Abstract
Aging is an important feature of animal biology characterized by progressive, degenerative changes in somatic and reproductive tissues. The rate of age-related degeneration is genetically controlled, since genes that influence lifespan have been identified. However, little is known about genes that affect reproductive aging or aging of specific somatic tissues. To identify genes that are important for controlling these degenerative changes, we used chemical mutagenesis to perform forward genetic screens in Caenorhabditis elegans. By conducting a screen focused on somatic aging, we identified mutant hermaphrodites that displayed extended periods of pharyngeal pumping, body movement, or survival. One of these mutations is a novel allele of the age-1 gene. age-1 encodes a phosphatidylinositol-3-kinase (PI3K) that functions in the insulin/insulin-like growth factor-1 (IGF-1) signaling pathway. age-1(am88) creates a missense change in the conserved PIK domain and causes dramatic extensions of the pharyngeal pumping and body movement spans, as well as a twofold extension of the lifespan. By conducting screens focused on reproductive aging in mated hermaphrodites, we identified mutants that displayed increased progeny production late in life. To characterize these mutations, we developed quantitative measurements of age-related morphological changes in the gonad. The am117 mutation delayed age-related declines in progeny production and morphological changes in the gonad. These studies provide new insights into the genetic regulation of age-related degenerative changes in somatic and reproductive tissues.
AGING is an important aspect of animal biology. As adult animals advance in chronological age, they display progressive degenerative changes that compromise the function of a wide range of organ systems. These changes affect neuromuscular activities associated with perception and motility, reproductive organs that mediate progeny production, and many other systems. Age-related degenerative changes in life support systems ultimately result in death. Human aging has enormous medical significance, since age-related degenerative changes contribute to functional impairments and mortality in elderly people (Vijg and Campisi 2008). In human females, reproductive aging is an important medical issue because the age-related decline in oocyte quality results in increased birth defects and decreased fertility that culminates in reproductive cessation at menopause (Hartge 2009).
The soil nematode Caenorhabditis elegans has emerged as an important model organism for studies of aging (Guarente and Kenyon 2000; Braeckman et al. 2001; Gershon and Gershon 2002; Olsen et al. 2006). C. elegans has a relatively short adult lifespan of ∼2 weeks, and a number of age-related degenerative changes have been characterized (Collins et al. 2007; Pincus and Slack 2010). Sophisticated genetic techniques have contributed to the identification of a large number of genes that modulate lifespan, and these studies have made important contributions to understanding the genetic control of longevity (Kenyon 2005). These studies have demonstrated that insulin/IGF-1 signaling, mitochondrial function, chemosensory function, dietary intake, and the sir-2 gene are important modulators of adult lifespan.
Many studies of C. elegans aging use lifespan as the primary measurement of aging. Because aging is characterized by widespread degenerative changes, including changes in the reproductive system that may not affect longevity, genes that are important for aging in specific organ systems may not have been identified using lifespan as an assay. To characterize the genetic control of age-related degenerative changes, we have focused on the somatic and reproductive tissues of C. elegans (Huang et al. 2004; Hughes et al. 2007). Two somatic tissues are well suited to analyzing age-related degeneration. The pharynx is the feeding organ of the animal, and the rhythmic contractions of this organ can be easily visualized and display progressive age-related declines (Croll et al. 1977; Hosono et al. 1980). Similarly, body movement can be easily visualized and displays age-related degeneration (Bolanowski et al. 1981; Johnson 1987; Herndon et al. 2002; Huang et al. 2004). To characterize aging in the reproductive system, we have focused on changes in the hermaphrodite gonad, which produces fertilized eggs (Garigan et al. 2002; Hughes et al. 2007; Luo et al. 2009, 2010). Reproductive aging has special significance for evolutionary biology, since reproduction is the purpose of animal life and a critical aspect of fitness. Furthermore, prominent theories postulate that reproduction is a cause of aging. The disposable soma theory proposes a metabolic tradeoff between reproduction and aging, whereas the antagonistic pleiotropy theory proposes a genetic tradeoff between early and late reproduction (Williams 1957; Kirkwood 1977). Studies of reproductive aging have been used to test predictions of these theories. For example, selection experiments for enhanced late reproduction in Drosophila resulted in lower levels of early reproduction, which is interpreted as supporting the antagonistic pleiotropy theory (Rose and Charlesworth 1981).
Here we describe the identification of new mutations that were discovered by screening for mutant animals with delayed functional declines in processes mediated by specific somatic or reproductive tissues. By screening for animals with extended pharyngeal pumping or body movement, we identified a novel mutation in the age-1 gene that encodes a phosphatidylinositol-3-kinase. age-1 is an important mediator of the insulin/IGF-1 signaling pathway that controls longevity in C. elegans and other animals including vertebrates (Kenyon 2005). Three mutations were identified that influence reproductive aging and extend the mated reproductive period. We developed quantitative measurements of age-related changes in the gonad and demonstrated that the am117 mutation delays these morphological changes in addition to extending the mated reproductive period. These studies provide new insights into the genetic regulation of tissue-specific aging.
Materials and Methods
General methods and strains
C. elegans strains were cultured at 20° unless noted otherwise on 6-cm Petri dishes containing nematode growth media (NGM) agar and a lawn of Escherichia coli strain OP50 (Brenner 1974). The wild-type strain was N2. The following mutations that affect lifespan and Dauer development were used: daf-2(e1370P1465S) is a partial loss-of-function mutation that affects the kinase domain of the DAF-2 receptor tyrosine kinase (Kimura et al. 1997); age-1(hx546P806S) is a partial loss-of-function mutation that affects the AGE-1 PI3K (Morris et al. 1996; Ayyadevara et al. 2008); age-1(am88E725K), am115, am116, and am117 are described here. The following mutations on chromosome I were used for three-factor mapping experiments and are described by Riddle et al. (1997): dpy-5(e61) 0.0, bli-4(e937) +1.0, unc-37(e262) +1.3, unc-29(e193) +3.3, dpy-24(s71) +4.7, unc-75(e950) +9.5, and unc-101(m1) +13.2.
Analyses of aging phenotypes in self-fertilized hermaphrodites
Aging phenotypes of self-fertilized hermaphrodites were analyzed as described by Huang et al. (2004) unless noted otherwise. Briefly, studies of aging were begun on day 0 by placing one L4 hermaphrodite on a Petri dish. Each hermaphrodite was transferred to a fresh Petri dish about every 2 days during the reproductive period (approximately the first 7 days) to eliminate self-progeny and transferred as necessary thereafter. Each hermaphrodite was examined every 1–2 days using a dissecting microscope for the following phenotypes: (1) progeny production was determined by the presence of eggs or larvae; (2) body movement was observed for 10–30 sec and classified as fast if an animal displayed continuous and well-coordinated sinusoidal movement and not fast if an animal displayed discontinuous or sluggish movement; (3) pharyngeal pumping was observed for 10 sec and categorized as fast (≥25 contractions), slow (1–24 contractions), or none (0 contractions); and (4) survival was determined by spontaneous movement or movement in response to prodding with a pick. For each hermaphrodite, we calculated the time period in days between the start of the experiment (L4) and the last day of progeny production, fast pharyngeal pumping, fast body movement, and pharyngeal pumping. These periods are defined as the self-fertile reproductive span (RS), fast pharyngeal pumping span (FPS), fast body movement span (MS), and pharyngeal pumping span (PS). Lifespan (LS) was defined as the time period in days between L4 and the last day of survival.
Genetic screen for mutations that delay age-related degenerative changes in self-fertilized hermaphrodites
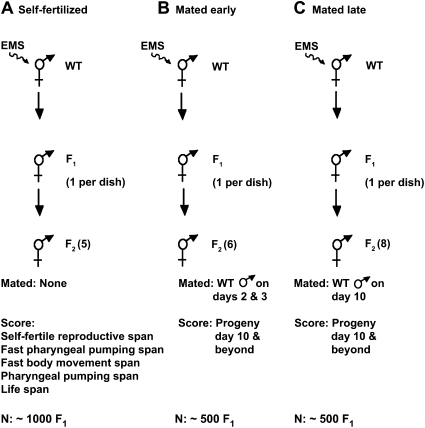
We used methanesulfonic acid ethyl ester (EMS) to mutagenize wild-type worms as previously described (Brenner 1974). For the screen of self-fertilized hermaphrodites, we mutagenized P0 hermaphrodites at the fourth larval stage (L4) and placed ∼1000 F1 self-progeny on individual plates (Figure 1A). F2 self-progeny hermaphrodites were synchronized by using a dissecting microscope to select animals at the L4 stage, on the basis of the appearance of the vulva as a dark half circle. Five F2 self-progeny from each F1 animal were placed one per Petri dish and monitored for self-fertile reproductive span, fast pharyngeal pumping span, fast body movement span, pharyngeal pumping span, and lifespan. F2 animals that displayed an extension of one or more spans were classified as preliminary mutants. Strains derived from self-fertilization of preliminary mutants were analyzed in secondary and tertiary experiments using progressively larger numbers of animals. We focused on measuring the span that was extended in the preliminary mutant, although sometimes additional spans were also analyzed. Forty-five mutant strains displayed a reproducible extension of one or more aging spans and were further analyzed.
Figure 1 .
Design of genetic screens. Wild-type hermaphrodites were mutagenized with EMS, F1 self-progeny were cultured individually, and the indicated number of F2 self-progeny was analyzed. F2 hermaphrodites were self-fertilized (A) or mated to wild-type males on the indicated days (B and C). F2 animals were analyzed for aging spans (A) or cumulative progeny production from day 10 until cessation (B and C). N indicates the number of F1 animals analyzed.
Backcrossing mutant strains to the wild-type strain was a challenge because it required extensive tests to assign genotypes with confidence. Because genetically identical individuals display substantial variation in aging spans, the analysis of one hermaphrodite cannot be used to reliably infer the genotype of that animal. Rather, it is necessary to analyze multiple progeny of one hermaphrodite and determine the mean value for this population to infer the genotype of the parent hermaphrodite. We mated one P0 mutant hermaphrodite with four to six wild-type males on a single Petri dish, transferred these animals daily to a fresh Petri dish, and isolated outcrossed F1 L4 hermaphrodites from Petri dishes containing about half male progeny. The F1 hermaphrodite was allowed to generate F2 self-progeny, and 12 randomly selected F2 eggs were placed one per Petri dish. F2 animals were selected as eggs to minimize bias against slow growing mutants, and 25% (3/12) of these F2 animals were predicted to have the genotype mutant/mutant. To identify mutant/mutant F2 animals, we allowed F2 eggs to mature and produce F3 self-progeny. Twenty-five F3 self-progeny from each F2 animal were analyzed for the aging phenotype. An F2 animal was considered a candidate to have the genotype mutant/mutant when the population mean of the F3 progeny displayed a significantly extended aging span. To confirm that the F2 animal was mutant/mutant and not mutant/+, we further chose 10 F3 progeny and analyzed 10 F4 progeny from each. The F2 animal was determined to be mutant/mutant if these 10 F4 populations consistently displayed the extended aging span.
To backcross the am88 allele, we mated am88 hermaphrodites to wild-type males, picked outcrossed F1 hermaphrodites to separate Petri dishes, and picked F2 self-progeny eggs to separate Petri dishes. After F2 animals matured and generated self-progeny, we extracted DNA from F2 animals and used DNA sequencing to genotype the am88 missense change in the age-1 locus (described below).
Genetic screen for mutations that delay reproductive aging in mated hermaphrodites
To screen for mutations that influence reproductive aging of mated hermaphrodites, we mutagenized wild-type N2 hermaphrodites at the L4 stage with EMS (Brenner 1974) and picked F1 self-progeny to individual Petri dishes. For the early-mating screen protocol shown in Figure 1B, six F2 hermaphrodites from an individual F1 were picked at the L4 stage to individual Petri dishes (day 1). The hermaphrodites were mated to approximately seven wild-type males on days 2 and 3 in sibling groups of three hermaphrodites. The day 1 Petri dishes were used for recovery of candidate mutations. Hermaphrodites were transferred to fresh Petri dishes every other day until day 10 and then placed on individual Petri dishes to score progeny production. Worms were transferred every 2 days thereafter if progeny were present. The number of progeny was scored ∼2 days after transfer. Worms were monitored until at least 4 days of no progeny production after day 10 or the death of the animal. We analyzed the F2 descendants of ∼500 F1 animals. Our criteria for an F2 animal with a candidate mutation was either (1) the production on day 10 and subsequent days of >10 progeny in total or (2) the production on day 14 and subsequent days of >0 progeny in total.
For the late-mating screen protocol shown in Figure 1C, we picked to individual Petri dishes eight F2 hermaphrodites at the L4 stage that were self-progeny of one F1 hermaphrodite (day 1). The day-1 dishes were used to recover candidate mutations. F2 hermaphrodites were transferred to fresh dishes in groups of four siblings on days 3, 5, 7, and 9. F2 hermaphrodites were cultured individually with three males for ∼1 day beginning on day 10 and then transferred to a fresh dish on day 11 and every 2 days thereafter if progeny were present. The number of progeny was scored ∼2 days after transfer. We analyzed the F2 descendants of ∼500 F1 animals, which were distinct from the F1 animals analyzed in the early-mating protocol. We used the criteria described above for a candidate mutation.
When a mated F2 hermaphrodite met our criteria for increased progeny production, then we recovered the candidate mutation by analyzing about eight L4 self-progeny hermaphrodites derived from each potential day-1 plate. Our criteria for further analyzing the strain was that at least half of these eight worms displayed increased progeny production on day 10 and subsequent days. Candidate strains were then analyzed by testing ∼20 hermaphrodites followed by testing ∼50 hermaphrodites using the initial screening conditions. Seven candidate strains displayed a statistically significant increase in late progeny production in these tests and were further analyzed.
To backcross candidate mutations, we mated mutant hermaphrodites to wild-type males, picked F1 cross-progeny hermaphrodites, and picked 12 F2 self-progeny as eggs or L4 larvae to individual plates. To identify F2 animals that were homozygous for the new mutation, we assayed 25 F3 self-progeny hermaphrodites from each F2 animal for late progeny production. F3 populations that displayed statistically significant increases in late progeny production compared to wild type (P < 0.05) were considered successfully backcrossed.
To confirm that a mutation was homozygous after a backcross, we picked 10 animals from the population to separate Petri dishes as eggs or L4 larvae and analyzed 10 self-progeny from each animal for late progeny production. We concluded that a strain was homozygous for the mutation if all 10 of the original populations displayed the late progeny production phenotype. We used these procedures to backcross am115 one time, am116 two times, and am117 two times. am117 was backcrossed one additional time by scoring the scrawny phenotype and then using these procedures to confirm that the strain was also homozygous for the mutation that increases late progeny production.
Analyses of Dauer formation
Dauer formation was assayed as described by Kimura et al. (1997). Briefly, we collected eggs from hermaphrodites cultured at 20°, transferred the eggs to 27° with ample food for 72 hr, and examined hatched animals. Animals were classified as non-Dauer (including adults and non-Dauer larvae) or Dauer on the basis of morphological criteria (Riddle et al. 1997).
Complementation tests with am88 were performed using the Dauer formation assay. To generate heterozygous animals, we mated one L4 hermaphrodite with four to eight males on a single Petri dish at 20° and transferred the animals to a fresh Petri dish daily. On the third day, the mated hermaphrodite was allowed to lay eggs on a fresh Petri dish for ∼4 hr. These eggs were then transferred to 27° and assayed for Dauer formation. We confirmed that the hermaphrodites mated successfully by observing the production of male cross-progeny both before and after day 3. Mating experiments with daf-2(e1370) males were conducted at 15° since these males mate poorly at 20°.
Analyses of additional phenotypes in reproductive aging mutants
To analyze the rate of egg hatching, larval viability, and adult sterility, we picked 200 eggs from self-fertilized hermaphrodites to individual Petri dishes and monitored development to adulthood. We recorded the number of eggs that failed to hatch, the number of larvae that failed to develop to adulthood, and the number of adults that did not lay eggs. To determine whether these processes were temperature sensitive, we cultured animals at 15°, 20°, or 25°.
To measure the schedule of progeny production, we picked L4 hermaphrodites to individual Petri dishes and transferred these animals to fresh dishes daily until death or at least 4 days without progeny production. Progeny were counted ∼2 days after the transfer. For analysis of mated hermaphrodites, three young, wild-type males were added to the dish for the first 2 days. Fertile animals that died during the experiment were included in the data until the day of death, and N values are the number of animals at the start of the experiment.
The pharyngeal pumping rate was monitored at the L4 stage using a dissecting microscope by counting the number of pharyngeal pumps in a 10-sec interval.
Determining the map position of am117
The am117 mutation was mapped in three stages. First, we used a low-density, genome-wide map of single nucleotide polymorphisms (SNPs) that differ between the N2 (Bristol) and the CB4856 (Hawaiian) strains as described by Bruinsma et al. (2008). am117 males were mated to CB4856 hermaphrodites, F1 cross-progeny were isolated, and F2 self-progeny were picked as eggs to individual Petri dishes. The F2 adults and their self-progeny were scored for the am117 scrawny phenotype, and SNPs were scored using pyrosequencing or standard sequencing techniques. am117 displayed linkage of 86% to SNP marker amP20 in cosmid C09D4 positioned at the center of chromosome I (−0.1), and am117 did not display significant linkage to SNP markers located at the center of the other five chromosomes. An analysis of six SNP markers on chromosome I showed that am117 displayed the highest linkage to SNP markers in cosmid C36B1 located at +3.0 (89%) and in cosmid T22A3 located at +5.0 (88%). Second, the position of am117 was determined relative to mutations that cause visible phenotypes using standard 3-factor mapping techniques. From dpy-5unc-101/am117 hermaphrodites, 6/47 Unc non-Dpy and 11/25 Dpy non-Unc self-progeny segregated am117 on the basis of the scrawny phenotype, indicating that am117 is between dpy-5 and unc-101. Third, we used a local, high-density SNP map to refine the position of am117 as described by Jakubowski and Kornfeld (1999). The triple mutant dpy-5(e61) am117 unc-101(m1) was constructed using standard genetic techniques. We crossed this mutant to CB4856, selected F1 cross-progeny, selected F2 self-progeny that displayed a Dpy non-Unc or Unc non-Dpy phenotype, and selected self-progeny homozygous for the recombinant chromosome. Because the am117 scrawny phenotype of these strains was difficult to score in the presence of the Unc or Dpy phenotype, we performed a complementation test by mating to am117 males and scoring the scrawny phenotype in cross-progeny. SNP markers were scored by pyrosequencing or standard DNA sequencing. For Dpy non-Unc strains, 3/86 with a crossover event left of a SNP in cosmid F32B4 segregated am117 and 25/53 with a crossover event right of a SNP in cosmid F32B4 segregated am117. For Unc non-Dpy strains, 8/34 with a crossover event right of a SNP in cosmid F32B4 segregated am117 and 1/69 with a crossover event left of a SNP in cosmid F32B4 segregated am117. These results indicate that am117 is positioned in a 4.3-map-unit interval between F32B4 at +9.0 and unc-101 at +13.3.
Development of morphological markers of reproductive aging
To analyze age-related changes in the morphology of the reproductive system, we selected hermaphrodites at the L4 stage (day 1) and transferred the hermaphrodites to a fresh Petri dish every 2 days. Aged hermaphrodites were placed on a slide with a coverslip and examined using Nomarski optics on a Zeiss axioplan microscope. dpy-5(e61) worms were placed on the slide to prevent the coverslip from killing older, more fragile hermaphrodites.
DNA sequencing and alignment of am88
Unless otherwise noted, molecular biology techniques were performed as described by Sambrook et al. (1989). Genomic DNA was prepared, PCR amplified, and sequenced as described by Williams et al. (1992). DNA sequence data were analyzed using the Sequencher software (version 4.1.2; Genes Codes Corporation).
The accession numbers of the human and mouse PI3 kinase delta polypeptides are NP_005017 and AAN05615, respectively. The multiple sequence alignment in Figure 3 was performed using ClustalW (v1.4) with the following parameters: open gap penalty= 10.0, extend gap penalty= 0.1, delay divergent= 40%, gap distance= 8, and similarity matrix was blosum (MecVector software version 7.2; Accelrys).
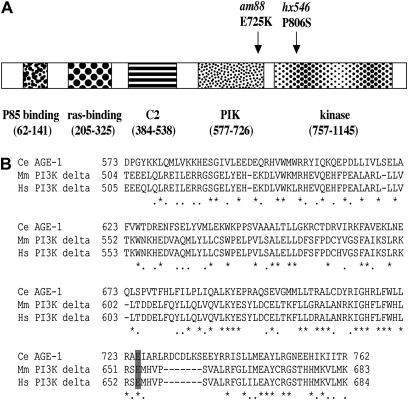
Figure 3 .
The am88 mutation affects a conserved residue in the AGE-1 PI3 kinase. (A) The full-length AGE-1 PI3 kinase is schematized, and conserved domains are shown as stippled boxes and labeled below. The positions of the am88 mutation (E-to-K substitution in the PIK domain) and the hx546 mutation are indicated. (B) The amino acid sequence of the PIK domain of the C. elegans AGE-1 protein is aligned with that of mouse PI3 kinase delta (Chantry et al. 1997; Okkenhaug et al. 2002) and human PI3 kinase delta (Chantry et al. 1997; Vanhaesebroeck et al. 1997). Identical residues are marked by asterisks, similar residues are marked by dots, and gaps are represented by dashes. The am88 change affects the conserved glutamate at residue 725 (shaded gray).
Statistical analyses
The statistical comparison of two different strains was carried out by using the Student’s t-test when the standard deviations of the two strains were not significantly different from each other. When they were significantly different from each other, the alternate Welch test was used instead. All of these analyses were carried out by using the software Instat for Macintosh version 2.03 (GraphPad software).
Results
Identification of mutations that delay age-related degenerative changes of self-fertilized hermaphrodites
To analyze age-related degeneration in C. elegans, we previously described four measurements of age-related changes of physiological functions (Huang et al. 2004). These measurements are quantitative, so they can be used to compare two populations, and noninvasive, so that serial measurements can be made on the same animal. The self-fertile reproductive span is defined as the time from the L4 larval stage to the last day of self-fertile progeny production. This measures the duration of self-fertile reproduction, which depends on the number of sperm generated by the hermaphrodite during development and the rate of self-sperm utilization (Hughes et al. 2007). The fast pharyngeal pumping span is defined as the time from L4 to the last day of fast pharyngeal pumping, defined as ≥150 contractions/minute. This measures the duration of vigorous pharyngeal pumping. The fast body movement span is defined as the time from L4 to the last day of fast body movement, defined as continuous and well-coordinated sinusoidal movement. This measures the duration of vigorous body movement. The pharyngeal pumping span is defined as the time from L4 to the last day of detectable pharyngeal pumping. Together with lifespan, the standard measurement used to infer the rate of aging, these measurements quantitatively describe the decline of physiological functions during adulthood of self-fertilized hermaphrodites (Huang et al. 2004).
Wild-type hermaphrodites cultured with abundant food at 20° displayed a self-fertile reproductive span of 5.8 days, a fast pharyngeal pumping span of 8.1 days, a fast body movement span of 8.2 days, a pharyngeal pumping span of 11.8 days, and a lifespan of 15.2 days (Table 1; Huang et al. 2004). To identify genes that modulate the rate of aging, we conducted a forward genetic screen for mutations that extend one or more of these spans. A clonal screen was conducted by mutagenizing wild-type P0 worms with the chemical mutagen EMS and measuring the five spans in multiple F2 self-progeny that were cultured separately (Figure 1A). We screened ∼2000 haploid genomes and identified 45 mutant strains that displayed a significant extension of one or more of the spans. Of these strains, 22 primarily extended lifespan, 14 primarily extended fast body movement span, four primarily extended self-fertile reproductive span, three primarily extended fast pharyngeal pumping span, and two primarily extended pharyngeal pumping span (data not shown).
Table 1 . age-1 (am88) delays aging.
| Genotype | Self-fertile reproductive spana | Fast pharyngeal pumping spana | Fast body movement spana | Pharyngeal pumping spana | Lifespana | Nb |
|---|---|---|---|---|---|---|
| WT | 5.8 ± 0.15 | 8.1 ± 0.16 | 8.2 ± 0.13 | 11.8 ± 0.22 | 15.2 ± 0.27 | 180 |
| am88 | 5.6 ± 0.16 | 10.8 ± 0.3* | 15.5 ± 0.56* | 21.4 ± 0.71* | 29.8 ± 0.92* | 105 |
*P-value ≤ 0.0001 compared to WT. WT data are from Huang et al. (2004).
Values are mean number of days and standard error of the mean for self-fertile hermaphrodites.
Number of hermaphrodites analyzed.
The am88 mutation substantially delays multiple age-related changes
To analyze newly identified strains, we first backcrossed the mutations to wild-type animals. We devised methods to demonstrate that newly identified mutations were homozygous following the backcross (see Materials and Methods). However, these methods were laborious, and if a mutant strain displayed small or moderate extensions of an aging span, then it was difficult to successfully conduct the backcross experiments. Therefore, we focused on the mutation am88, since am88 mutants displayed the largest span extensions among the newly identified strains. am88 was initially identified on the basis of lifespan extension. Compared to wild type, the lifespan increased 100% to 29.8 days, the fast pharyngeal pumping span increased 30% to 10.8 days, the fast body movement span increased 90% to 15.5 days, and the pharyngeal pumping span increased 80% to 21.4 days. am88 did not significantly affect the self-fertile reproductive span (Table 1, Figure 2).
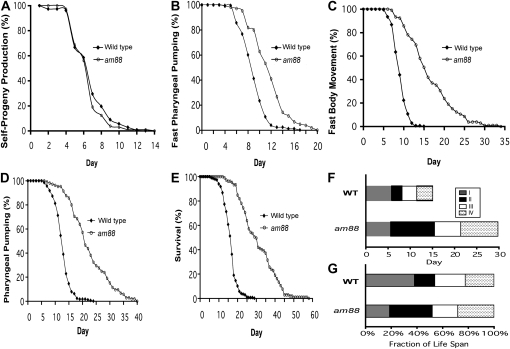
Figure 2 .
The am88 mutation delays multiple age-related degenerative changes. Wild-type (black diamonds) and am88 (open circles) hermaphrodites at the L4 stage were placed on individual Petri dishes and monitored for self-progeny production (A), fast pharyngeal pumping (B), fast body movement (C), any pharyngeal pumping (D), and survival (E). The percentage of animals in the population that displayed the phenotype is graphed vs. age in days (see Table 1 for summary statistics). Total number of animals analyzed was 180 for wild type and 105 for am88. (F) Horizontal bars represent the time in days spent in stages I (gray), II (black), III (white), and IV (stippled) (described in the text). (G) Horizontal bars represent the fraction of the lifespan occupied by each stage, calculated by setting the lifespan of each strain equal to 100%. WT data are from Huang et al. (2004).
We analyzed the aging stage patterns of the am88 animals using the staging system described in Huang et al. (2004). Briefly, stage I is defined as L4 to the end of the self-fertile reproductive span, stage II is defined as the end of stage I to the end of the fast body movement span, stage III is defined as the end of stage II to the end of the pharyngeal pumping span, and stage IV is defined as the end of stage III to the end of the lifespan. Figure 2F shows the amount of time in days of each stage, while Figure 2G shows the proportion of the adult lifespan for each stage. am88 extended the adult lifespan almost 100% and dramatically changed the pattern of the four stages. Specifically, am88 expanded stages II (33% compared to 16% of wild type) and IV (28% compared to 22% of wild type) and reduced stage I (19% compared to 38% of wild type) as a fraction of the lifespan.
am88 is a novel mutation in the age-1 gene
We previously used this staging system to analyze mutations that increase lifespan by affecting the insulin/IGF-1 signaling pathway, caloric intake, and mitochondrial function; mutations from these classes produced characteristic changes in aging stage patterns (Huang et al. 2004). The changes in patterns caused by am88 were similar to the changes in patterns caused by mutations affecting the insulin/IGF-1 signaling pathway such as age-1(hx546) and daf-2(e1370) (Huang et al. 2004), indicating that am88 might affect the insulin/IGF-1 signaling pathway. Furthermore, the extremely long lifespan of am88 mutants was also similar to the extremely long lifespans of age-1 and daf-2 mutants (Klass 1983; Friedman and Johnson 1988; Kenyon et al. 1993).
To test the hypothesis that am88 affects a gene in the DAF-2 pathway, we determined whether am88 causes a Dauer constitutive (Daf-c) phenotype as displayed by age-1 and daf-2 mutants (Riddle et al. 1997). Whereas wild-type worms cultured at 27° with abundant food formed no Dauer larvae, am88 mutants formed 8% Dauer larvae, indicating that am88 causes a Daf-c phenotype (Table 2, lines 1 and 2). To test the hypothesis that am88 is a mutation in the daf-2 or age-1 gene, we performed complementation tests. The Daf-c phenotype caused by am88 was recessive (Table 2, lines 3 and 4) and failed to complement the recessive Daf-c phenotype caused by age-1(hx546) (Table 2, line 9). By contrast, am88 complemented the semidominant Daf-c phenotype caused by daf-2(e1370) (Table 2, lines 8, 10, and 11). These results indicate that am88 affects the age-1 gene.
Table 2 . am88 affects Dauer development.
| Genotype (paternal/maternal) | Dauer (%) | N |
|---|---|---|
| WT | 0 | 466 |
| am88 | 8 | 1341 |
| am88/+ | 0 | 32 |
| +/am88 | 1 | 71 |
| age-1(hx546) | 56 | 324 |
| +/age-1(hx546) | 0 | 94 |
| daf-2(e1370) | 87 | 199 |
| +/daf-2(e1370) | 13 | 31 |
| am88/age-1(hx546) | 42 | 45 |
| daf-2(e1370)/am88 | 12 | 43 |
| daf-2(e1370)/age-1(hx546) | 8 | 39 |
Percentage of Dauer larvae formed at 27°. N, total number of worms examined.
We used genomic DNA sequencing to identify the am88 molecular lesion. The age-1 gene consists of nine exons (Morris et al. 1996). We sequenced all of the exons as well as the intron/exon boundaries using DNA from the am88 strain and the parental wild-type strain. We detected only one base change in am88 mutants, a G-to-A change at nucleotide 2289 in exon 4 (numbered according to cDNA accession number NM_064061) that is predicted to change codon 725 from glutamate to lysine. This nonconservative missense change substitutes a basic amino acid for an acidic residue. Furthermore, Glu 725 is a highly conserved residue in the PIK domain of the AGE-1 protein, which is a homolog of the vertebrate PI3 kinase (Flanagan et al. 1993; Morris et al. 1996; Domin and Waterfield 1997) (Figure 3). This mutation segregated with the lifespan extension phenotype after four rounds of backcrossing (data not shown), supporting the conclusion that the missense change resulting in the E725K substitution is the am88 mutation, and am88 is a novel allele of age-1.
Identification of mutations that delay reproductive aging of mated hermaphrodites
There are important differences between the age-related declines in progeny production displayed by self-fertilized and mated hermaphrodites. Hermaphrodites produce ∼300 self-sperm at the start of gametogenesis before switching to oocyte production (Ward and Carrel 1979). For self-fertilized hermaphrodites, the duration of live progeny production and the total number of progeny generated is limited by the number of self-sperm. First, the number of self-progeny corresponds closely to the number of self-sperm, indicating that nearly every sperm is utilized to produce a zygote (Ward and Carrel 1979). By contrast, hermaphrodites produce oocytes in excess, and unfertilized oocytes begin to be laid as sperm is depleted. Second, hermaphrodites that are mated to males and have abundant sperm generate substantially >300 progeny (Hodgkin and Barnes 1991). Third, mated hermaphrodites produce progeny for a longer time period, since the mean mated reproductive span is ∼30% longer than the mean self-fertile reproductive span (Hughes et al. 2007). These observations indicate that live progeny production at the end of the reproductive period is an accurate measure of reproductive capacity only for mated hermaphrodites that continually have sperm.
The genetic screen described above was designed to identify mutations that extended the reproductive period of self-fertilized hermaphrodites. Mutations that increase the self-fertile reproductive span are predicted to either increase the number of self-sperm generated or decrease the rate of self-sperm utilization. Indeed, the mutations that we identified on the basis of an extended self-fertile reproductive span reduced the rate of sperm utilization, as measured by the rate of self-progeny production (data not shown).
To identify genes that influence the rate of reproductive aging, we conducted a forward genetic screen for mutations that increase progeny production late in life in mated hermaphrodites that have abundant sperm. The analysis of mated hermaphrodites overcomes the issue of sperm limitation, and mutations that increase progeny production by mated hermaphrodites late in life are likely to delay age-related degenerative changes in the reproductive system. To design the screen, we analyzed the number of progeny generated daily by wild-type hermaphrodites mated early in life or late in life as previously determined by Hughes et al. (2007). Wild-type hermaphrodites mated late (day 10 of adulthood) generated 3.6 progeny during their remaining lifespan (Table 3, line 1), indicating that mating is still possible at this late stage but there is little remaining capacity for progeny production. Wild-type hermaphrodites mated early (day 1 or 2 of adulthood) produced an average of 1.8 progeny on day 10 and subsequent days (Table 3, line 1), indicating that reproductive senescence is almost complete by day 10. On the basis of these observations, we developed two strategies to screen for EMS-induced mutations that increase progeny production late in life. In the early-mating protocol, mutant hermaphrodites of the F2 generation were mated early in adulthood (days 2 and 3), and progeny production of individual hermaphrodites was measured on day 10 and subsequent days (Figure 1B). In the late-mating protocol, mutant hermaphrodites of the F2 generation were mated on day 10 of adulthood, and progeny production of individual hermaphrodites was measured on day 10 and subsequent days (Figure 1C, see Materials and Methods). In both protocols, new mutations were recovered from self-progeny generated by hermaphrodites before mating to males. We screened ∼1000 mutagenized haploid genomes using each protocol. If a mutant F2 hermaphrodite generated significantly more progeny late in life than wild type, then we propagated a strain from self-progeny and retested multiple descendants. Seven candidate strains consistently generated more late progeny than wild type: six were identified using the late-mating protocol and one was identified using the early-mating protocol.
Table 3 . Late reproduction by hermaphrodites.
| Genotype | Mated on day 10, progeny day 10, and beyonda | Numberb | P-valuec | Mated on days 1 and 2, progeny day 10, and beyondd | Numberb | P-valuec |
|---|---|---|---|---|---|---|
| WT | 3.6 ± 0.56 | 99 | 1.8 ± 0.48 | 86 | ||
| am115 | 5.6 ± 0.97 | 83 | 0.0781 | ND | ||
| am116 | 9.7 ± 1.4 | 103 | <0.0001 | 7.9 ± 2.7 | 18 | 0.0406 |
| am117 | 30.6 ± 3.4 | 102 | <0.0001 | 25.2 ± 2.4 | 92 | <0.0001 |
Hermaphrodites that survived until day 10 as self-fertile animals were mated on day 10 for ∼24 hr and scored for progeny production. Values are mean number of total progeny generated on day 10 and subsequent days with standard error of the mean. WT data are from Hughes et al. (2007).
Number of hermaphrodites analyzed.
P-value is compared to WT (line 1).
Hermaphrodites that were mated on day 1 or 2 for ∼48 hr and that survived until day 10 were scored for progeny production.
Phenotypic analysis of mutations that delay reproductive aging of mated hermaphrodites
Three of the mutant strains were successfully backcrossed to wild type; these experiments indicated that increased progeny production late in life was caused by a single mutation, and we designated these mutations am115, am116, and am117. am115 hermaphrodites mated late in life produced 5.6 progeny on day 10 and subsequent days, an increase of ∼60% compared to wild type (Table 3, line 2). Because this increase was of marginal statistical significance, we did not further analyze am115.
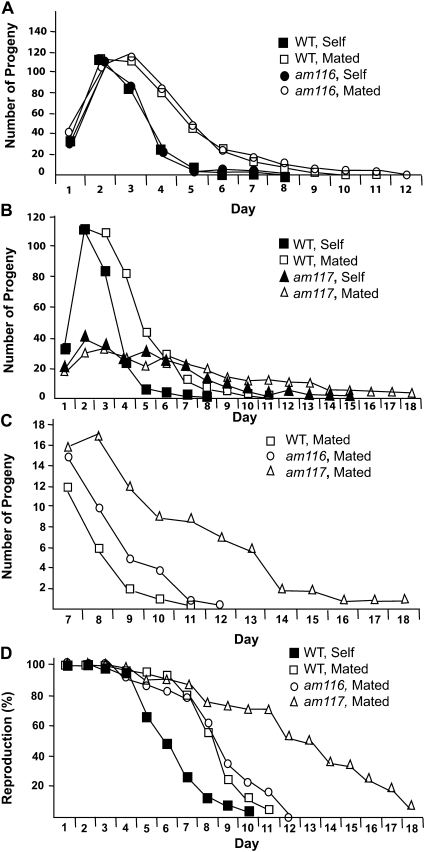
The mutation am116 significantly increased late progeny production of hermaphrodites mated early or late in life. am116 hermaphrodites mated late in life produced 9.7 progeny on day 10 and subsequent days, an increase of ∼170% compared to wild type (Table 3, line 3). am116 hermaphrodites mated early in life produced 7.9 progeny on day 10 and subsequent days, an increase of ∼340% compared to wild type (Table 3, line 2; Figure 4C). am116 hermaphrodites displayed high levels of egg hatching and low levels of larval lethality and adult sterility at 15°, 20°, and 25° that were similar to wild type (data not shown). To analyze somatic aging, we measured lifespan of self-fertilized am116 hermaphrodites. The am116 mutants displayed an increase in mean lifespan of ∼19% to 19.1 ± 1.2 days (N= 19, ± SEM) compared to 16.1 ± 0.7 days for wild type (N= 59, P= 0.05).
Figure 4 .
Progeny production of am116 and am117 mutants. (A–C) Average daily progeny production of live hermaphrodites containing the am116 or am117 mutation compared to wild type (WT). Hermaphrodites were self-fertilized (self) or mated for days 1 and 2 to three wild-type males (mated). (D) Percentage of hermaphrodites producing progeny. See Table 4 for values and summary statistics. Number of hermaphrodites at the start of the experiment: WT self = 76, WT mated = 65, am116 self = 25, am116 mated = 19, am117 self = 50, and am117 mated = 24. WT data are from Hughes et al. (2007).
The eat-2(ad465) mutation causes caloric restriction resulting in extended lifespan and increased progeny production by mated hermaphrodites late in life (Lakowski and Hekimi 1998; Hughes et al. 2007). To determine whether am116 causes caloric restriction, we monitored daily progeny production, since caloric restriction causes significant decreases in progeny production early in life (Hughes et al. 2007). Daily progeny production before day 9 for self-fertilized or mated am116 hermaphrodites was similar to self-fertilized or mated wild-type hermaphrodites, respectively (Table 4, Figure 4A). These observations suggest that am116 does not cause dietary restriction. To determine whether am116 affects insulin/IGF-1 signaling, we monitored formation of Dauer larvae, since mutations that decrease insulin/IGF-1 signaling typically cause a Dauer constitutive phenotype (Riddle et al. 1997). am116 mutants did not display a Dauer constitutive phenotype (data not shown), suggesting that the am116 mutation does not affect a major component of the insulin/ IGF-1 signaling pathway.
Table 4 . Progeny production and reproductive span of mated hermaphrodites.
| Daily progeny productionc | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotypea | Reproductive span (days)b | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 |
| WT | 7.7 ± 0.20 (65) | 34 ± 1.7 (65) | 114 ± 3.1 (65) | 112 ± 3.3 (65) | 83 ± 4 (64) | 45 ± 3.3 (64) | 26 ± 2.1 (64) | 12 ± 1.4 (64) | 6 ± 0.9 (61) | 2 ± 0.6 (51) | 0.4 ± 0.1 (45) | 0.1 ± 0.1 (45) | |||||||
| am116 | 7.8 ± 0.6 (19) | 42 ± 2.3* (19) | 107 ± 6.2 (19) | 117 ± 6.4 (19) | 86 ± 5.8 (17) | 48 ± 5.6 (17) | 25 ± 3.3 (16) | 15 ± 2.6 (15) | 10 ± 1.9 (13) | 5 ± 1.6* (10) | 4 ± 1.7* (9) | 1 ± 0.8* (6) | 0 (4) | ||||||
| am117 | 12 ± 0.9* (24) | 21 ± 1.3* (23) | 36 ± 1.8* (24) | 34 ± 2.2* (24) | 25 ± 1.7* (23) | 23 ± 1.5* (22) | 21 ± 3.4 (22) | 16 ± 2.0 (21) | 17 ± 2.4* (21) | 12 ± 2.6* (21) | 9 ± 2.0* (21) | 9 ± 2.4* (21) | 7 ± 1.8 (20) | 6 ± 1.8 (19) | 2 ± 0.7 (17) | 2 ± 0.7 (17) | 1 ± 0.3 (16) | 1 ± 0.5 (14) | 1 ± 0.9 (11) |
*P-value ≤ 0.05 compared to WT.
Hermaphrodites were mated to wild-type males on days 1 and 2.
Values are mean number of days and standard error of the mean.
Values are mean number of total progeny generated on a single day and standard error of the mean. Number of hermaphrodites analyzed is shown in parentheses.
The mutation am117 was identified using the early-mating protocol, and am117 mutants displayed the largest increases in progeny production late in life among the mutant strains that we identified. am117 hermaphrodites mated late in life produced 30.6 progeny on day 10 and subsequent days, an increase of ∼750% compared to wild type (Table 3, line 4). am117 hermaphrodites mated early in life produced 25.2 progeny on day 10 and subsequent days, an increase of ∼1300% compared to wild type (Table 3, line 3; Figure 4C). We measured daily progeny production of self-fertilized and mated am117 hermaphrodites (Table 4, Figure 4B). Self-fertilized am117 hermaphrodites displayed decreased early progeny production and an extended reproductive period, suggesting that the am117 mutation delays the rate of self-sperm utilization. Mated am117 hermaphrodites displayed significantly decreased early progeny production (days 1–5), significantly increased late progeny production (days 8–11), and a significantly increased reproductive span of 12 days compared to 7.7 days for wild type (Figure 4D). am117 hermaphrodites mated early in life produced a total of 225 ± 85 progeny, a decrease of about 48% compared to the 432 ± 103 progeny produced by wild-type hermaphrodites mated early in life (Hughes et al. 2007).
am117 mutants are thinner and smaller than wild-type worms, and we refer to this body morphology phenotype as “scrawny.” The development of am117 mutants from egg to adult was examined for hermaphrodites cultured at 15°, 20°, and 25°. Compared to wild-type animals, am117 mutants displayed a slightly higher level of eggs that failed to hatch and a similar level of death during larval development and adult sterility (data not shown). This small reduction in the rate of egg hatching might be caused by the am117 mutation or additional changes resulting from mutagenesis of this strain. To analyze somatic aging, we determined that the mean lifespan of self-fertilized am117 hermaphrodites was increased 19% to 19.1 ± 1.0 days (N= 30), a significant difference compared to the self-fertilized wild-type hermaphrodite value of 16.1 ± 0.7 days (N= 59, P= 0.03). The extended lifespan suggests that the am117 mutation delays somatic aging as well as reproductive aging.
A scrawny phenotype similar to that displayed by am117 mutants is characteristic of worms that experience dietary restriction (Avery 1993). Additional phenotypes associated with dietary restriction include decreased early progeny production, decreased total progeny production, increased late progeny production, and an increased lifespan (Lakowski and Hekimi 1998; Hughes et al. 2007). One class of mutations that cause dietary restriction in C. elegans are the eat mutations, which cause defects in pharyngeal pumping (Avery 1993). To determine whether am117 causes dietary restriction due to a defect in pharyngeal pumping, we monitored the rate of pharyngeal pumping in self-fertilized am117 hermaphrodites. Young adult am117 hermaphrodites displayed over 250 pharyngeal pumps per minute, a value that is similar to young adult wild-type worms (data not shown). This suggests that gross defects in pharyngeal pumping do not cause the am117 phenotype, although subtle defects may not have been detected in this analysis. To determine whether the am117 mutation affects the insulin/IGF-1 signaling pathway, we analyzed the am117 strain for defects in Dauer formation. am117 worms cultured at 27° did not display measurable levels of Dauer larvae (data not shown), suggesting that am117 does not affect the insulin/ IGF-1 signaling pathway.
The am117 mutation is positioned on the right arm of chromosome I
After the am117 strain was backcrossed twice to wild-type animals, the strain maintained the scrawny body morphology. This suggests that the scrawny phenotype is caused by am117 or a linked mutation. To investigate these possibilities, we crossed the am117 mutant strain to wild type, selected F2 hermaphrodites that displayed the scrawny phenotype or the wild-type phenotype, and analyzed late progeny production in strains derived from these F2 animals. Strains that displayed the scrawny phenotype also displayed increased late progeny production, whereas strains that displayed wild-type body morphology displayed wild-type levels of late progeny production (data not shown). These observations suggest that either the am117 mutation causes the scrawny phenotype or the am117 strain contains a tightly linked mutation that causes the scrawny phenotype.
To identify the position of the am117 mutation, we took advantage of the scrawny body morphology phenotype because it can be readily scored. To determine the chromosomal linkage of am117, we mated homozygous am117 mutants to the polymorphic CB4856 strain isolated in Hawaii and selected F2 worms that displayed the scrawny phenotype. We used the method of DNA pyrosequncing to analyze single nucleotide polymorphisms (SNPs) that differ between the am117 strain and the CB4856 strain (Bruinsma et al. 2008). am117 displayed highest linkage to a SNP marker positioned on the right arm of chromosome I (Materials and Methods). To confirm this position, we used the independent method of 3-factor mapping relative to mutations that cause visible phenotypes; am117 was positioned right of dpy-24 located at +4.7, left of unc-101 located at +13.3, and close to unc-75 located at +9.3. To further refine the position of am117, we used a local SNP map (Jakubowski and Kornfeld 1999). We constructed a dpy-5(e61) am117 unc-101(m1) triple mutant, crossed the strain to CB4856 to generate F1 heterozygotes, and selected 139 Dpy non-Unc and 103 Unc non-Dpy F2 recombinants. These strains were scored for SNP markers and the scrawny phenotype, and the results indicated that am117 is positioned right of a SNP marker positioned in cosmid F32B4 located at +9.0. These results show that the am117 mutation is in an interval of ∼4.3 map units between cosmid F32B4 and unc-101.
Identification of morphological markers of reproductive aging: the am117 mutation delays age-related degenerative changes
Differential interference contrast (DIC) microscopy has been used to visualize age-related morphological changes in the reproductive system. Garigan et al. (2002) showed that the distal gonad displays age-related increases in cavities and grainy material and decreases in the number of nuclei, and these changes were analyzed by assigning scores from 1 to 5 on the basis of the degree of deterioration. Luo et al. (2010) reported a similar analysis of the distal gonad and an analysis of the proximal gonad that showed age-related decreases in oocyte size and increases in cavities and cluster formation. These changes were analyzed by assigning scores of normal, mild, or severe. Our goal was to identify age-related morphological changes that could be directly quantified to compare the rates of age-related changes in mutant and wild-type worms. Hermaphrodites between the ages of day 1 and days 6–7 usually displayed well-organized germlines; the distal gonad contained many germ cell nuclei, and the proximal gonad contained multiple late-stage oocytes and eggs (Figure 5A). Self-fertilized hermaphrodites often displayed changes by day 7 as a result of sperm depletion. Many late-stage oocytes were “stacked” or accumulated between the spermatheca and the loop region due to the lack of the ovulation signal from sperm (Greenstein 2005) (Figure 5B). In addition, unfertilized eggs, rather than fertilized eggs, were typically observed near the vulva (Figure 5C). The eggs and oocytes appeared to be healthy in these animals, and if a hermaphrodite at this stage receives sperm by mating to a male, then the hermaphrodite can resume producing fertilized eggs (Hughes et al. 2007).
Figure 5 .
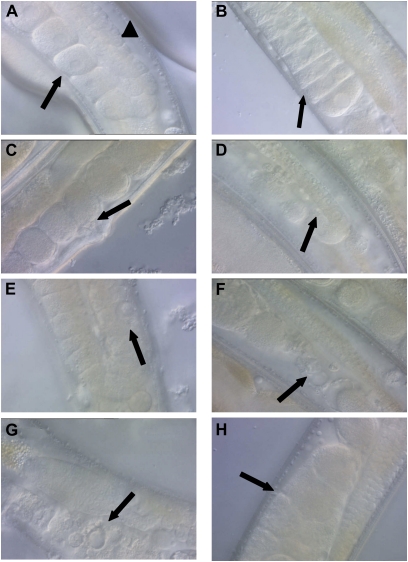
Age-related morphological changes in the hermaphrodite germline. Wild-type hermaphrodites were isolated at the L4 larval stage (day 1), allowed to mature until the indicated day, and visualized by differential interference contrast (DIC) microscopy at ×400 magnification. Images are oriented with ventral below and dorsal above. (A) Day 7, well-organized late-stage oocytes in the proximal gonad (arrow, ventral) and many developing germ cell nuclei in the distal gonad (arrowhead, dorsal). (B) Day 7, late-stage oocytes display “stacking” in the proximal gonad (arrow, ventral), a result of sperm depletion in a self-fertilized hermaphrodite. (C) Day 7, unfertilized eggs in the proximal gonad (arrow, ventral), a result of sperm depletion in a self-fertilized hermaphrodite. (D) Day 8, a narrow distal gonad containing a reduced number of developing germ cells (arrow, dorsal). (E) Day 9, a swollen nucleus in the distal gonad (arrow, dorsal). (F) Day 8, debris in the proximal gonad (arrow, ventral). (G) Day 9, a vacuole-like structure in the proximal gonad (arrow, ventral). (H) Day 8, an opaque mass in the proximal gonad (arrow, ventral).
By day 8, some hermaphrodites began displaying detectable and even extreme degenerative changes in the gonad, as noted in previous studies (Garigan et al. 2002; Herndon et al. 2002; Luo et al. 2010). The distal gonad displayed a decreased diameter and contained fewer germ cell nuclei (Figure 5D). In some hermaphrodites, the distal gonad was contracted into a small clump or the somatic gonad appeared to pucker around gaps in the distal gonad rather than appearing straight and smooth. The distal gonad occasionally contained large nuclei that appeared swollen (Figure 5E). As hermaphrodites become progressively older, the proximal gonad displayed a decrease in the number of large oocytes and eggs and an increase in the number of disorganized structures. The disorganized structures included debris (Figure 5F), vacuoles (Figure 5G), and large spaces lacking oocytes or eggs. A dramatic change in the proximal gonad was the appearance of large, dark masses between the spermatheca and vulva that appeared to be composed of a fusion of many eggs (Figure 5H). These masses correspond to the clusters described by Luo et al. (2010). Unfertilized oocytes undergo endoreduplication (Golden et al. 2007), and it is possible that these masses are related to this process. We observed similar abnormalities in mated hermaphrodites (data not shown), suggesting that mating and the subsequent increased progeny production on days 5–10 has little effect on the deterioration of the proximal gonad.
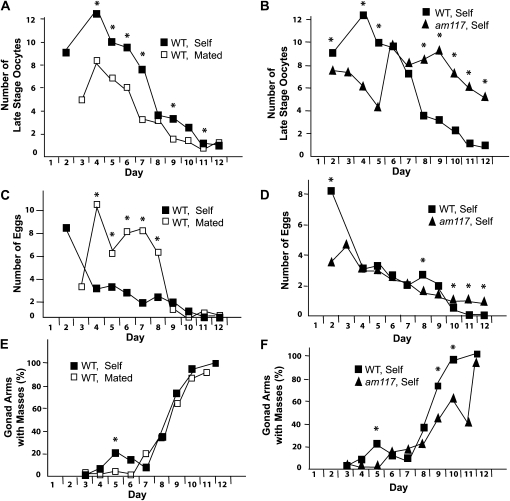
To quantify age-related degenerative changes in the gonad, we focused on the disappearance of normal structures and the appearance of abnormal structures that can be readily identified and quantified. While a variety of normal structures are modified with age, two that can be readily scored are late-stage oocytes and eggs. We defined late-stage oocytes as large oocytes that were located between the reflex in the gonad arm and the spermatheca. We defined eggs as fertilized and unfertilized eggs located between the spermatheca and the vulva. The number of late-stage oocytes and eggs in the proximal gonad of self-fertilized wild-type hermaphrodites steadily declined with age (Figure 6, A and C). To determine how these age-related changes are affected by increased progeny production, we analyzed mated hermaphrodites. Prior to days 8–9, mated hermaphrodites displayed fewer oocytes and more eggs, which may reflect the greater progeny production of mated hermaphrodites during these days (Figure 6, A and C). After days 8–9, mated and self-fertilized hermaphrodites displayed more similar declines.
Figure 6 .
Quantification of age-related changes in the proximal germline of hermaphrodites. Wild-type (WT) hermaphrodites were self fertilized (self) or mated to males (mated), and am117 mutant hermaphrodites were self-fertilized. Ages of adult hermaphrodites raised at 20° are shown in days: the L4 stage is day 1. (A and B) Average number per gonad arm of large oocytes located between the reflex in the gonad arm and the spermatheca. Compared to WT self, WT mated was significantly different on days 4–7, 9, and 11, and am117 self was significantly different on days 2, 4, 5, and 8–12 (*P ≤ 0.05; N = 10–68, 17–64, and 12–68 gonad arms for WT self, WT mated, and am117 self, respectively). (C and D) Average number per gonad arm of unfertilized and fertilized eggs located between the spermatheca and the vulva. Compared to WT self, WT mated was significantly different on days 4–8, and am117 self was significantly different on days 2, 8, and 10–12 (*P ≤ 0.05; N = 10–64, 24–68, and 12–69 gonad arms for WT self, WT mated, and am117 self, respectively). (E and F) Percentage of gonad arms with one or more masses between the spermatheca and the vulva. Compared to WT self, WT mated was significantly different on day 5, and am117 self was significantly different on days 5, 9, and 10 (*P ≤ 0.05, see Table 5).
These assays were used to characterize am117 mutant hermaphrodites. Compared to wild-type hermaphrodites, self-fertilized am117 hermaphrodites displayed significantly more late stage oocytes than on days 8–12 (Figure 6B) and eggs on days 10–12 (Figure 6D). These observations demonstrate that the am117 mutation delays the age-related decline in the number of late-stage oocytes and eggs. Since am117 mutant hermaphrodites also displayed extended reproduction compared to wild type, these studies indicate that the number of late-stage oocytes and eggs is a positive indicator of reproductive function.
To quantify the accumulation of degenerative changes, we chose to monitor masses that appear in the proximal gonad. These masses accumulated steadily with age in both self-fertilized and mated hermaphrodites until almost 100% of hermaphrodite gonad arms possessed such a mass (Table 5, Figure 6E). These masses might interfere with the entry of sperm following mating, which would limit progeny production late in life. The masses might also interfere with the exit of fertilized eggs, which would result in the internal hatching of progeny or “bagging.” Related to this possible consequence of the masses, mated hermaphrodites produce more progeny late in life when the masses are accumulating and internal hatching of progeny increases substantially in mated hermaphrodites compared to self-fertilized hermaphrodites (data not shown). The am117 mutation delayed the accumulation of masses, since self-fertilized am117 hermaphrodites displayed significantly fewer masses than self-fertilized wild-type hermaphrodites on days 9–10 (Table 5, Figure 6F).
Table 5 . Gonad arms with masses.
| Daya | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| WT, self | 0 | 5 | 23.5 | 13.3 | 6.7 | 36.7 | 74.5 | 92.6 | ND | 100 |
| (33) | (20) | (39) | (15) | (15) | (30) | (41) | (14) | (14) | ||
| WT, mated | 2.5 | 0 | 3.8* | 0 | 21.4 | 36.7 | 67.6 | 92 | 95.2 | ND |
| (46) | (7) | (27) | (11) | (14) | (30) | (34) | (23) | (21) | ||
| am117, self | 0 | 0 | 0* | 14.9 | 17.7 | 25.9 | 48* | 64.1* | 44.4 | 92.5 |
| (22) | (34) | (31) | (67) | (41) | (58) | (49) | (39) | (27) | (40) | |
Number of gonad arms examined is shown in parentheses. ND, not determined. *P-value ≤ 0.05 compared to WT, self (line 1).
Values are the percentage of gonad arms that contained at least one mass.
Discussion
Identification of mutations that delay somatic aging
The identification of mutations that extend adult lifespan is the basis for many recent and exciting advances in the genetic analysis of aging. C. elegans is a leading model system in this field, and two basic approaches have been used to identify genes that can be manipulated to extend worm lifespan. One approach is determining whether candidate mutations that were identified in genetic screens for nonaging phenotypes also affect lifespan. For example, eat-2 mutations were identified on the basis of pharyngeal pumping defects and subsequently demonstrated to cause lifespan extensions and represent a genetic model of caloric restriction (Lakowski and Hekimi 1998). Lifespan extending daf-2 mutations that affect the insulin/IGF-1 signaling pathway were identified on the basis of Dauer defective phenotypes (Kenyon et al. 1993). Lifespan extending clk-1 mutations that affect mitochondrial function were identified on the basis of delays of rhythmic behaviors (Lakowski and Hekimi 1996). Lifespan extending che-3 mutations that affect sensory perception were identified on the basis of defective chemosensation (Apfeld and Kenyon 1999). More recently, screens using chemical mutagenesis or RNAi have been conducted for surrogate phenotypes, and candidates were then analyzed for lifespan extension. Surrogate phenotypes include resistance to heat stress (Sampayo et al. 2000; Munoz and Riddle 2003), resistance to oxidative stress (De Castro et al. 2004; Kim and Sun 2007), and lethality during development (Chen et al. 2007; Curran and Ruvkun 2007).
A second approach is to directly perform screens for mutants that display extended longevity. The advantage of this approach is that it is relatively unbiased and many or all genes can be analyzed. The first such screen in C. elegans was reported by Klass (1983) and utilized chemical mutagenesis and extended lifespan as an assay. This approach resulted in the discovery of the age-1(hx546) allele (Friedman and Johnson 1988). age-1 was subsequently demonstrated to encode a PI3 kinase that functions in the insulin/IGF-1 signaling pathway (Morris et al. 1996). Additional screens have used the method of feeding RNAi to search for genes that can extend longevity when gene activity is reduced (Dillin et al. 2002; Lee et al. 2003; Hamilton et al. 2005; Hansen et al. 2005; Pan et al. 2007). RNAi screens have identified new genes involved in lifespan determination and demonstrated that many genes necessary for mitochondrial function modulate lifespan.
Here we report a novel approach for identifying mutations that delay age-related degenerative changes. We developed quantitative measurements of age-related declines in pharyngeal pumping, body movement, and mated reproductive output and screened for mutants that displayed extended performance in these measurements of youthful vigor. A chemical mutagen was used to generate alleles that would be useful and enduring genetic reagents. A large collection of mutations was identified on the basis of the extension of one or more age-related degenerative changes. We focused on the analysis of the age-1(am88) mutation that caused the most substantial extension of youthful vigor. Genetic and molecular studies demonstrated that am88 is a missense mutation of the age-1 gene. It is intriguing that the only other well-characterized mutation identified in a chemical mutagenisis screen for longevity phenotypes also affects the age-1 gene: age-1(hx546) (Klass 1983; Friedman and Johnson 1988). Whereas age-1(am88) changes glutamate 725 to lysine and affects the PIK domain, age-1(hx546) changes proline 806 to serine and affects the kinase domain (Morris et al. 1996; Ayyadevara et al. 2008). Both mutations cause a partial loss of function, since null mutations in age-1 cause lethality. Drosophila is an important model system for studies of aging, and flies display many age-related degenerative changes, including progressive impairment of locomotor activity (Jones and Grotewiel 2011). A forward genetic screen for delayed locomotor activity in flies identified phosphoinositide-dependent kinase 1 (PDK1), and candidate gene approaches confirmed that reducing the activity of additional components of the insulin/IGF-1 signaling pathway, such as PI3K, also delayed locomotor decline (Jones et al. 2009). These findings are consistent with our results and indicate that the insulin/IGF-1 signaling pathway plays a conserved role in modulating age-related functional declines in animals.
It is interesting to compare the two age-1 alleles identified in worms on the basis of delayed aging. age-1(hx546) hermaphrodites display a movement span of 10.8 days (30% increase), a pumping span of 15.2 days (30% increase), and a lifespan of 24.8 days (60% increase) (Huang et al. 2004); age-1(am88) hermaphrodites displayed a movement span of 15.5 days (90% increase), a pumping span of 21.4 days (80% increase), and a lifespan of 29.8 days (100% increase). Thus, age-1(am88) caused more substantial delays of age-related degenerative changes. By contrast, age-1(hx546) caused 56% Dauer constitutive phenotype, whereas age-1(am88) caused only 8% Dauer constitutive. These findings indicate that these mutations do not form a simple allelic series and suggest that different aspects of age-1 function may regulate Dauer formation and adult lifespan. Furthermore, age-1(am88) is an example of an allele that would have been difficult to isolate using the Dauer constitutive phenotype as a surrogate. The identification of age-1(am88) is important because it identifies a unique residue of the AGE-1 PI3K that is functionally significant, providing new structure-function insights into this important signaling protein. The age-1(am88) mutation causes dramatic extensions of vigorous pharyngeal pumping and body movement and doubles the lifespan; it will be a useful reagent for future studies of the age-1 gene and the insulin/IGF-1 signaling pathway during aging.
Identification of mutations that delay reproductive aging
Aging of the reproductive system is a fascinating and important topic. Compared to aging of life support systems that ultimately determine lifespan, aging of the reproductive system has received little attention. In human females, reproductive aging causes age-related increases in birth defects and declines in fertility, culminating in reproductive cessation at menopause (Hartge 2009). Age-related infertility is a major medical issue, and while some treatments have been developed that enhance fertility in older women, many of the causes of age-related declines in fertility remain to be elucidated. Reproductive aging is also a critical aspect of the evolutionary biology of aging (Partridge et al. 2005). Because progeny production is the purpose of animal life and a critical aspect of fitness, reproductive aging is likely to be subject to strong selection. However, the relationships between selective forces in the environment that promote accelerated or delayed reproductive aging and the genetic mechanisms that control reproductive aging have not been determined.
Here we describe a novel genetic screen for C. elegans mutants with delayed reproductive aging. Because progeny production is an effective measure of reproductive aging in mated but not self-fertilized hermaphrodites (Ward and Carrel 1979; Hughes et al. 2007), an important aspect of these screens was the use of mated hermaphrodites. Hermaphrodites generate ∼300 self-sperm that are used efficiently to fertilize oocytes. When these sperm are depleted, self-fertilized hermaphrodites cease progeny production. By contrast, mated hermaphrodites produce substantially more progeny over an extended period of time, and mated hermaphrodites cease progeny production as a result of reproductive aging, not sperm depletion. We identified two mutations that significantly extended mated reproduction, am116 and am117. To characterize the am117 mutation, we developed quantitative measurements of age-related changes in the morphology of the proximal gonad. Several age-related changes in gonad morphology have been described (Garigan et al. 2002; Herndon et al. 2002; Luo et al. 2010). We extended these studies by focusing on three features that change dramatically over the time period of reproductive cessation and can be readily quantified: number of late-stage oocytes, number of eggs, and number of gonad arms that contain masses. We have not used longitudinal studies to determine how these age-related morphological changes correlate with age-related changes in somatic or reproductive function, since scoring these morphological changes requires sacrificing the animal. Interestingly, am117 delayed both the age-related declines of reproductive function and the age-related morphological changes in the gonad. These findings suggest that age-related changes in gonad morphology may cause functional declines in progeny production, and the am117 mutation may delay the functional decline by delaying these morphological changes. Alternatively, the morphological changes and the functional declines may be independent events that are both influenced by the am117 mutation.
The identification of mutations that delay reproductive aging using an unbiased forward genetic screen is an important advance, since all the genes previously demonstrated to delay reproductive aging were identified using candidate gene approaches. We previously used candidate approaches to show that an eat-2 mutation that causes caloric restriction and a daf-2 mutation that causes reduced insulin/IGF-1 signaling can delay reproductive aging (Hughes et al. 2007). Recently, candidate gene approaches were used to show that genes in the TGF-β Sma/Mab pathway such as sma-2 (Luo et al. 2009, 2010) and egl-1 mutations that influence programmed cell death (Andux and Ellis 2008) can extend the mated reproductive span. Not all mutations that cause extended longevity increase the reproductive span, since mutations that decrease mitochondrial function such as clk-1 and isp-1 do not extend mated reproduction (Hughes et al. 2007). The genes affected by the am116 and am117 mutations have not been identified, and it is possible that these genes may function in one of the pathways previously implicated in controlling reproductive aging or in a novel pathway.
It is important to elucidate the relationships between reproductive and somatic aging, and the findings presented here shed new light on this issue. Our studies of long-lived mutants demonstrated that some but not all of these strains display delayed reproductive aging (Hughes et al. 2007). Furthermore, mutations in the TGF-β Sma/Mab pathway cause substantial extensions of the reproductive span with relatively minor lifespan extension, indicating that somatic and reproductive aging can be independently controlled (Luo et al. 2009). Interestingly, am116 and am117 both caused ∼20% extensions of mean lifespan, indicating that the genes affected by these mutations influence somatic and reproductive aging. Thus, a genetic screen that selected for mutants with delayed reproductive aging resulted in the identification of mutations that also delayed somatic aging. An important goal of future studies is to identify the genes affected by these mutations and elucidate their role in aging.
Acknowledgments
We thank Luke Schneider and John Murphy for assistance with genetic experiments and data analyses, Barbara Scott and Mike Crowder for sequencing primers, Paul Goodfellow for suggestions with statistics, and Tim Schedl for advice. Some strains were provided by the Caenorhabditis Genetics Center (St. Paul, MN), which is funded by the National Institutes of Health (NIH) National Center for Research Resources. This work was supported by a predoctoral fellowship from the Howard Hughes Medical Institute and a Glenn/American Federation for Aging Research (AFAR) scholarship from AFAR (C.H.), a predoctoral fellowship from the National Science Foundation (S.H.), an Ellison Medical Foundation senior scholar award (K.K.), and grants from the National Science Foundation (IOBO446240) and the NIH (RO1AG02656101A1) (K.K.).
Literature Cited
- Andux S., Ellis R. E., 2008. Apoptosis maintains oocyte quality in aging Caenorhabditis elegans females. PLoS Genet. 4: e10000295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apfeld J., Kenyon C., 1999. Regulation of lifespan by sensory perception in Caenorhabditis elegans. Nature 402: 804–809 [DOI] [PubMed] [Google Scholar]
- Avery L., 1993. The genetics of feeding in Caenorhabditis elegans. Genetics 133: 897–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyadevara S., Alla R., Thaden J. J., Shmookler Reis R. J., 2008. Remarkable longevity and stress resistance of nematode PI3K-null mutants. Aging Cell 7: 13–22 [DOI] [PubMed] [Google Scholar]
- Bolanowski M. A., Russell R. L., Jacobson L. A., 1981. Quantitative measures of aging in the nematode Caenorhabditis elegans. I. Population and longitudinal studies of two behavioral parameters. Mech. Ageing Dev. 15: 279–295 [DOI] [PubMed] [Google Scholar]
- Braeckman B. P., Houthoofd K., Vanfleteren J. R., 2001. Insulin-like signaling, metabolism, stress resistance and ageing in Caenorhabditis elegans. Mech. Ageing Dev. 122: 673–693 [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruinsma J. J., Schneider D., Davis D., Kornfeld K., 2008. Identification of mutations in Caenorhabditis elegans that cause resistance to high levels of dietary zinc and analysis using a genome-wide map of single-nucleotide polymorphisms scored by pyrosequencing. Genetics 179: 811–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantry D., Vojtek A., Kashishian A., Holtzman D. A., Wood C., et al. , 1997. p110δ, a novel phosphatidylinositol 3-kinase catalytic subunit that associates with p85 and is expressed predominantly in leukocytes. J. Biol. Chem. 272: 19236–19241 [DOI] [PubMed] [Google Scholar]
- Chen D., Pan K. Z., Palter J. E., Kapahi P., 2007. Longevity determined by developmental arrest genes in Caenorhabditis elegans. Aging Cell 6: 525–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J. J., Huang C., Hughes S., Kornfeld K., 2007. The measurement and analysis of age-related changes in Caenorhabditis elegans. WormBook, ed. The C. elegans Research Community, WormBook, http://www.wormbook.org [DOI] [PMC free article] [PubMed]
- Croll N. A., Smith J. M., Zuckerman B. M., 1977. The aging process of the nematode Caenorhabditis elegans in bacterial and axenic culture. Exp. Aging Res. 3: 175–189 [DOI] [PubMed] [Google Scholar]
- Curran S. P., Ruvkun G., 2007. Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 3: e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Castro E., De Castro S. H., Johnson T. E., 2004. Isolation of long-lived mutants in Caenorhabditis elegans using selection for resistance to juglone. Free Radic. Biol. Med. 37: 139–145 [DOI] [PubMed] [Google Scholar]
- Dillin A., Hsu A.-L., Arentes-Oliveira N., Lehrer-Graiwer J., Hsin H., et al. , 2002. Rates of behavior and aging specified by mitochondrial function during development. Science 298: 2398–2401 [DOI] [PubMed] [Google Scholar]
- Domin J., Waterfield M. D., 1997. Using structure to define the function of phosphoinositide 3-kinase family members. FEBS Lett. 410: 91–95 [DOI] [PubMed] [Google Scholar]
- Flanagan C. A., Schnieders E. A., Emerick A. W., Kunisawa R., Admon A., et al. , 1993. Phosphatidylinositol 4-kinase: gene structure and requirement for yeast cell viability. Science 262: 1444–1448 [DOI] [PubMed] [Google Scholar]
- Friedman D. B., Johnson T. E., 1988. A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics 118: 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garigan D., Hsu A. L., Fraser A. G., Kamath R. S., Ahringer J., et al. , 2002. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics 161: 1101–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershon H., Gershon D., 2002. Caenorhabditis elegans: a paradigm for aging research: advantages and limitations. Mech. Ageing Dev. 123: 261–274 [DOI] [PubMed] [Google Scholar]
- Golden T. R., Beckman K. B., Lee A. H., Dudek N., Hubbard A., et al. , 2007. Dramatic age-related changes in nuclear and genome copy number in the nematode Caenorhabditis elegans. Aging Cell 2: 179–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenstein D., 2005. Control of oocyte meiotic maturation and fertilization. WormBook, ed. The C. elegans Research Community, WormBook, http://www.wormbook.org [DOI] [PMC free article] [PubMed]
- Guarente L., Kenyon C., 2000. Genetic pathways that regulate ageing in model organisms. Nature 408: 255–262 [DOI] [PubMed] [Google Scholar]
- Hamilton B., Dong Y., Shindo M., Liu W., Odell I., et al. , 2005. A systematic RNAi screen for longevity genes in C. elegans. Genes Dev. 19: 1544–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M., Hsu A. L., Dillin A., Kenyon C., 2005. New genes tied to endocrine, metabolic and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 1: e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartge P., 2009. Genetics of reproductive lifespan. Nat. Genet. 41: 637–638 [DOI] [PubMed] [Google Scholar]
- Herndon L. A., Schmeissner P. J., Dudaronek J. M., Brown P. A., Listner K. M., et al. , 2002. Stochastic and genetic factors influence tissue-specific declines in ageing C. elegans. Nature 419: 808–814 [DOI] [PubMed] [Google Scholar]
- Hodgkin J., Barnes T. M., 1991. More is not better: brood size and population growth in a self-fertilizing nematode. Proc. R. Soc. Lond. B Biol. Sci. 246: 19–24 [DOI] [PubMed] [Google Scholar]
- Hosono R., Sato Y., Aizawa S.-I., Mitsui Y., 1980. Age-dependent changes in mobility and separation of the nematode Caenorhabditis elegans. Exp. Gerontol. 15: 285–289 [DOI] [PubMed] [Google Scholar]
- Huang C., Xiong C., Kornfeld K., 2004. Measurements of age-related changes of physiological processes that predict life span of Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 101: 8084–8089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S., Evason K., Xiong C., Kornfeld K., 2007. Genetic and pharmacological factors that influence reproductive aging in nematodes. PLoS Genet. 3: 254–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowski J., Kornfeld K., 1999. A local, high-density, single-nucleotide polymorphism map used to clone Caenorhabditis elegans cdf-1. Genetics 153: 743–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T. E., 1987. Aging can be genetically dissected into component processes using long-lived lines of Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 84: 3777–3781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. A., Gargano J. W., Rhodenizer D., Martin I., Bhandari P., et al. , 2009. A forward genetic screen in Drosophila implicates insulin signaling in age-related locomoter impairment. Exp. Gerontol. 44: 532–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. A., Grotewiel M., 2011. Drosophila as a model for age-related impairment in locomoter and other behaviors. Exp. Gerontol. 46: 320–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C., Chang J., Gensch E., Rudner A., Tabtiang R., 1993. A C. elegans mutant that lives twice as long as wild type. Nature 366: 461–464 [DOI] [PubMed] [Google Scholar]
- Kenyon C., 2005. The plasticity of aging: insights from long-lived mutants. Cell 120: 449–460 [DOI] [PubMed] [Google Scholar]
- Kim Y., Sun H., 2007. Functional genomic approach to identify novel genes involved in the regulation of oxidative stress resistance and animal lifespan. Aging Cell 6: 489–503 [DOI] [PubMed] [Google Scholar]
- Kimura K. D., Tissenbaum H. A., Liu Y., Ruvkun G., 1997. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277: 942–946 [DOI] [PubMed] [Google Scholar]
- Kirkwood T. B., 1977. Evolution of aging. Nature 270: 301–304 [DOI] [PubMed] [Google Scholar]
- Klass M. R., 1983. A method for the isolation of longevity mutants in the nematode Caenorhabditis elegans and initial results. Mech. Ageing Dev. 22: 279–286 [DOI] [PubMed] [Google Scholar]
- Lakowski B., Hekimi S., 1996. Determination of life-span in Caenorhabditis elegans by four clock genes. Science 272: 1010–1013 [DOI] [PubMed] [Google Scholar]
- Lakowski B., Hekimi S., 1998. The genetics of caloric restriction in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 95: 13091–13096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. S., Lee R. Y. N., Fraser A. G., Kamath R. S., Ahringer J., et al. , 2003. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat. Genet. 33: 40–48 [DOI] [PubMed] [Google Scholar]
- Luo S., Shaw W. M., Ashraf J., Murphy C. T., 2009. TGF-B Sma/Mab signaling mutations uncouple reproductive aging from somatic aging. PLoS Genet. 5: e1000789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S., Kleemann G. A., Ashraf J. M., Shaw W. M., Murphy C. T., 2010. TGF-B and insulin signaling regulate reproductive aging via oocyte and germline quality maintenance. Cell 143: 299–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J. Z., Tissenbaum H. A., Ruvkun G., 1996. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature 382: 536–539 [DOI] [PubMed] [Google Scholar]
- Munoz M. J., Riddle D. L., 2003. Positive selection of Caenorhabditis elegans mutants with increased stress resistance and longevity. Genetics 163: 171–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkenhaug K., Bilancio A., Farjot G., Priddle H., Sancho S., et al. , 2002. Impaired B and T cell antigen receptor signaling in p110delta PI 3-kinase mutant mice. Science 297: 1031–1034 [DOI] [PubMed] [Google Scholar]
- Olsen A., Vantipalli M. C., Lithgow G. J., 2006. Using Caenorhabditis elegans as a model for aging and age-related diseases. Ann. N. Y. Acad. Sci. 1067: 120–128 [DOI] [PubMed] [Google Scholar]
- Pan K. Z., Palter J. E., Rogers A. N., Olsen A., Chen D., et al. , 2007. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell 6: 111–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge L., Gems D., Withers D. J., 2005. Sex and death: What is the connection? Cell 120: 461–472 [DOI] [PubMed] [Google Scholar]
- Pincus Z., Slack F. J., 2010. Developmental biomarkers of aging in Caenorhabditis elegans. Dev. Dyn. 239: 1306–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle D. L., Blumenthal T., Meyer B. J., Priess J. R., 1997. C. elegans II. Cold Spring Harbor Laboratory Press, Plainview, NY: [PubMed] [Google Scholar]
- Rose M. R., Charlesworth B., 1981. Genetics of life history in Drosophila. II. Exploratory selection experiments. Genetics 97: 187–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T., 1989. Molecular Cloning. Cold Spring Harbor Laboratory Press, Plainview, NY [Google Scholar]
- Sampayo J. N., Jenkins N. L., Lithgow G. J., 2000. Using stress resistance to isolate novel longevity mutations in Caenorhabditis elegans. Ann. N. Y. Acad. Sci. 908: 324–326 [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B., Welham M. J., Kotani K., Stein R., Warne P. H., et al. , 1997. P110delta, a novel phosphoinositide 3-kinase in leukocytes. Proc. Natl. Acad. Sci. USA 94: 4330–4335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijg J., Campisi J., 2008. Puzzles, promises and a cure for ageing. Nature 454: 1065–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S., Carrel J. S., 1979. Fertilization and sperm competition in the nematode Caenorhabditis elegans. Dev. Biol. 73: 304–321 [DOI] [PubMed] [Google Scholar]
- Williams B. D., Schrank B., Huynh C., Shownkeen R., Waterston R. H., 1992. A genetic mapping system in Caenorhabditis elegans based on polymorphic sequence-tagged sites. Genetics 131: 609–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams G., 1957. Pleiotropy, natural selection, and the evolution of senescence. Evolution 11: 398–411 [Google Scholar]