Abstract
Hypoacetylated H4 is present at regional centromeres; however, its role in kinetochore function is poorly understood. We characterized H4 acetylation at point centromeres in Saccharomyces cerevisiae and determined the consequences of altered H4 acetylation on chromosome segregation. We observed low levels of tetra-acetylated and K16 acetylated histone H4 (H4K16Ac) at centromeres. Low levels of H4K16Ac were also observed at noncentromeric regions associated with Cse4p. Inhibition of histone deacetylases (HDAC) using nicotinamide (NAM) caused lethality in cse4 and hhf1-20 kinetochore mutants and increased centromeric H4K16Ac. Overexpression of Sas2-mediated H4K16 acetylation activity in wild-type cells led to increased rates of chromosome loss and synthetic dosage lethality in kinetochore mutants. Consistent with increased H4K16 acetylation as a cause of the phenotypes, deletion of the H4K16 deacetylase SIR2 or a sir2-H364Y catalytic mutant resulted in higher rates of chromosome loss compared to wild-type cells. Moreover, H4K16Q acetylmimic mutants displayed increased rates of chromosome loss compared to H4K16R nonacetylatable mutants and wild-type cells. Our work shows that hypoacetylated centromeric H4 is conserved across eukaryotic centromeres and hypoacetylation of H4K16 at centromeres plays an important role in accurate chromosome segregation.
CENTROMERES serve as the chromosomal regions on which the kinetochores (i.e., centromeric DNA and associated proteins) are assembled, multisubunit complexes essential for mediating chromosome transmission fidelity (ctf) during cell division. Eukaryotes, such as budding yeast, have “point” centromeres, whereas fission yeast, fruit flies, and humans have “regional” centromeres. The point centromeres are small (∼125 bp) and consist of CDE I–III, DNA elements that are highly conserved between each centromere. By contrast, regional centromeres are composed of repeated arrays of α-satellite DNA that range in size from ∼0.1 to 5 Mb (Karpen and Allshire 1997). Despite the lack of sequence conservation, centromeres in all eukaryotes are assembled into a specialized chromatin that contains the centromere specific histone H3 variant (CenH3; Cse4p in budding yeast) (Ekwall 2007).
Several reports indicate that a unique chromatin structure is assembled at the centromere and pericentromeric regions in budding yeast. Work by Bloom and Carbon (1982) reported a specialized chromatin that extended several kilobases beyond the centromere sequence on both sides. Studies using fluorescence microscopy and chromosome conformation capture (3C) assays, revealed that the pericentromeric chromatin can form an intramolecular cruciform structure (pericentromeric loop) that has been proposed to be an important component of the mitotic spindle, balancing the forces between the kinetochore and spindle pole body (Bouck et al. 2008; Yeh et al. 2008; Anderson et al. 2009). In turn, disruption of nucleosomes by histone depletion (Saunders et al. 1990; Bouck and Bloom 2007), deletion of both CAC1 and HIR1 (Sharp et al. 2002), or deletion of SPT4 (Crotti and Basrai 2004) result in declustering of kinetochores and compromised centromeric structure. Moreover, deletion of a single copy of histone H2A leads to polyploidy and mutants in the Hda1 deacetylase complex can suppress polyploidization (Kanta et al. 2006).
N-terminal covalent histone modifications modulate virtually all DNA-templated processes (Strahl and Allis 2000) and combinations of various histone modifications correlate with functional chromatin domains and activities (Millar and Grunstein 2006; Wang et al. 2009). Studies of regional centromeres showed that centromeric histones H3 and H4 are hypoacetylated, flanked by pericentromeric heterochromatin containing hypoacetylated H3 and H4 (Sullivan and Karpen 2004). How histone H4 modifications affect kinetochore assembly and function is still not fully understood. Regional centromeres are composed of CenH3 nucleosomes interspersed with H3 nucleosomes that contain repetitive DNA, making it difficult to delineate the precise role of centromeric histone modifications. The point centromeres in Saccharomyces cerevisiae with a defined sequence and single centromeric nucleosome (Furuyama and Biggins 2007) provide a highly tractable system that bypasses these challenges.
Our work shows that there is a low level of acetylated histone H4 at the S. cerevisiae point centromere. Maintaining hypoacetylated H4 at the centromere is important for kinetochore function, as deregulated histone H4K16 acetylation causes growth defects in kinetochore mutants and chromosome missegregation. Taken together, we propose that hypoacetylated H4K16 is required for an optimal chromatin environment to ensure that the kinetochore mediates faithful chromosome segregation.
Materials and Methods
Strains and plasmids
Please refer to supporting information, Table S1 for a description of all yeast strains used. pRS316 (CEN6URA3), pRS314 (CEN6TRP1), pRS425 (GAL1 2μ LEU2), and pRS426 (GAL1 2μ URA3) were gifts from P. Hieter, University of British Columbia, British Columbia, Canada (Sikorski and Hieter 1989). pLG39 (GAL1/10-HHF1 2μ URA3) and pLG41 (GAL1/10-HHT1 2μ URA3) were provided by M. M. Smith, University of Virginia, Charlottesville, VA (Glowczewski et al. 2000). pSB816 (GAL1/10-MYC-CSE4 2µ URA3) was a gift from S. Biggins, Fred Hutchinson Cancer Center, Seattle, WA. pLP1197 (GST-ESA1 in pRS425) was a gift from L. Pillus, UC-San Diego, La Jolla, CA. pMB1193 (GAL1 2μ SCM3LEU2) was generated by cloning SCM3 in a pRS425 vector. pS116 (pESC/URA/6XHIS-SAS2-FLAG) and pS117 (pESC/URA/6XHIS-sas2m1-FLAG) were donated by J. L. Workman, Stowers Institute for Medical Research, Kansas City, MO (Osada et al. 2001). pDM607 (sir2-H364Y) and pDM608 (SIR2) were provided by D. Moazed, Harvard Medical School, Boston, MA (Tanny et al. 2004). pMB1434 and pMB1435 were generated by PCR-amplifying SAS2 and SAS2m1 using pS116 and pS117 as templates with primers including HindIII/EcoRI restriction sites and subcloning the respective fragments into pRS426 cut with the same enzymes. Cloning was verified by digestion and sequencing. For experiments, yeast strains were transformed with the aforementioned plasmids or the pBG1805 plasmid (GAL1URA3 His6X-3C-Protein A “zz” domain) containing the desired mORF (SAS2, SAS4, and SAS5) obtained from Open Biosystems (Gelperin et al. 2005).
Growth assays
Strains grown on selective or rich media were used to prepare fivefold serial dilutions in water and 3.5 µl of each dilution was spotted on selective media with either 2% glucose or 2% raffinose and 2% galactose. Nicotinamide (NAM; Fluka Analytical, St. Louis, MO, CAS: 72340) was added at the indicated concentration where designated. Plates were incubated at the specified temperatures for 3–5 days and three independent transformants were tested for each strain.
Chromatin immunoprecipitation (ChIP) experiments
Cultures of each strain, starting OD ∼0.2, were grown until OD ∼0.8–1.0, typically at 30°, and cross-linked with formaldehyde for 30 min and processed essentially as described in Au et al. (2008). In experiments with cse4-1, cells were grown at 35° in 0 mM or 30 mM NAM from a starting OD ∼0.2 and cross-linked with formaldehyde for 30 min when cells reached OD ∼0.8. Glycine was added to a final concentration of 0.325 M, washed with 1× PBS, pelleted, frozen on dry ice, and stored. Pellets were resuspended in FA buffer (50 mM Hepes-NaOH pH 7.6, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-Deoxycholate) with Roche Protease inhibitor cocktail, 1 mM PMSF, 30 mM sodium butyrate. An equal volume of glass beads was added and samples were vortexed at 4° until >90% of cells were lysed as determined by microscopic inspection. Cells were separated from glass beads and sonicated to obtain sheared chromatin. Magnetic beads containing protein A or sheep antirabbit (Invitrogen) were preblocked with BSA, treated with antibodies to Myc (Santa Cruz), tAcH4 (Millipore), H4Pan (Millipore), H4K16 (ChIP grade from Active Motif), then added to chromatin samples for immunoprecipitation overnight at 4°. Beads were washed three times sequentially with FA buffer, HS-FA buffer (same as FA except with 500 ml NaCl), and two times with RIPA (10 mM Tris-Cl pH 8.0, 250 mM LiCl, 0.5% NP-40, 0.5% Na-Deoxycholate, 1 mM EDTA), and TE (Tris-Cl pH 8.0, 1 mM EDTA) before elution buffer (25 mM Tris-Cl pH 7.6, 10 mM EDTA, 0.5% SDS) was added to resuspend the beads and incubated overnight at 65° to reverse cross-links. Beads were removed and the samples treated with proteinase K at 55° for ∼4 hr, phenol chloroform extracted or a Qiagen PCR clean up kit used to isolate DNA. We used the relative quantification method of qPCR with Sybr Green (ABI) and calculated enrichment from 2-(CT(IP)-CT(mock)) (Figures 1, 3, S1, S3). Alternatively, we analyzed ChIP DNA samples with standard PCR and 1.5% agarose gel electrophoresis containing EtBr. Gels were imaged and DNA products were quantified using GeneSnap and GeneTools from SynGene, respectively. The ratio of H4K16Ac and/or tAcH4 products to pan H4 products were plotted (Figures 3C, 4B, S2, S3, S4, S5). Primer pairs to each region assayed were designed to amplify DNA fragments ∼200–300 bp in length. Primer sequences are available upon request.
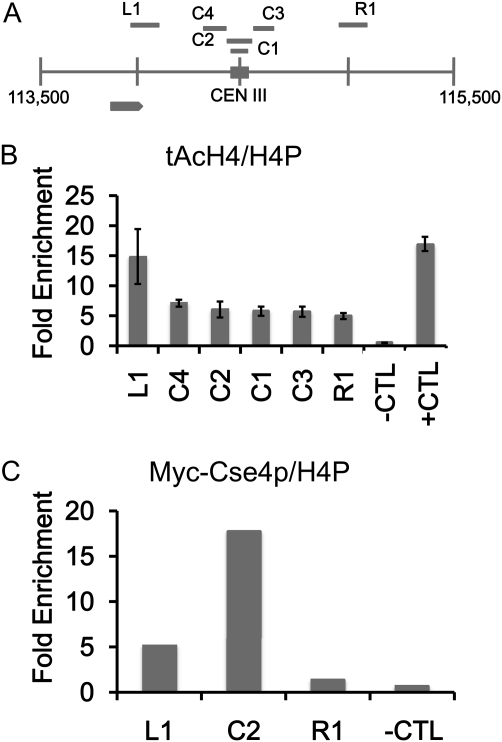
Figure 1 .
Low levels of acetylated H4 are observed at the centromere. Chromatin immunoprecipitation (ChIP) experiments were performed with asynchronously grown wild-type strains expressing Myc-Cse4p (YMB6955). (A) Diagram of centromere (CEN) (C1–4) and pericentromeric regions (L1 and R1) on chromosome III analyzed by ChIP. Solid square represents the CEN III sequence and each vertical line from chromosomal coordinate 113,500 to 115,500 denotes 500-bp increments. Gray arrow represents an uncharacterized open reading frame (YCL001W-B). (B and C) ChIP with antibodies that recognize acetylated isoforms of acetylated histone H4 (tAcH4) to measure the levels of H4 acetylation at centromeric (C1–4) regions or to Myc to measure levels of Myc-Cse4p at centromeric C2 region and pericentromeric (L1 and R1) regions by quantitative PCR. HML serves as a hypoacetylated H4 control (−CTL) region and SNR189 (SNR) (Chr III: 178729–178589) is a hyperacetylated H4 control (+CTL) region. ChIP for total H4 was performed with antibodies to pan H4 (H4P) that recognize modified and unmodified forms. Bar graphs in B show mean fold enrichment for tAcH4 normalized to total H4 from three biological replicates with error bars showing standard error of the mean. Bar graphs in C show mean fold enrichment for Myc-Cse4p normalized to total H4 from two biological replicates.
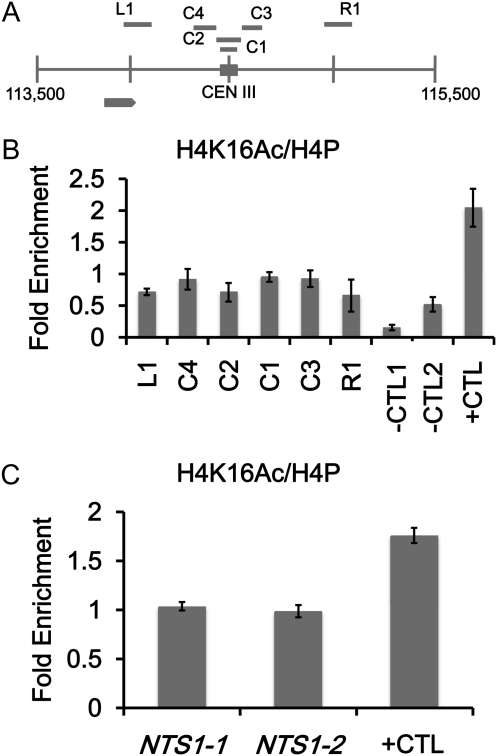
Figure 3 .
Low levels of H4K16Ac are present at the centromere and at noncentromeric loci enriched for Cse4p. ChIP experiments were performed with asynchronously grown wild-type strains (YMB6955 in B and YMB8077 in C). Immunoprecipitated DNA was subjected to quantitative PCR to determine enrichment for the indicated regions. (A) Diagram of CEN III (C1–4) and pericentromeric regions (L1 and R1) analyzed by ChIP. (B) ChIP experiments with antibodies to acetylated H4K16 (H4K16Ac) and pan H4 to measure levels of H4K16Ac and total H4 at C1–4 and at L1 and R1 or in C at NTS1-1 and NTS1-2 rDNA regions that contain Cse4p. HML (−CTL1) and SPS22 (−CTL2) (Chr III: 42437–42630) serve as hypoacetylated and ARE1 (+CTL) (Chr III: 212230–212450) serves as hyperacetylated H4K16 controls. Bar graphs show mean fold enrichment for H4K16Ac normalized to total H4 from three biological replicates with error bars showing standard error of the mean.
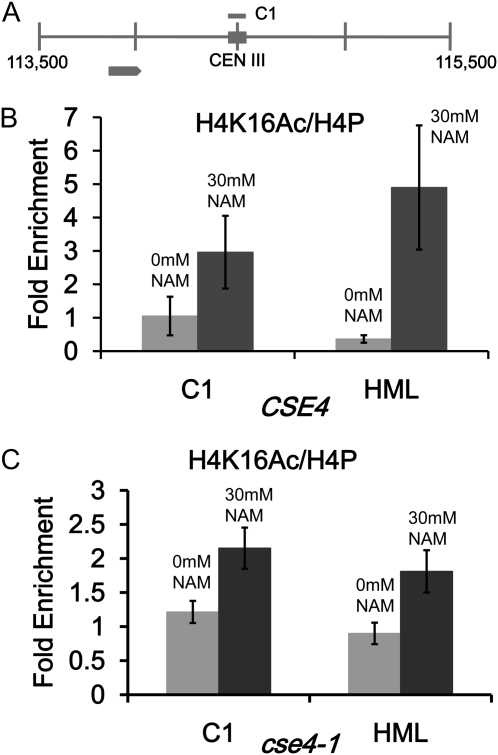
Figure 4 .
NAM causes an increase in centromeric H4K16Ac in wild-type and cse4-1 strains. ChIP experiments were performed with CSE4 (RC154) and cse4-1 strains (RC147) grown in 0 mM or 30 mM NAM. Immunoprecipitated DNA was subjected to quantitative PCR to determine enrichment for H4K16Ac. (A) Diagram of CEN III and pericentromeric regions. (B) ChIP experiments with antibodies to H4K16Ac and pan H4 to measure levels of H4K16Ac at C1 in CSE4 strains. (C) Same as B except in cse4-1 strains. HML serves as a hypoacetylated H4K16 control region. Bar graphs show mean fold enrichment for H4K16Ac normalized to total H4 from three biological replicates with error bars showing standard error of the mean.
Chromosome loss assays
Assays were performed essentially as previously described in Hieter et al. (1985). Strains containing the nonessential chromosome fragment (CF) were grown on medium selective for the CF and the selectable marker on the plasmid that was being assayed. Cells were plated on SC media with limiting adenine and incubated at 30° for 5–7 days. Chromosome loss was determined by counting colonies that were at least half-sectored, indicating loss of the CF during the first cell division. Three transformants for each strain were assayed; ∼1–2000 colonies were counted.
Results
Low levels of acetylated histone H4 are present at centromeric chromatin
A distinct pattern of histone H3 and H4 modifications has been reported for regional centromeres (Hayashi et al. 2004; Sullivan and Karpen 2004). Considering the role of H4 acetylation in chromatin structure, we explored the possibility that there were acetylated H4 isoforms at centromeric (∼200 bp centered on the CDE I–III sequences) chromatin in S. cerevisiae. Using chromatin immunoprecipitation (ChIP) with an antibody that recognizes all forms of N-terminal acetylated histone H4 (tAcH4), we observed that the centromere region of chromosome III (CEN III) contains lower levels of tAcH4 (∼3-fold less) compared to SNR189 (+CTL), a region with hyperacetylated histone H4 (Liu et al. 2005) (Figure 1). Our results for tAcH4 are derived from four different primer pairs that amplify the core centromere sequence (C1 and C2) and the 200-bp regions immediately upstream and downstream of the centromere (C3 and C4). As a negative control, we analyzed the HML locus (−CTL), which is known to contain hypoacetylated H4 (Rusche et al. 2003). As expected, we found virtually no tAcH4 at HML, indicated by <1-fold enrichment (Figure 1B). Low levels of tAcH4 were also present in the pericentromeric region ∼300 bp downstream of the centromere (R1) compared to the region ∼300 bp upstream of the centromere (L1). The latter region overlaps with an uncharacterized open reading frame (YCL001W-B) and this may contribute to the higher level of tAcH4. We also observed a low level of tAcH4 (∼3-fold less than SNR189) at the core centromere of chromosome VI (Figure S1). All regions analyzed were normalized to total histone H4 (modified and unmodified) using an antipan H4 antibody to account for any differences in nucleosome positioning or potential differences in ChIP efficiencies at the centromere compared to noncentromeric regions as previously described (Camahort et al. 2009). We have provided examples of the differences in the percentage of input for pan-H4 at CEN III and VI compared to noncentromeric regions (Figure S2). We refer to the lower levels of tAcH4 at the centromere region as hypoacetylated. To confirm that the domain of hypoacetylated H4 overlapped with the centromere region, we performed ChIP assays for Cse4p (Furuyama and Biggins 2007). Our results showed that Myc-Cse4p was highly enriched (on average ∼20-fold enrichment) in the same C2 region of CEN III containing hypoacetylated H4 but not in the flanking regions L1 and R1 or at HML (−CTL) (Figure 1C).
We wanted to determine whether chromosomal sequences were important for the observed hypoacetylated histone H4 at the centromere or whether the centromere sequence alone was sufficient. Thus, we examined tAcH4 at a minimal centromere on a pBluescript-based plasmid (pRS316) with heterologous sequences on both sides. An ARS element and the URA3 marker are downstream, and bacterial sequences are upstream of the centromeric sequence (CEN VI) (Figure S3). We introduced pRS316 into a S. cerevisiae strain in which the endogenous CEN VI is replaced with CEN XI and performed ChIP experiments to determine the level of tAcH4 at the plasmid-based CEN VI. As in Figure 1 we used two different primer pairs that amplify the core centromere sequence (pC1 and pC2) and two different primer pairs that amplify the 200-bp regions immediately upstream and downstream of the centromere (pL and pR). Consistent with a centromere-dependent pattern, we observed that there were ∼2-fold lower levels of tAcH4 at CEN VI than at SNR189 (+CTL) (Figure S3), similar to that observed for endogenous CEN III (Figure 1B).
Mutants in CSE4 and HHF1 are sensitized to nicotinamide (NAM)
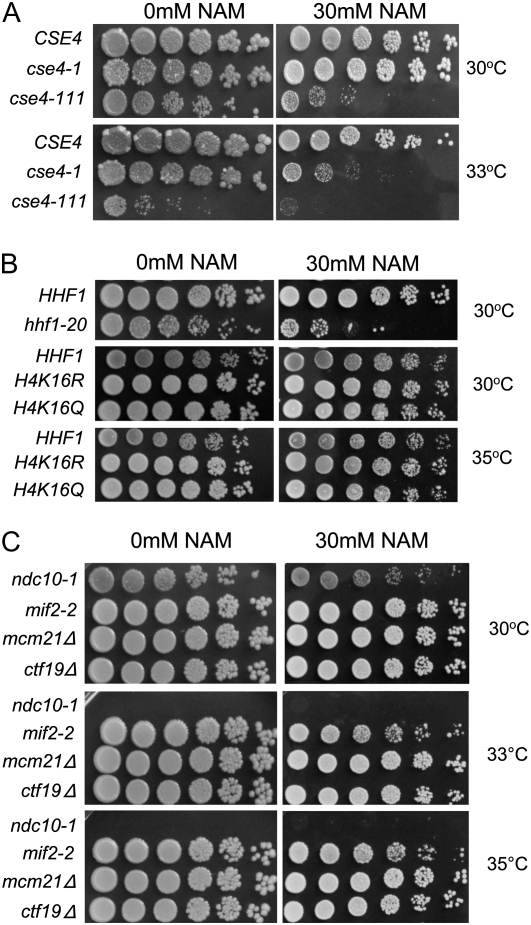
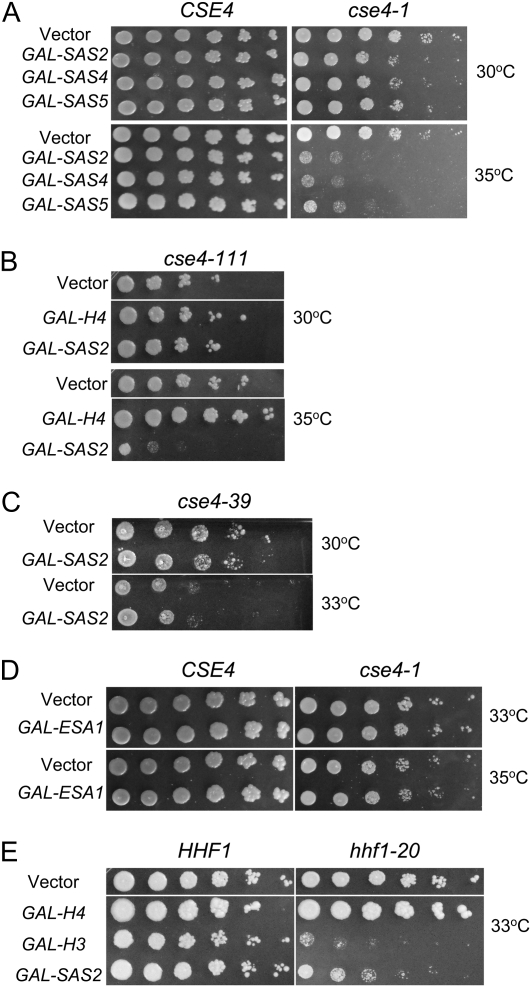
The results of our ChIP experiments for tAcH4 suggested that similar to regional centromeres, histone H4 at the centromere in S. cerevisiae is hypoacetylated. Acetylation of specific lysine residues on H3 and H4 can lead to different functional outcomes (Millar and Grunstein 2006; Wang et al. 2009). Levels of K16 acetylation on H4 is a major determinant in regulating chromatin structure in yeast and metazoans (Shogren-Knaak and Peterson 2006). Therefore, we investigated whether altering H4K16 acetylation might exhibit synthetic interactions with kinetochore mutants. Several studies have shown that NAM inhibits NAD-dependent HDACs, including Sir2p (Sanders et al. 2007), and leads to higher levels of acetylated H4K16 (H4K16Ac) (Dang et al. 2009). The centromeric nucleosome contains Cse4p and H4, and the C-terminal histone fold domain of Cse4p has been shown to be important for interaction with H4 (Smith et al. 1996; Glowczewski et al. 2000). Temperature-sensitive (ts) alleles of cse4-1, cse4-111, and hhf1-20 have chromosome segregation defects. The cse4-1 and cse4-111 strains have mutations in the histone fold domain and the hhf1-20 strain contains mutations near the DNA binding region and helix 3 of the histone fold domain that are thought to weaken interactions with Cse4p (Stoler et al. 1995; Smith et al. 1996; Glowczewski et al. 2000). If low levels of H4K16Ac are important for kinetochore function, we predict that cse4 and hhf1-20 mutants treated with NAM would result in enhanced growth defects. We found that cse4-1 and cse4-111 displayed an increase in temperature sensitivity to NAM at 33° and 30°, respectively, but wild-type cells remained unaffected at both temperatures (Figure 2A). Similar to cse4 mutants, the histone H4 mutant hhf1-20 was also sensitive to NAM at 30° (Figure 2B). If the growth defects observed in cse4 and hhf1-20 mutants were mediated through modulating H4K16 acetylation, then we would predict that mutants in which lysine 16 is changed to glutamine (H4K16Q), an acetylmimic or an arginine (H4K16R) that is nonacetylatable, should not result in decreased fitness. On the other hand, if the NAM effects were a result of H4 independent pathway(s) then H4K16Q and H4K16R mutants might both show enhanced growth defects in the presence of NAM. Consistent with NAM inihibition acting on H4K16 acetylation, we observed no growth defects for these mutants when grown on NAM at 30° or 35° (Figure 2B). To determine whether the growth defects in cse4-1, cse4-111, and hhf1-20 strains were a general toxicity of kinetochore mutants to NAM, we tested the kinetochore mutants, ndc10-1, mif2-3, mcm21Δ, and ctf19Δ. A mild growth defect was observed in ndc10-1 strain at 30° (note that the ndc10-1 strain is inviable above 30°) but no growth defects were observed for mif2-3, ctf19∆, or mcm21∆ to NAM at 30°, 33° and 35° (Figure 2C).
Figure 2 .
Kinetochore mutants are sensitized to nicotinamide (NAM). Strains were serially diluted fivefold and spotted onto YPD plates with 0 mM or 30 mM NAM, incubated at the indicated temperatures, and photographed after 3 days. (A) Wild-type CSE4 (RC154) and temperature-sensitive C-terminal cse4 mutants, cse4-1 (RC147) and cse4-111 (MSY1520). (B) Wild-type HHF1 (MSY559) and temperature-sensitive chromosome segregation histone H4 mutant hhf1-20 (MSY554) (top), and strains expressing wild-type histone H4 (YMB8155), nonacetylatable H4K16R (YMB8156), and acetylmimic H4K16Q (YMB8157) (middle and bottom). (C) Temperature-sensitive kinetochore mutants, ndc10-1 (JK421), mif2-2 (6849-10-1), and nonessential kinetochore mutants, mcm21∆ (YPH1715) and ctf19∆ (YPH1713). Note the ndc10-1 mutant is inviable above 30°.
Low levels of H4K16Ac are present at the centromere and noncentromeric regions enriched for Cse4p
The low level of tAcH4 at the centromere together with the sensitivity of cse4 and hhf1-20 mutants to NAM prompted us to measure the levels of H4K16Ac at the centromere (Shahbazian and Grunstein 2007). We performed ChIP assays with an antibody that recognizes acetylated histone H4K16 and four different primer pairs for CEN III (C1–C4) as described in Figure 1. We determined that all four regions corresponding to the centromere region of chromosome III and the upstream (L1) and downstream regions (R1) contain lower levels of H4K16Ac (∼twofold less) similar to hypoacetylated HML (−CTL1) and SPS22 (−CTL2) regions, previously reported to have low H4K16Ac (Liu et al. 2005), compared to ARE1 (+CTL) (Figure 3B), a region with high levels of H4K16Ac (Liu et al. 2005). Similarly, we also observed low levels of H4K16Ac (approximately twofold less) at CEN VI compared to ARE1 (+CTL) (Figure S4). Moreover, H4K16Ac levels are higher in the regions immediately upstream (∼50 bp) and downstream (∼400 bp) of CEN VI, which contains the DEG1 and LOC1 genes, respectively (Figure S5). These results show that H4K16 is hypoacetylated at centromeric chromatin and is not affected by the acetylation of neighboring genes. We note that while the SNR189 region contains hyperacetylated H4, the level of acetylated H4K16 is lower compared to ARE1 (Liu et al. 2005) and therefore, we have used ARE1 as a positive control for H4K16 hyperacetylation rather than SNR189.
We have observed low levels of acetylated H4 at centromeres in the context of the endogenous chromosome and on a plasmid. The low level of acetylated centromeric H4 may be regulated by the kinetochore or associated proteins. To test this possibility, we sought to determine whether the hypoacetylated state of H4K16 is associated with noncentromeric regions containing Cse4p. In a recent report by Camahort et al. (2009) they found approximately four- to sixfold higher levels of Cse4p at rDNA (NTS1-1 and NTS1-2) regions compared to loci where Cse4p is absent. We performed ChIP experiments to measure the levels of H4K16Ac at NTS1-1 and NTS1-2 and observed slightly less than twofold lower H4K16Ac compared to ARE1 (Figure 3C), similar to the difference observed at CEN III (Figure 3B). We also observed enrichment for Cse4p at NTS1-1 and NTS1-2 consistent with that reported by Camahort et al. (2009) (Figure S6). These results suggest that acetylation of H4 may be dictated by association with Cse4p, or Cse4p tends to associate with genomic regions in which histone H4K16 is hypoacetylated.
Centromeric H4K16 acetylation is increased in NAM treated cells
The increased sensitivity of hhf1-20 and cse4 mutants to NAM prompted us to examine whether NAM treatment affected the level of H4K16Ac at the centromere. We performed ChIP experiments with wild-type and cse4-1 strains grown in 0 mM or 30 mM NAM. We observed an approximately twofold increase in centromeric H4K16Ac in wild-type and cse4-1 strains grown in NAM compared to untreated cells (Figure 4, B and C). In addition, we also observed an increase in H4K16Ac at HML, which likely reflects a more general effect of NAM-mediated inhibition of NAD-dependent H4K16 deacetylation activity. These results show that NAM treatment causes an increase in H4K16 acetylation at the centromere and in combination with a kinetochore mutant (cse4 and hhf1-20), results in growth defects.
Overexpression of SAS subunits is synthetic dosage lethal (SDL) in CSE4 and HHF1 mutants
Our ChIP experiments revealed that centromeric chromatin had lower levels of acetylated H4K16 and inhibiting HDACs with NAM led to loss of viability in kinetochore mutants. On the basis of these observations, we predict that increasing H4K16 histone acetyltransferase activity would exacerbate the growth defects in kinetochore mutants. To test this possibility, we determined whether overexpression of Sas2p (GAL-SAS2), an H4K16 histone acetyltransferase (HAT) (Shia et al. 2005) and Esa1p, an essential HAT that acetylates all four N-terminal histone H4 lysines (Smith et al. 1998; Allard et al. 1999), displayed genetic interactions in cse4 and hhf1 mutants. Indeed, we found that cse4 mutants (cse4-1 and cse4-111) with mutations in the C-terminal histone fold domain were sensitized to GAL-SAS2 with the strongest effect at 35° and 33°, respectively (Figure 5, A and B). Moreover, overexpression of the Sas2p partners, Sas4p and Sas5p, also exhibited a SDL in cse4-1 at 35° (Figure 5A). As previously reported, we also observed that overexpression of histone H4 suppresses the growth defect in cse4-111 mutants (Figure 5B) (Glowczewski et al. 2000). However, there was no growth defect conferred by GAL-SAS2 in cse4-39 mutants, which have a mutation at the N-terminal domain of Cse4p (Figure 5C). We then tested the phenotypes of ESA1 overexpression at 33° and 35° in cse4-1 and observed no growth defect (Figure 5D). This is likely not due to a dominant negative effect, as we found no obvious differences in acetylated H4K16 protein levels in the presence or absence of ESA1 overexpression in wild-type or cse4-1 strains (Figure S7).
Figure 5 .
Overexpression of SAS subunits leads to SDL in kinetochore mutants. Strains were transformed with vector (vector) or vector containing the indicated gene under the Gal1 promoter (GAL-). Fivefold serial dilutions of each strain were spotted on SC −Ura plates containing 2% galactose and 2% raffinose, incubated at the indicated temperatures, and photographed after 3–5 days. (A) Wild-type CSE4 (RC154) and C-terminal mutant cse4-1 (RC147). (B) C-terminal mutant cse4-111 (MSY1520). (C) N-terminal mutant cse4-39 (YC190). (D) Wild-type CSE4 (RC154) and C-terminal mutant cse4-1 (RC147). (E) Wild-type HHF1 (MSY559) and temperature-sensitive chromosome segregation mutant hhf1-20 (MSY554) strains. Three or more independent transformants were tested for each experiment.
Considering that Cse4p binds to H4 to form the core centromeric nucleosome (Smith et al. 1996; Glowczewski et al. 2000; Furuyama and Biggins 2007), we tested whether hhf1-20 was sensitized to GAL-SAS2. Consistent with previously published observations, we found that overexpression of histone H3 is SDL in hhf1-20 (Figure 5E) (Glowczewski et al. 2000). Similar to the results in cse4 mutants, GAL-SAS2 exhibited SDL in an hhf1-20 strain at 33°, compared to vector control (Figure 5E).
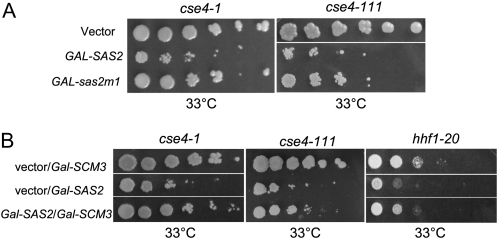
The SDL conferred by GAL-SAS2 may be mediated by its catalytic or noncatalytic activity. To distinguish between these two possibilities, we expressed a SAS2 catalytic mutant (Gal-sas2m1) in cse4-1 and cse4-111 mutants. The sas2m1 allele encodes for a mutant protein that replaces residues 219–221 from GLG to AAA within the active site (Osada et al. 2001; Shia et al. 2005). Previous results have shown that the catalytic mutant, sas2m1p, displays a sixfold decrease in activity and is unable to complement a loss of silencing defect in sas2Δ mutants (Osada et al. 2001; Shia et al. 2005). Compared to wild-type SAS2, which causes a marked loss in viability in cse4-1 and cse4-111 at 33°, GAL-sas2m1 had a reduced effect in these strains (Figure 6A).
Figure 6 .
Catalytic activity of Sas2p contributes to SDL and GAL-SCM3 suppresses the GAL-SAS2 SDL. Strains were transformed with vector (vector) or vector containing the indicated gene under the Gal1 promoter (GAL-). Fivefold serial dilutions of each strain were spotted on SC −Ura plates containing 2% galactose and 2% raffinose, incubated at the indicated temperatures, and photographed after 3–5 days. (A) C-terminal mutants cse4-1 (RC147) and cse4-111 (MSY1520) strains, carrying vector, GAL-SAS2, or the catalytic mutant GAL-sas2m1. (B) C-terminal mutant cse4-1, cse4-111, and histone H4 mutant hhf1-20 (MSY554) strains, carrying vector and GAL-SCM3 or GAL-SAS2 or carrying both GAL-SAS2 and GAL-SCM3. At least three independent transformants were tested for each experiment.
Several reports have demonstrated that Scm3p binds to Cse4p and is required to maintain Cse4p at the centromere (Camahort et al. 2007; Mizuguchi et al. 2007; Stoler et al. 2007). If the SAS2 SDL is associated with disruption of the Cse4p-H4 complex then increasing the dosage of SCM3 might suppress the loss in viability. Consistent with this prediction, we found that the SDL conferred by SAS2 overexpression in cse4-1, cse4-111, and hhf1-20 mutants was suppressed by overexpression of SCM3 (Figure 6B). Taken together, these results suggest that SAS activity enhances the kinetochore defects in cse4 C-terminal mutants and hhf1-20 mutants.
Altered acetylation of histone H4 affects chromosome segregation
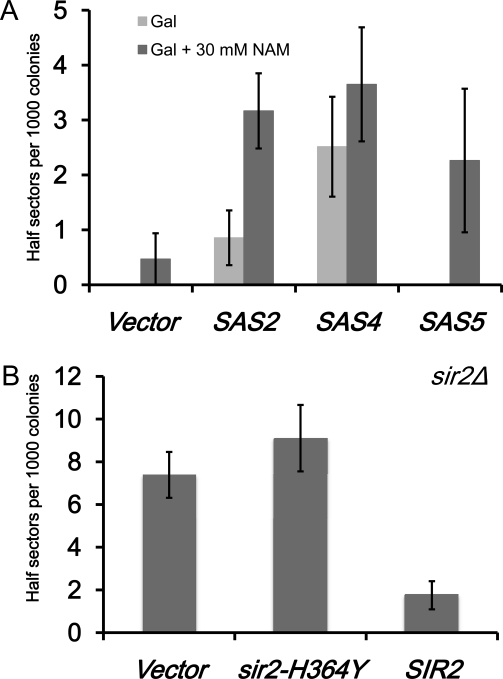
The observed lethality in kinetochore mutants overexpressing SAS subunits along with the reduced SDL displayed by the SAS2 catalytic mutant suggest that increased H4K16 acetylation may lead to chromosome missegregation. To test this possibility, we determined the rate of chromosome loss in a reporter strain overexpressing SAS2, SAS4, and SAS5. Cells with a chromosome segregation defect lose the minichromosome and give rise to white colonies with red sectors (Hieter et al. 1985). Half-sectored colonies reflect chromosome loss within the first cell division. For example, a deletion of the spindle assembly checkpoint gene MAD1 results in approximately eight half-sectored colonies per 1000 examined (Kastenmayer et al. 2005). We found an absence of half-sectored colonies for wild-type strains with vector alone or Gal-SAS5 in ∼1000 colonies, but observed half-sectored colonies for those expressing GAL-SAS2 or GAL-SAS4 (∼1 or ∼2.5 per 1000 colonies, respectively) (Figure 7A). We propose that the increased chromosome loss in GAL-SAS4 might reflect the importance of this subunit for Sas2p activity as previously reported (Sutton et al. 2003).
Figure 7 .
Altered H4K16Ac affects chromosome segregation. Chromosome loss assays were done with wild-type strains carrying a nonessential minichromosome (YPH1015) with vector or SAS2, SAS4, or SAS5 expressed under the GAL1 promoter. Loss of the minichromosome within the first cell division gives rise to colonies that are half red and half white. We measured colonies that were at least half red. (A) Bar graph showing the number of half-sectored colonies per 1000 for each strain on galactose media without (Gal) or with NAM (Gal + 30 mM NAM). (B) Bar graph shows the number of half-sectored colonies per 1000 for SIR2 deletion strain (YMB7911) carrying pRS314 (vector), sir2-H364Y (catalytic mutant), or SIR2. Error bars show standard error of the mean from three biological replicates.
The NAD-dependent deacetylases have been reported to deacetylate H4K16 to mediate silencing at the telomeres and the silent mating loci (Krebs 2007). Previous studies have shown that NAM inhibits Sir2p both in vitro and in vivo (Sanders et al. 2007). We predicted that inhibiting H4K16 deacetylation with NAM in combination with overexpression of SAS2, SAS4, and SAS5 might enhance the rate of chromosome missegregation. Indeed, we found that addition of NAM resulted in increased chromosome loss in wild-type strains expressing GAL-SAS2, GAL-SAS4, and GAL-SAS5 when compared to the vector control (∼3, ∼3.5, and ∼2 per 1000 colonies, respectively) (Figure 7A).
If the enhanced chromosome loss defect conferred by NAM is in part through inhibition of SIR2, then deletion of SIR2 or a sir2-H364Y catalytic mutant may lead to similar defects in chromosome segregation. We deleted SIR2 in the reporter strain and transformed the strain with vector or vector containing either sir2-H364Y or wild-type SIR2 and measured the number of half-sectored colonies as described for Figure 7A. We observed that the sir2∆ strain with either vector or sir2-H364Y displayed an increase in chromosome loss when compared to the strain with the complementing SIR2 plasmid (∼7–8 vs. <2 per 1000 colonies, respectively) (Figure 7B). These results show that the catalytic activity of Sir2p contributes to accurate chromosome segregation and that deregulation of histone H4 acetylation leads to chromosome loss.
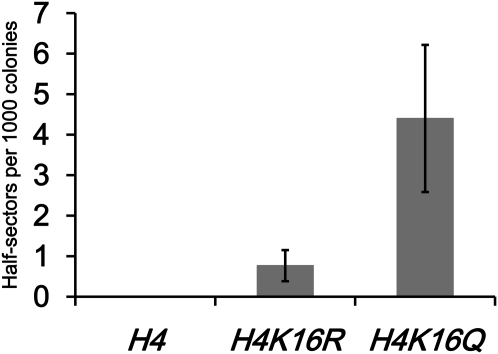
Histone H4K16Q acetylmimic mutant exhibits a chromosome loss phenotype
The observed growth and chromosome loss defects associated with altered H4 acetylation activity suggest that deregulated H4K16 acetylation may be the underlying cause for these phenotypes. To directly test this possibility, we created reporter strains that express either wild-type H4, H4K16R, which is nonacetylatable, or H4K16Q, an acetylmimic as the only forms of histone H4 and measured the rate of chromosome loss. We observed on average <1 half-sectored colony per 1000 for wild-type H4 and H4K16R strains (Figure 8). In contrast, H4K16Q mutants showed increased chromosome loss with an average ∼4 half-sectored colonies per 1000 (Figure 8). Taken together, our results are consistent with the conclusion that acetylated lysine 16 on histone H4 may underlie the SAS-mediated SDL and sensitivity to NAM in cse4-1 and hhf1-20 strains and increased rate of chromosome loss in sir2Δ, sir2-H364Y, and wild-type cells treated with NAM.
Figure 8 .
An H4K16Q acetylmimic mutant shows increased chromosome loss. Chromosome loss assays were performed with strains carrying a nonessential minichromosome that expresses wild-type H4 (YMB8155), H4K16R (YMB8156), or H4K16Q (YMB8157). Loss of the minichromosome within the first cell division gives rise to colonies that are half red and half white. We measured colonies that were at least half red. Bar graph shows the number of half-sectored colonies per 1000. Shown are results for three independent experiments and error bars show standard error of the mean.
Discussion
The function of histone H4 hypoacetylation in regional centromeres has remained an open question in chromosome biology. In this report, we show that hypoacetylated H4 is present at point centromeres in S. cerevisiae and that deregulation of H4 acetylation affects kinetochore function. We propose that hypoacetylated H4K16 is important for maintaining the integrity of the kinetochore and accurate chromosome segregation. That there were low levels of tAcH4, even at a minimal centromere flanked by heterologous sequences on a plasmid suggests that the kinetochore may contribute to the acetylation state of centromeric H4. On the basis of our results, we propose that H4K16 acetylation is a key modification that impacts kinetochore function and chromosome segregation. Our conclusion is supported by the following: (a) sensitivity of cse4 and hhf1 strains to NAM and SAS overexpression, (b) higher levels of centromeric H4K16Ac in wild-type and cse4-1 strains treated with NAM, (c) increased chromosome loss in wild-type strains overexpressing subunits of the SAS complex, (d) increased chromosome loss in a SIR2 deletion and sir2-H364Y catalytic mutant, and (e) increased chromosome loss in mutants expressing H4K16Q acetylmimic but not in H4K16R nonacetylatable mutants.
We hypothesize that the Cse4p-H4 interactions within the histone fold domain of Cse4p plays a major role in protecting cells from the effects of high levels of histone acetylation. When these interactions are compromised, as in the cse4 and hhf1-20 mutants, a hypoacetylated N-terminal H4 becomes essential to maintain kinetochore integrity. This is supported by our results that showed both wild-type and cse4-1 strains had an increase in centromeric acetylation of H4K16 when treated with NAM but unlike cse4-1 and hhf1-20 mutants, wild-type strains were not sensitive to NAM. Moreover, several kinetochore mutants (ndc10-1, mif2-2, mcm21, and ctf19) did not display growth defects when treated with NAM further supporting the hypothesis that the effects of NAM originate at the Cse4p-H4 complex. Similar to the results with NAM treatment, we propose that the phenotypes conferred by overexpression of SAS subunits are consistent with defects that originate at the Cse4p-H4 centromeric nucleosome. First, SAS2 exhibited SDL in C-terminal cse4 mutants (cse4-1 and cse4-111) with defects in interactions with histone H4, but not an N-terminal cse4 mutant (cse4-39). Second, overexpression of SAS2 results in SDL in hhf1-20 mutants that are predicted to have weakened interactions with Cse4p (Smith et al. 1996). Third, overexpression of SCM3 suppressed the SAS2 induced SDL in cse4 and hhf1-20 mutants. Scm3p, which coimmunoprecipitates with Cse4p, is required for kinetochore function and its overexpression suppresses temperature sensitivity of cse4-1 strains (Camahort et al. 2007; Mizuguchi et al. 2007; Stoler et al. 2007). Suppression of the SAS SDL in cse4 and hhf1-20 by SCM3 overexpression suggests that Scm3p might interact with Cse4p-H4 such that the nucleosome becomes protected from SAS-mediated acetylation, help recruit an H4 deacetylase to counteract the effects of SAS overexpression, or stabilizes mutant Cse4p interactions with H4.
A role for hypoacetylated histone H4 in chromosome segregation is further supported by our observations that sir2Δ and a sir2-H364Y catalytically inactive mutant exhibit increased chromosome loss rates. Previously, Sir2p was shown to be important for assembly of silent telomeric chromatin by deacetylation of H4K16 and Sas2p was necessary to restrict the spread of silenced chromatin into euchromatin (Kimura et al. 2002; Suka et al. 2002; Shia et al. 2006). In the absence of SIR2, SAS2-mediated acetylation of H4K16 occurs inappropriately at telomeres, thereby altering their chromatin structure. Moreover, overexpression of SIR2 has been reported to cause an increase in chromosome loss (Holmes et al. 1997). However, the mechanism of this defect is not understood. The phenotypes we observed for overexpression of SAS2 might also reflect an imbalance with Sir2p activity leading to higher levels of acetylated H4K16. We hypothesize that Sir2p might act as the deacetylase that maintains low levels of centromeric acetylated H4K16. However, ChIP experiments revealed that Sir1p but not Sir2p, Sir3p, or Sir4p was present at the kinetochore (Sharp et al. 2003). Perhaps the association of Sir2p is weak and cannot be detected by ChIP or Sir2p might act on the Cse4p-H4 complex prior to centromeric assembly. Biochemical work demonstrated that Sir2p can deacetylate histone octamers and free histones, lending support for the latter possibility (Parsons et al. 2003). Sir2p has been shown to be important for transcriptional silencing within regional centromeric chromatin in Drosophila, but its effect on centromeric H4 and chromosome segregation has not been explored (Rosenberg and Parkhurst 2002). Given the role for SIR2 at telomeres, it is possible that the chromosome loss phenotype of the sir2 mutants may be due to telomeric dysfunction, which in turn leads to chromosome missegregation. While we cannot rule this out completely, we propose that our genetic analysis of cse4 and hhf1-20 mutants are consistent with centromeric-based defects due to altered histone acetylation.
Our genetic results with the catalytic mutant, sas2m1, the SDL conferred by overexpression of each of the SAS complex subunits, the loss of viability in cse4 and hhf1-20 mutants treated with NAM are all effects that support an H4K16 acetylation-based mechanism. To address this directly, we measured chromosome loss rates in H4K16R and H4K16Q mutants, in which K16 is nonacetylatable or mimics acetylation, respectively. Consistent with our hypothesis, we observed higher levels of chromosome loss in the H4K16Q strains compared to the H4K16R and wild-type H4 strains. We also showed a correlation between low levels of H4K16Ac and the presence of Cse4p at noncentromeric regions, suggesting that Cse4p either prefers to assemble with hypoacetylated H4 or the Cse4p-H4 complex is a substrate of deacetylases before or after nucleosome assembly. This is perhaps reminiscent of trans-histone modification pathways previously reported for canonical histones (Fingerman et al. 2007) and raises the possibility that a similar pathway may regulate modification of centromeric histone H4.
That we find wild-type cells remain relatively unaffected by altered HAT activity, while there is a severe loss in viability in kinetochore mutants, provides an exciting avenue for further research. Cancer therapies based on antimitotic drugs are being actively investigated (Jackson et al. 2007; Carpinelli and Moll 2009; Wood et al. 2010). On the basis of our work, combining HDAC inhibitors with drugs that compromise kinetochore function may provide a more efficacious approach to treat cancers with minimal effect on normal cells. Given the vast genetic tools in budding yeast, a comprehensive analysis of mitotic pathways that are vulnerable to altered H4 acetylation by small molecule HDAC inhibitors will facilitate the identification of potential targets for cancer therapy.
In summary, our work establishes that the point centromere of S. cerevisiae contains hypoacetylated H4. We show that hypoacetylated H4 at the centromere is a conserved feature in eukaryotes, despite having highly divergent centromeric sequences. Furthermore, our studies suggest that hypoacetylated H4K16 is important for kinetochore function and accurate chromosome segregation. While CenH3 alone cannot establish an active kinetochore in any system studied thus far, our results show a correlation between the acetylation state of H4 and regions of Cse4p association. Future studies will help understand whether the role for hypoacetylation of H4K16 in chromosome segregation is a conserved feature across all eukaryotic centromeres.
Acknowledgments
The authors thank S. Biggins, C. Chan, Danesh Moazed, J. Gerton, P. Megee, V. Measday, M. Smith, L. Pillus, and J. Workman for strains and plasmids. We are grateful to members of the Basrai lab, R. Baker, and D. Clarke for critical discussions and comments on the manuscript. This research was supported by the Intramural Research Program of the National Cancer Institute, National Institutes of Health.
Literature Cited
- Allard S., Utley R. T., Savard J., Clarke A., Grant P., et al. , 1999. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 18: 5108–5119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson M., Haase J., Yeh E., Bloom K., 2009. Function and assembly of DNA looping, clustering, and microtubule attachment complexes within a eukaryotic kinetochore. Mol. Biol. Cell 20: 4131–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au W. C., Crisp M. J., DeLuca S. Z., Rando O. J., Basrai M. A., 2008. Altered dosage and mislocalization of histone H3 and Cse4p lead to chromosome loss in Saccharomyces cerevisiae. Genetics 179: 263–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom K. S., Carbon J., 1982. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell 29: 305–317 [DOI] [PubMed] [Google Scholar]
- Bouck D. C., Bloom K., 2007. Pericentric chromatin is an elastic component of the mitotic spindle. Curr. Biol. 17: 741–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouck D. C., Joglekar A. P., Bloom K. S., 2008. Design features of a mitotic spindle: balancing tension and compression at a single microtubule kinetochore interface in budding yeast. Annu. Rev. Genet. 42: 335–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camahort R., Li B., Florens L., Swanson S. K., Washburn M. P., et al. , 2007. Scm3 is essential to recruit the histone H3 variant Cse4 to centromeres and to maintain a functional kinetochore. Mol. Cell 26: 853–865 [DOI] [PubMed] [Google Scholar]
- Camahort R., Shivaraju M., Mattingly M., Li B., Nakanishi S., et al. , 2009. Cse4 is part of an octameric nucleosome in budding yeast. Mol. Cell 35: 794–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpinelli P., Moll J., 2009. Is there a future for Aurora kinase inhibitors for anticancer therapy? Curr. Opin. Drug Discov. Devel. 12: 533–542 [PubMed] [Google Scholar]
- Crotti L. B., Basrai M. A., 2004. Functional roles for evolutionarily conserved Spt4p at centromeres and heterochromatin in Saccharomyces cerevisiae. EMBO J. 23: 1804–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang W., Steffen K. K., Perry R., Dorsey J. A., Johnson F. B., et al. 2009. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 459: 802–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall K., 2007. Epigenetic control of centromere behavior. Annu. Rev. Genet. 41: 63–81 [DOI] [PubMed] [Google Scholar]
- Fingerman I. M., Li H. C., Briggs S. D., 2007. A charge-based interaction between histone H4 and Dot1 is required for H3K79 methylation and telomere silencing: identification of a new trans-histone pathway. Genes Dev. 21: 2018–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama S., Biggins S., 2007. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc. Natl. Acad. Sci. USA 104: 14706–14711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelperin D. M., White M. A., Wilkinson M. L., Kon Y., Kung L. A., et al. 2005. Biochemical and genetic analysis of the yeast proteome with movable ORF collection. Genes Dev. 23: 2816–2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowczewski L., Yang P., Kalashnikova T., Santisteban M. S., Smith M. M., 2000. Histone-histone interactions and centromere function. Mol. Cell. Biol. 20: 5700–5711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T., Fujita Y., Iwasaki O., Adachi Y., Takahashi K., et al. , 2004. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell 118: 715–729 [DOI] [PubMed] [Google Scholar]
- Hieter P., Mann C., Snyder M., Davis R. W., 1985. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell 40: 381–392 [DOI] [PubMed] [Google Scholar]
- Holmes S. G., Rose A. B., Steuerle K., Saez E., Sayegh S., et al. , 1997. Hyperactivation of the silencing proteins, Sir2p and Sir3p, causes chromosome loss. Genetics 145: 605–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson J. R., Patrick D. R., Dar M. M., Huang P. S., 2007. Targeted anti-mitotic therapies: Can we improve on tubulin agents? Nat. Rev. Cancer 7: 107–117 [DOI] [PubMed] [Google Scholar]
- Kanta H., Laprade L., Almutairi A., Pinto I., 2006. Suppressor analysis of a histone defect identifies a new function for the Hda1 complex in chromosome segregation. Genetics 173: 435–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen G. H., Allshire R. C., 1997. The case for epigenetic effects on centromere identity and function. Trends Genet. 13: 489–496 [DOI] [PubMed] [Google Scholar]
- Kastenmayer J. P., Lee M. S., Hong A. L., Spencer F. A., Basrai M. A., 2005. The C-terminal half of Saccharomyces cerevisiae Mad1p mediates spindle checkpoint function, chromosome transmission fidelity and CEN association. Genetics 170: 509–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A., Umehara T., Horikoshi M., 2002. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat. Genet. 32: 370–377 [DOI] [PubMed] [Google Scholar]
- Krebs J. E., 2007. Moving marks: dynamic histone modifications in yeast. Mol. Biosyst. 3: 590–597 [DOI] [PubMed] [Google Scholar]
- Liu C. L., Kaplan T., Kim M., Buratowski S., Schreiber S. L., et al. , 2005. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 3: e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar C. B., Grunstein M., 2006. Genome-wide patterns of histone modifications in yeast. Nat. Rev. Mol. Cell Biol. 7: 657–666 [DOI] [PubMed] [Google Scholar]
- Mizuguchi G., Xiao H., Wisniewski J., Smith M. M., Wu C., 2007. Nonhistone Scm3 and histones CenH3–H4 assemble the core of centromere-specific nucleosomes. Cell 129: 1153–1164 [DOI] [PubMed] [Google Scholar]
- Osada S., Sutton A., Muster N., Brown C. E., Yates J. R., 3rd, et al. , 2001. The yeast SAS (something about silencing) protein complex contains a MYST-type putative acetyltransferase and functions with chromatin assembly factor Asf1. Genes Dev. 15: 3155–3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons X. H., Garcia S. N., Pillus L., Kadonaga J. T., 2003. Histone deacetylation by Sir2 generates a transcriptionally repressed nucleoprotein complex. Proc. Natl. Acad. Sci. USA 100: 1609–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M. I., Parkhurst S. M., 2002. Drosophila Sir2 is required for heterochromatic silencing and by euchromatic Hairy/E(Spl) bHLH repressors in segmentation and sex determination. Cell 109: 447–458 [DOI] [PubMed] [Google Scholar]
- Rusche L. N., Kirchmaier A. L., Rine J., 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72: 481–516 [DOI] [PubMed] [Google Scholar]
- Sanders B. D., Zhao K., Slama J. T., Marmorstein R., 2007. Structural basis for nicotinamide inhibition and base exchange in Sir2 enzymes. Mol. Cell 25: 463–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders M.J., Yeh E., Grunstein M., Bloom K., 1990. Nucleosome depletion alters the chromatin structure of Saccharomyces cerevisiae centromeres. Mol. Cell. Biol. 10: 5721–5727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian M. D., Grunstein M., 2007. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76: 75–100 [DOI] [PubMed] [Google Scholar]
- Sharp J. A., Franco A. A., Osley M. A., Kaufman P. D., 2002. Chromatin assembly factor I and Hir proteins contribute to building functional kinetochores in S. cerevisiae. Genes Dev. 16: 85–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp J. A., Krawitz D. C., Gardner K. A., Fox C. A., Kaufman P. D., 2003. The budding yeast silencing protein Sir1 is a functional component of centromeric chromatin. Genes Dev. 17: 2356–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shia W. J., Osada S., Florens L., Swanson S. K., Washburn M. P., et al. , 2005. Characterization of the yeast trimeric-SAS acetyltransferase complex. J. Biol. Chem. 280: 11987–11994 [DOI] [PubMed] [Google Scholar]
- Shia W. J., Li B., Workman J. L., 2006. SAS-mediated acetylation of histone H4 Lys 16 is required for H2A.Z incorporation at subtelomeric regions in Saccharomyces cerevisiae. Genes Dev. 20: 2507–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shogren-Knaak M., Peterson C. L., 2006. Switching on chromatin: mechanistic role of histone H4–K16 acetylation. Cell Cycle 5: 1361–1365 [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P., 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae Genetics 122: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E. R., Eisen A., Gu W., Sattah M., Pannuti A., et al. , 1998. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc. Natl. Acad. Sci. USA 95: 3561–3565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. M., Yang P., Santisteban M. S., Boone P. W., Goldstein A. T., et al. , 1996. A novel histone H4 mutant defective in nuclear division and mitotic chromosome transmission. Mol. Cell. Biol. 16: 1017–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler S., Keith K. C., Curnick K. E., Fitzgerald-Hayes M., 1995. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 9: 573–586 [DOI] [PubMed] [Google Scholar]
- Stoler S., Rogers K., Weitze S., Morey L., Fitzgerald-Hayes M., et al. , 2007. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc. Natl. Acad. Sci. USA 104: 10571–10576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl B. D., Allis C. D., 2000. The language of covalent histone modifications. Nature 403: 41–45 [DOI] [PubMed] [Google Scholar]
- Suka N., Luo K., Grunstein M., 2002. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat. Genet. 32: 378–383 [DOI] [PubMed] [Google Scholar]
- Sullivan B. A., Karpen G. H., 2004. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat. Struct. Mol. Biol. 11: 1076–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton A., Shia W. J., Band D., Kaufman P. D., Osada S., et al. , 2003. Sas4 and Sas5 are required for the histone acetyltransferase activity of Sas2 in the SAS complex. J. Biol. Chem. 278: 16887–16892 [DOI] [PubMed] [Google Scholar]
- Tanny J. C., Kirkpatrick D. S., Gerber S. A., Gygi S. P., Moazed D., 2004. Budding yeast silencing complexes and regulation of Sir2 activity by protein-protein interactions. Mol. Cell Biol. 16: 6931–6946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Schones D. E., Zhao K., 2009. Characterization of human epigenomes. Curr. Opin. Genet. Dev. 19: 127–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood K. W., Lad L., Luo L., Qian X., Knight S. D., et al. , 2010. Antitumor activity of an allosteric inhibitor of centromere-associated protein-E. Proc. Natl. Acad. Sci. USA 107: 5839–5844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E., Haase J., Paliulis L. V., Joglekar A., Bond L., et al. , 2008. Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr. Biol. 18: 81–90 [DOI] [PMC free article] [PubMed] [Google Scholar]