Abstract
Regulation of alternative mRNA processing by ELAV (embryonic lethal abnormal visual system)/Hu proteins is mediated by binding to AU-rich elements of low complexity. Since such sequences diverge very rapidly during evolution, it has not been clear if ELAV regulation is maintained over extended phylogenetic distances. The transcription factor Erect wing (Ewg) is a major target of ELAV in Drosophila melanogaster and coordinates metabolic gene expression with regulation of synaptic plasticity. Here, we demonstrate evolutionary conservation of ELAV regulation of ewg despite massive degeneration of its binding site and of associated elements in the regulated intronic 3′-end processing site in distantly related Drosophila virilis. In this species, the RNA-binding part of ELAV protein is identical to D. melanogaster. ELAV expression as well as expression and regulation of ewg are also conserved. Using in vitro binding assays and in vivo transgene analysis, we demonstrate, however, that the ELAV-binding site of D. virilis is fully functional in regulating alternative splicing of ewg intron 6 in D. melanogaster. Known features of the ELAV-binding site, such as the requirement of multiple poly(U) motifs spread over an extended binding site of ∼150 nt and a higher affinity to the 3′ part of the binding site, are conserved. We further show that the 135-bp ELAV-binding site from D. melanogaster is sufficient for ELAV recruitment in vivo. Hence, our data suggest that ELAV/Hu protein-regulated alternative RNA processing is more conserved than anticipated from the alignment of degenerate low-complexity sequences.
ALTERNATIVE mRNA processing is major mechanism to generate molecular diversity and organismal complexity from the limited number of genes present in higher eukaryotes. Through alternative splicing and polyadenylation, more than one mRNA can be generated from a single gene that differs in the encoded protein and/or alters expression or localization of the encoded protein post-transcriptionally (Matlin et al. 2005; Soller 2006; Chen and Manley 2009; Licatalosi and Darnell 2010). In humans, alternative splicing and polyadenylation occur in 92–94% and in >50% of genes, respectively, and are particularly abundant in the brain (Licatalosi and Darnell 2006; Li et al. 2007; Wang et al. 2008; Neilson and Sandberg 2010). Our understanding of the regulation of alternative mRNA processing, however, is limited. Since RNA-binding regulators are generally well conserved, but noncoding parts of pre-mRNAs that harbor regulatory sequences for its processing diverge very rapidly during evolution, it is not clear if and how evolutionary conservation is maintained at the sequence level.
ELAV (embryonic lethal abnormal visual system)/Hu family proteins are prominent regulators of alternative mRNA processing in the brain and are widely used neuronal markers (Soller and White 2004; Hinman and Lou 2008). ELAV/Hu proteins are proto-type RNA-binding proteins that contain three RNA recognition motifs (RRMs). The RRMs of ELAV/Hu proteins, as in many other RRM-containing proteins, are highly conserved with identities of 52–82% between human Hu and Drosophila ELAV. Despite the high sequence conservation of ELAV/Hu family proteins, however, their number varies between different clades, suggesting either highly dynamic functions or redundancy among individual family members (Samson 2008). The founding member of this family of proteins, neuron-specific ELAV from Drosophila, has been shown to affect gene-specific alternative pre-mRNA processing of erect wing (ewg), neuroglian (nrg), and armadillo (arm) (Koushika et al. 2000). The neuronally expressed human homologs HuB-C and the ubiquitously expressed HuR were initially assigned cytoplasmic functions, but also regulate pre-mRNA processing (Antic et al. 1999; Kasashima et al. 1999; Brennan and Steitz 2001; Zhu et al. 2007, 2008). ELAV/Hu proteins preferentially bind to U-rich sequences that are abundant in introns and untranslated regions. Furthermore, ELAV/Hu proteins have been shown to multimerize, suggesting an important role of this feature in achieving target specificity in a complex cellular environment (Kasashima et al. 2002; Soller and White 2005; David et al. 2007; Toba and White 2008).
The ewg gene encoding a transcriptional regulator homologous to human NRF-1 is a major target of ELAV in Drosophila as ewg transgenes can rescue post-embryonic development and viability of elav mutants (Haussmann et al. 2008). ELAV is required for splicing of the last intron 6 of ewg that results in EWG protein expression (Soller et al. 2008). In ewg intron 6, ELAV binds distal of a poly(A) site and inhibits 3′-end processing in vitro and in vivo (Soller and White 2003). At this regulated poly(A) site, ELAV-binding requires a number of short poly(U) motifs spread over an extended binding site of ∼135 nt (Soller and White 2005). ELAV forms a defined dodecameric complex in vitro, and the importance of complex formation for ewg intron 6 splicing is indicated by the requirement of multiple poly(U) motifs. These poly(U) motifs, however, can be variably positioned in the ELAV-binding site as indicated by the presence of deletions in very closely related species. Also, introduction of spacer sequences minimally affects ELAV regulation of ewg intron 6 splicing. These features make it unlikely that target-specific binding depends upon the formation of a higher-order RNA structure encompassing the ELAV-binding site (Soller and White 2005; Soller et al. 2010).
The ewg gene integrates multiple signaling pathways (e.g., Notch, Wnt/wingless, TGF-β, and AP-1) in coordinating neuronal metabolism and synaptic plasticity (Haussmann et al. 2008; Haussmann and Soller 2010). Given the importance of ELAV-mediated regulation of ewg in Drosophila melanogaster, we were wondering if the mechanism of ELAV-dependent splicing of ewg intron 6 is evolutionarily conserved in the distantly related Drosophila virilis that separated ∼40–60 MYA, a phylogentic distance similar to mice and humans. Since the RNA-binding part of ELAV protein is identical in D. virilis ELAV (Yao and White 1991), we further anticipated gaining insights into the evolution of RNA-processing signals and the underlying mechanism governing gene-specific target RNA recognition by ELAV. Our analysis of ELAV and EWG expression shows evolutionary conservation in D. virilis since both ELAV and EWG proteins are restricted to neurons and ewg transcripts are broadly expressed as in D. melanogaster. Furthermore, we identified a functional poly(A) site in D. virilis at a similar position in the regulated intron 6 as in D. melanogaster and demonstrate binding of ELAV to the vicinity of this poly(A) site. By using reporter transgenes in D. melanogaster, we show that a 600-nt-long region containing the regulated poly(A) site from D. virilis provides full functionality and is regulated in an ELAV-dependent manner in neurons. Intriguingly, however, the ELAV-binding site in ewg intron 6 diverged such that it is not recognized with sequence alignment algorithms in a genomic context due to its low complexity. The ELAV-binding site in D. virilis ewg intron 6 extends over ∼150 nt, and as for the D. melanogaster sequence, multiple and spaced poly(U) motifs are required for binding and regulation of intron 6 splicing. Within this sequence the importance of the 3′ part for ELAV binding in vitro and splicing regulation in vivo is also conserved. In addition, our analysis demonstrates the flexibility of regulatory elements involved in 3′-end processing since the distance of the cleavage site relative to the poly(A) site recognition element (AAUAAA) is not conserved. Given the massive sequence degeneration of the ELAV-binding site and the redundancy of ELAV-binding motifs at a genomic scale, it has not been clear if the 135-bp sequence identified from D. melanogaster is sufficient to recruit ELAV for binding in the natural context of Drosophila neurons. Using a number of reporter transgenes, we demonstrate that the choice of the promoter has no role in the recruitment of ELAV to its binding site in ewg and that the 135-bp sequence from D. melanogaster is sufficient for ELAV recruitment in a heterologous context. Our data demonstrate evolutionary conservation of pre-mRNA processing by ELAV/Hu proteins that are mediated by degenerate low-complexity sequences.
Materials and Methods
Fly genetics and recombinant DNA technology
Fly breeding, genetics and P-element-mediated transformation and recombinant DNA technology were according to standard procedures as described (Soller and White 2005). For phiC31 transformation, tcgERv and tcgRm derivative constructs were injected into embryos of the following genotype: y1 w* M{vas-int.Dm}ZH-2A; PBac{y+-attP-3B}VK00002 and y1 w* M{vas-int.Dm}ZH-2A, M{3xP3-RFP.attP}ZH-64A with insertion sites at 28E and 64A, respectively. Insertion of tcgERv and tcgRm constructs yielded comparable rescue levels as obtained previously by P-element-mediated transformation. The RNA null allele of ewg, ewg∆, was generated by FLP/FRT-mediated recombination between two transposon insertion lines, PBac{WH}CG3777f05779 and P{XP}d08061 (Parks et al. 2004; Thibault et al. 2004).
Accession numbers of D. melanogaster and D. virilis genomic ewg sequences are AF135590 and HM746707, respectively. The D. virilis ELAV-binding site (with mutations and flanking sequences indicated by capital letters) was cloned with three oligonuclotides, leaving a 6- and a 5-nt 5′ overhang: CTAGAttctttgttgttgtgattttataatCtcaaCtCtccctttctGttGGtCtCAGAaaatactctatttaattg, gtcatatcatCtGctGtctCagcaggtcatcaatCtCgtaataggtctacaaatGtc, and cttgctCtGCtCtatttacctgatCtGtaagtaagtatgatCtatctGcatCtGCtCtCgactgtgtacaaGC into a modified pB SK+ cut with XbaI and NotI, adding the following vector sequence to the ewg substrate (GGGCGAATTGGGTACGCGATCCTCTAGA-ewg-GCGGCCGCCACCGCGGTGGAG) when transcribing the Ecl136II (Fermentas) linearized plasmid with T7 RNA polymerase. To add the mutant ELAV-binding site to the fly transfomation vector, the sequence up to exon J was cloned into the previous vector using the BsrGI and NotI sites with primers virF12 KpnIMfeI (agtGGTACCaattgttttagagcaaattttaattacgtgtaaacca) and ewg 6R3 SacII (CCGCcaCCGCGGTctatacatgcgatgactagatgg). This fragment was then PCR-amplified using primers ewg vir F16 (AAttctttgttgttgtgattttataatttcaattttccc) or ewg vir F16 (AAttctttgttgttgtgattttataatCtcaaCtCtccc) and ewg vir J R NheI (atcggtgtagctagcTTGCTCCATTATGATTGTGTCCTCGGCCT) and cloned into a pBS SK+ containing the NotI–SpeI fragment of the final construct with XmnI and NheI in a three-way ligation and then swapped into a modified pCaSpeR transformation vector where the attB site had been added at the end of the polylinker.
Primers to amplify promoters of endoA (1.73 kb) were gacctcgagacCTGTGCACTGATGCAGGCAATGCTG and ccagtGGCGCGCCTGCTGCTGTCTCTTCTGGTTTTCTTCC; of nwk (1.67 kb), gaCCTCGAGAAGTTCTGTTCGCTTTTGGCCAGTTC and ccagtGGCGCGCCttggggctttttctacgacaatcgggtcactc; and of Nrx-1 (2.41 kb), gacctcgagCACACGACGCCTTGTAAAGTGCACTTG and ccagtGGCGCGCCtcacacgcacggtggcactcgggcttac. Promoters were cloned with XhoI and AscI into a modified pBS KS+ containing the ewg fragment of the tcgERm construct and an attB site. Transformants were identified by rescue of the lethal ewgl1allele.
The elav-SP construct was made by three-way ligation of PCR fragments amplified with SP F3 Not (GGCTCGAGGCGGCCGCCGATTAGCTTGAATGTCGGTG) and SP R5 Bgl (CTAGATCTATATCTTAACATCTTCCACCCCAG), cut with NotI and BglII, and SP F5 BglNhe (GGAGATCTAGGCTAGCCTGGGGTGGAAGATGTTAAGATATGAATATTTGAGCTTAATATAAAATAAACCCAC) and M13rev, cut with BglII and SpeI from a vector containing the SP gene and an attB site and a modified pCaSpeR containing the elav promoter cut with NotI and SpeI. In this construct, BglII and NheI sites are introduced in the 3′ UTR of the sex-peptide gene. The ewg pA2 poly(A) site was PCR-amplified with primers F6i2 Bgl (CGGAGATCTGCCAAGTCAATTGCAAAAGAGGGAGAATGAAAAAGCAAC) and ewgPyextNhe (CCGCTAGCTTAAAAGAAAAGAACATAAAGTATAAAATTATAAGATAAAATGTATAATAGC) and cloned with BglII and NheI into elav-SP. RNA in situ hybridizations were done according to the Berkeley Drosophila Genome Project (BDGP) protocol or obtained from the BDGP web site (http://www.fruitfly.org).
Sequence alignments were done using ClustalW (DNAstar), CHAOS/DIALIGN (http://dialign.gobics.de) (Brudno et al. 2004), Lagan (http://lagan.stanford.edu) (Brudno et al. 2003), Vista (http://genome.lbl.gov/vista/index.shtml) (Mayor et al. 2000), the University of California at Santa Cruz (UCSC) genome browser (http://genome.ucsc.edu) (Rhead et al. 2010), and EvoPrinter (http://evoprinter.ninds.nih.gov) (Odenwald et al. 2005).
In vitro and in vivo binding of ELAV, RT-PCR, 3′ RACE, antibody stainings, and RNase protection
Production of recombinant proteins, 32P-labeled in vitro transcripts, and electrophoretic mobility shift assays (EMSAs) were done as described (Soller and White 2005). RNA extraction and RT-PCR for the analysis of ewg in D. melanogaster and D. virilis and ewg rescue constructs were done as described (Soller and White 2005). Polyacrylamide gels from the analysis of 32P-labeled PCR products were dried, exposed to phosphoimager screens (BioRad), and quantified with QuantityOne (BioRad). cDNAs from all ewg transcripts were amplified with primers 4F and 5R, eeF and eeR, 6F and 6R, or VSV-R (Koushika et al. 1999; Soller and White 2003). Primers for the amplification of elav were elavFhinge (CTAAGCTTGGGCAGCACCAGTAAGATCATCCAG) and elavBamR (GTGGGATCCTTGACAATCTTTACCG). D. virilis primers for exons 4 and 5 were vF4 (CAGGTGGATCCTAACAATCCGATC) and vR52 (CACGGTGCCATCACTATTCGTCTG) and for intron 6 were vF6 (ATATCCGGTCTCAGTGAGCAAT) and vR6 (GCCTGGCGAAACGGTAATGG). 3′ RACE of cDNAs from D. virilis was done with nested primers vF4 and vF6, vF13 (GTATGCATAAAATTGAATTGCCAAGTTCCTAAAACAC) and vF14 (GTCGCGTTCGTGCTTCAATCCAAAATG), and vF10 (CCCGCCCACATGCCATTGGAGCTAG) and vF11 (GGACACCATATTTAAAAGAATATCTAAATAG) using the return primer AUAP in two reactions on AP reverse-transcribed RNA as described (Soller and White 2003). Antibody stainings were done as described (Haussmann et al. 2008).
To analyze in vivo binding of ELAV, 14- to 18-hr-old embryos were first dechorionated and then fixed in heptane containing 5% formaldehyde (10 ml heptane, 1.75 ml 37% formaldehyde, and 1.3 ml PBS equilibrated for 30 min) for 10 min with vigorous shaking. Embryo extracts were then prepared in RIPA buffer (150 mM NaCl, 50 mM Tris–HCL, pH 7.5, 1% NP-40, 0.5% Na-deoxycholate, 0.05% SDS) in a 1-ml Dounce homogenizer. After 20–40 strokes with the tight pestil, 1 vol of immuno-precipitation (IP) buffer was added (150 mM NaCl, 50 mM Tris–HCL, pH 7.5, 0.05% NP-40). The extract was then sonified (Misonic XL2020) using a small tip in an Eppendorf vial three times for 20 sec (setting 3, ∼20% output) and cleared by centrifugation for 15 sec. IPs were done with the monoclonal anti-ELAV antibody 7D and protein A/G beads (SantaCruz) in IP buffer containing 7 mM CaCl2, 40 U of RNase inhibitor (Roche), 2 U of TurboDNase (Ambion), and 15% of extract for 2 hr at room temperature. After washing and Proteinase K digestion (0.5 mg/ml in 150 mM NaCl, 100 mM Tris–HCl, pH 7.5, 10 mM EDTA, 0.25% SDS) for 30 min at 37°, RNA was isolated by phenol/chloroform extraction and ethanol precipitation in the presence of glycogen. After DNase I treatment, the RNA was then reverse-transcribed with primer ewgPyext with Superscript II (Invitrogen) according to the manufacturer’s instructions and PCR-amplified (30 sec at 94°, 45 sec at 56°, and 45 sec at 72° for 40 cycles with 1 min initial denaturation and 4 min final extension). Primers used were SP F3 (GGCTCGAGGCGGCCGCCGATTAGCTTGAATGTCGGTG), F6i (CGCGGAGAAATGAGTTTACGAG), and GR3 (TTTATTTAGCATTTCAGTTTACAAAATGTACAAGC).
Results
ELAV expression is conserved in D. virilis
Comparison of ELAV from D. melanogaster and D. virilis revealed that the RNA-binding part consisting of the three RRMs is identical (Yao and White 1991). This part of the ELAV protein has previously been termed RBD60 and fully rescues the viability of lethal elav mutants, as does D. virilis ELAV (Yao and White 1991; Yao et al. 1993). RBD60 also binds ewg RNA in vitro with the same affinity as wild-type ELAV (Soller and White 2005). Consistent with its wide use as a neuronal marker, ELAV expression is also restricted to neurons in both D. melanogaster and D. virilis (Figure 1, A and B).
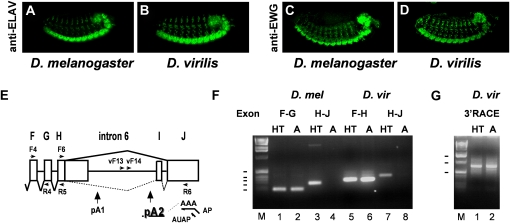
Figure 1 .
ELAV and EWG expression is conserved in D. virilis. (A–D) Expression of ELAV (A and B) and EWG (C and D) in D. melanogaster embryos (A and C) and D. virilis embryos (B and D). (E) Schematic of ewg intron 6 from D. melanogaster. ELAV-regulated splicing of intron 6 is shown as a solid line, and non-neuronal splicing of introns 6a and 6b is shown by a dashed line. The ELAV-regulated poly(A) site is underlined. Schematic position of primers used in both species for PCR or for 3′ RACE in D. virilis (vF13 and vF14) are indicated by arrows. (F and G) RT-PCR of ewg in D. melanogaster and D. virilis. Splicing of intron 6 is confined to the neuron-rich head/thorax (HT) and reduced in the neuron-poor abdomens (“A” in F and G) in both species as shown in F. ewg cDNAs from exons F and G in D. melanogaster, exons F and H in D. virilis, and exons H–J in both species were amplified with primers F4 and R4 or R5, and F6 and R6, respectively. 3′ RACE confirms a single intronic poly(A) site (pA) in intron 6 in D. virilis as shown in G. 3′ RACE of cDNAs in D. virilis was done with nested primers vF13 and vF14, and the return primer AUAP in two reactions on AP reverse-transcribed RNA. Molecular weight markers (M) are indicated on the left in F (100–500 bp) and G (0.5, 1, and 1.5 kb).
EWG expression is conserved in D. virilis
We next analyzed expression of EWG in D. virilis. EWG expression is also conserved and restricted to neurons (Figure 1, C and D). In D. melanogaster, ewg RNA is broadly expressed, but EWG protein expression is restricted to neurons by ELAV-regulated splicing of the last intron 6 (Figure 1E) (Soller et al. 2008). We therefore analyzed the expression of ewg RNA and splicing of intron 6 in neuron-rich heads and thorax (“HT” in Figure 1F) and compared it with the expression in neuron-poor abdomens (“A” in Figure 1F). Again, expression of ewg is conserved in D. virilis as the body of the RNA is expressed broadly (Figure 1F; compare lanes 1 and 2 with lanes 5 and 6), but splicing of intron 6 is restricted to neuron-rich tissue (Figure 1F; compare lanes 3 and 4 with lanes 7 and 8). We also noted that exon I is absent in D. virilis (Figure 1F). In D. melanogaster, exon I is prominently included in wing discs (Koushika et al. 2000), but exon I could not be detected in wing discs of D. virilis by the analysis of PCR products on agarose gels or by sequencing (data not shown).
D. virilis ELAV-binding site of ewg is fully functional despite massive degeneracy
Comparative analysis of intron 6 in D. melanogaster and D. virilis revealed a considerable size difference (1722 and 2420 nt, respectively). Although seven consensus AAUAAA poly(A) signals (nt 547, 722, 1404, 1605, 1884, 2071, and 2277 relative to the start of intron 6) are present in D. virilis compared to two in D. melanogaster, 3′ RACE and sequencing identified only a single 3′-end processing site in the 3′ part of intron 6 (nt 2071, termed virpA, Figure 1G and Figure 2A) compared to two 3′-end processing sites in D. melanogaster (pA1 and pA2, Figures 1E and 2A). Intriguingly, the distance of the cleavage site relative to the AAUAAA poly(A) signal was not conserved and increased considerably from 17 nt in D. melanogaster to 28 nt in D. virilis. No sequence conservation in D. virilis was found at the position of exon I in D. melanogaster (Figure 2A). Conserved features of ewg intron 6 are the extension of the ORF into intron 6 (172 aa in D. melanogaster and 241 aa in D. virilis from the start of intron 6), but without amino acid sequence conservation. In addition, an extensive stem loop structure at the end of the ORF terminating in intron 6 is conserved in D. virilis (nt 450–507 in D. melanogaster and nt 531–596 in D. virilis from the start of intron 6) (data not shown).
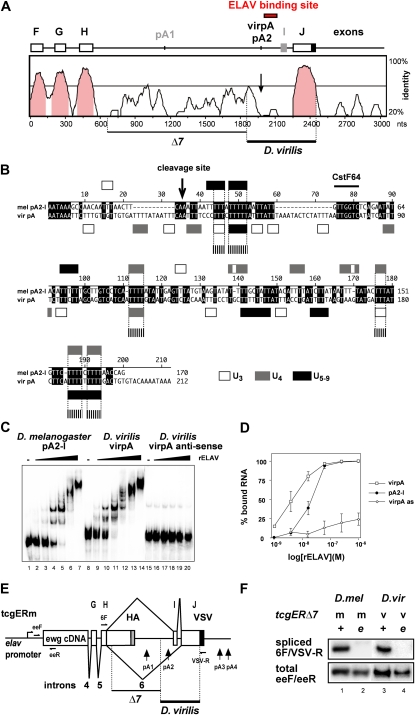
Figure 2 .
The ELAV-binding site in ewg intron 6 from D. virilis is functional in D. melanogaster, but not identified by sequence alignment. (A) Sequence alignment of the 3′ part of the ewg gene from D. melanogaster and D. virilis. A summarized exon/intron structure and the ELAV-binding site of both species are shown at the top and features absent in D. virilis are shown in gray. D. melanogaster pA2 corresponds to D. virilis virpA according to a similar position in intron 6. Sequence identity is indicated on the right and identity >50% is shown in pink. The deleted part in the tcgERm transgene not required for ELAV-dependent regulation (∆7) and the substituted part from D. virilis are indicated at the bottom. (B) Manual sequence alignment according to mapped features in the ewg 3′-end processing site. Poly(U) motifs are open (three Us), shaded (four Us), or solid (five to nine Us) boxes, and aligned poly(U) motifs between the two species longer than three nucleotides are indicated at the bottom with dashed boxes. (C). EMSA gel with RNAs from the ELAV-binding site of D. melanogaster (pA2-I) and D. virilis (virpA) and, as control, antisense RNA from D. virilis (virpAas) with recombinant ELAV from D. melanogaster. Uniformly 32P-labeled RNAs (100 pM) were incubated with recombinant ELAV (1, 3.9, 15.6, and 62.5 nM and 0.25 and 1 µM for pA2-I and virpA, and 3.9, 15.6, and 62.5 nM and 0.25 and 1 µM for virpAas) and separated on 4% native polyacrylamide gels. (D) Graphic representation of EMSA data from C as means with standard error from three experiments. The percentage of bound RNA (input RNA-unbound RNA/input RNA × 100) is plotted against the concentration of recombinant ELAV (in mol/Liter, M) presented as log. (E) Schematic of the tcgERm reporter construct with primers used in F depicted at the top. (F) Semiquantitative RT-PCR of intron 6 splicing using 32P-labeled forward primers from tcgER∆7m and tcgER∆7v transgenes in eye discs from wild type (+) and elavedr (e) that have reduced ELAV levels in photoreceptor neurons using primers 6F and VSV-R (cycle 26) compared to total expression levels of tcgER∆7m and v (primers eeF and eeR, cycle 28) analyzed on 8% polyacrylamide gels.
Next, we analyzed the genomic sequence of the 3′ part of the ewg gene of the two species by alignment with various algorithms (ClustalW, CHAOS/DIALIGN, Lagan, and Vista). As illustrated by the alignment and visualization with Vista, these algorithms did not detect the ELAV-binding site in the genomic context (Figure 2A). Recently, sequences from 12 Drosophila species became available (Clark et al. 2007). Reanalysis with multi-alignment tools such as the UCSC browser or EvoPrinter also did not identify the ELAV-binding site (data not shown). Motif-finder algorithms to identify degenerate U-rich motifs involved in ELAV binding are not applicable due to the abundance of these motifs in introns and UTRs. Manual curation of the ELAV-binding site in D. virilis on the basis of the functional elements involved in 3′-end processing (cleavage site and GU-rich CstF64-binding site) revealed ∼50% conservation, but with only marginal overlap in short poly(U) motifs (Figure 2B). Although ELAV binds to multiple motifs in the ewg-binding site, no repetitive elements aligned. Although some additional sequence conservation upstream of the ELAV-regulated poly(A) site is detected in intron 6, this part is not relevant for ELAV-dependent splicing regulation and can be deleted (Figure 2A) (Soller and White 2003).
To test if the sequence 3′ of the poly(A) site in D. virilis binds ELAV with high affinity, we employed in vitro binding assays using EMSAs and the D. melanogaster RNA pA2-I (encompassing the sequence from pA2 to exon I) and the D. virilis RNA virpA (encoding the sequence between the sixth and the seventh AAUAAA of intron 6). Both the pA2-I and the virpA substrate RNAs from D. melanogaster and D. virilis, respectively, bind recombinant ELAV, and a similar multimeric complex is formed (Figure 2C). We also noted that ELAV binds the virpA RNA from D. virilis with a higher affinity [Figure 2D; dissociation constant (Kd), 4.5 nM compared to 17 nM for pA2-I from D. melanogaster]. In addition, the slope for binding to pA2-I is higher, suggesting higher co-operativity in binding, which has been observed previously for binding at lower affinity (Soller and White 2003, 2005). Binding specificity is indicated by the very low affinity of ELAV to the A-rich antisense transcript of virpA, virpAas (Figure 2, C and D).
To test if ELAV regulation of intron 6 splicing is mediated by the divergent sequences from D. virilis, we used the tcgER∆7m reporter rescue construct, which expresses EWG under the control of the elav promoter and which includes the genomic part of the regulated intron (Figure 2E), but contains a deletion of sequences in intron 6 not required for ELAV-dependent regulation (∆7 in Figure 2, A and E) (Soller and White 2003, p. 2527). Since splicing generally dominates over 3′ processing, we reasoned that additional sequences in exon J are likely required to silence the 3′ splice site of exon J such that 3′ processing at pA2 is favored in the absence of ELAV. We therefore included additional sequences from D. virilis up to the end of exon J and replaced the D. melanogaster sequence with the D. virilis sequence in tcgER∆7m to generate tcgER∆7v. Transgenes with the tcgER∆7v construct were fully functional; e.g., the level of splicing was indistinguishable from the tcgER∆7m construct in the presence of ELAV in photoreceptor neurons and dramatically reduced when ELAV levels were reduced in the elavedr mutants (Figure 2F). tcgER∆7v transgenes also fully rescued viability of ewgl1 mutants (98%). Hence, the 600-nt sequence of D. virilis containing the ELAV-regulated poly(A) site provides fully functional regulation despite massive sequence degeneracy in the ELAV-binding site.
Multiple short poly(U) motifs in the D. virilis ELAV-binding site contribute to ELAV-dependent regulation of ewg in vivo
Poly(U) motifs important for ELAV binding in D. virilis ewg do not align with the D. melanogaster sequence (Figure 3A). We therefore wanted to test if multiple short poly(U) motifs are also required in the D. virilis ELAV-binding site for ELAV-dependent regulation. For these experiments the D. virilis ELAV-binding site was divided into three parts (v1–3, Figure 3A), which are analogous to our previous experiments with the D. melanogaster ELAV-binding site (m1–3) (Soller and White 2005, p. 7581). Mutations introduced were U-to-C substitutions, and since these were not very effective in disrupting ELAV binding in vitro to the pA2-I substrate, we also included U-to-G substitutions (see Materials and Methods for details). In addition, the sequence after the cleavage site in D. virilis does not contain a clear GU auxiliary element involved in 3′-end processing, and we therefore replaced this U-rich sequence with the GU auxiliary element from D. melanogaster to disturb this U-rich sequence after the cleavage site.
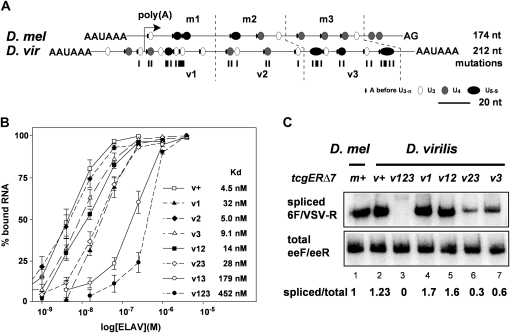
Figure 3 .
Requirement of multiple short poly(U) motives for ewg intron 6 splicing is conserved in D. virilis. (A) Schematic of the ELAV-binding site at pA2 from D. melanogaster and at virpA from D. virilis centered at the cleavage site. The ELAV-binding site is divided into three parts for each species: m1–3 for D. melanogaster and v1–3 for D. virilis. Poly(U) motifs are indicated as circles according to their length, starting with three Us, and A in front of the poly(U) motifs is indicated as a line. Mutations, mostly U-to-C substitutions, introduced into the D. virilis ELAV-binding site are indicated below as lines. (B) Graphic representation of EMSA data using recombinant D. melanogaster ELAV and virpA substrate RNAs containing mutations in the parts depicted in A as means with standard error from three to five experiments. EMSAs were done by incubating uniformly 32P-labeled RNAs (100 pM) with recombinant ELAV in five to six concentrations over the binding range (1, 3.9, 15.6, and 62.5 nM and 0.25, 1, and 4 µM) separated on 4% native polyacrylamide gels. The percentage of bound RNA (input RNA-unbound RNA/input RNA × 100) is plotted against the concentration of recombinant ELAV (in morgans) presented as log. (C) Semiquantitative RT-PCR of intron 6 splicing using 32P-labeled forward primers from tcgER∆7m and v transgenes containing mutated parts of the D. virilis ELAV-binding site as depicted in A in third instar larval brains using primers 6F and VSV-R (cycle 26) compared to total expression levels of tcgER∆7 (primers eeF and eeR, cycle 28) analyzed on 8% polyacrylamide gels. Quantification of three experiments is shown at the bottom.
In in vitro binding assays using EMSAs, mutations in one or two elements of the virpA substrate RNA had only a minimal effect on ELAV-binding affinity with the exception of mutations in v1 and v3, which greatly reduced in vitro binding (Figure 3B). The most dramatic decrease in ELAV binding was observed when mutations in all three poly(U) motif elements were combined (Figure 3B), which is analogous to the effect of mutations in the ELAV-binding site in D. melanogaster (Soller and White 2005).
Next, we introduced these mutations into the tcgER∆7v construct and generated transgenes with identical insertion sites by phiC31-mediated transformation to test for in vivo regulation of ewg intron 6 splicing. Mutations in all three elements, as in the tcgER∆7v123 transgene, reduced splicing dramatically, while mutations in v3 and v23 had weaker effects that increased with the number of mutations (Figure 3C), as had been observed for the cumulative effect of mutations in the ELAV-binding site in D. melanogaster (Soller and White 2005). Mutations in the v1 and v12 elements of the tcgER∆7v construct resulted in increased splicing, suggesting that these mutations in the v1 element, which overlap the cleavage site, weakened the strength of the 3′-end processing site (Figure 3C; compare lane 2 with lanes 4 and 5). Since the combination of mutations in the v1, v2, and v3 elements resulted in a dramatic decrease of intron 6 splicing, these effects on the strength of the 3′-end processing site were overruled (Figure 3C; compare lane 3 with lanes 4 and 5).
ELAV regulation of ewg intron 6 splicing is promoter-independent
Since the ELAV-binding site in D. virilis is massively degenerate compared to the analogous site in D. melanogaster, sequence-specific recruitment of ELAV to the regulated poly(A) site could be mediated by other mechanisms. Since ELAV autoregulates (Samson 1998) and since we had used the elav promoter in our tcgERm reporter constructs (Soller and White 2003), sequence-specific binding of ELAV could be achieved through recruitment at the promoter and deposition onto the nascent RNA. We had previously shown that, within the genomic part of tcgER∆7m, only a single ELAV-binding site is present and that ELAV regulation is not mediated via the 3′ UTR (Koushika et al. 2000; Soller and White 2003). Such recruitment of ELAV protein to target genes could explain the low sequence complexity needed for target-specific regulation.
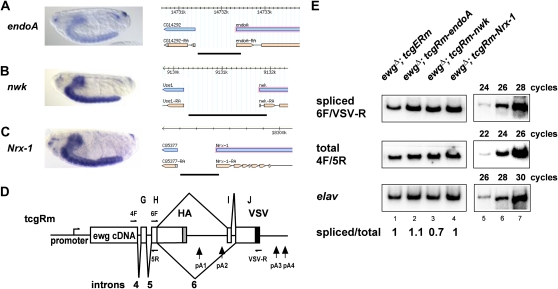
To test if promoters are important for ELAV gene-specific regulation, we searched for genes in FlyBase with the same expression pattern as ELAV and a simple gene structure. From the list of FlyBase genes we chose the endophilin A (endoA, CG14296), nervous wreck (nwk, CG4684), and Neurexin-1 (Nrx-1, CG7050) genes, which are not differentially regulated in ewgl1 and elav mutants (Haussmann et al. 2008) and thus are independent from ELAV regulation (Figure 4, A–C). From these three genes, we cloned the promoters into the tcgRm construct (Figure 4D) to generate tcgRm-endoA, tcgRm-nwk, and tcgRm-Nrx-1 constructs and established transgenic lines by phiC31-mediated transformation, which can be directly compared due to their exact same insertion site (Venken et al. 2006; Bischof et al. 2007).
Figure 4 .
ELAV-dependent regulation of ewg intron 6 is promoter-independent. (A–C) Expression of endoA, nwk, and Nrx-1 genes in embryos is confined to the ventral nerve cord as shown by RNA in situ. (Right) The promoter fragment used to drive expression of the tcgR construct is shown as a bold line at the bottom of the genomic organization. (D) Schematic of the tcgRm reporter construct with primers used in E depicted at the top. (E) Semiquantitative RT-PCR of intron 6 splicing using 32P-labeled forward primers from tcgERm compared to tcgRm-endoA, tcgRm-nwk, and tcgRm-NRX-1 transgenes in ewg∆, an RNA null allele using primers 6F and VSV-R (cycle 26). Expression levels of transgenes was determined with primers 4F and 5R (cycle 24) and expression levels of elav from primers elavFhinge/elavBamR (cycle 28) are shown as standard. The linear amplification range is indicated on the right for the control (lanes 5–7). PCR products were analyzed on 8% polyacrylamide gels. Quantification of three experiments is shown at the bottom.
It had come to our attention that transcripts of the oscar gene exert functions that are required for proper localization and expression of Oscar protein (Hachet and Ephrussi 2004; Jenny et al. 2006). To exclude that broadly expressed RNAs from the ewg gene are involved in processing of ewg intron 6, we generated an RNA null allele of ewg by deleting the genomic region, ewg∆. Viability and synaptic growth defects of ewg∆ mutants are fully rescued by ewg transgenes, and no difference of splicing of intron 6 is found in tcgERm transgenes compared to the protein null allele ewgl1 (data not shown).
Analysis of intron 6 splicing of transgenes from tcgRm-endoA, tcgRm-nwk, and tcgRm-Nrx-1 in ewg∆ revealed no difference of ewg intron 6 splicing compared to tcgERm transgenes (Figure 4E). In addition, transgenes from these constructs fully rescued viability of the lethal RNA null allele ewg∆ (88, 100, and 97%, respectively). Hence, regulation of ewg intron 6 is independent of the promoter and does not involve additional transcripts from the ewg gene, strongly arguing that the single ELAV-binding site after pA2 in D. melanogaster is sufficient for ELAV-dependent regulation.
The 135-bp ELAV-binding site of D. melanogaster is sufficient for ELAV recruitment in vivo
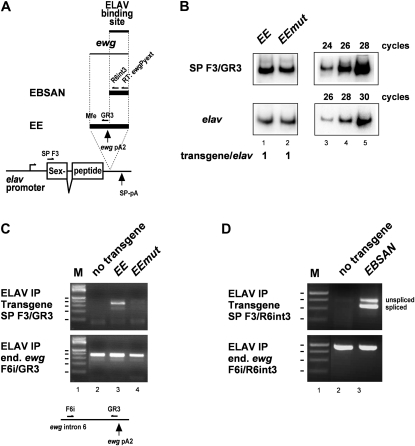
Although we had previously shown that ewg intron 6 regulation is mediated by a single ELAV-binding site, the body of the ewg gene could contribute to ELAV recruitment (Koushika et al. 2000; Soller and White 2003). To exclude this possibility and to identify the minimal element required for suppression of 3′-end processing at ewg pA2, we exchanged the ewg sequences with the gene coding for Sex-peptide (SP) that is not expressed in neurons (Saudan et al. 2002). In this construct, elav-SP, we placed the ELAV-regulated pA site sufficient for inhibition of cleavage in vitro (“EE,” Figure 5A), or a version with the mutated ELAV-binding site (“EEmut,” Figure 5B), or the ELAV-binding site pA-I (“EBSAN,” Figure 5A) into the 3′ UTR of this reporter (Soller and White 2003) and generated transgenes by phiC31-mediated transformation at the same insertion site. In D. melanogaster, ELAV does not bind to sequences beyond exon I (Soller and White 2003). Since the EEmut construct differs by mutations from the EE construct, we analyzed expression levels in these transgenes. Semiquantitative RT-PCR showed that transcript levels from the EEmut construct are not affected by the sequence variations present compared to transcript levels from the EE construct (Figure 5B).
Figure 5 .
ELAV binds in neurons to the minimal ELAV-dependent ewg pA2 processing site. (A) Schematic of the EE and EBSAN reporter constructs where the minimal ELAV-dependent ewg pA2 (EE) or the 135-bp ELAV-binding site (EBSAN) is included in the Sex-peptide gene expressed in neurons under the elav promoter. After reverse transcription with oligo(dT) (EE and EEmut) or ewgPyext (EBSAN), immunoprecipitated RNA was amplified with primers SP F3 and GR3 (EE and EEmut) or with primers SP F3 and R6int3 (EBSAN). ewg pA2 and SP-pA indicate the position of 3′-end processing sites. (B) Semiquantitative RT-PCR to determine transcript levels of EE and EEmut constructs using 32P-labeled forward primers from EE and EEmut transgenes with primers SP F3 and GR3 (cycle 26) compared to expression levels of elav (primers elavFhinge and elavBamR, cycle 28) analyzed on 8% polyacrylamide gels. The linear amplification range is indicated on the right for the control (lanes 3–5). Quantification of three experiments is shown at the bottom. (C) Amplification of ELAV-bound RNA after immunoprecipitation from embryonic extracts of transgenes indicated at the top with primers SP F3/GR3 for reporter-derived RNAs (Top) and with primers F6i/GR3 from endogenous ewg (Bottom). The schematic below the gels shows the position of primers around pA2. PCR products were separated on 3% agarose gels. Markers are shown in lane 1 with 100–500 bp indicated. (D) Amplification of ELAV-bound RNA after immunoprecipitation from embryonic extracts of the EBSAN transgene containing the pA2-I sequence from ewg with primers SP F3/R6int3 for reporter-derived RNAs (Top) and with primers F6i/R6int3 from endogenous ewg (Bottom). PCR products were separated on 3% agarose gels. Markers are shown in lane 1 with 200–500 bp indicated. Note that the pA2-I ELAV-binding site leads to increased amounts of unspliced RNA.
Immunoprecipitation of ELAV from embryonic extracts demonstrates in vivo binding of ELAV to RNA transcripts of the EE transgene, but not to the EEmut transgene with the mutated ELAV-binding site (Figure 5C). In vivo binding of ELAV could also be demonstrated when the shorter pA2-I sequence used in in vitro binding assays was inserted into the elav-SP transgene (“EBSAN,” Figure 5D). Intriguingly, in this context ELAV inhibited splicing of the upstream intron, suggesting that ELAV bound to RNA also interacts with spliceosomal components.
Discussion
ELAV/Hu family proteins are highly conserved like many other RNA-binding proteins. They bind gene-specifically to AU-rich sequences prominently found in introns and untranslated regions. A salient feature of such low-complexity sequences is their rapid divergence through evolution. Here we demonstrate that alternative splicing regulation of the ELAV target ewg is evolutionarily conserved in distantly related D. virilis despite massive degeneration of its binding site and of associated elements in the regulated intronic 3′-end processing site.
Although the ELAV-binding site from D. melanogaster is not recognized in D. virilis by sequence alignments in a genomic context, it is identified by functional analysis using transgenes. Furthermore, mutational analysis of the ELAV-binding site in D. virilis ewg revealed a number of conserved sequence features. First, the ELAV-binding site is in close distal proximity relative to the AAUAAA consensus sequence of the poly(A) site in ewg intron 6. Second, the ELAV-binding site in ewg extends over ∼150 nt, and both sequences have ∼50% U content. Third, multiple and spaced U-rich sequence motifs are important for in vitro binding and in vivo splicing regulation. Fourth, the 3′ part of the ELAV-binding site in ewg is more important for ewg splicing regulation in vivo and harbors a high U content. Therefore, the v3/m3 element might initiate ELAV complex assembly in vivo similar to sequences at the 3′ splice site of the HIV tat exon 3 involved in the hnRNP A1 complex assembly (Zhu et al. 2001; Okunola and Krainer 2009). Here, hnRNP A1 binds to a high-affinity site, and then the complex expands toward 5′ by recruiting additional hnRNP A1 proteins that bind neighboring sequences.
Potentially, more complicated scenarios of how ELAV comes into contact with its binding site in ewg could apply, e.g., recruitment at the promoter and deposition along the nascent transcript. Placing the ELAV-binding site of ewg in a heterologous reporter transgene clearly indicates that this sequence is sufficient for ELAV binding and that ELAV recognizes targets by diffusion. These results are in agreement with findings from the ELAV-regulated intron of nrg. Here, ectopic overexpression of ELAV in non-neuronal wing-disc cells is sufficient for neuronal splicing of the minimal ELAV-regulated intron of nrg overexpressed by a heterologous ubiquitin promoter (Toba and White 2008).
Given the degeneracy of the ELAV-binding site in ewg intron 6, differences therein and in its proximity also have evolved. Namely, the D. virilis site has an about fourfold higher affinity for binding ELAV. A likely contribution to this effect comes from two long U stretches of 8 and 9 nt in the 3′ part as deletion of this part in D. melanogaster results in a dramatic reduction of ELAV binding in EMSAs (Soller and White 2005). Also, the distance of the cleavage site to the highly conserved AAUAAA hexamer involved in poly(A) site recognition by the cleavage and polyadenylation specificity factor (CPSF) increased considerably (from 17 nt in D. melanogaster to 28 nt D. virilis). Furthermore, the distance of the AAUAAA hexamer to the 3′ splice site of intron 6 increased from 266 nt in D. melanogaster to 346 nt in D. virilis. In addition, the 5′ part of the D. virilis ELAV-binding site is more important for ELAV binding in vitro than is its complementary part in D. melanogaster (Soller and White 2005). Since this part is also directly involved in the recruitment of the cleavage stimulatory factor (CstF), the in vivo role of this element is not readily separable from affecting 3′-end processing efficiency as the mutations introduced weakened the poly(A) site. Interestingly, exon I is not present in D. virilis, and the sequence diverged completely.
To allow for evolutionary conservation in a degenerate sequence context, compensatory mechanisms must exist to allow for a large degree of flexibility in the positioning of short binding motifs. A prominent feature of ELAV/Hu proteins is multimerization upon binding to RNA (Kasashima et al. 2002; Soller and White 2005; David et al. 2007; Toba and White 2008). We demonstrated that individual mutations in the ELAV-binding site of ewg have little effect on RNA binding as well as on splicing regulation in vivo. Thus, ELAV multimerization can compensate for mutations in the binding site and allow it to diversify such that ELAV-binding sites can become refractory to detection by comparative genomics. This could also explain the divergence of the ELAV-binding sites found in other ELAV targets, e.g., in nrg and elav itself, which have U-rich motifs but do not align with the ELAV-binding site in ewg (Lisbin et al. 2001; Borgeson and Samson 2005). Since most long noncoding RNAs also show a low degree of evolutionary conservation by sequence alignment, combinatorial binding of RNA-binding proteins in a similar way as ELAV binds to ewg could be conserved (Prasanth and Spector 2007; Mattick 2009; Ponting et al. 2009).
Importance of multimerization has also been demonstrated for hnRNP A1 as well as for hnRNP F/H proteins in binding distantly localized binding sites resulting in looping out of the intervening sequence and alternative exclusion of the regulated exon (Martinez-Contreras et al. 2006). Multimerization is also involved in alternative splicing regulation of HIV-1 tat exon 3 (Zhu et al. 2001; Damgaard et al. 2002). A high-affinity hnRNP A1 binding site mediates recruitment of additional hnRNP A1 proteins, resulting in expansion of the complex preferentially in the 5′ direction and in antagonizing of splice site recognition by the spliceosome (Okunola and Krainer 2009). Since multimerization can lead to looping-out of intervening sequences, it might provide an efficient mechanism to protect splicing regulation against novel transposon inserts. In contrast to ewg, where only a single ELAV-binding site is present, ELAV has been shown to bind to multiple regions spread through the entire 3.2 kb of the regulated intron of nrg (Lisbin et al. 2001). Similar to the ELAV-binding site in ewg, mutations in multiple binding sites are required to reduce ELAV-mediated splicing of nrg, suggesting that multimerization of ELAV might be important for the regulation of this intron (Lisbin et al. 2001).
In conclusion, we demonstrate that ELAV regulation of ewg intron 6 splicing is conserved in distantly related D. virilis despite massive degeneracy of its binding site. Since multimerization is an inherent feature of ELAV/Hu proteins, our results indicate that ELAV/Hu regulated post-transcriptional gene regulation is likely more conserved than currently anticipated from genome alignments.
Acknowledgments
We thank the Bloomington and Exelixis/Harvard stock centers for fly lines, Dan Motola for initial analysis of the D. virilis intron 6, Declan Bostock for promoter analysis and cloning, and Chris Franklin for comments on the manuscript. We are indebted to Kalpana White for her support when this study was initiated. We acknowledge funding for this work from the Biotechnology and Biological Sciences Research Council.
Literature Cited
- Antic D., Lu N., Keene J. D., 1999. ELAV tumor antigen, hel-N1, increases translation of neurofilament M mRNA and induces formation of neurites in human teratocarcinoma cells. Genes Dev. 13: 449–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J., Maeda R. K., Hediger M., Karch F., Basler K., 2007. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA 104: 3312–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgeson C. D., Samson M. L., 2005. Shared RNA-binding sites for interacting members of the Drosophila ELAV family of neuronal proteins. Nucleic Acids Res. 33: 6372–6383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan C. M., Steitz J. A., 2001. HuR and mRNA stability. Cell. Mol. Life Sci. 58: 266–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudno M., Do C. B., Cooper G. M., Kim M. F., Davydov E., et al. , 2003. LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 13: 721–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudno M., Steinkamp R., Morgenstern B., 2004. The CHAOS/DIALIGN WWW server for multiple alignment of genomic sequences. Nucleic Acids Res. 32: W41–W44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Manley J. L., 2009. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 10: 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A. G., Eisen M. B., Smith D. R., Bergman C. M., Oliver B., et al. , 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450: 203–218 [DOI] [PubMed] [Google Scholar]
- Damgaard C. K., Tange T. O., Kjems J., 2002. hnRNP A1 controls HIV-1 mRNA splicing through cooperative binding to intron and exon splicing silencers in the context of a conserved secondary structure. RNA 8: 1401–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David P. S., Tanveer R., Port J. D., 2007. FRET-detectable interactions between the ARE binding proteins, HuR and p37AUF1. RNA 13: 1453–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachet O., Ephrussi A., 2004. Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature 428: 959–963 [DOI] [PubMed] [Google Scholar]
- Haussmann I. U., Soller M., 2010. Differential activity of EWG transcription factor isoforms identifies a subset of differentially regulated genes important for synaptic growth regulation. Dev. Biol. 348: 224–230 [DOI] [PubMed] [Google Scholar]
- Haussmann I. U., White K., Soller M., 2008. Erect wing regulates synaptic growth in Drosophila by integration of multiple signaling pathways. Genome Biol. 9: R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinman M. N., Lou H., 2008. Diverse molecular functions of Hu proteins. Cell. Mol. Life Sci. 65: 3168–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny A., Hachet O., Zavorszky P., Cyrklaff A., Weston M. D., et al. , 2006. A translation-independent role of oskar RNA in early Drosophila oogenesis. Development 133: 2827–2833 [DOI] [PubMed] [Google Scholar]
- Kasashima K., Terashima K., Yamamoto K., Sakashita E., Sakamoto H., 1999. Cytoplasmic localization is required for the mammalian ELAV-like protein HuD to induce neuronal differentiation. Genes Cells 4: 667–683 [DOI] [PubMed] [Google Scholar]
- Kasashima K., Sakashita E., Saito K., Sakamoto H., 2002. Complex formation of the neuron-specific ELAV-like Hu RNA-binding proteins. Nucleic Acids Res. 30: 4519–4526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koushika S. P., Soller M., DeSimone S. M., Daub D. M., White K., 1999. Differential and inefficient splicing of a broadly expressed Drosophila erect wing transcript results in tissue-specific enrichment of the vital EWG protein isoform. Mol. Cell. Biol. 19: 3998–4007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koushika S. P., Soller M., White K., 2000. The neuron-enriched splicing pattern of Drosophila erect wing is dependent on the presence of ELAV protein. Mol. Cell. Biol. 20: 1836–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Lee J. A., Black D. L., 2007. Neuronal regulation of alternative pre-mRNA splicing. Nat. Rev. Neurosci. 8: 819–831 [DOI] [PubMed] [Google Scholar]
- Licatalosi D. D., Darnell R. B., 2006. Splicing regulation in neurologic disease. Neuron 52: 93–101 [DOI] [PubMed] [Google Scholar]
- Licatalosi D. D., Darnell R. B., 2010. RNA processing and its regulation: global insights into biological networks. Nat. Rev. Genet. 11: 75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisbin M. J., Qiu J., White K., 2001. The neuron-specific RNA-binding protein ELAV regulates neuroglian alternative splicing in neurons and binds directly to its pre-mRNA. Genes Dev. 15: 2546–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Contreras R., Fisette J. F., Nasim F. U., Madden R., Cordeau M., et al. , 2006. Intronic binding sites for hnRNP A/B and hnRNP F/H proteins stimulate pre-mRNA splicing. PLoS Biol. 4: e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin A. J., Clark F., Smith C. W., 2005. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol. 6: 386–398 [DOI] [PubMed] [Google Scholar]
- Mattick J. S., 2009. The genetic signatures of noncoding RNAs. PLoS Genet. 5: e1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor C., Brudno M., Schwartz J. R., Poliakov A., Rubin E. M., et al. , 2000. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16: 1046–1047 [DOI] [PubMed] [Google Scholar]
- Neilson J. R., Sandberg R., 2010. Heterogeneity in mammalian RNA 3′ end formation. Exp. Cell Res. 316: 1357–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odenwald W. F., Rasband W., Kuzin A., Brody T., 2005. EVOPRINTER, a multigenomic comparative tool for rapid identification of functionally important DNA. Proc. Natl. Acad. Sci. USA 102: 14700–14705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okunola H. L., Krainer A. R., 2009. Cooperative-binding and splicing-repressive properties of hnRNP A1. Mol. Cell. Biol. 29: 5620–5631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks A. L., Cook K. R., Belvin M., Dompe N. A., Fawcett R., et al. , 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36: 288–292 [DOI] [PubMed] [Google Scholar]
- Ponting C. P., Oliver P. L., Reik W., 2009. Evolution and functions of long noncoding RNAs. Cell 136: 629–641 [DOI] [PubMed] [Google Scholar]
- Prasanth K. V., Spector D. L., 2007. Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum. Genes Dev. 21: 11–42 [DOI] [PubMed] [Google Scholar]
- Rhead B., Karolchik D., Kuhn R. M., Hinrichs A. S., Zweig A. S., et al. , 2010. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 38: D613–D619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson M. L., 1998. Evidence for 3′ untranslated region-dependent autoregulation of the Drosophila gene encoding the neuronal nuclear RNA-binding protein ELAV. Genetics 150: 723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson M. L., 2008. Rapid functional diversification in the structurally conserved ELAV family of neuronal RNA binding proteins. BMC Genomics 9: 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saudan P., Hauck K., Soller M., Choffat Y., Ottiger M., et al. , 2002. Ductus ejaculatorius peptide 99B (DUP99B), a novel Drosophila melanogaster sex-peptide pheromone. Eur. J. Biochem. 269: 989–997 [DOI] [PubMed] [Google Scholar]
- Soller M., 2006. Pre-messenger RNA processing and its regulation: a genomic perspective. Cell. Mol. Life Sci. 63: 796–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soller M., White K., 2003. ELAV inhibits 3′-end processing to promote neural splicing of ewg pre-mRNA. Genes Dev. 17: 2526–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soller M., White K., 2004. Elav. Curr. Biol. 14: R53. [DOI] [PubMed] [Google Scholar]
- Soller M., White K., 2005. ELAV multimerizes on conserved AU4–6 motifs important for ewg splicing regulation. Mol. Cell. Biol. 25: 7580–7591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soller M., Li M., Haussmann I. U., 2008. Regulation of the ELAV target ewg: insights from an evolutionary perspective. Biochem. Soc. Trans. 36: 502–504 [DOI] [PubMed] [Google Scholar]
- Soller M., Li M., Haussmann I. U., 2010. Determinants of ELAV gene-specific regulation. Biochem. Soc. Trans. 38: 1122–1124 [DOI] [PubMed] [Google Scholar]
- Thibault S. T., Singer M. A., Miyazaki W. Y., Milash B., Dompe N. A., et al. , 2004. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36: 283–287 [DOI] [PubMed] [Google Scholar]
- Toba G., White K., 2008. The third RNA recognition motif of Drosophila ELAV protein has a role in multimerization. Nucleic Acids Res. 36: 1390–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken K. J., He Y., Hoskins R. A., Bellen H. J., 2006. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314: 1747–1751 [DOI] [PubMed] [Google Scholar]
- Wang E. T., Sandberg R., Luo S., Khrebtukova I., Zhang L., et al. , 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao K. M., White K., 1991. Organizational analysis of elav gene and functional analysis of ELAV protein of Drosophila melanogaster and Drosophila virilis. Mol. Cell. Biol. 11: 2994–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao K.-M., Samson M.-L., Reeves R., White K., 1993. Gene elav of Drosophila melanogaster: a prototype for neuronal-specific RNA binding protein gene family that is conserved in flies and humans. J. Neurobiol. 24: 723–739 [DOI] [PubMed] [Google Scholar]
- Zhu H., Zhou H. L., Hasman R. A., Lou H., 2007. Hu proteins regulate polyadenylation by blocking sites containing U-rich sequences. J. Biol. Chem. 282: 2203–2210 [DOI] [PubMed] [Google Scholar]
- Zhu H., Hinman M. N., Hasman R. A., Mehta P., Lou H., 2008. Regulation of neuron-specific alternative splicing of neurofibromatosis type 1 pre-mRNA. Mol. Cell. Biol. 28: 1240–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Mayeda A., Krainer A. R., 2001. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol. Cell 8: 1351–1361 [DOI] [PubMed] [Google Scholar]