Abstract
Sequencing errors and random sampling of nucleotide types among sequencing reads at heterozygous sites present challenges for accurate, unbiased inference of single-nucleotide polymorphism genotypes from high-throughput sequence data. Here, we develop a maximum-likelihood approach to estimate the frequency distribution of the number of alleles in a sample of individuals (the site frequency spectrum), using high-throughput sequence data. Our method assumes binomial sampling of nucleotide types in heterozygotes and random sequencing error. By simulations, we show that close to unbiased estimates of the site frequency spectrum can be obtained if the error rate per base read does not exceed the population nucleotide diversity. We also show that these estimates are reasonably robust if errors are nonrandom. We then apply the method to infer site frequency spectra for zerofold degenerate, fourfold degenerate, and intronic sites of protein-coding genes using the low coverage human sequence data produced by the 1000 Genomes Project phase-one pilot. By fitting a model to the inferred site frequency spectra that estimates parameters of the distribution of fitness effects of new mutations, we find evidence for significant natural selection operating on fourfold sites. We also find that a model with variable effects of mutations at synonymous sites fits the data significantly better than a model with equal mutational effects. Under the variable effects model, we infer that 11% of synonymous mutations are subject to strong purifying selection.
WHOLE-GENOME resequencing of multiple individuals promises to significantly enhance progress in population and quantitative genetics. In principle, it should be possible to estimate single-nucleotide polymorphism (SNP) frequencies throughout the genome in large population samples, leading to more detailed information concerning several important processes, including demographic history and the impact of natural selection on nucleotide variation. To this end, the distribution of allele frequencies in a sample of individuals (the site frequency spectrum, SFS) is of fundamental interest. Within the SFS, alleles segregating at low frequencies may represent recent mutations, and these are particularly informative about the distribution of fitness effects of new mutations (Keightley and Eyre-Walker 2010). Therefore, for many purposes, it is preferable to estimate the frequencies of SNPs on a site-by-site basis rather than by incorporating information from linked sites (e.g., Le and Durbin 2011). Current high-throughput sequence data typically consist of multiple short reads, of the order of 100 bases, sampled more or less randomly from the genome. Each site is typically sequenced several times—the number of times that a site is sequenced within an individual is known as its depth of coverage. Depth of coverage tends to be limited by a trade-off between the number of individuals sequenced and the sequencing effort per individual. Sequence reads are usually aligned to a reference genome by one of several software packages that have been written for this purpose.
In diploid species, two features of high-throughput sequence data cause difficulties for unambiguous assignment of SNP genotypes and for inference of the SFS. First, if a site is heterozygous, the number of reads of each nucleotide type is subject to sampling variation. One of the two nucleotides at a site may therefore be at a low frequency or may even be absent. Second, population genetic inference is susceptible to sequencing errors (Clark and Whittam 1992; Johnson and Slatkin 2008). Error rates in current high-throughput sequencing technologies are nontrivial: for example, even after filtering of low-quality data, empirical estimates of error rates per base sequenced by the Illumina high-throughput sequencing platform are ∼10−3 (Keightley et al. 2009; Ossowski et al. 2010). In a diploid, a true homozygote may therefore have reads containing a mixture of two or more nucleotides, which could be naively misclassified as a heterozygote. This problem is expected to be exacerbated if there is nonrandom error, such that nucleotide A is more likely to generate nucleotide B by error than nucleotide C or D. Errors can arise by sequencing error, if the wrong nucleotide is assigned at a site, or by mapping error, if a read is aligned to an incorrect location in the reference genome.
In this article, we develop an approach to estimate the SFS by maximum likelihood (ML), using high-throughput sequence data. We assume binomial sampling of nucleotide types at heterozygous sites and that errors affect bases uniformly. We assume that genotype frequencies at each site within the sample of individuals are in Hardy–Weinberg proportions and that sites are unlinked. Our approach builds on work of Lynch (2008, 2009) and Haubold et al. (2010), who have developed ML methods to estimate several fundamental population genetics parameters, including levels of nucleotide diversity and the allele frequency at a site. Similarly, Hellmann et al. (2008) and Liu et al. (2010) have developed composite-likelihood approaches to estimate population genetic parameters from shotgun sequence data of a population sample. We investigate the performance of our method in simulations and check its robustness to departures from random error.
We have applied our ML approach to low-coverage sequence data from 57 Yoruba individuals that were sequenced as part of the 1000 Genomes Project phase-one pilot (1000 Genomes Project Consortium 2010). We inferred SFSs for zerofold degenerate, fourfold degenerate, and intronic sites, and from these we infer levels of nucleotide diversity in each class of sites and use the method of Keightley and Eyre-Walker (2007) to estimate the distribution of fitness effects of new mutations. Consistent with previous studies (Eyre-Walker et al. 2006; Keightley and Eyre-Walker 2007; Boyko et al. 2008) we find evidence that most amino acid-changing mutations are subject to strong purifying selection and that a significant proportion of mutations are slightly deleterious. We also find significant evidence for selection operating on fourfold sites that varies in strength among sites.
Materials and Methods
ML method to infer the SFS
The data are assumed to consist of sequence reads at s nucleotide sites in a sample of n diploid individuals. The depth of sequencing coverage at site i in individual j is denoted dij. The first step of the analysis is to determine, for each site, the derived base (if the SFS is unfolded) or the majority base (if the SFS is folded) among the complete set of the reads in the n individuals. To simplify computations and for computational efficiency, the observed reads for an individual are then coded. Reads at a site that have a mixture of nucleotides are distinguished according to whether there is a singleton or a doubleton present or whether there are three kinds of base present. Table 1 shows the read codes and their definitions, along with an example of that read code for the case of dij = 6, under the assumption that the most frequent (major) base is A. Note that under this coding system, read codes at a site depend on read depth; for example, code 1 is not possible for d = 5. In cases where d > 6, mixtures of reads for which the least frequent (minor) base occurs three times or more are assigned code 1. Sites that have three kinds of nucleotide present are assigned read code 7. To simplify computations, sites that have four kinds of nucleotide (which must contain at least two errors according to our model assumptions) are also assigned read code 7.
Table 1. Read codes and their definitions and an example of each read code for a site sequenced at depth 6.
| Read code | Description | Example |
|---|---|---|
| 0 | All reads are of major type | AAAAAA |
| 1 | Reads consist of two types (excluding singletons/doubletons) | AAATTT |
| 2 | All reads are of minor type | TTTTTT |
| 3 | Singleton (of minor type) | AAAAAT |
| 4 | Doubleton (of minor type) | AAAATT |
| 5 | Doubleton (of major type) | AATTTT |
| 6 | Singleton (of major type) | ATTTTT |
| 7 | Reads comprised of >2 nucleotides | AAAACT |
Our inference procedure computes the likelihood of the data given the SFS, denoted φ(x). The SFS is a vector of frequencies, which are parameters of the model estimated by ML. Element x of the SFS is the probability of x derived or major alleles at a site among 2n alleles sampled from a population. Because , there are 2n d.f., and 2n frequencies need to be estimated for an unfolded SFS (n frequencies for a folded SFS). We also estimate a sequencing error rate parameter, ε.
Assumptions
Sequencing reads are assumed to be generated from nucleotide sites at which up to two alleles segregate. Genotype frequencies are assumed to be independent across sites, and within a site, frequencies are assumed to be in Hardy–Weinberg proportions across individuals. It is important to note that due to random sampling, the reads that we observe at a heterozygous site within an individual could consist of only one nucleotide type. Additionally, the reads observed at a homozygous site within an individual could contain two or more nucleotide types, if there are sequencing errors.
In our model we assume that sequencing errors occur with equal probabilities across sites and individuals. We assume that up to one and two errors can occur in the reads at a site generated from a single heterozygous and a homozygous individual, respectively. We allow more errors in homozygous individuals because the level of genetic variation is expected to be relatively low, such that the vast majority of individuals are homozygous at a given site. In our model, if we observe three different nucleotides among the reads at a site, then there must be at least one error.
Computation of likelihood
We assume independence across sites, so the overall likelihood (L) of the data is computed from the product across s sites,
| (1) |
where readsi is the vector of read codes observed for the n individuals at site i. The likelihood of readsi is the sum of the probabilities of observing readsi, enumerated over 2n + 1 numbers of derived alleles (for the unfolded SFS), weighted by each SFS element:
| (2) |
The probability of observing the vector of read codes readsi given that there are j derived alleles at the site is calculated as
| (3) |
where g is the number of unique combinations of genotypes that can be constructed for n diploid individuals, given that there are j derived alleles, and genotypesk is a vector defining one such combination. For example, nine unique genotype combinations can be constructed if two individuals are sequenced at a biallelic locus, and genotypes are coded 1 (homozygous wild type), 2 (heterozygote), and 3 (homozygous derived): {11}, {12}, {13}, {21}, {22}, {23}, {31}, {32}, and {33}. The term p(genotypesk|j) is the probability of one such genotype combination given that there are j derived alleles. This is computed from the number of ways in which genotypesk can be generated from j derived and 2n – j nonderived alleles divided by the total number of possible permutations of j derived and 2n – j nonderived alleles,
| (4) |
where h is the number of heterozygous individuals in the genotype combination. Note that j derived and 2n – j nonderived alleles can be arranged to form the same genotype 2h ways because each heterozygous individual doubles the number of possibilities. The probability of observing the vector of read codes readsi, given the genotype vector genotypesk is calculated from
| (5) |
where z is the number of read code combinations that can be generated from genotypesk, and cl is a candidate vector of read codes defining one such combination. The term δil(readsi, cl) takes the value 1 if the candidate vector of read codes cl matches the vector of observed read codes reads or takes the value 0 otherwise. The term p(cl|genotypesk) is the probability of the candidate read codes cl vector given that the true genotype vector is genotypesk. An example of the read codes that can be generated for the case of one individual sequenced at read depth ≥8 and their associated probabilities is provided in Table 2 for a folded SFS (see below). The term ex = Poisson(x, dε) is the Poisson probability of x errors, given an expected number of errors dε. The term by = bin(y, d, 0.5) is the binomial probability of observing y major type bases in a sample of d reads at a site, given that the site is heterozygous. For example, the probability of code 0 (i.e., all wild-type alleles) given that the individual is a wild-type homozygote (i.e., two wild-type alleles) is e0 + e2/(3d), which is the probability of no error (e0) plus the probability of two errors, the second of which reverts the first (e2/(3d)). To be a revertant, the second error can occur at only one of d reads and can be only one of three possible nucleotide changes. With more than one individual sequenced, p(yl|genotypesk) is the probability of a multi-individual genotype vector and is obtained by multiplying entries in Table 2 across individuals.
Table 2. Read codes and associated probabilities for depth of d ≥ 8 for one individual and the number of wild-type alleles specified for a folded SFS.
| No. of wild-type alleles in genotype | Read code | Probability |
|---|---|---|
| 2 or 0 | 0 | e0 + e2/(3d) |
| 1 | 0 | |
| 3 or 6 | e1 + 2e2/(3d) | |
| 4 or 5 | (d − 1)e2/(3d) | |
| 7 | 2(d − 1)e2/(3d) | |
| 1 | 0 | 2b0e0 + 2b1e1/(3d) |
| 1 | 2b2(d − 2)e1/(3d) + 2b3e0 + 2b3(d − 3)e1/(3d) + (1 – 2(b0 + b1 + b2 + b3))(e0 + e1/3) | |
| 3 or 6 | 2b0e1 + 2b1e0 + 4b1e1/(3d) + 4b2e1/(3d) | |
| 4 or 5 | 2b1(d − 1)e1/(3d) + 2b2e0 + 2b3e1/d | |
| 7 | 4b1(d − 1)e1/(3d) + 4b2e1/d + 4b3e1/3 + (1 – 2(b0 + b1 + b2 + b3))(2e1/3) |
The term ex = Poisson(x, dε) is the Poisson probability of x errors, given an expected number of errors dε. The term by = bin(y, d, 0.5) is the binomial probability of observing y major type bases in a sample of d reads at a site, given that the site is heterozygous.
Folding the site frequency spectrum
By analyzing the folded SFS, only the frequency of the most frequent allele (or the major allele) at a site in the sample of individuals is estimated. The folded SFS therefore contains n + 1 elements (rather than 2n + 1 as in the case of the unfolded SFS), so n SFS parameters and the error rate parameter need to be estimated. To infer the folded spectrum, both the data and the candidate genotype vectors are coded such that there are ≥n major alleles in the read combination. The probability of a read combination needs to be doubled if equivalent read codes can be generated by a given read combination.
Maximization of likelihood
We maximized log likelihood using the simplex algorithm (Nelder and Mead 1965). To explore the possibility of multiple maxima, we randomly sampled 10 independent sets of starting values for each data set analyzed. The maximum likelihood was assumed to be the highest likelihood found among these trials. We found that increasing the number of trials to 20 did not affect the outcome (results not shown).
Simulating the site frequency spectrum
To evaluate the ML inference procedure, we generated simulated SFSs for s sites at which 2n alleles have been sampled. Elements of the SFS were a function of the genetic diversity (θ) and a parameter, r, that specifies the shape of the SFS. More specifically, for values of r < 1, we assume that the density at each consecutive element of the SFS drops according to a geometric function ϕ(y) = ry−1x, where x = ϕ(1) is the density associated with SFS element 1. To solve for x, we start with the nucleotide diversity for a given s in a sequence of length L, which is defined by
| (6) |
where . The proportion of segregating sites in the sample is
| (7) |
and, therefore,
| (8) |
Generation of simulated data
For each site simulated, we sampled a number (z) of mutant alleles from the SFS with probability proportional to the density of each SFS element. A wild-type base (e.g., A) and a different mutant base (e.g., T, G, or C) were each sampled with uniform probability. The z mutant and 2n − z wild-type alleles were randomly allocated among the n diploid individuals to be simulated. This procedure generated the genotypes for the individuals sampled at each site. If the true genotype for an individual was homozygous mutant or homozygous wild type, d mutant or wild-type base reads were allocated to the site. If it was heterozygous, the number of mutant base reads was sampled from a binomial distribution for d trials with P = 0.5. The “true” reads at the site were then subjected to either uniform or nonuniform errors. In either case, we sampled a number of errors from a Poisson distribution parameter dε. To simulate uniform errors, base reads were randomly changed to a different base. To simulate nonuniform errors, we sampled base changes from an empirically determined matrix of error probabilities (supporting information, Table S1).
Human polymorphism data
We applied our ML inference procedure to sequence data from the 1000 Genomes Project phase-one pilot (1000 Genomes Project Consortium 2010). Specifically, we attempted to infer the SFS for three different classes of sites: zerofold and fourfold degenerate sites and intronic sites from protein-coding genes. To compile these data, we initially obtained a set of coordinates for all zerofold and fourfold sites and a random sample of 2.5% of all intronic sites from the whole genome (excluding the X and Y chromosomes and the mitochondrial genome). We included zerofold and fourfold sites from coding regions of genes where every overlapping exon was in the same reading frame and orientation. We defined intronic sites as any sites overlapping any annotated protein-coding transcript (using the genome annotation available from ftp.1000genomes.ebi.ac.uk), and, to exclude any site likely to be involved in splicing, we excluded sites within 30 bp of any annotated protein-coding exon. Because CpG dinucleotides are hypermutable and differ in frequency between these classes of sites, we restricted our analysis to sites defined as non-CpG prone (sites that are not preceded by a C or followed by a G, following Keightley et al. 2005). We obtained a total of 12,352,702, 1,811,238, and 18,648,553 zerofold, fourfold, and intronic non-CpG–prone sites, respectively.
We obtained “pileup” format files for each class of sites from the 57 Yoruba (YRI) individuals sequenced at low coverage as part of the 1000 Genomes Project phase-one pilot (1000 Genomes Project Consortium 2010). To analyze data that have a consistent error spectrum, we restricted our analysis to reads sequenced by the Illumina platform. Mean depth of coverage for these data ranges from ∼1.5 to 11 per individual (1000 Genomes Project Consortium 2010, figure S2). We trimmed any reads that had a base quality of <20 or a map quality of <20. We investigated the effect of using less stringent base and map quality cutoffs, and although the inferred error rate was higher, the results were qualitatively unaffected (data not shown). For each site, we then attempted to select 6, 8, or 10 individuals at random from those individuals that had five or more valid reads at that site after filtering reads on the basis of base and map quality scores. Sites were excluded in cases where there was an insufficient number of individuals with five valid reads. Although mean coverage was fairly low in most individuals, by using this method we were able to obtain data for a high proportion of sites of each site type (see Table S2). For all usable sites within each of the three different classes of sites, we randomly selected five reads from 6, 8, or 10 randomly selected individuals from those that had at least five reads and created a data file suitable for inputting into our SFS inference program.
Having inferred SFSs for each site class, we calculated two standard estimates of the level of diversity: nucleotide diversity (θπ, Tajima 1983) and Watterson’s θ (θW, Watterson 1975). We also estimated parameters of the distribution of homozygous fitness effects (DFE) of new mutations at zerofold and fourfold sites using the maximum-likelihood method of Keightley and Eyre-Walker (2007). This method infers the DFE assuming a model of equal fitness effects of new mutations or a gamma distribution of effects of new mutations, using the SFS of a set of sites assumed to be subject to selection together with the SFS of a set of sites at which mutations are assumed to be neutral. We also compared the fit of these models to a model with two classes of fitness effects and estimated the strength of selection operating on each class (s1 and s2) and the proportion of mutations in class 1 (p1). We assume that intronic sites are evolving neutrally, since it has previously been shown that intronic sites evolve only slightly slower than ancestral transposable elements (Eöry et al. 2010), which are themselves among the best candidates for neutrally evolving sites in the mammalian genome (Lunter et al. 2006). The method also simultaneously estimates the parameters of a simple demographic model, which is a step change in population size, from N1 to N2, at some time (t) in the past.
Results
Evaluation of the ML inference procedure by simulation
We focused on evaluating the SFS inference method for simulated data sets giving nucleotide diversity θ = 10−3. This is similar to silent-site nucleotide diversity in many human populations (Li and Sadler 1991) and should be a stringent test of the method, since the higher diversity in many other outbreeding species will reduce the relative contribution of error. We present results for simulations generated with the SFS shape parameter (r) set at 0.75, which implies that the density associated with consecutive elements of the SFS declines more steeply than typically seen for putatively neutral sites in real data sets. Again, this should present a stiffer test than many real data sets. We also examined the more extreme case of r = 0.5, but results are qualitatively similar (not shown). We simulated data sets with a large number of sites, i.e., 107, as would be generated for whole-genome sequencing of eukaryotes.
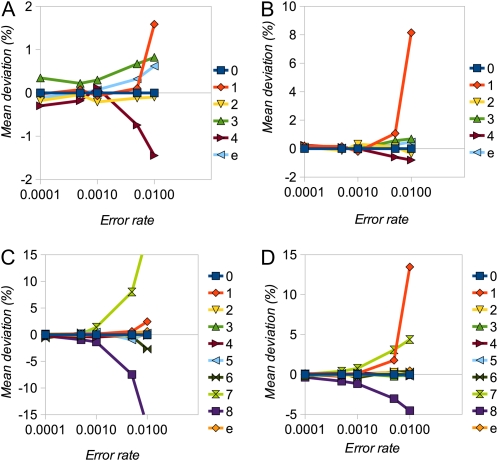
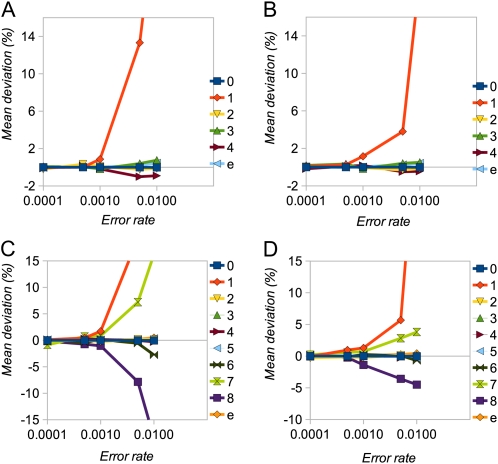
We assessed the performance of the inference procedure by estimating the mean deviations (as a percentage in 100 replicates) of each element of the estimated SFS and of the error parameter (ε) from their true values. Figure 1 shows percentage deviation statistics plotted against the true value of ε (ranging from 0.1θ to 10θ) for the case of random sequencing errors. We examined SFSs with 8 and 16 alleles and depths of coverage of 5 and 10. In all cases the procedure produces close to unbiased estimates of the SFS elements and ε, as long as the true error rate is not substantially larger than θ. As the true value of ε increases, estimates become increasingly biased. In particular, the frequency of the singleton class (one minor allele) becomes overestimated, presumably because it is not possible to distinguish multiple errors in one individual from heterozygotes. Bias tends to be more serious at a higher depth of coverage, presumably because there are more opportunities for error. With nonrandom errors (see Table S1 for parameterization), estimates remain close to unbiased, as long as ε ≤ θ, but bias becomes more serious for higher ε-values (Figure 2).
Figure 1 .
Simulation results for random errors. (A–D) Mean deviation (%) of SFS elements (labeled numerically in terms of number of minor alleles) and error rate (ε) for cases of n = 8 (A and B) and n = 16 (C and D) and d = 5 (A and C) and d = 10 (B and D). The shape parameter, r, of the simulated SFS was 0.75.
Figure 2 .
Simulation results for nonrandom errors. (A–D) Mean deviation (%) of SFS elements (labeled numerically in terms of number of minor alleles) and error rate (ε) for n = 8 (A and B) and n = 16 (C and D) and d = 5 (A and C) and d = 10 (B and D). The shape parameter, r, of the SFS was 0.75.
Human polymorphism data
We inferred the SFS and the error rate for three site classes (zerofold, fourfold, and intronic non-CpG–prone sites) in data sets generated by sampling 6, 8, or 10 individuals at each site from the low-coverage sequence data of Yoruba individuals produced as part of the 1000 Genomes Project phase-one pilot (1000 Genomes Project Consortium 2010). For the map- and base-quality filtering levels chosen, estimates of the error rate were relatively constant (ε ≈ 0.1%) across site classes and across different numbers of individuals sampled (Table S2). However, for data recompiled using lower-quality thresholds (by filtering base calls of base quality <10 and map quality <10), we inferred a higher error rate at all sites, as expected (Table S2).
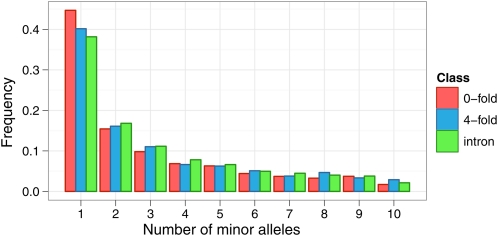
The inferred SFSs for zerofold, fourfold, and intronic sites are plotted in Figure 3. As expected, zerofold sites show a higher frequency of singletons than both fourfold and intronic sites, indicative of the action of purifying selection maintaining recently arisen deleterious mutations at low frequencies. Furthermore, fourfold degenerate sites also show a higher frequency of singletons than intronic sites, consistent with the presence of weak negative selection at synonymous sites in humans (Comeron 2006). We then calculated two standard measures of diversity, θπ and θW, from the inferred SFSs.
Figure 3 .
Inferred folded SFSs based on 20 alleles for zerofold, fourfold, and intronic non-CpG–prone sites. The SFSs were inferred using our ML approach applied to low-coverage whole-genome sequences for 57 Yoruba individuals sequenced as part of the 1000 Genomes Project phase one. Data suitable for analysis were generated by selecting five reads from each of 10 individuals chosen at random from sites that had at least 10 individuals with five or more reads after filtering reads with base quality <20 and map quality <40.
Zerofold sites have substantially lower diversity by both measures (θπ = 0.041%, θW = 0.045%) than both fourfold degenerate sites (θπ = 0.086%, θW = 0.092%) and intronic sites (θπ = 0.10%, θW = 0.11%). Furthermore, fourfold sites show lower diversity by both measures than intronic sites, a result that is also consistent with weak negative selection acting on fourfold sites.
We then estimated the strength of selection acting on zerofold and fourfold degenerate mutations, under the assumption that introns evolve neutrally, using a published method (Keightley and Eyre-Walker 2007). Introns have been shown to evolve at similar rates to ancestrally inserted transposable elements (Eöry et al. 2010), which themselves appear to be good candidates for neutrally evolving sequence (Lunter et al. 2006). Under a model with equal deleterious mutational effects, estimates of the strength of selection (Nes) acting on zerofold and fourfold mutations are 3.2 and 0.64, respectively (Table 3). For both zerofold and fourfold sites, a model that includes selection fits significantly better than one without (likelihood-ratio test, P < 0.001 in both cases). Interestingly, the estimated effect size for fourfold degenerate sites is remarkably consistent with the mean estimate produced by Comeron (2006, figure 4) of ∼0.5, using an independent approach based on the ratio of polymorphism to divergence of synonymous preferred and unpreferred mutations, also assuming that Nes is constant. A model with a gamma distribution of fitness effects fits significantly better than a model with equal effects for both zerofold and fourfold sites (likelihood-ratio test, P < 0.001 in both cases). Under this model, the estimated parameters imply that, for zerofold sites, the majority of mutations (53%) are strongly selected with Nes > 10 and 66% have an Nes > 1. On the other hand, only 24% evolve effectively neutrally (Nes < 0.1). Surprisingly, however, we find evidence for strong selection operating on a proportion of fourfold degenerate sites: our estimated parameter values suggest that 11% have an Nes > 10, and as many as 21% have an Nes > 1. For both zerofold and fourfold sites, fitting a model with two classes of effects gives higher log-likelihoods, although the increase is only marginal for fourfold sites. The Akaike information criterion (AIC), which penalizes models that have more parameters, suggests that the two-effects model is the best choice for zerofold sites and that the gamma distribution is (marginally) the best choice for fourfold sites (Table 3). The parameter estimates obtained under a two-effects model are in broad agreement with the results from a gamma distribution model. For zerofold sites we find that the majority (64%) of zerofold mutations are strongly deleterious (Nes = 34), whereas 36% have weak selection coefficients (Nes = 0.0013). For fourfold sites we infer that the majority of mutations (p1 = 0.82) are neutral (Nes1 = 0), while the remainder are strongly selected (Nes2 = 15).
Table 3. Demographic and strength of selection parameter estimates obtained by fitting models with a single selection coefficient fitted (equal-effects model), a two-selection–coefficient model (two-effects model), and a model that assumes a gamma distribution of mutational effects to the inferred SFSs for zerofold and fourfold degenerate sites.
| Class | Model | N2/N1 | t/N2 | b | Mean Nes | Nes1 | Nes2 | p1 | ΔLogL | ΔAIC |
|---|---|---|---|---|---|---|---|---|---|---|
| Zerofold | Equal effects | 1.58 | 0.40 | — | 2.4 | — | — | — | 860 | 1710 |
| Zerofold | Gamma | 2.31 | 0.52 | 0.14 | 490 | — | — | — | 12 | 22 |
| Zerofold | Two effects | 2.31 | 0.59 | — | 21 | 0.0013 | 33 | 0.36 | 0 | 0 |
| Fourfold | Equal effects | 2.31 | 0.57 | — | 0.62 | — | — | — | 9.8 | 16 |
| Fourfold | Gamma | 2.31 | 0.55 | 0.038 | 20 | — | — | — | 0.8 | 0 |
| Fourfold | Two effects | 2.31 | 0.56 | — | 2.8 | 0 | 15 | 0.82 | 0 | 0.4 |
We report estimates of the inferred change in population size (N2/N1, where N1 is the ancestral population size and N2 is the current population size), the inferred number of generations since the population size change in units of the current population size (t/N2), the inferred shape of the gamma distribution (b, where applicable), and the inferred mean scaled selection coefficient (Nes). For the two-effects model we also report the estimated scaled selection coefficient for each effects class (Nes1 and Nes2) and the proportion of mutations in the first class (p1). In the final columns we report the difference between the model’s log-likelihood and Akaike information criteria (AIC) from that of the best-fitting model for that data set (ΔLogL and ΔAIC, respectively).
Discussion
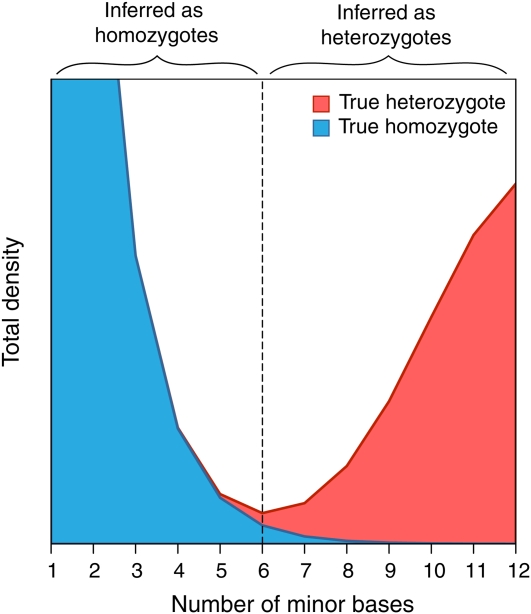
Genotype inference using high-throughput sequence data from diploids has two principal problems. First, due to sampling, reads from heterozygous sites could all come from a single allele, and therefore heterozygotes may be misclassified as homozygous. Second, reads from any site are subject to sequencing errors, so a true homozygous site could be misclassified as heterozygous. This problem is illustrated in Figure 4, which shows a cartoon example of the contribution of heterozygous and homozygous sites to the minor nucleotide frequency distribution, under the assumption of binomial sampling of the number of minor nucleotides in heterozygotes along with random error. A heuristic approach to distinguish homozygous from heterozygous sites could, for example, set a threshold above (below) which a site is classified as heterozygous (homozygous) at the point where incorrect calls of homozygotes as heterozygotes and vice versa are minimized. Ideally the total density at this point should be zero, but if the density is greater than zero then some proportion of both types of erroneous genotype calls will occur. The magnitude of this problem will increase with decreasing nucleotide diversity, because the number of base reads containing error increases relative to the frequency of heterozygotes. The problems associated with inferring the SFS from high-throughput sequence data therefore will tend to become more serious in species that have low nucleotide diversity (like humans) and are expected to be worse for sites that are under strong purifying selection, such as nonsynonymous sites of protein-coding genes and conserved noncoding elements.
Figure 4 .
Graphical representation of the frequency distribution of the number of minor class reads assuming binomial sampling of reads in heterozygotes and random sequencing error. The total density for each point has a contribution from two distributions: (1) binomial sampling of the number of minor class reads in heterozygotes (red) and (2) a geometric distribution of the number of minor class reads due to sequencing error (blue). Genotypes may be assigned by a simple heuristic approach where a minor allele frequency cutoff is chosen (represented as a dotted vertical line), above (below) which sites are inferred as being heterozygous (homozygous). Assigning genotypes becomes problematic when the two distributions substantially overlap, resulting in true homozygotes being called heterozygotes and vice versa.
Intuitively, we might expect that strong filtering on base and mapping quality should reduce the contribution of errors and allow more confident assignment of heterozygotes. We might also expect that by using sites with only high coverage, we should better distinguish between homozygotes subject to sequencing error and true heterozygotes. However, this is not necessarily the case for three reasons:
Reads in which a correct base call differs from the reference tend to have low mapping quality.
The subset of sites sequenced at the highest depth is more likely to have incorrectly mapped reads. Strong filtering on minimum depth of coverage therefore tends to increase the contribution from mapping errors.
Strong filtering on base quality reduces the number of usable base reads and therefore the number of usable sites, with the consequence that remaining sites come from the population of sites sequenced at high depth. The reads at these sites are more likely than average to be incorrectly mapped.
The interaction between these factors results in a trade-off between them. Less stringent filtering on base quality increases the contribution from sequencing errors, but too strong filtering removes most of the data, and the remaining sites are subject to an increased frequency of mismapped reads. Reduction of filtering on mapping quality includes more mismapped reads, but stronger filtering tends to remove reads showing genuine differences between the sequenced individual and the reference. Finally, reducing the minimum coverage threshold makes it harder to distinguish between the contribution from sequence errors and binomial sampling of heterozygotes, whereas considering only sites with a high depth of coverage increases the contribution from mismapped reads.
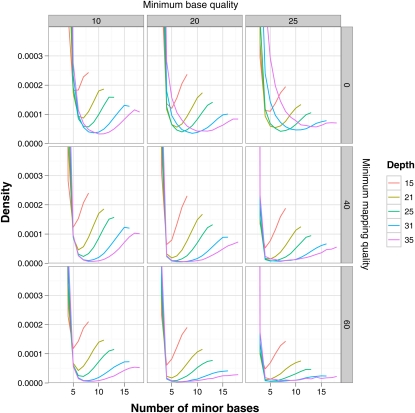
The interaction between these effects is illustrated in plots of the minor nucleotide frequency from a single individual sequenced at high coverage as part of the phase-two pilot of the 1000 Genomes Project (Figure 5). The data consist of sites from chromosome 1 from individual NA12891 (from population CEU of European ancestry), which has a mean depth of coverage of ∼27. We calculated the minor nucleotide frequency at each site after filtering the data on the basis of minimum base quality, minimum map quality, and minimum coverage and plotted the frequency distributions. As expected, in each plot there is a high frequency of sites where the minor nucleotide type is rare, presumably representing sequencing errors at homozygous sites, whereas there also appears to be a population of sites where the two nucleotides are present at nearly equal frequencies, presumably representing heterozygous sites. The distributions indicate that the best compromise for inference of an individual’s genotype from these particular data would be achieved by setting a minimum base quality of 20, a minimum mapping quality of 40, and a minimum coverage of 25. However, it is notable that the minima in all of the plotted distributions never reach zero, implying that errors will contribute to the inferred SFS, even if a cutoff for accepting/rejecting a site as heterozygous is set at this minimum. It is clear therefore that even with relatively high coverage, simple heuristic inference of genotypes will be error prone. Whereas the ultimate solution to the inference problem outlined above is increasing depth of coverage, problems will also be reduced by longer reads and paired-end reads, both of which will tend to reduce mapping errors.
Figure 5 .
Influence of minimum base quality, mapping quality, and read depth on the frequency distribution of the number of minor class nucleotides. The data are all sites from chromosome 1 from individual NA12891 (from population CEU) sequenced to high depth of coverage (mean ∼27×) as part of phase two of the 1000 Genomes Project (1000 Genomes Project Consortium 2010). Sequencing for these data was carried out by single (rather than paired-end) reads, using variable read lengths. In the presence of sequencing or mapping errors there can be more than two nucleotides at a site within an individual, and we therefore define the minor nucleotide as the second most frequent nucleotide; i.e., we assume that less common nucleotides are due to error. All plots are cropped at a maximum frequency of 0.0004.
Our ML approach to infer the SFS is best suited to low coverage (say up to a depth of 10) because higher coverage than this will tend to lead to multiple errors at sites within an individual, and we model only up to two errors in the case of sites that are truly heterozygous. However, the algorithm could be modified to allow higher numbers of errors at a cost of increased computing time. We have also assumed independent errors, but nonindependent errors would be difficult to model within the current framework. With high depth of coverage, problems of inference are expected to essentially disappear for the reasons we have stated above.
We applied our ML method to the low-coverage genome-wide data produced as part of the 1000 Genomes Project phase-one pilot (1000 Genomes Project Consortium 2010). We used our method to infer SFSs and from these obtained estimates for the nucleotide diversity (θπ) at zerofold and fourfold sites of 0.041% and 0.086%, respectively. The estimate for zerofold sites is somewhat higher than an estimate of ∼0.025% obtained for nonsynonymous sites by Torgerson et al. (2009) for a data set of African-Americans. This could be partly a consequence of sequencing error contributing to our estimates, since θπ < ε for zerofold sites. On the other hand, in the case of fourfold sites, θπ ≈ ε, so we would expect our estimates to be less biased. Indeed our estimate of 0.086% is close to the estimate of ∼0.08% obtained for synonymous sites by Torgerson et al. (2009). It should be noted, however, that these estimates are not directly comparable for two reasons: First, we analyze zerofold and fourfold degenerate sites whereas Torgerson et al. obtained estimates for nonsynonymous and synonymous sites, which both include contributions from twofold degenerate sites. Second, we obtained estimates for non-CpG–prone sites only, whereas Torgerson et al. obtained estimates using all sites. The inclusion of hypermutable CpG-prone sites will increase estimates of diversity. Indeed, if we estimate θπ for all fourfold sites (including CpG-prone sites), we obtain a higher estimate of θπ = 0.11%. Finally, Torgerson et al. obtained their estimates from a data set of African-Americans, which potentially has lower levels of diversity than the Yoruba individuals analyzed here.
We used the inferred SFSs at zerofold, fourfold, and intronic sites to infer the strength of selection operating on zerofold and fourfold mutations under the assumption that mutations at intronic sites are neutral. We find significant evidence for selection operating on both zerofold and fourfold sites. Furthermore, models that include variable selection coefficients fit significantly better than an equal-effects model for both zerofold and fourfold sites (likelihood-ratio test, P < 0.001 in all cases). Consistent with previous studies (Eyre-Walker et al. 2006; Keightley and Eyre-Walker 2007; Boyko et al. 2008) when we fit a gamma distribution to the DFE, we infer that the majority (54%) of mutations at zerofold degenerate sites are strongly deleterious, and only 24% are effectively neutral. Intriguingly, the same model applied to fourfold sites reveals that 11% of mutations are subject to strong purifying selection and that only 70% are effectively neutral. Possible causes of selection on synonymous sites have been reviewed elsewhere (Chamary et al. 2006). Further analysis may reveal whether likely mechanisms, including the presence of subsets of sites important for mRNA stability and to control the removal of introns, are implicated.
Acknowledgments
We thank Thanasis Kousathanas for help in analyzing the two-effects model and Adam Eyre-Walker, Rob Ness, and two reviewers for helpful comments. D.L.H. and P.D.K. acknowledge support from a Biotechnology and Biological Sciences Research Council grant.
Literature Cited
- Boyko A. R., Williamson S. H., Indap A. R., Degenhardt J. D., Hernandez R. D., et al. , 2008. Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet. 4: e1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamary J. V., Parmley J. L., Hurst L. D., 2006. Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat. Rev. Genet. 7: 98–108 [DOI] [PubMed] [Google Scholar]
- Clark A. G., Whittam T. S., 1992. Sequencing errors and molecular evolutionary analysis. Mol. Biol. Evol. 9: 744–752 [DOI] [PubMed] [Google Scholar]
- Comeron J. M., 2006. Weak selection and recent mutational changes influence polymorphic synonymous mutations in humans. Proc. Natl. Acad. Sci. USA 103: 6940–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eöry L., Halligan D. L., Keightley P. D., 2010. Distributions of selectively constrained sites and deleterious mutation rates in the hominid and murid genomes. Mol. Biol. Evol. 27: 177–192 [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A., Woolfit M., Phelps T., 2006. The distribution of fitness of new deleterious amino acid mutations in humans. Genetics 173: 891–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubold B., Pfaffelhuber P., Lynch M., 2010. mlRho a program for estimating the population mutation and recombination rates from shotgun-sequenced diploid genomes. Mol. Ecol. 19: 277–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann I., Mang Y., Gu Z., Li P., de la Vega F. M., et al. , 2008. Population genetic analysis of shotgun assemblies of genomic sequences from multiple individuals. Genome Res. 18: 1020–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P. L. F., Slatkin M., 2008. Accounting for bias from sequencing error in population genetic estimates. Mol. Biol. Evol. 25: 199–206 [DOI] [PubMed] [Google Scholar]
- Keightley P. D., Eyre-Walker A., 2007. Joint inference of the distribution of fitness effects of deleterious mutations and population demography based on nucleotide polymorphism frequencies. Genetics 177: 2251–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., Eyre-Walker A., 2010. What can we learn about the distribution of fitness effects of new mutations from DNA sequence data? Philos. Trans. R. Soc. B. 365: 1187–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., Kryukov G. V., Sunyaev S., Halligan D. L., Gaffney D. J., 2005. Evolutionary constraints in conserved nongenic sequences of mammals. Genome Res. 15: 1373–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., Trivedi U., Thomson M., Oliver F., Kumar S., et al. , 2009. Analysis of the genome sequences of three Drosophila melanogaster spontaneous mutation accumulation lines. Genome Res. 19: 1195–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le S. Q., Durbin R., 2011. SNP detection and genotyping from low-coverage sequencing data on multiple diploid samples. Genome Res. 21: 952–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.-H., Sadler L. A., 1991. Low nucleotide diversity in man. Genetics 129: 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Fu Y.-X., Maxwell T. J., Boerwinkle E., 2010. Estimating population genetic parameters and comparing model goodness-of-fit using DNA sequences with error. Genome Res. 20: 101–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunter G., Ponting C. P., Hein J., 2006. Genome-wide identification of human functional DNA using a neutral indel model. PLoS Comput. Biol. 2: e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., 2008. Estimation of nucleotide diversity, disequilibrium coefficients, and mutation rates from high-coverage genomes-sequencing projects. Mol. Biol. Evol. 25: 2421–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., 2009. Estimation of allele frequencies from high-coverage genome-sequencing projects. Genetics 182: 295–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelder J. A., Mead R., 1965. A simplex method for function minimization. Comput. J. 7: 308–313 [Google Scholar]
- 1000 Genomes Project Consortium, 2010. A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S., K., Schneeberger J. I., Lucas-Lledó N., Warthmann R. M., Clark, et al. , 2010. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327: 92–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F., 1983. Evolutionary relationship of DNA sequences in finite populations. Genetics 105: 437–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torgerson D. G., Boyko A. R., Hernandez R. D., Indap A., Hu X., et al. , 2009. Evolutionary processes acting on candidate cis-regulatory regions in humans inferred from patterns of polymorphism and divergence. PLoS Genet. 5: e1000592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson G. A., 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7: 256–276 [DOI] [PubMed] [Google Scholar]