Abstract
Reproductive isolation between species is often caused by deleterious interactions among loci in hybrids. Finding the genes involved in these incompatibilities provides insight into the mechanisms of speciation. With recently diverged subspecies, house mice provide a powerful system for understanding the genetics of reproductive isolation early in the speciation process. Although previous studies have yielded important clues about the genetics of hybrid male sterility in house mice, they have been restricted to F1 sterility or incompatibilities involving the X chromosome. To provide a more complete characterization of this key reproductive barrier, we conducted an F2 intercross between wild-derived inbred strains from two subspecies of house mice, Mus musculus musculus and Mus musculus domesticus. We identified a suite of autosomal and X-linked QTL that underlie measures of hybrid male sterility, including testis weight, sperm density, and sperm morphology. In many cases, the autosomal loci were unique to a specific sterility trait and exhibited an effect only when homozygous, underscoring the importance of examining reproductive barriers beyond the F1 generation. We also found novel two-locus incompatibilities between the M. m. musculus X chromosome and M. m. domesticus autosomal alleles. Our results reveal a complex genetic architecture for hybrid male sterility and suggest a prominent role for reproductive barriers in advanced generations in maintaining subspecies integrity in house mice.
EXPLAINING patterns of biodiversity requires a mechanistic understanding of how new species arise. Genetic dissection of reproductive barriers between nascent species is a powerful approach for revealing the causes of speciation. This strategy has been highly successful, particularly in Drosophila, where nine specific genes that cause reduced fitness in hybrids have been identified (Sawamura and Yamamoto 1997; Ting et al. 1998; Barbash et al. 2003; Presgraves et al. 2003; Brideau et al. 2006; Bayes and Malik 2009; Ferree and Barbash 2009; Phadnis and Orr 2009; Tang and Presgraves 2009). Despite this progress, several factors motivate additional genetic studies of postzygotic isolation. Although other species have provided important insights (e.g., Sweigart et al. 2006; Bomblies et al. 2007; Moyle 2007; Chen et al. 2008; Lee et al. 2008; Long et al. 2008; Kao et al. 2010; Martin and Willis 2010), current knowledge of the genetics of postzygotic isolation remains highly biased toward Drosophila. In addition, some of the best-studied species hybridize rarely or not at all in the wild, complicating attempts to connect reproductive barriers examined in the lab with gene flow in nature. Finally, there is a dearth of information on taxa in the process of speciating, where it is possible to find genetic changes responsible for the initial development of reproductive isolation.
House mice provide a powerful system for studying the genetics of reproductive isolation. Three recognizable subspecies diverged from a common ancestor only ∼500,000 generations ago (She et al. 1990; Boursot et al. 1996; Suzuki et al. 2004; Salcedo et al. 2007; Geraldes et al. 2008). Despite this short divergence time, several lines of evidence indicate reproductive isolation between two of the subspecies, Mus musculus musculus and Mus musculus domesticus. The subspecies meet in a well-studied zone of secondary contact that stretches across central Europe (Boursot et al. 1993; Sage et al. 1993), where diagnostic allele frequencies shift rapidly over short geographic distances (Boursot et al. 1993; Sage et al. 1993). Individual loci often exhibit marked reductions in gene flow (Vanlerberghe et al. 1986; Dod et al. 1993; Munclinger et al. 2002; Payseur et al. 2004; Dod et al. 2005; Payseur and Nachman 2005; Raufaste et al. 2005; Macholán et al. 2007, 2008; Teeter et al. 2008, 2010), as expected for genomic regions involved in reproductive isolation (Payseur 2010). Hybrids sampled from this zone show signs of reduced fitness, including increased parasite loads (Sage et al. 1986; Moulia et al. 1991, 1993). In addition, M. m. musculus and M. m. domesticus exhibit prezygotic isolation, including preferences for mates from the same subspecies (Laukaitis et al. 1997; Talley et al. 2001; Smadja and Ganem 2002, 2005; Smadja et al. 2004; Ganem et al. 2008) and higher fertilization rates by sperm from the same subspecies when females are multiply mated (Dean and Nachman 2009). The most direct evidence for reproductive isolation comes from laboratory crosses involving wild-derived inbred strains, where F1 hybrid male sterility is routinely observed, usually without hybrid female sterility (Iványi et al. 1969; Forejt and Iványi 1974; Storchová et al. 2004; Britton-Davidian et al. 2005; Vyskočilová et al. 2005, 2009; Good et al. 2008a,b).
As with many other cases of postzygotic isolation (Coyne and Orr 2004), hybrid male sterility in house mice is thought to be caused primarily by genetic changes that fail to interact properly in hybrids (“Dobzhansky–Muller incompatibilities”) (Bateson 1909; Dobzhansky 1936; Muller 1942). Substantial effort directed toward finding the incompatibilities that underlie hybrid male sterility between M. m. domesticus and M. m. musculus has revealed some genetic patterns. First, the X chromosome is an important contributor (Oka et al. 2004; Storchová et al. 2004; Britton-Davidian et al. 2005; Good et al. 2008a,b, 2010; Vyskočilová et al. 2009). F1’s from most crosses follow Haldane’s rule (Haldane 1922), with hybrid females showing few signs of sterility. In hybrid males, sterility is more common when M. m. musculus is the mother (Britton-Davidian et al. 2005; Good et al. 2008b; Vyskočilová et al. 2009). The role for the M. m. musculus X chromosome suggested by these observations has been confirmed through backcrosses (Storchová et al. 2004) and chromosomal introgression studies (Good et al. 2008a; Gregorová et al. 2008). Reduced gene flow of X-linked loci across the hybrid zone (Tucker et al. 1992; Dod et al. 1993; Munclinger et al. 2002; Payseur et al. 2004; Dod et al. 2005; Macholán et al. 2007) raises the possibility that this chromosome also confers hybrid male sterility in natural populations. Additional patterns involving the X chromosome suggest that hybrid male sterility in house mice is genetically complex. At least four regions of the M. m. musculus X chromosome cause sterile phenotypes when introgressed onto a M. m. domesticus genomic background (Good et al. 2008a). Furthermore, genetic mapping in multiple crosses suggests that interactions involving several segments of the X chromosome are required to generate hybrid male sterility (Oka et al. 2004; Storchová et al. 2004).
Although previous studies have focused on the role of the X chromosome, evidence exists for the involvement of autosomal loci in hybrid male sterility. The only gene known to cause hybrid sterility in vertebrates, Prdm9, resides on chromosome 17 in house mice (Forejt and Iványi 1974; Forejt 1996; Mihola et al. 2009). Consistent with a Dobzhansky–Muller incompatibility framework, the sterility effect of Prdm9 requires interaction with additional, unidentified loci (Forejt and Iványi 1974; Forejt 1996; Mihola et al. 2009). Backcrosses between M. m. domesticus and Mus musculus molossinus (a hybrid between M. m. musculus and Mus musculus castaneus) have identified several autosomal QTL that affect hybrid sterility when the entire M. m. molossinus X chromosome is present on the M. m. domesticus genetic background (Oka et al. 2007). Substituting M. m. musculus chromosomes 10 or 11 onto a mostly M. m. domesticus background (C57BL/6J inbred line) strongly reduces litter size (Gregorová et al. 2008). Finally, some autosomal loci show reduced introgression across the European hybrid zone (Macholán et al. 2007; Teeter et al. 2008, 2010).
The observations of Haldane’s rule, asymmetrical isolation, and the role of the X chromosome in hybrid sterility between M. m. musculus and M. m. domesticus have encouraged most investigators to focus attention on F1 sterility. Importantly, F1 reproductive barriers might not be representative of those in later generations. Both data from other species (Presgraves 2003) and theory (Muller 1942) suggest that incompatibilities involving recessive mutations will be the most numerous, and these disrupted interactions are completely hidden in F1’s. Remarkably, a standard F2 QTL study of hybrid male sterility between M. m. musculus and M. m. domesticus, which would sample a broader range of multi-locus genotype combinations, has not been reported. To characterize the genetic architecture of hybrid male sterility on a genome-wide scale, we conducted an F2 intercross between inbred lines of M. m. musculus and M. m. domesticus. We detected a large number of autosomal loci associated with different measures of hybrid male sterility, with each phenotype distinguished by a unique genetic architecture. In addition, we found several novel Dobzhansky–Muller interactions between the M. m. musculus X chromosome and homozygous M. m. domesticus QTL on the autosomes. Our results revealed a number of QTL with recessive effects, highlighting an important role for reproductive isolation beyond the F1 generation in house mice.
Materials and Methods
Animal husbandry and crossing design
Crosses were conducted using two wild-derived inbred strains purchased from The Jackson Laboratory (http://www.jax.org): M. m. domesticus (WSB/EiJ) and M. m. musculus (PWD/PhJ). WSB/EiJ was derived from a population in the eastern United States, and PWD/PhJ was derived from a population in the Czech Republic (Gregorová and Forejt 2000). Although WSB/EiJ originated in North America, allozyme studies suggest that M. m. domesticus animals from eastern North America are genetically similar to European mice closer to the ancestral subspecies range (Selander et al. 1969; Selander and Yang 1969).
Parents were crossed in reciprocal directions (M. m. domesticus × M. m. musculus and M. m. musculus × M. m. domesticus) to generate F1 hybrids. F1’s were bred with siblings according to the following scheme: (M. m. domesticus × M. m. musculus)F1 × (M. m. domesticus × M. m. musculus)F1 and (M. m. musculus × M. m. domesticus)F1 × (M. m. musculus × M. m. domesticus)F1. All crosses occurred within the University of Wisconsin School of Medicine and Public Health mouse facility according to animal care protocols approved by the University of Wisconsin Animal Care and Use Committee. Mice were provided with food and water ad libitum. Pups were weaned into same-sex sibling groups at 21 days, and males were separated into individual cages at ∼56 days. Males were killed for phenotyping at 70 days of age (±5 days) using carbon dioxide.
Quantification of male fertility phenotypes
We quantified five diagnostic measures of subfertility and sterility in house mice: testis weight (Iványi et al. 1969; Forejt and Iványi 1974), sperm density (Searle and Beechey 1974; Storchová et al. 2004; Vyskočilová et al. 2005), sperm head morphology (Oka et al. 2004; Storchová et al. 2004; Kawai et al. 2006), proportion of abnormal sperm (Kawai et al. 2006), and cross-sectional area of seminiferous tubules. Although we did not perform additional matings to directly examine the fertility of each F2 male, the measured phenotypes allowed a fine-scale dissection of different components of hybrid sterility. Testes were weighed fresh upon dissection, fixed overnight in Bouin’s, and washed in an ethanol series of 25, 50, and 75%. The right testis was embedded in paraffin, sectioned at 6 μm, and stained with hematoxylin and eosin according to standard procedures. Testis weight was positively correlated with body weight in F2 males (Pearson’s r = 0.306, P < 0.001). To account for this correlation, we divided testis weight by body weight prior to QTL analyses. QTL mapping was also conducted using the residual trait scores from a least-squares regression of testis weight on body weight and using absolute testis weights. Because mapping results were similar for all procedures, we focus on results using ratios. All genetic analyses of testis weight focused on the right testis to avoid effects of size differences between the testes.
Sperm density was estimated using a Makler counting chamber (Sefi-Medical Instruments) at ×200 magnification under a light microscope. The left and right cauda epididymides were dissected and coarsely chopped in 500 μl of sperm media solution (bovine gamete medium 3) (Vredenburgh-Wilberg and Parrish 1995) without calcium chloride, normally required for capacitation. Sperm were allowed to passively diffuse into the media over 20 min, and 5 μl of sperm suspension was loaded in the Makler counting chamber. Each male was measured three times, and the average density was used as the final estimate.
Sperm head morphology was measured in samples collected from the epididymides. Sperm suspension was air dried on a glass side, fixed in 5% acetic acid:95% ethanol for 3 min, stained with 1% Eosin-Y for 4 min, washed three times in 70% ethanol, air dried, and then mounted in mounting medium (Richard-Allan Scientific). Sperm were measured at ×630 magnification using differential interference contrast (DIC) imaging. Five sperm heads were randomly selected per male and traced using the lasso tool in Adobe Photoshop. Traced sperm heads were converted to binary images and quantified using a standardized elliptic Fourier descriptor as implemented in the software package SHAPE (Iwata and Ukai 2002; http:/cse.naro.affrc.go.jp/iwatah/shape/). Sperm head morphologies were more similar within males than between males for the first 12 harmonics of the elliptic Fourier descriptors (ANOVA, P < 0.05). This indicated that genetic factors contributed to variance in sperm head morphology. One elliptic Fourier descriptor was calculated for each male by averaging the coefficients from the five individual sperm head descriptors. Within the F2 intercross, the major sources of shape variation were identified using a principal component analysis. The principal component scores were transformed to normal quantiles to improve the fit to normality.
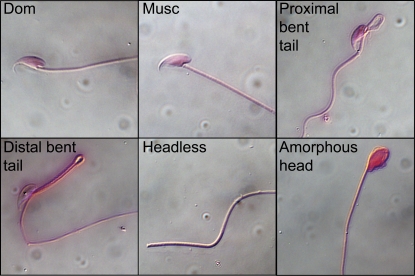
The proportion of abnormal sperm in each male was estimated by randomly counting 100 sperm from DIC images taken at ×200 magnification. Sperm were categorized on the basis of four primary abnormalities identified within the F2 intercross: proximal bent tail, distal bent tail, missing head or missing tail, and severely amorphous head (Figure 1). Sperm could be assigned to multiple categories, but the majority of sperm fit into a single category. As a result, the sum of sperm from the four abnormal categories was used as a close approximation to the total number of abnormal sperm. The proportions for each type of abnormal sperm were transformed using a modified arcsine square-root transformation (Freeman and Tukey 1950) to improve the fit to normality.
Figure 1 .
The most frequent abnormal sperm types observed in F2 males. Normal morphologies are depicted for both parents, M. m. domesticusWSB (Dom.) and M. m. musculusPWD (Musc.).
A cross-sectional area of seminiferous tubules was measured only in stage VII tubules (Russell et al. 1990) to control for variance across stages of spermatogenesis. In each male, the major and minor axes of 10 stage VII seminiferous tubules were measured using Spectrum software (Aperio Technologies). Elliptical area was calculated for each seminiferous tubule, and the average of the 10 tubule areas was used for QTL mapping. In males with <10 stage VII tubules, averages were taken over the number of tubules present. A cross-sectional area of seminiferous tubules was positively correlated with testis weight (Pearson’s r = 0.561, P < 0.001). To account for this correlation, QTL analyses were conducted using the residual trait scores from a least-squares regression of seminiferous tubule area on testis weight.
Genotyping
Genomic DNA was extracted from liver tissue with the Wizard Genomic DNA Purification Kit (Promega) following manufacturer-recommended protocols. Single nucleotide polymorphism (SNP) markers were designed from the Perlegen phase 4 release of the mouse resequencing project (Frazer et al. 2007). A total of 331 SNP markers were genotyped using the Sequenom iPLEX MassARRAY system (San Diego) as previously described (Gabriel et al. 2009).
Quality control procedures
Several quality control measures were applied to ensure accurate QTL mapping. SNPs that failed parental and F1 controls, significantly deviated from an expected autosomal 1:2:1 segregation ratio, or were missing >20% of SNP calls, were removed (Dumont et al. 2011). Animals missing genotypes at ≥20% of SNPs were removed before constructing the genetic map (Dumont et al. 2011; these animals were included in QTL analyses). After these conservative filtering steps, there were 198 SNPs across the autosomes, X chromosome, Y chromosome, and mitochondrion. Genotypes were compared for all pairs of males to search for possible duplicate samples. With one exception, genotypic similarity fell within the bounds of 15.5 and 71.1%. Males were also examined across autosomal markers for high levels of heterozygosity, which could suggest poor DNA quality or contaminated DNA samples. The two most genotypically similar males also contained the highest fraction of heterozygous SNPs (84.6 and 75.8%) and were removed from the data set. This quality control procedure retained 310 F2 males. Only 3.2% of genotypes were missing in the entire data matrix.
QTL analyses
A genetic map was estimated from a total of 553 males and females with the est.map function of R/qtl (Broman et al. 2003; Broman and Sen 2009), assuming a genotyping error rate of zero (the stringent filtering scheme for markers ensured few genotyping errors) (Dumont et al. 2011) and a Carter–Falconer mapping function (Carter and Falconer 1951; Broman et al. 2002). SNP order on the linkage map matched that in the reference genome sequence (Mouse Genome Sequencing Consortium et al. 2002), suggesting no large chromosomal rearrangements between the two inbred strains. SNPs were spaced at an average distance of 7.03 cM. Physical positions were interpolated between markers using the physical and genetic map positions of flanking markers.
Standard interval mapping was implemented using the scanone function in R/qtl (Lander and Botstein 1989; Broman and Sen 2009). Genotype probabilities between markers were calculated at a grid size of 2 cM and with a genotyping error rate of 0.001. This slightly higher genotyping error rate was used because males removed during the initial SNP-filtering scheme were added back to maximize the number of animals used for QTL mapping. All phenotypes were analyzed using standard interval mapping except the abnormal sperm types, which were analyzed using the extended Haley–Knott method (Feenstra et al. 2006). Genome-wide significance thresholds were calculated from 1000 permutations of the autosomes and separate permutations for the X chromosome (Churchill and Doerge 1994; Broman et al. 2006; Broman and Sen 2009). For phenotypes that did not follow a normal distribution, results from several alternative mapping procedures were compared, including nonparametric interval mapping, interval mapping for binary traits, and a two-part model for phenotypes that exhibited a spike near zero in the distribution (Broman 2003; Broman and Sen 2009).
Joint analyses of multiple QTL were implemented using two methods. First, two-dimensional, two-QTL scans were conducted with the scantwo function in R/qtl using standard interval mapping (Sen and Churchill 2001; Broman and Sen 2009). For sperm head morphology and abnormal sperm types, mapping was performed using Haley–Knott regression (Haley and Knott 1992). Genotype probabilities were calculated at a grid size of 2 cM and with a genotyping error rate of 0.001. Significance thresholds were calculated from 10,000 permutations. Second, models incorporating multiple QTL were fit using the stepwiseqtl function, which uses a forward/backward stepwise search algorithm to select the best-fitting model (Manichaikul et al. 2009; Arends et al. 2010). Models were compared using a penalized LOD score with thresholds calculated from the 10,000 scantwo permutations. For sperm head morphology and abnormal sperm types, genotype probabilities were calculated every 3 cM with a genotyping error rate of 0.001 and mapped using Haley–Knott regression. For all other phenotypes, genotypes were imputed between markers every 3 cM using 600 draws with a genotyping error rate of 0.001 and mapped using multiple imputations. Six hundred draws were sufficient to produce consistent results between independent runs.
Results
F1 hybrid sterility
F1 hybrid male sterility is polymorphic among strains of M. m. musculus and M. m. domesticus (Vyskočilová et al. 2005, 2009; Good et al. 2008b; Piálek et al. 2008) and has not been evaluated in crosses between the wild-derived inbred strains PWD/PhJ (M. m. musculusPWD) and WSB/EiJ (M. m. domesticusWSB). To determine if F1 males were sterile in crosses between these strains, we quantified a range of phenotypes in 70-day-old F1 and parental males. Both parental strains were fertile in all crosses, although M. m. musculusPWD had a significantly lower relative right testis weight and sperm density than M. m. domesticusWSB (Table 1). There was strong, asymmetric F1 sterility between M. m. musculusPWD and M. m. domesticusWSB. Males with M. m. musculusPWD mothers consistently displayed higher levels of sterility for every trait measured (significantly lower relative right testis weight, sperm density, and seminiferous tubule area and significantly higher levels of each type of abnormal sperm) (Table 1; Figure 1). Sperm head morphology and overall testis histology also showed marked differences from the parental males. The apical hook was clearly reduced in the heads of the low number of sperm present (Figure 2A). All seminiferous tubules were characterized by a large reduction in or complete absence of post-meiotic round spermatids (Figure 3). In contrast, males with M. m. domesticusWSB mothers had phenotypic averages that were within or exceeded parental values. Sperm heads also exhibited clearly defined apical hooks, and seminiferous tubules had large populations of post-meiotic cells. These males had some degree of subfertility in seminiferous tubule area, with values that were significantly lower than parental means, but the areas were still significantly higher than the reciprocal F1 males.
Table 1 . Mean phenotypic values for parents and F1’s.
| Phenotype | M. m. musculusa | M. m. domesticusa | musculus × domesticusa | domesticus × musculusa |
|---|---|---|---|---|
| Right testis weight (mg) | 57.04 ± 4.43b (25) | 66.36 ± 11.51c (19) | 49.26 ± 7.75d (23) | 65.59 ± 8.13c (35) |
| Relative right testis weight (mg/g) | 3.57 ± 0.30b (25) | 3.95 ± 0.57c (19) | 2.63 ± 0.38d (23) | 3.61 ± 0.56b,c (35) |
| Sperm density (millions/ml) | 9.33 ± 3.37e (25) | 12.37 ± 4.08f (17) | 1.77 ± 3.46g (22) | 12.59 ± 4.80f (33) |
| Seminiferous tubule area (µm2) | 36,362.70 ± 2,206.02b (6) | 34,583.45 ± 5,328.86b (6) | 20,751.01 ± 4,021.83c (5) | 28,583.69 ± 1,260.76d (6) |
| Proximal bent tailh | 0.026 ± 0.031e (10) | 0.042 ± 0.022e (10) | 0.12 ± 0.064f (10) | 0.032 ± 0.057e (10) |
| Distal bent tailh | 0.027 ± 0.023e (10) | 0.011 ± 0.012e,f (10) | 0.029 ± 0.022e (10) | 0.0030 ± 0.0048f (10) |
| Headless/tailless spermh | 0.11 ± 0.058e (10) | 0.072 ± 0.036e (10) | 0.38 ± 0.10f (10) | 0.089 ± 0.036e (10) |
| Amorphous sperm headh | 0.0070 ± 0.0095e (10) | 0.013 ± 0.013e (10) | 0.10 ± 0.038f (10) | 0.0090 ± 0.0057e (10) |
| Total abnormal spermh | 0.17 ± 0.074e (10) | 0.14 ± 0.052e (10) | 0.63 ± 0.060f (10) | 0.13 ± 0.049e (10) |
Mean ± SD. N is within parentheses.
Groups significantly different by t-test, P < 0.05.
Groups significantly different by Mann–Whitney U-test, P < 0.05.
Proportion of abnormal sperm.
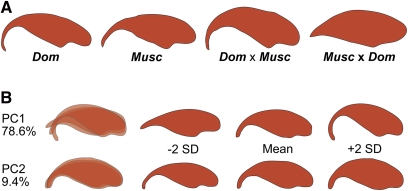
Figure 2 .
Epididymal sperm head morphologies. (A) M. m. domesticusWSB (Dom.), M. m. musculusPWD (Musc.), and the F1 hybrids. The sterile F1 direction (Musc. × Dom.) has a severely reduced apical hook. (B) F2 variation among epididymal sperm head morphology was characterized by two main principal components. The first principal component largely explained changes in the apical hook, whereas the second principal component characterized a change in sperm head width.
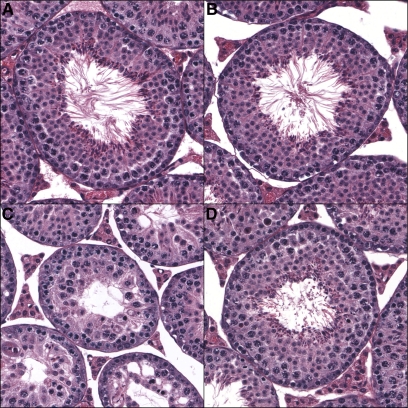
Figure 3 .
Stage VII seminiferous tubules. Cross sections from (A) M. m. domesticusWSB, (B) M. m. musculusPWD, (C) (M. m. musculusPWD × M. m. domesticusWSB)F1, and (D) (M. m. domesticusWSB × M. m. musculusPWD)F1 males. In the sterile F1 direction (M. m. musculusPWD × M. m. domesticusWSB)F1, there is a large reduction in post-meiotic cells, reducing the overall area of the tubule. All images are at ×200 magnification.
We paired F1 males with F1 females from the same parental cross direction to directly assess whether males were sterile. (M. m. domesticusWSB × M. m. musculusPWD)F1 males produced litters with (M. m. domesticusWSB × M. m. musculusPWD)F1 females in every intercross attempted (11 total crosses). Pairings between (M. m. musculusPWD × M. m. domesticusWSB)F1 males and (M. m. musculusPWD × M. m. domesticusWSB)F1 females resulted in no litters (4 total crosses), except in one case where small, male-biased litters were produced after 5 months of pairing (8 litters, 12 total males, 2 total females). To test whether the sterility that we observed in the (M. m. musculusPWD × M. m. domesticusWSB)F1 intercross was due to male rather than female sterility, we backcrossed F1 females to M. m. domesticusWSB males (2 total crosses). Both of these crosses regularly produced litters. The lack of offspring from (M. m. musculusPWD × M. m. domesticusWSB)F1 males suggests that these males were effectively sterile, and the phenotypes that we measured accurately reflected this sterility.
F2 hybrid sterility
To study the genetic architecture of hybrid male sterility, we measured phenotypes across 310 F2 males. Only 12 males were from the M. m. musculusPWD × M. m. domesticusWSB intercross direction, which limited our ability to detect sterility associated with the Y chromosome. We observed a wide expansion in the variance for all phenotypes in the F2’s, indicating that multiple loci contribute to hybrid male sterility (supporting information, Figure S1). There were significant correlations between most pairs of phenotypes (Table 2), raising the possibility that some loci contribute to multiple sterility measures. We calculated broad-sense heritability for each phenotype using the average phenotypic variance within parental strains and within F1’s as an estimate of environmental variance. For all phenotypes, we observed high broad-sense heritabilities (relative right testis weight: 0.838; sperm density: 0.718; seminiferous tubule area: 0.631; proximal bent tail: 0.517; distal bent tail: 0.789; headless/tailless sperm: 0.702; amorphous sperm heads: 0.846; and total number of abnormal sperm: 0.927), revealing a strong genetic component to the phenotypic variance. Broad-sense heritability was not calculated for the principal components of sperm head morphology because the variance estimated from the F2 is not directly comparable to the variance among parental and F1 shapes.
Table 2 . Spearman’s rank correlation coefficients between hybrid sterility phenotypes.
| Relative right testis | Sperm density | Sperm head PC1 | Sperm head PC2 | Seminiferous tubule area | Proximal bent tail | Distal bent tail | Headless/tailless | Amorphous head | Total abnormal | |
|---|---|---|---|---|---|---|---|---|---|---|
| Relative right testis | — | 0.37 | −0.01 | −0.18 | 0.40 | −0.18 | −0.14 | −0.25 | −0.21 | −0.26 |
| Sperm density | <0.001 | — | 0.18 | −0.18 | 0.24 | −0.21 | −0.10 | −0.14 | −0.28 | −0.24 |
| Sperm head PC1 | 0.850 | 0.002 | — | 0 | 0.18 | −0.45 | -0.29 | −0.32 | −0.42 | −0.51 |
| Sperm head PC2 | 0.002 | 0.002 | 1 | — | −0.11 | 0.05 | 0.12 | 0.05 | 0.08 | 0.06 |
| Seminiferous tubule area | <0.001 | <0.001 | 0.002 | 0.069 | — | −0.17 | −0.19 | −0.22 | −0.19 | −0.26 |
| Proximal bent tail | 0.002 | <0.001 | <0.001 | 0.358 | 0.003 | — | 0.36 | 0.27 | 0.44 | 0.66 |
| Distal bent tail | 0.019 | 0.070 | <0.001 | 0.037 | 0.001 | <0.001 | — | 0.30 | 0.31 | 0.55 |
| Headless/tailless | <0.001 | 0.013 | <0.001 | 0.412 | <0.001 | <0.001 | <0.001 | — | 0.33 | 0.80 |
| Amorphous head | <0.001 | <0.001 | <0.001 | 0.181 | 0.001 | <0.001 | <0.001 | <0.001 | — | 0.61 |
| Total abnormal | <0.001 | <0.001 | <0.001 | 0.147 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | — |
Upper diagonal: Spearman’s ρ with significant correlations in boldface type. Lower diagonal: P values.
We used the average trait values from the (M. m. musculusPWD × M. m. domesticusWSB)F1 males to set approximate sterility thresholds for F2 phenotypic distributions. For each phenotype, a percentage of the males fell within the sterile range (relative right testis weight: 9.4%; sperm density: 6.5%; seminiferous tubule area: 1.6%; proximal bent tail: 8.7%; distal bent tail: 27.7%; headless/tailless sperm: 2.6%; amorphous sperm head: 9.0%; total sperm abnormalities: 5.5%), suggesting that hybrid sterility has a prominent effect in generations beyond the F1.
We also observed a large amount of phenotypic variance in sperm head morphology. We applied a principal component analysis to identify the major axes of shape variation among the F2’s. The first two principal components explained 88.0% of the shape variation in the elliptic Fourier descriptors (Figure 2B). Principal component 1 (PC1; 78.6% of the phenotypic variance) tracked the curvature of the apical hook on the sperm head, whereas principal component 2 (PC2; 9.4% of the phenotypic variance) reflected changes largely in overall sperm head width. Lower values of PC1 were associated with a considerable reduction in the apical hook, consistent with the shape observed in the (M. m. musculusPWD × M. m. domesticusWSB)F1 sterile males.
Single QTL interval mapping
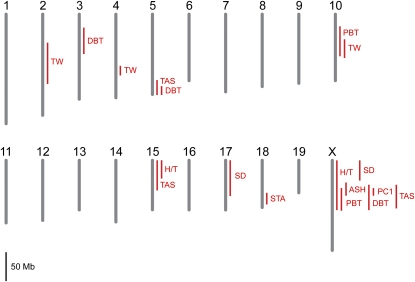
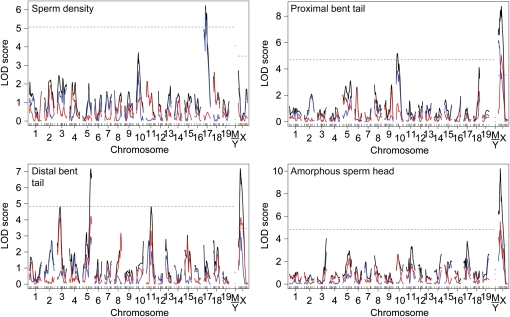
Standard interval mapping (Lander and Botstein 1989; Broman and Sen 2009) detected QTL for each hybrid sterility phenotype (Table 3; Figure 4; Figure S2). We identified QTL on chromosomes 2, 4, and 10 for relative right testis weight and on chromosome 18 for the residual trait score of seminiferous tubule area regressed on testis weight. Phenotypic distributions for other hybrid sterility measures exhibited strong deviations from normality. For each of these phenotypes, we applied parametric and nonparametric mapping methods to evaluate the robustness of the results. Except for a few cases, QTL remained significant regardless of the method used (Table S1).
Table 3 . Single QTL mapping.
| Phenotype | Chromosome | Position (cM) | LOD scorea | Position (Mb) | 1.5-LOD interval (Mb) | DDb | DMb | MMb |
|---|---|---|---|---|---|---|---|---|
| Relative right testis weight | 2 | 30 | 5.41 | 96.0 | 52.2–123.9 | 3.64 ± 0.13 | 4.40 ± 0.09 | 4.43 ± 0.14 |
| 4 | 45.9 | 8.66 | 108.9 | 98.8–115.3 | 4.68 ± 0.12 | 4.28 ± 0.08 | 3.49 ± 0.14 | |
| 10 | 20 | 6.36 | 69.3 | 55.1–87.4 | 3.81 ± 0.12 | 4.20 ± 0.09 | 4.80 ± 0.13 | |
| Sperm density | X | 0 | 3.28 | 10.2 | 10.2–45.1 | 13.82 ± 0.62 | — | 10.81 ± 0.58 |
| Sperm density (binary) | 17 | 13.3 | 6.00 | 30.0 | 3.1–65.2 | 0.85 ± 0.04 | 0.77 ± 0.03 | 1.00 ± 0.05 |
| Sperm head PC1c | X | 18 | 50.95 | 68.9 | 58.3–71.3 | 0.061 ± 0.004 | — | −0.056 ± 0.004 |
| Seminiferous tubule aread | 18 | 34 | 4.36 | 69.6 | 60.3–80.0 | 2681.17 ± 880.56 | −859.17 ± 486.73 | −610.82 ± 638.18 |
| Proximal bent taile | 10 | 10 | 4.20 | 47.6 | 32.6–84.3 | 0.24 ± 0.02 | 0.18 ± 0.01 | 0.15 ± 0.02 |
| X | 24.5 | 8.77 | 90.8 | 58.3–97.4 | 0.14 ± 0.01 | — | 0.23 ± 0.01 | |
| Distal bent taile | 3 | 22 | 4.34 | 51.4 | 30.0–75.9 | 0.18 ± 0.01 | 0.13 ± 0.01 | 0.12 ± 0.01 |
| 5 | 70 | 7.20 | 146.4 | 133.0–148.4 | 0.20 ± 0.01 | 0.13 ± 0.01 | 0.12 ± 0.01 | |
| X | 16 | 7.23 | 67.8 | 52.5–96.4 | 0.11 ± 0.01 | — | 0.17 ± 0.01 | |
| Headless/taillesse | 15 | 0 | 3.67 | 16.5 | 16.5–48.2 | 0.33 ± 0.02 | 0.34 ± 0.01 | 0.41 ± 0.02 |
| X | 2 | 3.20 | 18.7 | 10.2–96.4 | 0.32 ± 0.01 | — | 0.38 ± 0.01 | |
| Amorphous sperm heade | X | 15.9 | 10.28 | 67.7 | 48.6–71.3 | 0.13 ± 0.01 | — | 0.21 ± 0.01 |
| Total abnormal sperme | 5 | 64 | 4.25 | 138.2 | 122.4–148.4 | 0.60 ± 0.03 | 0.45 ± 0.02 | 0.47 ± 0.03 |
| 15 | 4 | 3.85 | 27.1 | 16.5–77.2 | 0.46 ± 0.03 | 0.45 ± 0.02 | 0.59 ± 0.03 | |
| X | 16 | 9.73 | 67.8 | 52.5–93.4 | 0.40 ± 0.017 | — | 0.57 ± 0.016 |
All QTL are significant at a 5% significance threshold.
Phenotype means of each genotype (±SE). D, M. m. domesticusWSB; M, M. m. musculusPWD.
Lower values of PC1 are correlated with higher levels of sterility.
Residual trait scores of seminiferous tubule area regressed on testis weight. Lower values are correlated with higher levels of sterility.
Arcsine square-root-transformed.
Figure 4 .
Single QTL scan for hybrid male sterility. The 1.5-LOD support intervals for QTL that exceed a genome-wide 5% significance threshold are shown (TW: relative right testis weight; SD: sperm density and sperm density binary; PC1: sperm head morphology PC1; STA: seminiferous tubule area; PBT: proximal bent tail; DBT: distal bent tail; H/T: headless/tailless; ASH: amorphous sperm head; TAS: total abnormal sperm). No QTL mapped to the mitochondrion or Y chromosome.
One QTL on the proximal end of the X chromosome was detected when the raw values of sperm density were used for nonparametric mapping. We also found a QTL linked to chromosome 17 when sperm density was reanalyzed as a binary trait to accommodate its spiked phenotypic distribution (Table 3; Figure 4; Figure S2). To empirically determine the best sperm density to split the two phenotypic categories, we repeated the single QTL scan using a range of densities. The LOD score for the QTL on chromosome 17 was maximized when a density of 5 million sperm per milliliter was used. We also applied a third method, which analyzed sperm density using two separate models. One model treated the phenotype as a binary trait (above or below 5 million sperm per milliliter) and a second model analyzed sperm density in the normal range as a quantitative trait, including only those animals above the cutoff (Broman 2003; Broman and Sen 2009). The two-part model detected the same QTL linked to chromosome 17, but this QTL received no support from the quantitative portion of the distribution (Figure 5). This indicated that the primary effect of the QTL was to severely reduce sperm density below 5 million/ml rather than to shape variation in sperm density within the normal range. The X-linked QTL contributed to both severe reduction of sperm density and variation within the normal range.
Figure 5 .
Single QTL scans with two-part models. The models evaluate support for QTL associations from the presence/absence of the trait (blue line), the normal portion of the distribution (red line), and the combined distribution (black line). Genome-wide significance thresholds are indicated by the dashed lines and were derived independently for the autosomes and the X chromosome from 1000 permutations of the combined distribution model (black line) (α = 0.05).
We transformed the first two principal components of sperm head morphology and mapped the phenotypes parametrically. There was a strong QTL on the X chromosome associated with PC1 (LOD score 50.95; Table 3; Figure 4; Figure S2). Although PC2 was significantly correlated with other sterility phenotypes, we did not detect QTL linked to this component and do not consider it further here.
Each of the transformed abnormal sperm types were affected by X-linked and autosomal QTL when mapped in a parametric framework (Table 3; Figure 4; Figure S2). Three of the abnormal sperm types (proximal bent tail, distal bent tail, and amorphous sperm head) had spikes in their distributions at zero. We applied a two-part model to these phenotypes (as described for sperm density) to differentiate the spike from the quantitative portion of the distribution. For all three phenotypes, the QTL on chromosome X had mixed effects on presence or absence of the abnormal sperm type (the binary trait) as well as the degree of severity of the abnormal type (the quantitative trait) (Figure 5). For proximal bent tail, the QTL on chromosome 10 affected largely the presence or absence of this defect. For distal bent tail, the QTL on chromosome 3 controlled only the degree of severity, whereas the QTL on chromosome 5 had mixed effects.
Consistent with results from previous studies using other strains of M. m. musculus and M. m. domesticus (Forejt 1996; Storchová et al. 2004; Britton-Davidian et al. 2005; Good et al. 2008a,b), the asymmetry in F1 sterility that we observed suggests an important role for the M. m. musculus X chromosome in hybrid male sterility (Forejt 1996; Storchová et al. 2004; Britton-Davidian et al. 2005; Good et al. 2008a,b). Every X-linked QTL that we identified supported this conclusion, with a large reduction in fertility associated with the M. m. musculusPWD genotype (Table 3). Furthermore, X-linked QTL affecting the different sperm phenotypes exhibited overlapping 1.5-LOD confidence intervals, which could indicate a common genetic mechanism for sperm head morphology and all abnormal sperm types.
In contrast, infertility was associated with both M. m. domesticusWSB and M. m. musculusPWD alleles at autosomal QTL. For 6 of the 11 QTL, the allele causing hybrid male sterility was recessive and hidden in F1 hybrids (Table 3). We observed only one case of underdominance, where the least-fertile genotype was associated with heterozygous males (sperm density: chromosome 17). This QTL overlaps with Prdm9, which also causes hybrid male sterility when heterozygous (Forejt et al. 1991; Gregorová et al. 1996; Trachtulec et al. 2008; Mihola et al. 2009).
Two-dimensional, two-QTL mapping
Joint consideration of multiple QTL allows the detection of epistatic interactions and can improve power to identify additional loci by reducing residual variance. We first used approaches that consider two-locus genotypes. We analyzed all phenotypes using parametric models, except for sperm density, which was analyzed as a binary trait. With a genome-wide 5% significance threshold, we found several pairs of QTL affecting relative right testis weight (chromosomes 2 and 4, 2 and 10, 4 and 4, 4 and 10, 10 and 17), sperm head morphology PC1 (chromosomes 4 and X, 11 and X, X and X), and seminiferous tubule area (chromosomes 14 and 16) in an additive manner. Although two-QTL models provided statistical improvements over their component one-QTL models, we observed little evidence of epistatic interactions among these QTL.
Multiple QTL mapping
We also considered models that jointly fit any number of QTL using an automated forward/backward stepwise search algorithm. Several additional QTL were found beyond those identified in two-dimensional QTL scans (Table 4). The percentage of the phenotypic variance explained by these full models varied among traits (relative right testis weight: 43.9%; sperm density (binary): 5.9%; sperm head morphology PC1: 74.0%; seminiferous tubule area: 21.1%; proximal bent tail: 17.5%; distal bent tail: 22.4%; headless/tailless sperm: 5.4%; amorphous sperm head: 14.4%; total abnormal sperm: 19.4%). Similar to the single QTL scans, alleles from both subspecies caused hybrid male sterility with effects ranging from recessive to dominant. Two QTL on chromosome 17 that overlap with Prdm9 (relative testis weight and sperm density) and one QTL on chromosome 7 (sperm head morphology PC1) exhibited underdominance. Only one phenotype included a significant epistatic effect. For relative right testis weight, an interaction between QTL on chromosomes 3 and 13 (LOD score: 6.1; 5.4% of the phenotypic variance) resulted in significantly lower weight when the genotypes at both QTL were homozygous for M. m. musculusPWD and therefore did not appear to represent a simple two-locus Dobzhansky–Muller interaction.
Table 4 . Multiple QTL mapping.
| Phenotype | Chromosome | Position (cM) | LOD scorea | Position (Mb) | 1.5-LOD interval (Mb) | % phenotype varianceb | Additivec | Dominanced | Effecte |
|---|---|---|---|---|---|---|---|---|---|
| Relative right testis weight | 2 | 33 | 6.06 | 105.9 | 52.2–128.5 | 5.32 | 0.35 ± 0.0 | 0.26 ± 0.12 | Dom. recessive |
| 3 | 54 | 8.06 | 129.8 | 108.5–143.7 | 7.19 | −0.16 ± 0.08 | −0.15 ± 0.11 | Musc. dominant | |
| 4 | 45.9 | 14.40 | 108.9 | 98.8–115.3 | 13.49 | −0.92 ± 0.12 | 0.070 ± 0.13 | Musc. additive | |
| 4 | 60 | 5.40 | 134.5 | 121.8–148.0 | 5.40 | 0.54 ± 0.12 | −0.15 ± 0.14 | Dom. additive | |
| 10 | 21 | 9.33 | 72.2 | 51.3–82.7 | 9.33 | 0.48 ± 0.08 | −0.08 ± 0.11 | Dom. additive | |
| 13 | 3 | 10.54 | 18.6 | 12.2–24.0 | 9.58 | −0.31 ± 0.08 | 0.03 ± 0.11 | Musc. additive | |
| 17 | 24 | 3.74 | 57.9 | 3.1–73.2 | 3.23 | 0.05 ± 0.08 | −0.44 ± 0.12 | Underdominant | |
| Sperm density (binary) | 17 | 13.3 | 4.04 | 30.0 | 3.1–64.5 | 5.86 | 0.07 ± 0.03 | −0.16 ± 0.04 | Underdominant |
| Sperm head PC1f | 7 | 6 | 3.74 | 23.4 | 13.7–44.3 | 1.90 | 0.01 ± 0.004 | −0.02 ± 0.01 | Underdominant |
| 11 | 39 | 4.22 | 57.3 | 44.6–73.2 | 2.15 | 0.01 ± 0.004 | 0.01 ± 0.01 | Dom. recessive | |
| X | 12 | 36.85 | 52.5 | 22.9–58.3 | 24.35 | −0.05 ± 0.003 | — | Musc. | |
| X | 45 | 18.46 | 110.0 | 98.6–128.2 | 10.51 | −0.03 ± 0.003 | — | Musc. | |
| Semin. tubule areag | 8 | 27.8 | 4.76 | 87.1 | 35.5–102.7 | 6.09 | 1711.39 ±372.67 | −185.52 ± 517.02 | Dom. additve |
| 14 | 16.1 | 4.35 | 61.6 | 25.6–63.2 | 5.55 | −1621.40 ± 373.64 | −289.60 ± 527.09 | Musc. additive | |
| 16 | 39 | 4.02 | 88.2 | 66.8–97.2 | 5.11 | −1147.66 ± 360.08 | −1490.79 ± 537.94 | Musc. dominant | |
| 18 | 33 | 4.62 | 68.5 | 58.0–78.1 | 6.00 | −1497.93 ± 404.78 | −1724.62 ± 554.91 | Musc. dominant | |
| Proximal bent tailh | 10 | 12 | 4.16 | 51.3 | 34.5–87.4 | 5.33 | −0.04 ± 0.10 | −0.02 ± 0.02 | Dom. additive |
| X | 24.5 | 8.89 | 90.8 | 58.5–97.1 | 11.81 | 0.044 ± 0.007 | — | Musc. | |
| Distal bent tailh | 3 | 24 | 3.90 | 54.7 | 28.2–72.1 | 4.71 | −0.023 ± 0.007 | −0.023 ± 0.009 | Dom. recessive |
| 5 | 65.6 | 5.85 | 139.6 | 132.2–148.4 | 7.14 | −0.03 ± 0.01 | −0.03 ± 0.01 | Dom. recessive | |
| X | 15 | 5.74 | 58.5 | 46.7–98.6 | 7.00 | 0.03 ± 0.01 | — | Musc. | |
| Headless/taillessh | 15 | 0 | 3.68 | 16.5 | 16.5–48.2 | 5.38 | 0.04 ± 0.01 | −0.03 ± 0.02 | Musc. recessive |
| Amorphous sperm headh | X | 15.9 | 10.30 | 67.7 | 52.5–70.7 | 14.36 | 0.04 ± 0.01 | — | Musc. |
| Total abnormal spermh | 15 | 6 | 4.35 | 32.3 | 16.5–68.7 | 5.45 | 0.06 ± 0.02 | −0.08 ± 0.03 | Musc. recessive |
| X | 18 | 10.63 | 68.9 | 52.5–92.7 | 13.97 | 0.09 ± 0.01 | — | Musc. |
All QTL are significant at a 5% genome-wide significance threshold.
Percentage of phenotypic variance explained by the QTL.
Additive effect: half the difference between the phenotype averages of the two homozygotes. Mean ± SD.
Dominance deviation: the difference between the phenotype average of the heterozygotes and the midpoint of the phenotype averages of the two homozygotes. Mean ± SD.
The subspecies allele associated with sterility and the effect of that allele (Dom., M. m. domesticusWSB; Musc., M. m. musculusPWD).
Lower values of PC1 are correlated with higher levels of sterility.
Residual trait scores of seminiferous tubule area regressed on testis weight. Lower values are correlated with higher levels of sterility.
Arcsine square-root-transformed.
Dobzhansky–Muller incompatibilities with the X chromosome
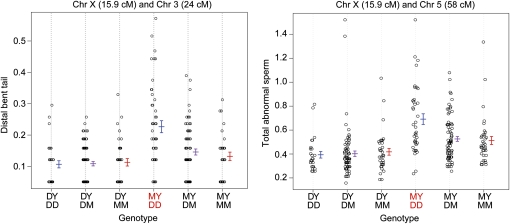
Although an F2 intercross with 310 animals is underpowered to identify epistatic loci (Mao and Da 2005), the Dobzhansky–Muller model suggests that the hybrid male sterility that we observed is indeed caused by multi-locus incompatibilities. To search further for these sets of loci, we focused on interactions involving the X chromosome because (i) it plays an important role in hybrid male sterility and (ii) its smaller number of genotypic classes (two) preserves power relative to autosomal loci (which have three genotypic classes). We conducted a single-QTL scan that was conditioned on either the M. m. musculusPWD or the M. m. domesticusWSB allele at each QTL on the X chromosome. All analyses conditioned on the M. m. domesticusWSB alleles yielded no QTL (as expected since this allele had no effect on sterility for any of the phenotypes). However, when we conditioned on the M. m. musculusPWD alleles, we found strong support (with LOD scores > 3.4) for five autosomal QTL interacting with the X chromosome (Figure 6; Table 5; Figure S3). For sperm head morphology PC1, we detected an interaction that reduced the apical hook of the sperm head when the M. m. musculusPWD allele on the X chromosome was combined with the homozygous M. m. domesticusWSB or heterozygous genotype on chromosome 7. For each abnormal sperm type category, there were higher proportions of abnormal sperm when the X-linked M. m. musculusPWD allele was combined with autosomal QTL homozygous for M. m. domesticusWSB.
Figure 6 .
Dobzhansky–Muller incompatibilities with the X chromosome. Phenotypic means are indicated by genotype for two of the six Dobzhansky–Muller interactions between the X chromosome and autosomal loci (see Figure S3 for the remaining plots). Red genotypes denote reduced fertility. Distal bent tail and total abnormal sperm are arcsine square-root-transformed proportions. Error bars are ±1 SE.
Table 5 . Dobzhansky–Muller incompatibilities with the X chromosome.
| Phenotype | Chromosome 1 | Position (cM) | Position (Mb) | Chromosome 2 | Position (cM) | LOD score | Position (Mb) | 1.5-LOD interval (Mb) |
|---|---|---|---|---|---|---|---|---|
| Sperm head PC1 | X | 18 | 68.9 | 7 | 8 | 3.64 | 26.6 | 13.7–44.3 |
| Proximal bent taila | X | 24.5 | 90.8 | 10 | 16 | 3.41 | 58.9 | 32.6–96.7 |
| Distal bent taila | X | 15.9 | 67.7 | 3 | 24 | 5.21 | 54.7 | 33.5–67.8 |
| X | 15.9 | 67.7 | 5 | 70 | 4.13 | 146.4 | 130.0–148.4 | |
| Headless/taillessa | X | 2 | 18.7 | 7 | 53.5 | 3.92 | 128.1 | 61.5–128.1 |
| Total abnormal sperma | X | 16 | 67.8 | 5 | 60 | 4.18 | 134.8 | 122.4–148.4 |
Arcsine square-root-transformed.
Discussion
Genetic mapping of hybrid male sterility in crosses between M. m. musculusPWD and M. m. domesticusWSB demonstrated contributions from multiple autosomal loci, the M. m. musculusPWD X chromosome, and incompatibilities between the X and the autosomes. These results provide a genetic portrait of a key reproductive barrier between two lineages in the early stages of speciation.
F1 hybrid male sterility
In addition to reduced testis weight and sperm density, F1 hybrid males from M. m. musculusPWD mothers presented abnormal sperm types similar to those previously connected with sterility. Abnormal sperm head shapes often arise as by-products of aneuploidy (Prisant et al. 2007; Perrin et al. 2008; Revay et al. 2009). In rodents, alterations of the sperm hook are correlated with decreases in fertilization success (Immler et al. 2007; Firman and Simmons 2009), and severely amorphous sperm heads are inefficient at penetrating the ova (Krzanowska and Lorenc 1983; Oka et al. 2007; Styrna 2008). Severity of these phenotypes also varies widely among the classical inbred strains of house mice, where the incidence of abnormal sperm is negatively correlated with fertilization success (Kawai et al. 2006). Consistent with observed defects in spermatogenesis, we also found a reduction in the area of seminiferous tubules, a change tied to decreases in the number of round spermatids in crosses between these subspecies (Britton-Davidian et al. 2005) that likely reflects failure during meiosis (Oka et al. 2010).
Hybrid male sterility QTL
Some of the QTL that we identified coincide with those found in previous studies of male reproductive traits in house mice. Testis weight QTL on chromosomes 4, 10, and 13 overlap with those mapped in crosses between two classical inbred strains that differ substantially in this trait (Le Roy et al. 2001; Bolor et al. 2006). The chromosome 11 sperm head morphology QTL maps near a locus that grossly alters head width and hook shape in crosses between C57BL/10 and MOLF/EiJ (a strain of M. m. molossinus; Oka et al. 2007). In addition, chromosome substitution lines that carry either chromosome 10 or 11 from M. m. musculusPWD on the genomic background of a classical inbred strain derived primarily from M. m. domesticus (C57BL/6J) show reduced fertility (Gregorová et al. 2008).
Two of the QTL that we found lie in the proximal region of chromosome 17, where the only gene known to cause hybrid sterility in vertebrates—Prdm9—resides (Mihola et al. 2009). Together with other loci that remain to be identified, this gene causes spermatogenic failure in F1’s between M. m. musculusPWD and C57BL/10 when heterozygous at this locus (Mihola et al. 2009). This genetic architecture is consistent with the chromosome 17 QTL for relative right testis weight and sperm density (binary) that acted underdominantly. Although the molecular mechanism responsible for hybrid sterility is unclear, divergence in the number of zinc fingers in the PRDM9 protein (Mihola et al. 2009) could affect its ability to methylate histones, thereby changing the transcription of target genes (Oliver et al. 2009). M. m. musculusPWD and M. m. domesticusWSB also differ in the number of zinc fingers at Prdm9 (Parvanov et al. 2010). Additional research will be required to determine whether this gene underlies the QTL that we found.
As might be expected from the observed asymmetry in F1 hybrid male sterility, several reproductive phenotypes showed strong linkage to the M. m. musculusPWD X chromosome. However, testis weight and seminiferous tubule area were not associated with the X chromosome. This result is surprising in light of previous studies that detected X-linked QTL for testis weight (Oka et al. 2004; Storchová et al. 2004; Good et al. 2008a).
Several factors could be responsible for this discrepancy. First, asymmetry in the F1 generation may be due to interactions between dominant autosomal loci and the M. m. domesticusWSB Y chromosome or M. m. musculusPWD mitochondrion rather than interactions with the M. m. musculusPWD X chromosome. Deletions of loci within the Y chromosome have been shown to severely alter sperm head morphology in mice (Styrna et al. 1991, 2002; Styrna and Krzanowska 1995). Because only 12 F2 males carried the necessary combination of chromosomes, our power to detect such an effect was severely limited. Second, X-linked sterility could require multiple loci on the X chromosome. In other crosses, M. m. musculus alleles at two X-linked loci were needed to observe sterility (Storchová et al. 2004), and testis weight was negatively correlated with the fraction of the M. m. musculus X chromosome introgressed on a M. m. domesticus background (Good et al. 2008a). Comparing reproductive phenotypes between F2’s with nonrecombinant and recombinant M. m. musculus X chromosomes in our study yielded no significant differences (Student’s t-test; P > 0.05 for relative right testis weight and seminiferous tubule area), providing no support for this explanation. A third possibility is that reductions in testis weight and seminiferous tubule area could result from many independent, small-effect interactions between homozygous M. m. domesticus autosomal loci and the M. m. musculus X chromosome. An F2 intercross would likely produce a small proportion of males with a sufficient number of these interactions. Consistent with this model, previous studies have noted fully fertile males with nonrecombinant M. m. musculus X chromosomes in early backcross generations (Oka et al. 2004; Storchová et al. 2004; Good et al. 2008a). As the autosomes approached complete homozygosity in later generations, all males developed sterility.
Dobzhansky–Muller incompatibilities with the X chromosome
By conditioning on the X-linked M. m. musculusPWD allele that generated sterile phenotypes, we identified several Dobzhansky–Muller interactions with autosomal loci. Across sperm phenotypes, we uncovered five recessive interactions and one dominant interaction involving M. m. domesticusWSB alleles, despite comparatively lower power to identify recessive incompatibilities. These observations indicate that most incompatibilities likely involve recessive partners, mirroring results from Drosophila (Presgraves 2003). The interacting autosomal locus that we identified for sperm head morphology PC1 (chromosome 7) differs from two previously identified M. m. domesticus loci (chromosomes 1 and 11) that cause defects when a M. m. molossinus X chromosome is introgressed into a largely M. m. domesticus genomic background (Oka et al. 2007). Although only two studies have attempted to map incompatibilities involving the X chromosome in house mice, the lack of overlap between them suggests that hybrid male sterility in generations beyond the F1 has a complex genetic basis.
Additional lines of evidence indicate that F2 hybrid male sterility between M. m. musculusPWD and M. m. domesticusWSB is caused by a large number of incompatibilities. Five of the six interactions with the X chromosome mapped to unique autosomal loci despite significant correlations between the abnormal sperm phenotypes. Furthermore, these interactions accounted for only a fraction of the reduced fertility caused by the X-linked M. m. musculusPWD allele. Each abnormal sperm phenotype may therefore be the result of multiple or higher-order Dobzhanksy–Muller incompatibilities with effects that are difficult to detect using our sample size.
Our results also indicate that M. m. musculusPWD loci on the X chromosome do not require an interaction with the M. m. domesticusWSB Y chromosome to cause hybrid male sterility. Previous studies have mapped hybrid male sterility between these subspecies only by backcrossing F1 females to M. m. domesticus males (Storchová et al. 2004; Good et al. 2008a). This crossing scheme does not separate the effects of the M. m. musculus X chromosome from the M. m. domesticus Y chromosome. In our study, nearly all males possessed a M. m. musculusPWD Y chromosome and still exhibited X-linked sterility from M. m. musculusPWD alleles.
Our study assumes that the genetics of hybrid male sterility between PWD and WSB accurately represents subspecific barriers between M. m. musculus and M. m. domesticus. The observation of certain patterns, including a large role for the M. m. musculus X chromosome, that agree with those seen in crosses between other strains, supports this assumption. However, there are reasons for caution. Some of the QTL that we identified might not be fixed differences between subspecies; under this scenario, the resulting incompatibilities would only have the potential to contribute to reproductive barriers in part of the subspecies range. Intra-subspecific variation in inter-subspecific hybrid male sterility has been documented in both M. m. musculus and M. m. domesticus (Good et al. 2008b). In addition, the large effective population size and short divergence time among subspecies have left a clear signature of incomplete lineage sorting across the genome (Geraldes et al. 2008; White et al. 2009). Therefore, some QTL might be found in genomic regions where phylogenies do not yet match the subspecies history. These considerations are inevitable consequences of studying reproductive barriers as they evolve and should motivate additional QTL studies across an array of wild-derived strains to assess the generality of the patterns described here.
Candidate regions
Existing data and genetic tools could help narrow the hybrid sterility QTL that we identified to specific genes in the near future. Patterns of differential introgression in natural hybrid zones allow identification of genomic regions that could affect reproductive isolation (Payseur 2010). Due to high levels of historical recombination, the mapping resolution of this approach is expected to exceed that achieved in laboratory crosses. A genomic analysis of two separate transects through the M. m. musculus–M. m. domesticus hybrid zone in central Europe identified five SNPs with narrow cline widths in both transects (Teeter et al. 2010), three of which are located within the 95% confidence intervals of QTL that we identified (testis weight: chromosome 4; seminiferous tubule area: chromosome 8; sperm head morphology PC1: chromosome X). If the same genes cause hybrid sterility in the lab and in nature, fine-scale mapping of these genes through comparative introgression in the hybrid zone might be feasible.
Other mouse strains also provide useful resources to localize the positions of genetic variants that cause hybrid male sterility. First, a large number of mutant mouse models have been generated to study abnormalities in male reproductive phenotypes (reviewed in Matzuk and Lamb 2002, 2008). This extensive list may contain genes that maintain reproductive barriers between subspecies. As an example, we queried this database for mutations that reside under the most significant QTL peak for testis weight (chromosome 4). The 95% confidence interval of this QTL spans 16.5 Mb of the chromosome. Within this interval, there are three mutations that cause partial or complete sterility in males (Dhcr24: Bilbao et al. 1998; Lepr: Bonache et al. 2007; Lrp8: Dam et al. 2007). Second, classical inbred mouse strains are genetic mosaics of the three subspecies. Patterns of genetic variation among these strains were shaped by selective removal of multi-locus, hetero-subspecific (M. m. musculus/M. m. domesticus) combinations of alleles (Payseur and Hoekstra 2005; Payseur and Place 2007). These genomic regions overlap with hybrid male sterility QTL on chromosomes 2, 7, 8, 13, 15, 17, 18, and X. Because the spatial scale of linkage disequilibrium is lower across the panel of inbred strains than in our intercross, this genomic overlap could be used for fine mapping of QTL. Finally, a large panel of recombinant inbred lines derived from crosses between eight strains—the Collaborative Cross—is being developed as a powerful resource for the genetic dissection of complex traits (Churchill et al. 2004; Chesler et al. 2008; Iraqi et al. 2008; Aylor et al. 2011). The eight founder strains include representatives of the two subspecies M. m. musculus and M. m. domesticus, raising the possibility of using this resource to localize hybrid sterility QTL and determine their epistatic partners.
Acknowledgments
We thank Maria Stubbings, John Bak, and Jenny Wagner for extensive help with measuring sterility traits and Denise Schwahn for characterizing sterility in the testis histology samples. We thank Karl Broman for assistance with QTL mapping. We acknowledge Leslie Turner for useful discussions and Jeff Good for comments on previous versions of this manuscript. This research was funded by National Science Foundation grant DEB 0918000. M.A.W. was supported by an National Library of Medicine (NLM) training grant in Computation and Informatics in Biology and Medicine to the University of Wisconsin (NLM 2T15LM007359).
Literature Cited
- Arends D., Prins P., Jansen R. C., Broman K. W., 2010. R/qtl: high-throughput multiple QTL mapping. Bioinformatics 26: 2990–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylor D. L., Valdar W., Foulds-Mathes W., Buus R. J., Verdugo R. A., et al. , 2011. Genetic analysis of complex traits in the emerging collaborative cross. Genome Res. 21: 1213–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbash D. A., Siino D. F., Tarone A. M., Roote J., 2003. A rapidly evolving MYB-related protein causes species isolation in Drosophila. Proc. Natl. Acad. Sci. USA 100: 5302–5307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateson W., 1909. Heredity and Variation in Modern Lights. Cambridge University Press, Cambridge, UK [Google Scholar]
- Bayes J. J., Malik H. S., 2009. Altered heterochromatin binding by a hybrid sterility protein in Drosophila sibling species. Science 326: 1538–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilbao J. R., Loridan L., Audí L., Gonzalo E., Castaño L., 1998. A novel missense (R80W) mutation in 17-beta-hydroxysteroid dehydrogenase type 3 gene associated with male pseudohermaphroditism. Eur. J. Endocrinol. 139: 330–333 [DOI] [PubMed] [Google Scholar]
- Bolor H., Wakasui N., Zhao W., Ishikawa A., 2006. Detection of quantitative trait loci causing abnormal spermatogenesis and reduced testis weight in the small testis (Smt) mutant mouse. Exp. Anim. 55: 97–108 [DOI] [PubMed] [Google Scholar]
- Bomblies K., Lempe J., Epple P., Warthmann N., Lanz C., et al. , 2007. Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol. 5: e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonache S., Martínez J., Fernández M., Bassas L., Larriba S., 2007. Single nucleotide polymorphisms in succinate dehydrogenase subunits and citrate synthase genes: association results for impaired spermatogenesis. Int. J. Androl. 30: 144–152 [DOI] [PubMed] [Google Scholar]
- Boursot P., Auffray J. C., Britton-Davidian J., Bonhomme F., 1993. The evolution of house mice. Annu. Rev. Ecol. Syst. 24: 119–152 [Google Scholar]
- Boursot P., Din W., Anand R., Darviche D., Dod B., et al. , 1996. Origin and radiation of the house mouse: mitochondrial DNA phylogeny. J. Evol. Biol. 9: 391–415 [Google Scholar]
- Brideau N. J., Flores H. A., Wang J., Maheshwari S., Wang X., et al. , 2006. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science 314: 1292–1295 [DOI] [PubMed] [Google Scholar]
- Britton-Davidian J., Fel-Clair F., Lopez J., Alibert P., Boursot P., 2005. Postzygotic isolation between the two European subspecies of the house mouse: estimates from fertility patterns in wild and laboratory-bred hybrids. Biol. J. Linn. Soc. Lond. 84: 379–393 [Google Scholar]
- Broman K. W., 2003. Mapping quantitative trait loci in the case of a spike in the phenotype distribution. Genetics 163: 1169–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K. W., Sen S., 2009. A Guide to QTL Mapping With R/qtl. Springer, New York [Google Scholar]
- Broman K. W., Rowe L. B., Churchill G. A., Paigen K., 2002. Crossover interference in the mouse. Genetics 160: 1123–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K. W., Wu H., Sen S., Churchill G. A., 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890 [DOI] [PubMed] [Google Scholar]
- Broman K. W., Sen S., Owens S. E., Manichaikul A., Southard-Smith E. M., et al. , 2006. The X chromosome in quantitative trait locus mapping. Genetics 174: 2151–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter T. C., Falconer D. S., 1951. Stocks for detecting linkage in the mouse, and the theory of their design. J. Genet. 50: 307–323 [DOI] [PubMed] [Google Scholar]
- Chen J., Ding J., Ouyang Y., Du H., Yang J., et al. , 2008. A triallelic system of S5 is a major regulator of the reproductive barrier and compatibility of indica-japonica hybrids in rice. Proc. Natl. Acad. Sci. USA 105: 11436–11441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler E. J., Miller D. R., Branstetter L. R., Galloway L. D., Jackson B. L., et al. , 2008. The Collaborative Cross at Oak Ridge National Laboratory: developing a powerful resource for systems genetics. Mamm. Genome 19: 382–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill G. A., Doerge R. W., 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill G. A., Airey D. C., Allayee H., Angel J. M., Attie A. D., et al. , 2004. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 36: 1133–1137 [DOI] [PubMed] [Google Scholar]
- Coyne J. A., Orr H. A., 2004. Speciation. Sinauer Associates, Sunderland, MA [Google Scholar]
- Dam A. H., Koscinski I., Kremer J. A. M., Moutou C., Jaeger A.-S., et al. , 2007. Homozygous mutation in SPATA16 is associated with male infertility in human globozoospermia. Am. J. Hum. Genet. 81: 813–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean M. D., Nachman M. W., 2009. Faster fertilization rate in conspecific vs. heterospecific matings in house mice. Evolution 63: 20–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T., 1936. Studies on hybrid sterility. II. Localization of sterility factors in Drosophila pseudoobscura hybrids. Genetics 21: 113–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dod B., Jermin L. S., Boursot P., Chapman V. H., Nielsen J. T., et al. , 1993. Counterselection on sex-chromosomes in the mus-musculus European hybrid zone. J. Evol. Biol. 6: 529–546 [Google Scholar]
- Dod B., Smadja C., Karn R., Boursot P., 2005. Testing for selection on the androgen-binding protein in the Danish mouse hybrid zone. Biol. J. Linn. Soc. Lond. 84: 447–459 [Google Scholar]
- Dumont B. L., White M. A., Steffy B., Wiltshire T., Payseur B. A., 2011. Extensive recombination rate variation in the house mouse species complex inferred from genetic linkage maps. Genome Res. 21: 114–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feenstra B., Skovgaard I. M., Broman K. W., 2006. Mapping quantitative trait loci by an extension of the Haley-Knott regression method using estimating equations. Genetics 173: 2269–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferree P. M., Barbash D. A., 2009. Species-specific heterochromatin prevents mitotic chromosome segregation to cause hybrid lethality in Drosophila. PLoS Biol. 7: e1000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firman R. C., Simmons L. W., 2009. Sperm competition and the evolution of the sperm hook in house mice. J. Evol. Biol. 22: 2505–2511 [DOI] [PubMed] [Google Scholar]
- Forejt J., 1996. Hybrid sterility in the mouse. Trends Genet. 12: 412–417 [DOI] [PubMed] [Google Scholar]
- Forejt J., Iványi P., 1974. Genetic studies on male sterility of hybrids between laboratory and wild mice (Mus musculus L.). Genet. Res. 24: 189–206 [DOI] [PubMed] [Google Scholar]
- Forejt J., Vincek V., Klein J., Lehrach H., Loudová-Micková M., 1991. Genetic mapping of the t-complex region on mouse chromosome 17 including the hybrid sterility-1 gene. Mamm. Genome 1: 84–91 [DOI] [PubMed] [Google Scholar]
- Frazer K. A., Eskin E., Kang H. M., Bogue M. A., Hinds D. A., et al. , 2007. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature 448: 1050–1053 [DOI] [PubMed] [Google Scholar]
- Freeman M., Tukey J., 1950. Transformations related to the angular and the square root. Ann. Math. Stat. 21: 607–611 [Google Scholar]
- Gabriel S., Ziaugra L., Tabbaa D., 2009. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr. Protoc. Hum. Genet; Chapter 2: Unit 2.12 [DOI] [PubMed] [Google Scholar]
- Ganem G., Litel C., Lenormand T., 2008. Variation in mate preference across a house mouse hybrid zone. Heredity 100: 594–601 [DOI] [PubMed] [Google Scholar]
- Geraldes A., Basset P., Gibson B., Smith K. L., Harr B., et al. , 2008. Inferring the history of speciation in house mice from autosomal, X-linked, Y-linked and mitochondrial genes. Mol. Ecol. 17: 5349–5363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good J. M., Dean M. D., Nachman M. W., 2008a. A complex genetic basis to X-linked hybrid male sterility between two species of house mice. Genetics 179: 2213–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good J. M., Handel M. A., Nachman M. W., 2008b. Asymmetry and polymorphism of hybrid male sterility during the early stages of speciation in house mice. Evolution 62: 50–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good J. M., Giger T., Dean M. D., Nachman M. W., 2010. Widespread over-expression of the X chromosome in sterile F1 hybrid mice. PLoS Genet. 6: e1001148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorová S., Forejt J., 2000. PWD/Ph and PWK/Ph inbred mouse strains of Mus m. musculus subspecies: a valuable resource of phenotypic variations and genomic polymorphisms. Folia Biol. (Praha) 46: 31–41 [PubMed] [Google Scholar]
- Gregorová S., Mnuková-Fajdelová M., Trachtulec Z., Capková J., Loudová M., et al. , 1996. Sub-milliMorgan map of the proximal part of mouse chromosome 17 including the hybrid sterility 1 gene. Mamm. Genome 7: 107–113 [DOI] [PubMed] [Google Scholar]
- Gregorová S., Divina P., Storchova R., Trachtulec Z., Fotopulosova V., et al. , 2008. Mouse consomic strains: exploiting genetic divergence between Mus m. musculus and Mus m. domesticus subspecies. Genome Res. 18: 509–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane J., 1922. Sex ratio and unisexual sterility in hybrid animals. J. Genet. 12: 101–109 [Google Scholar]
- Haley C. S., Knott S. A., 1992. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity 69: 315–324 [DOI] [PubMed] [Google Scholar]
- Immler S., Moore H. D. M., Breed W. G., Birkhead T. R., 2007. By hook or by crook? Morphometry, competition and cooperation in rodent sperm. PLoS ONE 2: e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iraqi F. A., Churchill G., Mott R., 2008. The Collaborative Cross, developing a resource for mammalian systems genetics: a status report of the Wellcome Trust cohort. Mamm. Genome 19: 379–381 [DOI] [PubMed] [Google Scholar]
- Iványi P., Vojtísková M., Démant P., Micková M., 1969. Genetic factors in the ninth linkage group influencing reproductive performance in male mice. Folia Biol. (Praha) 15: 401–421 [PubMed] [Google Scholar]
- Iwata H., Ukai Y., 2002. SHAPE: a computer program package for quantitative evaluation of biological shapes based on elliptic Fourier descriptors. J. Hered. 93: 384–385 [DOI] [PubMed] [Google Scholar]
- Kao K. C., Schwartz K., Sherlock G., 2010. A genome-wide analysis reveals no nuclear Dobzhansky-Muller pairs of determinants of speciation between S. cerevisiae and S. paradoxus, but suggests more complex incompatibilities. PLoS Genet. 6: e1001038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai Y., Hata T., Suzuki O., Matsuda J., 2006. The relationship between sperm morphology and in vitro fertilization ability in mice. J. Reprod. Dev. 52: 561–568 [DOI] [PubMed] [Google Scholar]
- Krzanowska H., Lorenc E., 1983. Influence of egg investments on in-vitro penetration of mouse eggs by misshapen spermatozoa. J. Reprod. Fertil. 68: 57–62 [DOI] [PubMed] [Google Scholar]
- Lander E. S., Botstein D., 1989. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laukaitis C., Critser E., Karn R., 1997. Salivary androgen-binding protein (ABP) mediates sexual isolation in Mus musculus. Evolution 51: 2000–2005 [DOI] [PubMed] [Google Scholar]
- Lee H.-Y., Chou J.-Y., Cheong L., Chang N.-H., Yang S.-Y., et al. , 2008. Incompatibility of nuclear and mitochondrial genomes causes hybrid sterility between two yeast species. Cell 135: 1065–1073 [DOI] [PubMed] [Google Scholar]
- Le Roy I., Tordjman S., Migliore-Samour D., Degrelle H., Roubertoux P., 2001. Genetic architecture of testis and seminal vesicle weights in mice. Genetics 158: 333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Y., Zhao L., Niu B., Su J., Wu H., et al. , 2008. Hybrid male sterility in rice controlled by interaction between divergent alleles of two adjacent genes. Proc. Natl. Acad. Sci. USA 105: 18871–18876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macholán M., Munclinger P., Sugerková M., Dufková P., Bímová B., et al. , 2007. Genetic analysis of autosomal and X-linked markers across a mouse hybrid zone. Evolution 61: 746–771 [DOI] [PubMed] [Google Scholar]
- Macholán M., Baird S. J. E., Munclinger P., Dufková P., Bímová B., et al. , 2008. Genetic conflict outweighs heterogametic incompatibility in the mouse hybrid zone? BMC Evol. Biol. 8: 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichaikul A., Moon J. Y., Sen S., Yandell B. S., Broman K. W., 2009. A model selection approach for the identification of quantitative trait loci in experimental crosses, allowing epistasis. Genetics 181: 1077–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y., Da Y., 2005. Statistical power for detecting epistasis QTL effects under the F-2 design. Genet. Sel. Evol. 37: 129–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin N. H., Willis J. H., 2010. Geographical variation in postzygotic isolation and its genetic basis within and between two Mimulus species. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 2469–2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzuk M. M., Lamb D. J., 2002. Genetic dissection of mammalian fertility pathways. Nat. Cell Biol. 4: s41–s49 [DOI] [PubMed] [Google Scholar]
- Matzuk M. M., Lamb D. J., 2008. The biology of infertility: research advances and clinical challenges. Nat. Med. 14: 1197–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihola O., Trachtulec Z., Vlcek C., Schimenti J. C., Forejt J., 2009. A mouse speciation gene encodes a meiotic histone h3 methyltransferase. Science 323: 373–375 [DOI] [PubMed] [Google Scholar]
- Moulia C., Aussel J., Bonhomme F., 1991. Wormy mice in a hybrid zone: a genetic control of susceptibility to parasite infection. J. Evol. Biol. 4: 679–687 [Google Scholar]
- Moulia C., Le Brun N., Dallas J., Orth A., Renaud F., 1993. Experimental evidence of genetic determinism in high susceptibility to intestinal pinworm infection in mice: a hybrid zone model. Parasitology 106: 387–393 [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium, Waterston R. H., Lindblad-Toh K., Birney E., Rogers J., et al. , 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562 [DOI] [PubMed] [Google Scholar]
- Moyle L. C., 2007. Comparative genetics of potential prezygotic and postzygotic isolating barriers in a Lycopersicon species cross. J. Hered. 98: 123–135 [DOI] [PubMed] [Google Scholar]
- Muller H. J., 1942. Isolating mechanisms, evolution, and temperature. Biol. Symp. 6: 71–125 [Google Scholar]
- Munclinger P., Bozikova E., Sugerkova M., Pialek J., Macholan M., 2002. Genetic variation in house mice (Mus, muridae, rodentia) from the Czech and Slovak republics. Folia Zool. (Brno) 51: 81–92 [Google Scholar]
- Oka A., Mita A., Sakurai-Yamatani N., Yamamoto H., Takagi N., et al. , 2004. Hybrid breakdown caused by substitution of the X chromosome between two mouse subspecies. Genetics 166: 913–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka A., Aoto T., Totsuka Y., Takahashi R., Ueda M., et al. , 2007. Disruption of genetic interaction between two autosomal regions and the X chromosome causes reproductive isolation between mouse strains derived from different subspecies. Genetics 175: 185–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka A., Mita A., Takada Y., Koseki H., Shiroishi T., 2010. Reproductive isolation in hybrid mice due to spermatogenesis defects at three meiotic stages. Genetics 186: 339–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver P. L., Goodstadt L., Bayes J. J., Birtle Z., Roach K. C., et al. , 2009. Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS Genet. 5: e1000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvanov E. D., Petkov P. M., Paigen K., 2010. Prdm9 controls activation of mammalian recombination hotspots. Science 327: 835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payseur B. A., 2010. Using differential introgression in hybrid zones to identify genomic regions involved in speciation. Mol. Ecol. Resour. 10: 806–820 [DOI] [PubMed] [Google Scholar]
- Payseur B. A., Hoekstra H. E., 2005. Signatures of reproductive isolation in patterns of single nucleotide diversity across inbred strains of mice. Genetics 171: 1905–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payseur B., Nachman M., 2005. The genomics of speciation: investigating the molecular correlates of X chromosome introgression across the hybrid zone between Mus domesticus and Mus musculus. Biol. J. Linn. Soc. Lond. 84: 523–534 [Google Scholar]
- Payseur B. A., Place M., 2007. Searching the genomes of inbred mouse strains for incompatibilities that reproductively isolate their wild relatives. J. Hered. 98: 115–122 [DOI] [PubMed] [Google Scholar]
- Payseur B. A., Krenz J. G., Nachman M. W., 2004. Differential patterns of introgression across the X chromosome in a hybrid zone between two species of house mice. Evolution 58: 2064–2078 [DOI] [PubMed] [Google Scholar]
- Perrin A., Morel F., Moy L., Colleu D., Amice V., et al. , 2008. Study of aneuploidy in large-headed, multiple-tailed spermatozoa: case report and review of the literature. Fertil. Steril. 90: 1201e1213–1207 [DOI] [PubMed] [Google Scholar]
- Phadnis N., Orr H. A., 2009. A single gene causes both male sterility and segregation distortion in Drosophila hybrids. Science 323: 376–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piálek J., Vyskočilová M., Bímová B., Havelková D., Piálková J., et al. , 2008. Development of unique house mouse resources suitable for evolutionary studies of speciation. J. Hered. 99: 34–44 [DOI] [PubMed] [Google Scholar]
- Presgraves D. C., 2003. A fine-scale genetic analysis of hybrid incompatibilities in Drosophila. Genetics 163: 955–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves D. C., Balagopalan L., Abmayr S. M., Orr H. A., 2003. Adaptive evolution drives divergence of a hybrid inviability gene between two species of Drosophila. Nature 423: 715–719 [DOI] [PubMed] [Google Scholar]
- Prisant N., Escalier D., Soufir J.-C., Morillon M., Schoevaert D., et al. , 2007. Ultrastructural nuclear defects and increased chromosome aneuploidies in spermatozoa with elongated heads. Hum. Reprod. 22: 1052–1059 [DOI] [PubMed] [Google Scholar]
- Raufaste N., Orth A., Belkhir K., Senet D., Smadja C., et al. , 2005. Inferences of selection and migration in the Danish house mouse hybrid zone. Biol. J. Linn. Soc. Lond. 84: 593–616 [Google Scholar]
- Revay T., Nagy S., Kopp C., Flyckt A., Rens W., et al. , 2009. Macrocephaly in bull spermatozoa is associated with nuclear vacuoles, diploidy and alteration of chromatin condensation. Cytogenet. Genome Res. 126: 202–209 [DOI] [PubMed] [Google Scholar]
- Russell L. D., Ettlin R., Sinha Hikin A., Clegg E., 1990. Histological and Histopathological Evaluation of the Testis. Cache River Press, Clearwater, FL [Google Scholar]
- Sage R. D., Heyneman D., Lim K. C., Wilson A. C., 1986. Wormy mice in a hybrid zone. Nature 324: 60–63 [DOI] [PubMed] [Google Scholar]
- Sage R. D., Atchley W. R., Capanna E., 1993. House mice as models in systematic biology. Syst. Biol. 42: 523–561 [Google Scholar]
- Salcedo T., Geraldes A., Nachman M. W., 2007. Nucleotide variation in wild and inbred mice. Genetics 177: 2277–2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamura K., Yamamoto M., 1997. Characterization of a reproductive isolation gene, zygotic hybrid rescue, of Drosophila melanogaster by using minichromosomes. Heredity 79: 97–103 [Google Scholar]
- Searle A. G., Beechey C. V., 1974. Sperm-count, egg-fertilization and dominant lethality after X-irradiation of mice. Mutat. Res. 22: 63–72 [DOI] [PubMed] [Google Scholar]
- Selander R. K., Yang S. Y., 1969. Protein polymorphism and genic heterozygosity in a wild population of the house mouse (Mus musculus). Genetics 63: 653–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selander R., Hunt W., Yang S., 1969. Protein polymorphism and genic heterozygosity in two European subspecies of the house mouse. Evolution 23: 379–390 [DOI] [PubMed] [Google Scholar]
- Sen S., Churchill G. A., 2001. A statistical framework for quantitative trait mapping. Genetics 159: 371–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She J. X., Bonhomme F., Boursot P., Thaler L., Catzeflis F., 1990. Molecular phylogenies in the genus Mus: comparative analysis of electrophoretic, scnDNA hybridization, and mtDNA RFLP data. Biol. J. Linn. Soc. Lond. 41: 83–103 [Google Scholar]
- Smadja C., Ganem G., 2002. Subspecies recognition in the house mouse: a study of two populations from the border of a hybrid zone. Behav. Ecol. 13: 312–320 [Google Scholar]
- Smadja C., Ganem G., 2005. Asymmetrical reproductive character displacement in the house mouse. J. Evol. Biol. 18: 1485–1493 [DOI] [PubMed] [Google Scholar]
- Smadja G., Catalan J., Ganem C., 2004. Strong premating divergence in a unimodal hybrid zone between two subspecies of the house mouse. J. Evol. Biol. 17: 165–176 [DOI] [PubMed] [Google Scholar]
- Storchová R., Gregorová S., Buckiová D., Kyselová V., Divina P., et al. , 2004. Genetic analysis of X-linked hybrid sterility in the house mouse. Mamm. Genome 15: 515–524 [DOI] [PubMed] [Google Scholar]
- Styrna J., 2008. Genetic control of gamete quality in the mouse: a tribute to Halina Krzanowska. Int. J. Dev. Biol. 52: 195–199 [DOI] [PubMed] [Google Scholar]
- Styrna J., Krzanowska H., 1995. Sperm select penetration test reveals differences in sperm quality in strains with different Y chromosome genotype in mice. Arch. Androl. 35: 111–118 [DOI] [PubMed] [Google Scholar]
- Styrna J., Klag J., Moriwaki K., 1991. Influence of partial deletion of the Y chromosome on mouse sperm phenotype. J. Reprod. Fertil. 92: 187–195 [DOI] [PubMed] [Google Scholar]
- Styrna J., Bilińska B., Krzanowskaa H., 2002. The effect of a partial Y chromosome deletion in B10.BR-Ydel mice on testis morphology, sperm quality and efficiency of fertilization. Reprod. Fertil. Dev. 14: 101–108 [DOI] [PubMed] [Google Scholar]
- Suzuki H., Shimada T., Terashima M., Tsuchiya K., Aplin K., 2004. Temporal, spatial, and ecological modes of evolution of Eurasian Mus based on mitochondrial and nuclear gene sequences. Mol. Phylogenet. Evol. 33: 626–646 [DOI] [PubMed] [Google Scholar]
- Sweigart A. L., Fishman L., Willis J. H., 2006. A simple genetic incompatibility causes hybrid male sterility in Mimulus. Genetics 172: 2465–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talley H. M., Laukaitis C. M., Karn R. C., 2001. Female preference for male saliva: implications for sexual isolation of Mus musculus subspecies. Evolution 55: 631–634 [DOI] [PubMed] [Google Scholar]
- Tang S., Presgraves D. C., 2009. Evolution of the Drosophila nuclear pore complex results in multiple hybrid incompatibilities. Science 323: 779–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeter K. C., Payseur B. A., Harris L. W., Bakewell M. A., Thibodeau L. M., et al. , 2008. Genome-wide patterns of gene flow across a house mouse hybrid zone. Genome Res. 18: 67–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeter K. C., Thibodeau L. M., Gompert Z., Buerkle C. A., Nachman M. W., et al. , 2010. The variable genomic architecture of isolation between hybridizing species of house mice. Evolution 64: 472–485 [DOI] [PubMed] [Google Scholar]
- Ting C.-T., Tsaur S.-C., Wu M.-L., Wu C.-I., 1998. A rapidly evolving homeobox at the site of a hybrid sterility gene. Science 282: 1501–1504 [DOI] [PubMed] [Google Scholar]
- Trachtulec Z., Vlcek C., Mihola O., Gregorova S., Fotopulosova V., et al. , 2008. Fine haplotype structure of a chromosome 17 region in the laboratory and wild mouse. Genetics 178: 1777–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker P. K., Sage R. D., Warner J., Wilson A. C., Eicher E., 1992. Abrupt cline for sex-chromosomes in a hybrid zone between two species of mice. Evolution 46: 1146–1163 [DOI] [PubMed] [Google Scholar]
- Vanlerberghe F., Dod B., Boursot P., Bellis M., Bonhomme F., 1986. Absence of Y-chromosome introgression across the hybrid zone between Mus musculus domesticus and Mus musculus musculus. Genet. Res. 48: 191–197 [DOI] [PubMed] [Google Scholar]
- Vredenburgh-Wilberg W. L., Parrish J. J., 1995. Intracellular pH of bovine sperm increases during capacitation. Mol. Reprod. Dev. 40: 490–502 [DOI] [PubMed] [Google Scholar]
- Vyskočilová M., Trachtulec Z., Forejt J., Pialek J., 2005. Does geography matter in hybrid sterility in house mice? Biol. J. Linn. Soc. Lond. 84: 663–674 [Google Scholar]
- Vyskočilová M., Pražanová G., Piálek J., 2009. Polymorphism in hybrid male sterility in wild-derived Mus musculus musculus strains on proximal chromosome 17. Mamm. Genome 20: 83–91 [DOI] [PubMed] [Google Scholar]
- White M. A., Ané C., Dewey C. N., Larget B. R., Payseur B. A., 2009. Fine-scale phylogenetic discordance across the house mouse genome. PLoS Genet. 5: e1000729. [DOI] [PMC free article] [PubMed] [Google Scholar]