution to each descendant to have a mean of 1 and measure the variance across descendants of this relative contribution. This variance averages <N2−t (Equation D11) and so quickly becomes small as t becomes large. This is because each descendant receives ancestry via very many routes through the pedigree ((2t/N)); the total contribution from any ancestor, summed over these many routes and renormalized by 2t, therefore clusters close to its mean.
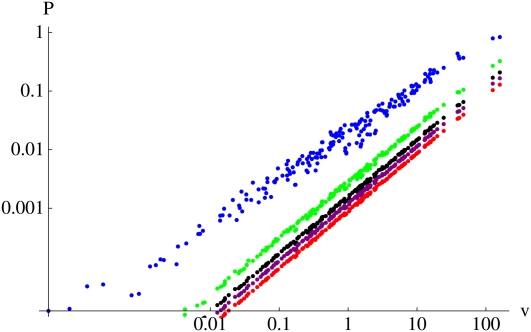
Probability of survival
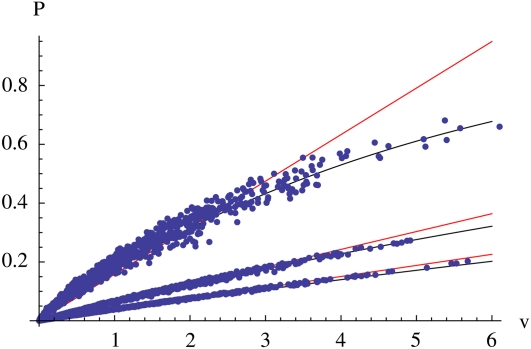
The chance that a neutral allele, initially present in a single copy, will survive for t generations becomes completely determined by the reproductive value of its initial bearer, as t becomes large. This is shown in Figure 3, which plots the survival probability against reproductive value for t = 10, 30, and 50 generations in a population of N = 1000. After ∼30 generations, the survival probability converges to vPN,t, where PN,t ≪ 1 is the probability of survival until time t of a neutral allele initially present in single copy in a haploid Wright–Fisher population of size 2N: this is shown by the tight fit of the points around the linear relationship, for t = 50. This is a strong result, which applies to single individuals, and not just to the population as a whole. Once we know an individual’s reproductive value, we know the chance that any one of its genes will survive for a given time. Indeed, this survival probability is in principle observable: unlinked genes are passed down independently through the pedigree, and so the fraction of an individual’s genes that survive to time t gives an estimate of the survival probability.
Figure 3 .
The relation between probability of survival, P, and reproductive value, v, at times t = 10 (top), t = 30 (middle), and t = 50 (bottom). For each time, there are 1000 dots, each representing a single ancestor. Each dot gives the probability that a single copy of a gene in the ancestor will survive to time t, plotted against the ancestor’s relative contribution to the pedigree up to time t. The straight lines show the linear relation vPN,t where PN,t is the probability of survival of a neutral allele in a branching process with growth rate λ = 1. The curves show the approximation is , where is an effective value determined by . This calculation is simplified by using the fact that is the generating function for v (Equation 5) evaluated at
For shorter times, there is more scatter: that is, the genes carried by individuals that make the same relative contribution to the pedigree, v, may have different chances of survival. This is not surprising, since the survival probability depends in a complex way on the structure of the pedigree, via Equations 2 and 3, and not just on the total number of descendants. For example, think of an individual that has two children and four grandchildren. If one child has all the grandchildren, ; if one has one and the other has three, ; and if both children have two grandchildren, What is remarkable is that for longer times, the fixation probability does depend only on the magnitude of an individual’s contribution, as measured by v.
The relation between survival probability, Pt, and relative contribution, v, cannot be precisely linear, if only because the probability cannot be >1. We show in Appendix E that, at least for a more accurate approximation is , where is an effective value determined by
This is shown by the curves in Figure 3. Note that is the generating function for the branching process with growth rate λ = 2, which can be calculated directly.
Distribution of numbers of copies
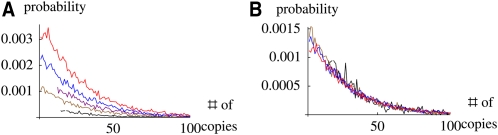
The generating function for the number of copies that survive to time t is given by the same recursion (Equation 5) as the probability of loss. Therefore, we expect that this distribution will also be determined entirely by the reproductive value and, moreover, can be approximated by assuming that each individual contributes an “effective number” v. Figure 4A shows the distribution of number of copies, after 50 generations, with individuals grouped by reproductive value; the spike at zero, representing the usual loss of the allele, is not shown. The distribution retains the same form, but its mass is proportional to the reproductive value. Thus, the distribution of copy number conditional on survival is independent of reproductive value (Figure 4B) and is the same for all individuals as t → ∞.
Figure 4 .
(A) The distribution of numbers of copies left by a single copy in an ancestor, after 50 generations in a population of 1000. The usual outcome—loss of the allele—is not shown. Ancestors are classified by their reproductive value (<0.5, 0.5−1, 1−1.5, 1.5−2, >2, bottom to top). (B) The distribution, conditional on survival, is almost independent of reproductive value. These distributions are for a single pedigree; they are estimated by simulating the flow of genes through that pedigree, using 1000 replicates. At the start of each replicate, every allele is labeled by a unique integer that denotes the individual that carries it. Thus, 1000 allele frequencies are estimated in each of the 1000 replicates.
In Appendix E, we show that, for sufficiently large populations, this conditional distribution should be approximately exponential. It would take a long time to calculate the full distribution for long times, but moments can easily be calculated. We know already, from results on survival probability, that the mean number, conditioned on survival, converges in this way. Figure 5 shows the coefficient of variation of the distribution, conditional on survival (i.e., the standard deviation divided by the mean), for t = 10, 30, and 50 generations. As expected, this converges to a constant value, for all individuals, by t = 50. This value is slightly <1, the coefficient of variation expected for an exponential distribution. Similar calculations for the generating function E[yn] over a range of values of y show similar convergence. This confirms that the distribution of copy number, conditional on survival, becomes close to an exponential, with mean 1/PN,t for all individuals.
Figure 5 .
The coefficient of variation of the distribution of numbers of copies, conditional on survival, plotted against relative contribution, v. This is calculated for a single pedigree, with N = 1000, at t = 10, 30, and 50 generations (bottom to top). Each dot represents one individual.
The probability of fixation of a favored allele
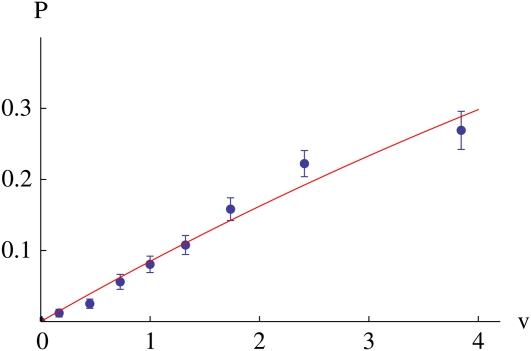
Figure 6 shows how the probability of fixation of an allele with advantage s = 0.05 depends on the reproductive value of the individual that carries it. It is not possible to calculate fixation probability on a given pedigree, because the selected allele itself influences the pedigree. Therefore, Figure 6 shows the average fixation probability for alleles that start in individuals within a range of reproductive values, averaged over many replicate simulations. Because the variance in reproductive value is much larger than the selection acting on the allele, we expect the distribution of numbers of copies left after the first few generations to be approximately independent of selection. This suggests that the probability that the allele will fix, given that it starts in an individual with reproductive value v, is 1 − (1 − P)v (where, as usual, P is the survival probability of a single copy of the allele). This fits closely with the simulations (Figure 6).
Figure 6 .
The probability of fixation of an allele with advantage s = 0.05 plotted against reproductive value, v. The curve gives the expected relationship, where is chosen such that the average fixation probability equals the standard value The fixation probability is estimated from 400 replicates, each starting with 10 favorable alleles in a population of N = 1000 and iterated for 30 generations. Each of the 4000 alleles is classified by the reproductive value of the individual that first carried it; the points show the mean reproductive value and the mean fixation probability for each class (±1 SE).
Population structure
So far we have considered the simplest Wright–Fisher model of diploid reproduction. However, the rate of convergence of the reproductive value is not greatly slowed under simple forms of structure. In Appendix F we consider two examples. First we take a population with partial selfing. We suppose that a proportion α of offspring are produced by self-fertilization and the remaining 1 − α by random mating. The change in relative contribution from generation to generation now decays at rate instead of the 2−t of the diploid Wright–Fisher model. This can be regarded as the simplest form of structure. We then extend to an island model. Reproduction is through random mating within demes, but a proportion of offspring are exchanged between demes before the next round of mating. Rohde et al. (2004) showed by simulation that mixing is rapid even with substructure and this is confirmed by our mathematical results. The variance in contribution from a particular ancestor to individuals within a given deme is rapidly whittled away through the same process as in a panmictic population, while migration works to eliminate variability between demes. This is quantified in Appendix F, The island model.
The effects of genome-wide selection; inherited variation in fitness at unlinked loci
We now use the infinitesimal model to describe the aggregate effects of selection on large numbers of unlinked loci. In Appendix B, we extend Equation 5 to find the generating function for the distribution of reproductive values under the infinitesimal model. If the mean log fitness is the mean reproductive value of individuals with log fitness z is which increases with the square of the fitness, ez. This can be understood from an argument first made by Robertson (1961): an individual with excess log fitness will have offspring that deviate by on average, grand-offspring that deviate by , and so on; the cumulative deviation in log fitness, summed over generations, is therefore and the net reproductive value is proportional to which is the square of the immediate relative log fitness. (The normalizing factor e−2V arises because, by definition, E[v] = 1.) The variance in reproductive value can be found explicitly (Equation B4); it is proportional to where β decreases from 2.65 for small V to 2.15 for V = 3. Averaging over z, the variance in reproductive value is just e4V, as can be seen by integrating Equation B3 over the distribution of z.
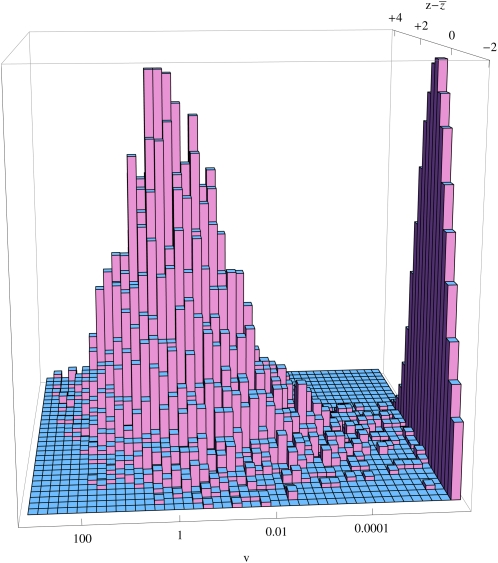
We give an example, in which the genetic variance in log fitness is V = 1. The heritable variance in fitness itself is varA(ez) = e2V − eV = 4.67. Since we assume a Poisson number of offspring, with mean averaging 2, the nongenetic variance in fitness is varE(ez) = 2, and so the heritability of fitness is h2 = varA(ez)/(varA(ez) + varE(ez)) = 70%. Now, there can be a very wide range of reproductive values: the variance in reproductive value has increased from 1 to 54.6 ∼ e4V, but fluctuates considerably from generation to generation, even with N = 1000. An individual’s reproductive value is correlated with its log fitness, z, but because the reproductive value is determined over several generations, not just one, the relationship is weak (r = 0.27; Figure 7).
Figure 7 .
The joint distribution of log relative fitness, and reproductive value, v, under the infinitesimal model. This distribution is taken from generations 60−100 of a simulated population of N = 1000 individuals, with genetic variance var(z) = 1. The variance in reproductive value is 73.45. Seventy-six percent of individuals have zero reproductive value; these are shown by the distribution at the right.
Just as is the case in the absence of selection, after a few tens of generations, the probability that an allele survives depends solely on an individual’s reproductive value (Figures 8 and 9), and not on the log fitness, z, or any other feature of the pedigree. Individuals with high values of z are expected to make a greater genetic contribution to distant generations, but this is mediated entirely by their increased reproductive value. Equations 2 and 3 still apply even when var(v) is greatly increased by selection.
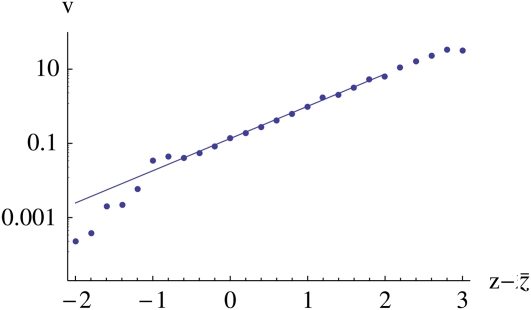
Figure 8 .
The mean reproductive value, as a function of log fitness, The line is the theoretical prediction exp Note that for very low z, the average reproductive value falls below the prediction. This is because almost all such individuals have zero reproductive value, so that the mean is determined by very rare individuals with high value and is correspondingly poorly estimated. These data are taken from the same simulation as in Figure 7 with var(z) = 1.
Figure 9 .
The probability of survival of a neutral allele, plotted against reproductive value, v, under the infinitesimal model (V = 1, N = 1000), for times t = 10, 20, 30, 40, and 50 (top to bottom).
Surviving blocks of ancestral genome
If we follow the descent of a single block of ancestral genome through a pedigree, large chunks will rapidly be lost. However, some small subblocks can be expected to persist for a long time. Baird et al. (2003) establish the full distribution of surviving block lengths by solving recursions, in much the same way as in Equations 2 and 3, but with the pedigree generated by a branching process. In Appendix G we show that for intermediate timescales, the first and second moment densities of the distribution of the numbers of surviving blocks of ancestral genome of given lengths both depend almost linearly on the reproductive value of the ancestor.
Discussion
The nature of the genetic contribution
Our central result is that the complex distribution of the genome that is passed down the pedigree differs between ancestors only through a single quantity: the reproductive value of the ancestral individual. Moreover, an individual’s reproductive value affects only the chance that it passes on some genes at a locus: given that it does, then at any time the distribution of the number of copies and the distribution of sizes of each small chunk of genome are the same for all ancestors, and conversely the contribution is (on average) the same to all descendants.
These strong results apply over timescales of a few tens of generations and require that selection be weak enough to have negligible effect during this time. They are due essentially to a difference in timescales: pedigrees mix rapidly, whereas genealogies drift over much longer timescales.
Equally, we could have formulated results backward in time. The genome of an individual alive in the present-day population can be thought of as a sequence of blocks of random lengths, each of which can be traced back to a specific ancestor. The distribution of lengths and positions of these blocks and of which subcollections of blocks have common ancestry at time t in the past is determined by the ancestral recombination graph. Over intermediate timescales, the only role of pedigrees in determining this complex distribution will be in assigning, to each collection of blocks that share an ancestor in this way, an ancestor that is sampled from the ancestral population with a weight proportional to reproductive value.
Population structure
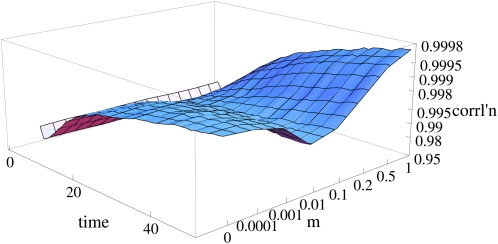
We have focused on a very simple model, in which parents are chosen at random, within a single panmictic pool. Plainly, our results do not hold if the population is strongly subdivided: individuals are more likely to descend from ancestors in the same deme. However, we believe that our results do apply to any well-mixed population and will be robust to moderate barriers (Figure 10). This is consistent with the examples checked in Appendix F and with Rohde et al.’s (2004) simulations, which showed that a population structure of the kind found in our own species does not greatly slow the mixing of the pedigree. This can be understood from the rapid growth in the numbers of descendants of ancestors in the pedigree, which double with every generation: once a single migrant establishes in a new deme, its descendants will double in number and quickly fill the deme.
Reproductive value as a measure of fitness
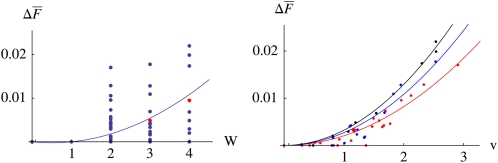
Of course, the reproductive value does not summarize every feature of the pedigree. For example, while the probability that two lineages coalesce in a given ancestor is approximately proportional to the square of the ancestor’s reproductive value, there is substantial scatter around this relationship: individuals with the same reproductive value may be more or less likely to propagate two independent lines of descent (Figure 11) and therefore to be the site of a coalescence event. Indeed, it is not clear what quantities can be completely determined by the reproductive value. One might think that the reproductive value—the expected number of copies of a gene that are passed on to distant generations—can determine only quantities that involve single lineages. Indeed, the contribution to inbreeding and coalescence involves a pair of lineages and is not completely determined by v. However, the variance in numbers of surviving copies is also a pairwise measure and is completely determined by v.
Figure 10 .
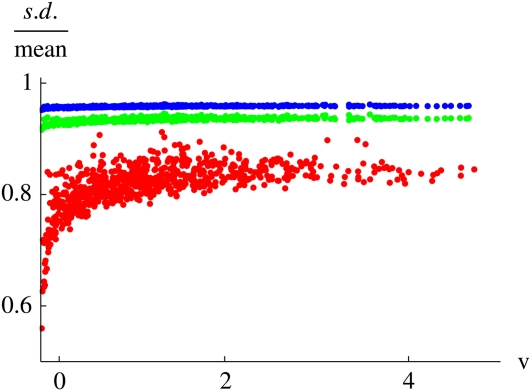
With intermediate migration rates, population structure slows the convergence of survival probability to strict dependence on reproductive value. The vertical axis shows the correlation between probability of survival to time t and the expected genetic contribution at time t; this correlation measures the tightness of the scatter in plots such as that in Figure 4. The correlation is plotted against time and against migration rate, m. There are 100 demes of 10 haploid individuals; parents are chosen from within the deme with probability 1 − m and randomly from the whole population with probability m.
Figure 11 .
The relation between an individual’s contribution to inbreeding, and its immediate fitness (W, left) or its reproductive value (v, right). is the average probability that two randomly chosen lineages will coalesce in a particular ancestor, 50 generations before; population size is N = 100 diploid individuals. The curve on the left is the best fit, The curves on the right are the best quadratic fits, for individuals with immediate fitness W = 2, 3, 4: 0.00205v2, 0.00271v2, and 0.00323v2, respectively.
Whether an individual will be the site of a coalescence event does not have any observable consequence at the time. In contrast, the contribution that deleterious recessive alleles carried by an individual will make to the future mutation load depends on the mean squared numbers of copies, E[n2], which in turn depends strictly on the reproductive value. Ballou (1997) defined a measure of ancestral inbreeding, which measures the chance that the genes carried by an individual have passed through a homozygote at some time in the past; presumably, the load of recessive deleterious mutations should be lower in individuals with higher levels of ancestral inbreeding (Suwanlee et al. 2007). If this is the case, then the future prospects of individuals with higher ancestral inbreeding should be higher. However, it is not clear how close is the relation between Ballou's measure and the mutation load: since deleterious alleles are mainly eliminated from outcrossing populations by their heterozygous effects (Wright et al. 1942; Charlesworth 1979), this effect may be small. Nevertheless, Ballou’s ancestral inbreeding may be a measure of the pedigree that complements the reproductive value by providing additional information about the extent of the mutation load carried by an individual.
The relation between selection and reproductive value
Typically, most individuals in a sexually reproducing population leave descendants, and all such individuals will be ancestors of every descendant; their relative contribution to the distant future is determined after a few tens of generations. This contrasts with the action of selection, which can fix a single gene, carried by a single individual, and acts over a long time: ∼(1/s)loge(4Ns) generations.
There is no paradox here: a favorable allele will gradually increase, through the slightly greater reproduction of individuals that carry it. Individuals carrying a set of unlinked alleles that collectively increase fitness by a factor W will have reproductive value greater by a factor W2 (Robertson 1961); the effect of a single allele on the reproductive value may be barely perceptible against the overall variance in reproductive value. The reproductive value of an individual that carries a single mutation that will ultimately fix will be substantially increased, but this is due primarily to the necessarily rapid increase of any allele that survives against the odds: the expected number of copies is est, and so the expected number conditioned on survival is est/P, where P is the probability of survival. For times less than ∼1/s this is dominated by the survival probability, P, which in turn depends on the individual’s reproductive value.
Does selection act on individuals or on genes? On the one hand, traits will evolve to maximize individual reproductive value; and natural selection must act through the reproduction of individuals. Moreover, we have shown here that the complete statistical distribution of the neutral alleles passed on by an individual is entirely determined by its reproductive value. On the other hand, an individual’s reproductive value tells us very little about the fates of the selected genes that it carries: even if an allele has an imperceptible effect on any one individual’s reproductive value, selection will ultimately determine its fate in the population as a whole.
Acknowledgments
N.H.B. was supported in part by European Research Council grant 250152. A.M.E. was supported in part by Engineering and Physical Sciences Research Council grant EP/E065945/1.
Appendix A
Matrix Representation of Recursions
We write Qt,j for the chance that an allele present in one copy in ancestor j alive t generations before the present will be lost by the current generation and for the chance that an allele present in two copies in individual j will be lost. Under the assumption that genes in different descendants are lost independently, these follow the recursions of Equations 2 and 3. To see this, first note that if an individual carrying a single gene mates with a different individual and has an offspring, then there is probability 1/2 of the gene not being passed on and Q(t−1),i/2 of it being passed on, but then lost, giving a factor of (1 + Q(t−1),i)/2 for each offspring produced by outcrossing (first term on right-hand side of Equation 2). If the offspring is produced by selfing, then 0, 1, or 2 copies of the gene present in the parent may be passed on, with probabilities in the ratio 1:2:1; the corresponding probabilities of loss are respectively. This gives the factor in Equation 2. If two copies are present in the parent, then an outcrossed progeny is certain to get one copy, and a selfed progeny is certain to get two, leading to the expression on the right-hand side of Equation 3. There is clearly some correlation between the fates of the two genes in a single individual, so that Q* > Q2; it is easy to see that Q* = Q2 does not satisfy the recursion. However, this correlation is surprisingly weak: numerical calculations show that Q* is close to Q2.
The variance in the number of copies left by ancestor j after t generations, Vt,j, is found by differentiating the generating function. Writing Q = E[ewn], the variance of n is
We know that for a neutral allele, the expected number of copies descended from a single ancestral copy is for all t; similarly, the expected number of copies produced by two copies in a homozygote is for all t. This leads to the recursion
| (A1) |
and
| (A2) |
The pedigree is represented by a series of matrices, Mt, that give the relationship between successive generations. Quantities such as the probability of survival, the distribution of numbers of gene copies, and the identity by descent can be calculated most efficiently in Mathematica by representing the recursions in matrix form.
Since 2M has entries with only value 0, 1, or 2, we can see that 4(1 − Mij)Mij is 1 iff 2Mij = 1 and Mij(2Mij − 1) is 1 iff Mij = 1. So to get efficient solutions, we rewrite the recursion as
| (A3) |
Here, logeV and exp(V) are threaded over each element of the vector V; V1V2 indicates element-by-element multiplication of two vectors and V1.V2 indicates a dot product of two vectors.
Appendix B
The Distribution of Reproductive Value With Selection
We consider the infinitesimal model in which the log fitness of each individual is assumed to be an additive trait, z, which is normally distributed with variance fixed at V. In the reproductive step, parents are chosen at random from the parental population. The probability that an individual with log fitness z is chosen as parent is proportional to ez. An offspring has normally distributed log fitness, with mean the average of the log fitness of its parents and variance V/2. Since by Fisher’s fundamental theorem the mean log fitness will advance by V in each generation, the mean log fitness t generations in the past is If population size is fixed, then the number of offspring of an individual of log fitness z alive t generations back will be approximately Poisson with parameter The generating function for the distribution of reproductive value is a function of the log fitness, gt[y, z] ≜ E[yv]. Extending Equation 5, we have
| (B1) |
Here is the generating function for the reproductive value of a single offspring of a parent with log fitness z in generation t. To identify the kernel ϕ, first note that the mean log fitness of the individuals chosen as parents from the tth generation back is Thus, the mean log fitness of an offspring of a parent with log fitness z in generation t is The variance of the log fitness of the offspring is the sum of contributions from within-family segregation (V/2) and from the random value of the mate (V/4). Thus the kernel ϕ is normal, with mean and variance 3V/4. The full recursion is of a complicated two-dimensional function. However, we can find recursions for the moments by differentiating gt[y, z]. The mean reproductive value, Mt(z), of an ancestor of log fitness z alive t generations in the past is given by
| (B2) |
which has the solution Similarly, the variance in reproductive value, is given by
Applying this to Equation B1 gives the recursion:
| (B3) |
This tends rapidly toward the steady-state value:
| (B4) |
The overall variance in reproductive value (obtained by averaging over z) is just e4V, as can be seen by integrating Equation B3 over the distribution of z.
Appendix C
Mathematical Notation for Asymptotic Behavior
For ease of reference, we record here the standard notation:
Appendix D
Rate of Convergence of Reproductive Value
Recall that the pedigree is determined by the sequence of (random) N × N matrices {Mt}t≥0 in which the ith row of Mt specifies the parents of individual i in generation t before the present. If the parents are distinct, then there is a in the corresponding positions in the vector (Mi1, Mi2, … , MiN). If the individual was produced by selfing, there is a single 1 in the corresponding position. All other entries in the matrix are zero. To see that the reproductive value of an ancestor settles down very quickly, we consider, for s < t,
Decreasing s corresponds to adding more generations of descendants. The sum of the entries in the jth column of Mst is just the relative contribution of the jth ancestor after t − s generations. The (ij)th entry is the contribution made by ancestor j to the descendant labeled i in generation s.
In the simplest Wright–Fisher model, each individual chooses its parents independently at random from the previous generation. Writing mj(t) for the sum of the entries in the jth column of Mt, the vector (2m1(t), 2m2(t), … , 2mN(t)) is determined by class sizes formed by multinomial sampling with 2N trials and equal weights on classes labeled 1, 2, … , N. For more general offspring distributions, the vector (2m1(t), 2m2(t), … , 2mN (t)) is an exchangeable random vector with E[mj(t)] = 1. Our first claim is that as t − s increases, the matrix Mst rapidly settles down into a fixed form in which the entries in each column are constant. This is made more precise by the following Lemma.
Lemma D.1. Suppose that the jth column of the matrix Mst is given by the vector (v1j(s), v2j(s), … , vNj(s))T (where T means transpose) and write vj(s) for the sum of the entries. Conditioned on this vector,
| (D1) |
| (D2) |
Remark D.2. The first assertion tells us how quickly the reproductive value settles down. The second controls the variance in the contribution that the jth ancestor at time t makes to each descendant alive in generation s before the present.
In the special case of a diploid Wright–Fisher model,
and
which yields
| (D3) |
and
| (D4) |
Proof of Lemma D.1. For the first statement, note that
| (D5) |
where to get from the second to the third line we have interchanged the order of summation in the double sum and relabeled the indexes in the single sum. Now observe that, using exchangeability of the columns of the matrix M(s−1),
By exchangeability, var(mk(s − 1)) = var(mj(s − 1)) for all k = 1, … , N. Substituting into (D5) and using that
| (D6) |
completes the proof of (D1).
To prove (D2), we again exploit exchangeability of the columns of M(s−1). First note that
| (D7) |
| (D8) |
Now we use the same technique as before to rewrite
| (D9) |
Moreover,
| (D10) |
To see this, the first term corresponds to which requires that exactly one of the 2ml(s − 1) times that the lth individual in generation s was chosen as a parent, it was by the ith individual in generation s − 1. The second term corresponds to which requires that offspring i picked the lth individual as parent twice. Equation D9 now allows us to unify the terms in (D7) into a single sum. First use (D6) and then (D10), substitute, and simplify to arrive at (D2). ▪
The right-hand side of Equation D2 converges to zero exponentially fast as we decrease s. Equation D1 then guarantees convergence of vj(s). The limiting value vj is the reproductive value of ancestor j. To quantify the variability in the contribution of ancestor j to different descendants, we renormalize so that the expected contribution to each descendant is one and calculate the variance in this quantity across descendants. Using Equation D2 we see that
| (D11) |
and so the variance in contributions to the current population of ancestors alive t generations in the past decays like N2−t. Substituting in (D1) tells us that
| (D12) |
and hence the extremely rapid convergence to a constant reproductive value, independent of population size.
Appendix E
Probability of Survival and Distribution of Copy Numbers
In this section, for concreteness, we concentrate on the diploid Wright–Fisher model, but it should be clear that our arguments carry over to more general offspring distributions. If we take an allele from individual i in the population at time s, then the chance that it is inherited from individual j in generation t before the present is the (i, j) entry in the matrix Mst. To see this, one can think of the ancestral lineage of the allele as following a random walk through the pedigree. At each step it is equally likely to be derived from either parent. Conditional on the pedigree, the transition matrix of the walk between generation s and s + 1 (backward in time) is then precisely Ms.
Lemma E.1. If then the probability that an individual in generation t before the present contributes any genetic material to the present population is approximately vPN,t, where v is its reproductive value and PN,t is the probability of a survival until time t of a single neutral allele in a haploid population of size 2N.
Proof. To estimate the probability that a particular ancestor contributes at least one copy of one of its alleles to the current population, we trace the ancestral lineages of all current alleles simultaneously. In the diploid Wright–Fisher model, these are described by a system of coalescing random walks that can be described as follows. We start a walk off from each of the 2N alleles in the population in the present. The two alleles in a given individual can be traced one to each of two parents chosen (independently and uniformly) at random from the previous generation. Given the parent, an allele is equally likely to be descended from each of the two alleles carried by that parent. If two walks choose the same allele in the same parent, then they coalesce. Our aim then is to estimate the probability that one of the walks alive at time t is in the jth individual. Note that the system of coalescing walks is precisely that describing the genealogy of a (haploid) Wright–Fisher model of size 2N.
Write U(s) for the number of walks alive at time s before the present. Since we are starting at time zero from the whole population, this number cannot be simply deduced from Kingman’s coalescent. However, using Möhle (2004) and Fu (2006), we see that U(s) is dominated, at least in expectation, by the corresponding number for the Kingman coalescent. Moreover, since once the exact coalescent for the Wright–Fisher model will only rarely experience multiple coalescences, the Kingman coalescent becomes an increasingly good approximation for U(s) as s increases.
Now note that the expected time for the Kingman coalescent (with coalescence rate 1/2N per pair of lineages) to decrease from 2N to n lineages has mean
and variance
for some constant C. Thus, for with high probability we have that and then the total coalescence rate after time s is ≪1/logeN. In particular, if t − s is (logeN) generations, then between times s and t, there will be no coalescence between the ancestral lineages that make up U(s). Since the only dependence between ancestral lineages arises when they coalesce, this implies that the error that we make by assuming that they evolve as independent random walks through the pedigree between times s and t will be negligible.
Without loss of generality we may suppose that the individuals in the population at time s before the present that carry the U(s) lineages ancestral to the present-day population are labeled 1, 2, … , U(s) and in our previous notation we write v1j(s), v2j(s), … , vU(s)j(s) for the first U(s) entries in the jth column of Mst. Then the probability that at least one lineage traces back to the jth individual is approximately that for independent random walks,
and using Lemma D.1 we see that if t − s = (logeN), then up to an error of order U(s)/N this is just vjU(s)/N, where vj is the reproductive value of the jth individual. Now observe that E[U(s)] = NPN,t−s, which, since t − s = (logeN) (whereas is ∼NPN,t. Finally then, averaging over the distribution of U(s), we obtain the desired result. ▪
Remark E.2. An improved approximation follows by observing that
We know that if we average over the distribution of vj, we should recover the survival probability of a single neutral allele and so we choose in such a way that
and approximate the survival probability of an allele in an individual with reproductive value v by
Lemma E.3. For the distribution of the number of alleles in the current population that are descended from an allele in individual j in generation t before the present, conditional on being nonzero is independent of the reproductive value vj. Moreover, it is approximately exponentially distributed with parameter
Proof. The first statement follows easily from our argument above. In our previous notation, conditional on at least one of the U(s) ancestral lineages being descended from ancestor j, the probability that at least two are descended from j is approximately
since vjU(s)/N is small. Thus, conditional on there being any genetic material derived from ancestor j in the current population, with high probability it is all descended from a single ancestral lineage from U(s). Since the population is neutral, the distribution of this number is independent of reproductive value.
For the second claim, since the population is neutral, until it becomes common in the population, the number of copies of a neutral allele conditioned on survival is close to that of a critical branching process conditioned on survival. Since as we argued above, conditional on survival all copies of the allele are with high probability descended from a single lineage from U(s) and we are considering times t ∼ s ≪ N, we are in precisely this regime: the growth of the neutral allele conditioned on being nonzero is approximately that of a critical Galton–Watson branching process with Poisson offspring distribution conditioned on nonextinction. The result then follows from Fisher (1930).▪
Remark E.4. For more general offspring distributions, the appropriate modification of Lemma E.3 follows from Theorem 10.1, Chap. 1 of Harris (1963). We then have that, conditional on survival, the expected proportion of the population descended from the allele is approximately
where Z1 is the number of copies of the allele in the first generation (before conditioning).
Appendix F
Structure
In this section we show that the rapid convergence of an individual’s reproductive value proved in Appendix D for a panmictic population is not greatly slowed by two simple forms of structure: partial selfing and subdivision.
Partial selfing:
Consider a population in which a fraction α of offspring in the population is produced by self-fertilization and the remaining 1 − α by random mating.
Lemma F.1. In the notation of Appendix D, under partial selfing
| (F1) |
and
| (F2) |
In particular,
Proof. Still using the notation of Appendix D, (2m1, … , 2mN) will once again be an exchangeable random vector. It is tedious but not difficult to check that
Substituting in our previous proof yields the result.▪
Apart from the initial behavior, Nordborg and Donnelly (1997) and Möhle (1996) show that for a population with partial selfing, under the Wright–Fisher model, the Kingman coalescent remains a valid model for the genealogy of a “small” sample, but the rate of coalescence is increased by a factor 2/(2 − α). For the exact coalescent for the Wright–Fisher model too, the effect of selfing will be to increase the rate of coalescence and so, using the notation of the proof of Lemma E.1, for with high probability and we can approximate the coalescent after that time by the (time-changed) Kingman coalescent. The proof of Lemma E.1 will then carry over to this setting.
The island model:
In this subsection we consider an island model in which the population is subdivided into D demes, each with N0 occupants. Mathematically, it is convenient to separate the steps of reproduction and migration. Thus in a reproductive step, each deme (separately) undergoes the diploid Wright–Fisher reproduction that we have seen above. Between reproductive steps a number of migration steps take place in which two demes are chosen at random and an individual from deme i is exchanged with one in deme j.
Again we trace the matrix Mst whose (i, j)th entry records the probability that a gene in individual i at time s is derived from one in individual j at time t in the past. It is convenient to label individuals so that labels 1, … , N0 lie in the first deme, N0 + 1, … , 2N0 lie in the second, and so on. In place of the matrix Mt we now have two sorts of matrix. The first, corresponding to reproduction, is block diagonal, with each block a copy of the Mt corresponding to the diploid Wright–Fisher model for a population of size N0. Premultiplication by the second type of matrix corresponds to exchanging two randomly chosen rows of Mst.
We examine the rate of decay of the variance of the entries in the first column to obtain the analog of Equation D4. We denote the entries of the first column of Mst by mij, where 1 ≤ i ≤ D refers to the number of the deme and 1 ≤ j ≤ N0 to the number of the individual within that deme. Write
We now write the variance of the entries in the first column of Est as
Now note that we can rewrite the second term as
The variance of the entries in the first column of our matrix then becomes
Let us write var1(s) and var2(s) for these two terms in the variance of the first column in Mst. In a reproduction event, by (D4), the term var1 is reduced by a factor The term var2 on the other hand can increase. Let us write Then by Equation D3,
(independently for each i) and E[εi] = 0. Thus E[var2(s − 1) − var2(s)] becomes
In a migration step involving the interchange of just two columns, the overall variance cannot change (we are merely shuffling the entries in the column, not changing them), but the expected value of the change in the second term is easily checked to be
Combining these, if a proportion m of offspring migrates immediately after each reproduction step, the change in variance over a whole cycle of reproduction and migration is
From this we see that
The first part of the variance is reduced by a factor of 2 in each cycle and, once this has been repeated often enough that var2(s) > var1(s), mass from var2(s) is transfered to var1(s − 1) so that it, in turn, can be reduced.
Appendix G
Introducing Recombination
Suppose now that we are interested in the descent of blocks of genome through our pedigree. For convenience we normalize so that the genome is represented by the unit interval [0, 1]. Each individual carries two chromosomes and for each chromosome a random subset U ⊆ [0, 1] is inherited from one parent with the complement being inherited from the other parent. As before, each individual chooses its two parents, independently at random from the previous generation. Note that our original model can be considered as a special case of this in which U is the empty set or the whole of [0, 1] with equal probability.
Now, conditional on a pedigree, instead of considering the matrices Ms, in which in the ith row there is at the position of each parent of the ith individual from generation s, we put 1U and in the two positions. Taking the products of these matrices as before, we see that each entry in the matrix M0t will be an indicator function (where 0 is the indicator function of the empty set). Each row of the matrix will sum to 1[0, 1], corresponding to the fact that the entries correspond to indicators of disjoint sets whose union is [0, 1]. The (i, j) entry of M0t is the indicator function of the portion of a chromosome chosen from the ith individual that is inherited from the jth individual from generation t before the present.
This, in principle, provides a route to the analysis of the whole distribution of the sizes and positions on the genome of the blocks of ancestral material inherited from the different ancestors in the population, just by superposing the descent of blocks of genome on the pedigree.
To prove a concrete result we specialize to the case of a single crossover, which we take to be uniformly distributed on [0, 1], in each individual in each generation. We consider the fate of a block of length y carried by a single ancestor in generation t before the present. For simplicity we do not specify the location of surviving blocks, but instead restrict our attention to the numbers of blocks of different sizes. Until blocks become common, with high probability, each diploid individual in the population carries blocks on at most one chromosome. Thus, if a parent carries a block of length z, then each offspring, independently, will carry a block of length z with probability (1 − z)/2, none of the block with probability (1 − z)/2, and with probability z a block of random length, uniformly distributed on (0, z).
Baird et al. (2003, Appendix B), establish the block size distribution in an infinite population. The first moments are obtained by multiplying the probability that a block of size z is passed down a single line of descent through the pedigree by the expected number of lines of descent. In particular, the total expected number of blocks in the population after t generations is shown to be 1 + yt. The number of lines of descent through the pedigree depends linearly on reproductive value and so conditioning on the pedigree, the expected number of blocks will be (1 + yt)v, where v is the reproductive value of the ancestor. The chance that some blocks survive until time t decays only logarithmically in t, and so even conditioned on survival, the total number of blocks in the population does not grow too quickly. In particular, since the number of blocks to survive in an infinite population is an upper bound for the number that survive in a finite population, over the intermediate timescales considered here, for moderate populations we may use the approximation that two different blocks never “meet” in the same diploid individual. Blocks of ancestral material are then simply eroded as they pass down the pedigree.
The block size distribution can be investigated through its moment densities. The first moment density, M1(t, y, dz) is defined through
Evidently M1(t, y, dz) will vanish for z > y. It will have a point mass at z = y, corresponding to the (strictly positive) probability that the whole ancestral block survives, and it will have a density for z < y. The second-order moment densities are defined in an analogous way,
and we use the notation M2[t, y, z1, z2] for the corresponding density (which has singularities at z1 = y and z2 = y), and so on for moments of all orders.Lemma G.1. The first and second moment densities of the distribution of numbers and sizes of blocks of genome descended from a given ancestor depend (approximately) linearly on the reproductive value of that ancestor.Proof. For the first moment density this is obvious. The expected number of blocks with lengths in the infinitesimal interval [z, z + dz) is the expected number of lines of descent through the pedigree times the probability, P1(t, y, dz) that each of them carries a block with length in [z, z + dz). Since the erosion of the ancestral block along a given lineage is independent of the pedigree, and the number of paths through the pedigree is proportional to the reproductive value of the ancestor, the result follows.
To calculate the second-order moment densities, we also superimpose the erosion of blocks by recombination on the pedigree. To do this we write Eτ for the expected number of pairs of genomes in the pedigree that share their most recent common ancestor at time t − τ. We now superimpose the erosion of the block of ancestral genome onto the pedigree. Suppose that a pair of genomes in the pedigree at time t had their most recent common ancestor (MRCA) at time t − τ. Conditional on knowing the length of the ancestral block carried by their MRCA, the recombination events down their two lines of descent between times t − τ and 0 are independent, and so, summing over all pairs of genomes in the pedigree at time t, we obtain for z1 ≠ z2
| (G1) |
For z1 = z2 we obtain half this quantity. (This formula holds regardless of the process that generates the pedigree.) Note that the expression under the integral is independent of the pedigree. Our claim will follow if Eτ is linear in v. This will not be true for very small values of t − τ, but once t − τ is large enough that the reproductive value has converged, the number of descendants of each individual in the population at time t − τ is independent of the reproductive value of our ancestor. The number of pairs of genomes with their MRCA at time t − τ depends on the reproductive value of the ancestor only through the number of potential ancestors at time t − τ and this is linear in reproductive value. Once t is big enough that the contribution to the sum in (G1) of the terms corresponding to t − τ less than say 10 is negligible, we see that the second moment density too is essentially a linear function of the reproductive value. ▪
We could continue indefinitely in this way, finding expressions for kth-order moment densities in terms of lower-order ones by first counting the number of k-tuples of genomes in the pedigree at time t with the MRCA at time τ, but the expressions rapidly become very cumbersome and for higher-order moments we expect the convergence to linear dependence on reproductive value to be slower (and so to require a larger population size for our assumptions to be valid).
Literature Cited
- Baird S. J. E., Barton N. H., Etheridge A. M., 2003. The distribution of the surviving blocks of an ancestral genome. Theor. Popul. Biol. 64: 451–471 [DOI] [PubMed] [Google Scholar]
- Ballou J. D., 1997. Ancestral inbreeding only minimally affects inbreeding depression in mammalian populations. J. Hered. 88: 169–178 [DOI] [PubMed] [Google Scholar]
- Bulmer M. G., 1971. The effect of selection on genetic variability. Am. Nat. 105: 201–211 [Google Scholar]
- Cannings C., Thompson E. A., Skolnick M., 1978. Probability functions on complex pedigrees. Adv. Appl. Probab. 10: 26–61 [Google Scholar]
- Caswell H., 1982. Optimal life histories and the maximization of reproductive value: a general theorem for complex life cycles. Ecology 63: 1218–1222 [Google Scholar]
- Chang J. T., 1999. Recent common ancestors of all present-day individuals. Adv. Appl. Probab. 31: 1002–1026 [Google Scholar]
- Chapman N. H., Thompson E. A., 2003. A model for the length of tracts of identity by descent in finite random mating populations. Theor. Popul. Biol. 64: 141–150 [DOI] [PubMed] [Google Scholar]
- Charlesworth B., 1979. Evidence against Fisher’s theory of dominance. Nature 278: 848–849 [Google Scholar]
- Derrida B., Manrubia S. C., Zanette D. H., 1999. Statistical properties of genealogical trees. Phys. Rev. Lett. 82: 1987–1990 [Google Scholar]
- Derrida B., Manrubia S. C., Zanette D. H., 2000. On the genealogy of a population of biparental individuals. J. Theor. Biol. 203: 303–315 [DOI] [PubMed] [Google Scholar]
- Edwards A. W. F., 1968. Simulation studies of genealogies. Heredity 23: 628 [Google Scholar]
- Fisher R. A., 1918. The correlation between relatives on the supposition of Mendelian inheritance. Proc. R. Soc. Edinb. 52: 399–433 [Google Scholar]
- Fisher R. A., 1930. The Genetical Theory of Natural Selection. Oxford University Press, Oxford [Google Scholar]
- Fu Y.-X., 2006. Exact coalescent for the Wright-Fisher model. Theor. Popul. Biol. 69: 385–394 [DOI] [PubMed] [Google Scholar]
- Grafen A., 2006. A theory of Fisher’s reproductive value. J. Math. Biol. 53: 15–60 [DOI] [PubMed] [Google Scholar]
- Harris T. E., 1963. The Theory of Branching Processes. Springer-Verlag, Berlin [Google Scholar]
- Huff C. D., Witherspoon D. J., Simonson T. S., Xing J., Watkins W. S., et al. 2011. Maximum-likelihood estimation of recent shared ancestry (ERSA). Genome Res. 21(5): 768–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCluer J. W., Vandeberg J. L., Read B., Ryder O. A., 1986. Pedigree analysis by computer simulation. Zoo Biol. 5: 147–160 [Google Scholar]
- Matsen F. A., Evans S. N., 2008. To what extent does genealogical ancestry imply genetic ancestry? Theor. Popul. Biol. 74: 182–190 [DOI] [PubMed] [Google Scholar]
- Möhle M., 1996. Coalescent results for diploid population models and the coalescent with selfing. Technical Report 433. Department of Statistics, University of Chicago, Chicago [Google Scholar]
- Möhle M., 2004. The time back to the most recent common ancestor in exchangeable population models. Adv. Appl. Probab. 36: 78–97 [Google Scholar]
- Nordborg M., Donnelly P., 1997. The coalescent process with selfing. Genetics 146: 1185–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson A., 1961. Inbreeding in artificial selection programmes. Genet. Res. 2: 189–194 [DOI] [PubMed] [Google Scholar]
- Rohde D. L. T., Olson S., Chang J. T., 2004. Modelling the recent common ancestry of all living humans. Nature 431: 562–566 [DOI] [PubMed] [Google Scholar]
- Smith C. A. B., 1976. The use of matrices in calculating Mendelian probabilities. Ann. Hum. Genet. 40: 37–54 [DOI] [PubMed] [Google Scholar]
- Suwanlee A., Baumung R., Solkner J., Curik I., 2007. Evaluation of ancestral inbreeding coefficients: Ballou’s formula vs. gene dropping. Conserv. Genet. 8: 489–495 [Google Scholar]
- Thompson E. A., 1979a. Extinction probabilities and pedigree structure. Adv. Appl. Probab. 11: 12–13 [Google Scholar]
- Thompson E. A., 1979b. Genealogical structure and correlations in gene extinction. Theor. Popul. Biol. 16: 191–222 [DOI] [PubMed] [Google Scholar]
- Thompson E. A., Cannings C., Skolnick M. H., 1978. Ancestral inference I. The problem and the method. Ann. Hum. Genet. 42: 95–108 [DOI] [PubMed] [Google Scholar]
- Vindenes Y., Lee A. M., Engen S., Saether B. E., 2009. Fixation of slightly beneficial mutations: effects of life history. Evolution 64: 1063–1075 [DOI] [PubMed] [Google Scholar]
- Wolfram S., 1991. Mathematica. Addison–Wesley, New York [Google Scholar]
- Wright S., Dobzhansky T., Hovanitz W., 1942. Genetics of natural populations VII. The allelism of lethals in the third chromosome of Drosophila pseudoobscura. Genetics 27: 363–394 [DOI] [PMC free article] [PubMed] [Google Scholar]