Abstract
In genetic studies, many interesting traits, including growth curves and skeletal shape, have temporal or spatial structure. They are better treated as curves or function-valued traits. Identification of genetic loci contributing to such traits is facilitated by specialized methods that explicitly address the function-valued nature of the data. Current methods for mapping function-valued traits are mostly likelihood-based, requiring specification of the distribution and error structure. However, such specification is difficult or impractical in many scenarios. We propose a general functional regression approach based on estimating equations that is robust to misspecification of the covariance structure. Estimation is based on a two-step least-squares algorithm, which is fast and applicable even when the number of time points exceeds the number of samples. It is also flexible due to a general linear functional model; changing the number of covariates does not necessitate a new set of formulas and programs. In addition, many meaningful extensions are straightforward. For example, we can accommodate incomplete genotype data, and the algorithm can be trivially parallelized. The framework is an attractive alternative to likelihood-based methods when the covariance structure of the data is not known. It provides a good compromise between model simplicity, statistical efficiency, and computational speed. We illustrate our method and its advantages using circadian mouse behavioral data.
MANY phenotypes (or traits) in genetic studies are quantitative and can be summarized by a single number; examples include bone mineral density and body weight. Other traits of interest to geneticists such as growth curves (Kramer et al. 1998), circadian rhythms (Shimomura et al. 2001), and morphology (Leamy et al. 2008) have spatial or temporal structure and cannot be reduced to a single number. Such traits may be viewed as curves or as function-valued (Pletcher and Geyer 1999). Quantitative genetics of such traits benefit from generalizations tailored to their function-valued nature (Kirkpatrick and Heckman 1989). In this article we present a new framework for identifying regions of the genome, quantitative trait loci (QTL) (Rapp 2000; Broman 2001), contributing to variation in function-valued traits.
Acknowledging the function-valued nature of a trait has several advantages. We can naturally treat features such as rates of growth or periodic fluctuations. We may also arrive at a more parsimonious representation of the data: a long sequence of correlated observations can often be described by a few values. From an evolutionary perspective, the function-valued nature of the data puts constraints on the patterns of variation possible and how selection might operate on the trait (Kingsolver et al. 2001). For these reasons, application of functional data analysis (FDA) (Ramsay and Silverman 2005) to genetic data has been fruitful. Wu and Lin (2006) called genetic mapping of function-valued traits functional mapping. Function-valued traits have been variously referred to as infinite-dimensional traits, longitudinal traits, repeated measures, functional traits, and dynamic traits (Kingsolver et al. 2001).
Most existing methods for mapping function-valued traits use a likelihood framework where the analyst must specify the mean function, the error distribution, and the error structure. When those features can be specified, one can use parametric functional mapping (Ma et al. 2002; Wu et al. 2004; Wu and Lin 2006). Here the analyst specifies the form of the mean function (say logistic, for modeling growth curves) and the error function (say autoregressive Gaussian errors). If there is not enough information about the form of the mean function, one may model the mean function nonparametrically using different basis function families: Legendre polynomials (Lin and Wu 2006), orthogonal polynomials (Yang et al. 2006), B-splines (Yang et al. 2009; Yap et al. 2009), and wavelets (Zhao et al. 2007) have all been used in the past. The approaches of Yang et al. (2009) and Yap et al. (2009) allow for an unstructured form of the variance–covariance matrix of the errors and use a multivariate Gaussian distribution. However, when the number of measurements per individual exceeds the number of samples and the empirical covariance matrix is thus singular, additional procedures such as wavelet dimension reduction (Zhao et al. 2007) or regularized estimation of the covariance matrix must be employed. For likelihood-based methods, incomplete genotype data are typically handled by the EM algorithm as in Lander and Botstein (1989). Bayesian formulations have been considered by Yang and Xu (2007) and Liu and Wu (2009).
In this report, we present an alternative framework for mapping function-valued traits on the basis of estimating equations. It is especially suited for scenarios where the error structure is unknown or poorly specified or when the number of time points exceeds the number of samples. Such data are becoming increasingly common with advances in automatic phenotyping. We present a mouse behavioral data set below that has these characteristics.
Our simulation studies indicate that likelihood functional regression models, when misspecified, may not have the desired false positive rates and can have lower power than our estimating equations approach. Generalized estimating equations have been used for mapping nonnormal traits (Lange and Whittaker 2001), but, to our knowledge, they have not been applied to mapping function-valued traits. Note that for genetic mapping, the covariance may be considered a nuisance parameter; for evolutionary studies and animal breeding, the covariance function is a key parameter of interest (Kingsolver et al. 2001; Mezey and Houle 2005).
Central to our approach is a general linear functional model that can accommodate any number of covariates and use a single set of computer programs without modification for different numbers of covariates. We use a simple two-step least-squares estimation procedure that is comparable in speed and memory to regular linear regression. This facilitates the use of commonly needed but computationally intensive procedures such as permutation tests for assessment of critical values or multiple testing correction, multiple imputation of incomplete genotypes, and model selection procedures.
Our article is organized as follows. The Mouse Behavioral Data section describes a mouse behavior data set that motivated our method. Then in Model and Estimation we outline the statistical theory underlying our method, including the model, estimation, and testing methods. We examine the method’s characteristics in simulations and on the mouse behavioral data in Simulation Studies and Data Analysis. We conclude with a summary in the Discussion.
Mouse Behavioral Data
The mouse experimental data come from a genetic analysis of mouse behavior using an automated home cage monitoring system. A backcross population was generated by crossing C57BL/6J and 129S1/SvImJ strains. The resulting F1 mice were then crossed back to the 129S1/SvImJ strain to generate the 89 N1 backcross mice that were phenotyped and genotyped. Genotyping was performed using Illumina’s low-density mouse SNP panel, which provided 233 informative (polymorphic) autosomal markers between these two strains. The average marker distance was 9.7 Mb (median 8.0 Mb, range 0.2–35 Mb). For phenotyping, mice were individually housed for 16 days in a home cage monitoring system that is described in detail elsewhere (Goulding et al. 2008). Briefly, this system allowed automated collection of feeding (detection of photobeam breaks when mice access food), drinking (detection of capacitance change with lick contacts at water spout), and movement (load cell position platform) data under a 12:12 light:dark cycle (lights on at 7 am and off at 7 pm).
Data were processed for errors in detection of feeding, drinking, and movement events as previously described (Goulding et al. 2008). Only mice with at least 4 days of complete data were used for subsequent analysis. Mice were allowed 4 days of acclimation to the monitoring cages and the final 12 days of data were used for analysis.
Mice in their home cages exhibit transitions between two major distinct states: an active state where the animals engage in bouts of feeding, drinking, and locomotion and an inactive state in which the animals engage in prolonged episodes of minimal movement at a discrete home base location (Goulding et al. 2008). Transitions between active and inactive states were identified with a resolution of 20 msec; the data then were aggregated into 6-min bins across all analysis days. Thus, each measurement represents the probability that an individual mouse was in the active state (active state probability, ASP) during any 6-min interval across all analysis days (Goulding et al. 2008).
As indicated earlier, the data showed several features that motivated the development of new methodology:
The number of measurements per mouse (220) exceeds the number of mice (89), which presents problems for many methods as the empirical covariance matrix is singular.
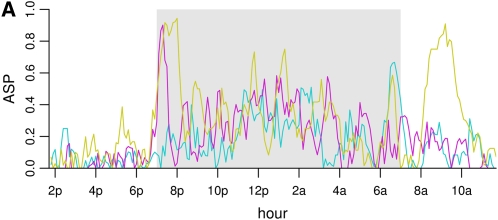
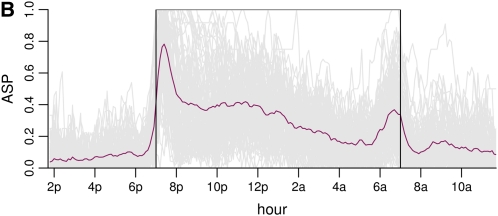
There is considerable variation in individual mouse active state probabilities (Figure 1, A and B).
The distribution is awkward to describe. There are mice with active state probabilities of zero or one at some time points.
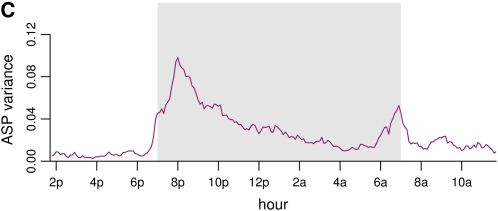
The variance between individuals as a function of time is smooth with two peaks (Figure 1C).
The average ASP pattern is also smooth with two peaks: one at 7–9 pm and one at 6–8 am. There is a third moderate peak between these two peaks. There is a minimum or nadir at night at 4–6 am. Note, however, that usual transformations of the data (such as the arcsine transformation) failed to stabilize the variance (results not shown).
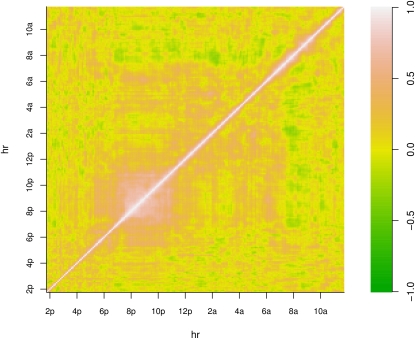
The correlation between time points was also smooth but without an easily articulated structure (Figure 2).
Figure 1 .
Mouse active state probability data trajectories, mean, and variance. (A) Raw trajectories of three randomly chosen individual mice. The dark period (7 pm–7 am) is shown in gray. (B) All individual trajectories are shown in gray, and the mean trajectory is overlaid in maroon. There is a peak in activity between 7 and 8 pm, which gradually dies out, followed by a second peak at ∼7 am. The dark period is shown in a box. (C) Variance of the trajectories as a function of time. The variance function shows roughly the same pattern as the mean function.
Figure 2 .
Correlation between time points for mouse active state probability data. There is some structure in the correlation matrix, although it does not fall into an easily classifiable category. There is high local correlation between 7 pm and 11 pm, and a negative correlation between 7–9 am and 7 pm–5 am.
Model and Estimation
We outline our method in this section. We start with the functional regression model. Using estimating equations, the parameter estimates and test statistics have a closed-form solution, assuming complete genotype data. Next, we present some extensions that broaden the method’s applicability and describe our strategy for handling incomplete genotypes. Finally, we discuss the computational requirements of our method, basis function choice, and shrinkage estimation of the covariance.
Model
When complete genotype data are available, we can express the trait data in terms of a regression model. For example, a linear regression model is traditionally used for mapping a quantitative trait,
where y is the quantitative trait of interest, and zk, k = 1, … , p, are covariates, βk, k = 1, … , p, are the effects of the covariates, and e is the random error. The covariates may include an intercept, the QTL genotypes, and any other factors such as age, sex, or body weight that may contribute to the trait. The focus of QTL mapping is to identify the genetic covariates contributing to the trait.
Analogously, we use a functional regression model for our function-valued trait when complete genotype data are available. Let y(t) be the function describing the observable trait as a function of t; in the remainder of our treatment we consider t to be time, but it can be extended to consider spatial position in one or more dimensions. We assume that the function can be represented as follows in terms of covariates z1, z2, … , zp (that may include genotypes),
| (1) |
where βk(t), k = 1, … , p, are unknown functions, and e(t) is random error. We assume that each of the βk(t) functions can be represented as a finite linear combination of a family of basis functions (say splines, polynomials, or Fourier series) as
where q is the number of basis functions. The number and nature of the covariates z1, z2, … , zp will depend on intended use. They may include a single QTL, multiple QTL with or without interaction effects, or nongenetic covariates such as age and sex.
Suppose there are n individuals with yi(t) denoting the function-valued trait for the ith individual. Further suppose that the trait is observed at m times, t1, t2, … , tm. If we denote by yij the trait value for the ith individual at tj, then we can represent the observed trait data as an n × m matrix,
Let zik denote the value of the kth covariate in the ith individual, ψjl = ψl(tj) be the value of the lth basis function at the jth time point tj, and let eij be the random error for the ith individual at the jth time point. In matrix notation, we write
Then, it can be shown that
| (2) |
Writing y = vec(YT), ε = vec(ET), β = vec(BT), and X = Z ⊗ Ψ, where ⊗ denotes the Kronecker product and vec is an operator that stacks columns of a matrix into a single, large column vector, the above equation can be written in the familiar form
If the n individuals are independent with common covariance matrix, then Λ = var(ε) = var(vec(E)) = In ⊗ Σ, where Σ is the covariance matrix of (e(t1), e(t2), … , e(tm))T. Note that Σ is, in general, unknown and must be estimated from the data.
Estimation
As stated earlier, without a distributional assumption for the random error, we cannot write out a likelihood function. Instead we choose B (or equivalently β) to minimize the residual sum of squares,
| (3) |
where is an estimate depending on . The criterion, Equation 3, is an approximation to the integrated squared error of the estimate
where [t*, t*] is the range of the interval we wish to consider. Assuming that the range of observed time points is close (or identical) to the range we wish to consider, the integrated squared error may be approximated by the sum
Optimizing this criterion is equivalent to solving a least-squares problem that has a closed-form solution,
| (4) |
This can be seen as a combination of two least squares: one on basis functions (the Ψ part) and the other on genotypes (the Z part). The least-squares equations are thus our estimating equations; the resulting solution is asymptotically unbiased (Liang and Zeger 1986). Further, a consistent estimate of the variance of the estimated coefficients is obtainable if a consistent estimate of the residual covariance matrix, Σ, is available. Note that estimation of the coefficients does not require knowledge or estimation of Σ.
The variance of the estimate is
| (5) |
The predictions are
In the context of statistical methods for longitudinal data, our approach corresponds to using estimating equations with a working independent correlation structure and assuming a Gaussian distribution. In general, these estimates will be less efficient compared to a correctly specified likelihood model (Godambe 1960). However, significant loss of efficiency seems to be the exception rather than the rule (Diggle et al. 2002; Chandler and Bate 2007; McCulloch et al. 2008). We explored this in our simulations (see Simulation Studies and Data Analysis).
Testing
A genome scan considers every locus on the genome and asks the following question: Is trait variation explained by genetic variation at this locus? The evidence is traditionally summarized by the LOD score, which is the logarithm (base 10) of a likelihood-ratio statistic. The likelihood ratio compares a trait regression model with and without the genetic locus under consideration. Since we do not specify a probability model for the data, we cannot calculate a traditional LOD score. Instead, we consider two related quantities as explained below. These can be used as test statistics to test the null hypothesis at each locus; i.e., genetic variation at that locus does not contribute to (function-valued) trait variation.
Consider the trait model in Equation 1. The standard null hypothesis is no effect; that is, some βk(t) are identically zero, which is same as the coefficients of their basis expansion being zero. Suppose the null hypothesis is B = 0 (testing select entries by replacing B with SB in Equation 4, where S is a selection matrix so that SB selects specific columns of B, and subsequent derivations would be based on modified estimation).
Wald statistic:
Let be the estimate of coefficients in vector form and be the estimated covariance matrix of . The quadratic form
| (6) |
follows a Hotelling’s T2 statistic with parameters d and r, where d is the degrees of freedom of the estimate of τ (Equation 5) and r is the length of vec(BT). For reasonably large sample sizes, this can be approximated by a χ2-statistic with r d.f.
The estimate of the covariance matrix τ is
where is an estimate of Σ. The simplest estimate one can consider is formed from the residuals:
This estimate is unbiased, but one may also want to consider biased estimates as discussed later in this section.
Residual error statistic:
An alternative statistic would be the difference in residual sum of squares between the model with the genetic locus and a null model corresponding to the null hypothesis. Thus, if denotes the fitted values from the null model, and denotes the fitted values from the model including the genetic locus under consideration, we would calculate and then use
as a test statistic. This statistic is closely connected to the proportion of the variance explained by the locus. The asymptotic null distribution of this statistic is a mixture of χ2-variables (Rotnitzky and Jewell 1990; Shen and Faraway 2004). The mixing proportions depend on the eigenvalues of a matrix depending on Ψ and Σ.
Assessing significance:
As stated earlier, the Wald statistic has an asymptotic χ2-distribution, while the distribution of the residual error statistic is more complicated. The genome-wide significance of either can be established by a permutation test (Churchill and Doerge 1994). For genome-wide association studies (GWAS), one may contemplate a Bonferroni correction or a false discovery rate correction on pointwise P-values. In this context the Wald statistic would be more convenient.
Incomplete genotypes
The functional regression model (Equation 1) assumed that we have complete genotypes; i.e., the genotype of every individual at every genomic location is known. In practice this is rarely the case, as there are gaps between typed markers, genotyping reactions for some individuals may fail, or selective genotyping may have been used. A key part of the QTL mapping problem is to accommodate incomplete genotype data. If typed markers are reasonably dense and no selective genotyping is used, one can use Haley–Knott regression (Haley and Knott 1992). Here we replace the indicator variables corresponding to possible genotypes at a locus by their probabilities conditional on typed markers. Then we use functional regression as if we had complete genotypes. This method is very fast, and easily parallelized, but is susceptible to bias when selective genotyping is used or when the marker spacing is big (Kao 2000; Sen et al. 2005). In such cases we can use multiple imputation (Sen and Churchill 2001). For both multiple imputation and Haley–Knott regression the analyst can contemplate different functional regression models for the trait without needing any additional computational machinery.
Computational considerations
The modularity of our algorithm also simplifies computation. If we assume n and m are greater than p and q, then a naive application of least squares to the y = Xβ + ε problem, where X ∼ mn × pq, would be in the order of O(p2q2nm + p3q3) (Golub and Van Loan 1996). In our method, we solve two smaller least-squares problems. In Equation 4, the two inverses are of order O(mq2 + q3 + np2 + p3), while other matrix multiplications add O(mnq + npq).
Furthermore, only the part involving Z needs to be recomputed for different loci, while the rest can be computed once and saved for later use. Likelihood-based methods employing the EM algorithm cannot take advantage of this; every instance of the EM algorithm has to be run separately. Suppose we are performing r permutations, and using s imputations, the complexity will be O(mq2 + q3 + mnq + lrs(np2 + p3 + npq)) for l loci. Use of the EM algorithm for maximum likelihood also has the disadvantage that it requires modification each time the function regression model is changed.
Basis functions
The choice of basis functions and their tuning parameters (such as knot position and number of splines) is vitally important in functional regression and an active research area. As this is beyond the scope of this article, we note only that the family and number of basis functions are key choices and can be flexibly accommodated with our method. We used B-splines for the mouse behavior data and natural splines for the simulations.
Shrinkage estimation of Σ
When the number of individuals is greater than the number of time points, the obvious estimate of Σ is the empirical covariance of residuals. It is unbiased, but may suffer from high variance (Kauermann and Carroll 2001). If the number of time points (m) is large relative to the number of individuals (n), the analyst may consider using a biased estimator such as the shrinkage estimator proposed by Ledoit and Wolf (2004) and adapted by Schafer and Strimmer (2005). The resulting covariance estimate is nonsingular (unlike the empirical estimate when the number of individuals is smaller than the number of time points). We evaluate this choice in a simulation study in the next section. Shrinkage estimation of the genetic covariance for function-valued data has been considered by Meyer and Kirkpatrick (2010).
Additional practical considerations
In our development we assumed that there were no missing trait data. If there is a modest amount of missing trait data, we can use local smoothing methods such as smoothing splines to fill in the missing data. Alternatively, one can incorporate estimation of missing data into the estimation procedure by using a basis expansion on the left-hand side of Equation 2 and replacing Y with the coefficients of the expansion.
Our estimating equations correspond to assuming homoscedasticity and independence between time points. We can easily accommodate heteroscedasticity by using weighted least squares instead of ordinary least squares. There can be weights on individuals and/or different time points. More generally, if the analyst has prior knowledge about the covariance between samples or time points, it can be used to increase efficiency, yet the method will remain robust to their misspecification.
Simulation Studies and Data Analysis
We conducted three simulation studies and analyzed the mouse behavioral data presented earlier, to evaluate our method. We studied its type-I error and power and compared it to other methods (functional and nonfunctional). In the first simulation study, we compared our method to a parametric likelihood-based functional mapping method based on Ma et al. (2002). We computed the type-I error under three covariance structures. In the second simulation study we compared our method to a likelihood-based method that estimates the covariance (Yap et al. 2009). In addition, we looked at the power of different methods. In the third simulation study, we compared our method with a nonfunctional method (a cross-sectional method), which compresses multiple observations within a sample into a summary statistic by averaging; this material is in Supporting Information, File S1 Figure S1, and Table S1 Finally, we applied both the Wald statistic and the integrated residual error statistic to our mouse behavioral data and compared the result to a cross-sectional method (taking means over time intervals) that is currently used to analyze such data (Nishi et al. 2010).
Simulation studies: Comparison with a parametric functional approach
Here we report simulation studies comparing our estimating equations approach to a likelihood-based parametric approach. Our objective was to compare the null distribution and power of our method against likelihood-based functional mapping under three conditions: when the likelihood model is correctly specified, the pointwise error distribution is incorrectly specified, and the covariance is misspecified.
Our simulations were loosely modeled after the poplar tree data in Ma et al. (2002). We assumed that 13 equally spaced measurements between times 0 and 6 were made on 200 individuals; 100 individuals each had one of two possible genotypes 0 and 1. The mean of the observations was assumed to follow a logistic curve as in Ma et al. (2002) with the functional form
| (7) |
where θ = (θ0, θ1, θ2) are parameters describing the mean curve, and ε(t) is a stationary stochastic process with mean 0 and point-wise variance σ2 = 0.01. The functional parameters of individuals with genotypes equal to i are described by θi, i = 0, 1. Under the alternative, θ0 = (1.00, 9.0, 1), and θ1 = (0.95, 8.5, 1). Under the null, θ0 = θ1 = (0.975, 8.75, 1). The plots of three mean functions are in Figure S2.
We considered three error structures:
Gaussian, autoregressive: The marginal (pointwise) and all finite-dimensional distributions of the error are Gaussian, and the correlation structure is autoregressive so that the covariance between measurements separated by a time t is described by the exponential correlation function ρ(t) = σ2rt. We evaluated performance under r = 0.61, 0.83, 0.94, which are the correlations between successive time points for the Matérn correlation function (with smoothness parameters 0.5, 1, and 2, respectively) considered below (see File S1 for definition of the covariance function and references).
t-distributed, autoregressive: The finite-dimensional distributions have a t-distribution with 4 d.f. (the smallest for which the third moment exists). The correlation function is autoregressive, as above.
Gaussian, Matérn: The finite-dimensional distributions are Gaussian, and the correlation function is Matérn with amplitude parameter σ2, scale parameter 1, and smoothness parameters 0.5, 1, and 2. The three correlation functions are shown in Figure S3.
We compared three functional approaches:
Likelihood (Lik): We used the likelihood-based method presented in Ma et al. (2002). This assumes that the mean function has a logistic form and that the errors are Gaussian with an autoregressive structure.
Estimating equations (EE): We used the Wald statistic without a shrinkage estimator for the error covariance matrix, Σ. The mean function was modeled using a natural spline basis with 7 d.f.
Estimating equations with shrinkage (EES): We used the Wald statistic with a natural spline basis with the Schafer and Strimmer (2005) method for shrinking the covariance matrix.
Note that the estimating equations approach is applied using no knowledge of the mean and correlation functions. The likelihood method is correctly specified for the first set of simulations (Gaussian, autoregressive). It misspecifies the finite-dimensional distributions in the second set (t-distributed, autoregressive), but the correlation is correctly specified. In the third set, the correlation functions are misspecified.
Table 1 gives tail probabilities of the test statistics under the null. It shows that the correctly specified likelihood-ratio statistic has χ2-tail probabilities as expected. This holds true when the marginal distribution of the errors has a t-distribution as well. However, when the correlation structure is misspecified, it no longer has a χ2-distribution and the type-I errors corresponding to all the critical values are higher than expected.
Table 1 . Null distribution P-values from parametric and estimating equation approaches under three error distributions using 1000 simulations each.
| Covariance structure | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gaussian, autoreg:Method | t, 4 d.f., autoreg:Method | Gaussian, Matérn:Method | |||||||
| Cutoff | Lik | EES | EE | Lik | EES | EE | Lik | EES | EE |
| Neighbor correlation 0.61 | |||||||||
| 0.1 | 0.11 | 0.11 | 0.11 | 0.11 | 0.12 | 0.12 | 0.42 | 0.077 | 0.11 |
| 0.05 | 0.057 | 0.053 | 0.054 | 0.064 | 0.067 | 0.061 | 0.32 | 0.032 | 0.052 |
| 0.01 | 0.013 | 0.011 | 0.011 | 0.016 | 0.014 | 0.010 | 0.19 | 0.007 | 0.010 |
| Neighbor correlation 0.83 | |||||||||
| 0.1 | 0.099 | 0.099 | 0.098 | 0.099 | 0.075 | 0.089 | 0.44 | 0.077 | 0.11 |
| 0.05 | 0.048 | 0.052 | 0.058 | 0.044 | 0.034 | 0.042 | 0.35 | 0.033 | 0.055 |
| 0.01 | 0.015 | 0.010 | 0.013 | 0.006 | 0.007 | 0.008 | 0.20 | 0.009 | 0.015 |
| Neighbor correlation 0.94 | |||||||||
| 0.1 | 0.11 | 0.084 | 0.11 | 0.092 | 0.067 | 0.10 | 0.45 | 0.070 | 0.11 |
| 0.05 | 0.050 | 0.039 | 0.054 | 0.051 | 0.034 | 0.056 | 0.34 | 0.038 | 0.055 |
| 0.01 | 0.012 | 0.006 | 0.011 | 0.013 | 0.004 | 0.008 | 0.18 | 0.009 | 0.012 |
We considered three different covariance structures: (a) Gaussian with autoregressive correlations, (b) t-distributed with 4 d.f. and an autocorrelated covariance, and (c) Gaussian with Matérn correlation function. We consider three correlation strengths as measured by the correlation between nearest neighbors: 0.61, 0.83, and 0.94. The parametric likelihood method assumes autocorrelated error (Lik) and has the desired type-I error for both Gaussian and t-distributed errors, when the correlation structure is correctly specified. However, it is substantially off target when the correlation is misspecified. The estimating equations approach with shrinkage (EES) has close to the desired type-I error, but is slightly off target. The estimating equations approach (EE) always has the correct type-I error.
The simulations indicate that the regular estimating equations approach has near the expected type-I error behavior under all circumstances. However, the estimating equations approach with shrinkage does not have this property with slightly too low type-I error rates, and thus its null distribution would need to be obtained empirically using permutations. The likelihood-ratio statistic has the expected χ2-distribution only when the correlation structure is correctly specified; otherwise it may be way off target. Thus, in practice, if the covariance is hard to specify, the likelihood-ratio statistic’s statistical significance should be established using a permutation distribution.
Since the null distribution of the statistics is not always as expected, we used the null distribution quantiles as the critical values to calculate power (Table 2). We find that the likelihood approach has the greatest power when the correlation structure is correctly specified (note that the likelihood method in this case also benefits from having the correct mean function). The estimating equations approaches have greater power when the correlation structure is incorrectly specified. In all situations, the estimating equations with shrinkage has slightly greater power than the approach without shrinkage. Thus, our simulations demonstrate that power of likelihood methods may be compromised if the correlation structure is misspecified (even if the type-I error has been recalibrated).
Table 2 . Power of parametric and estimating equations approaches under three error distributions: Gaussian, autoregressive (GA); t, 4 d.f., autoregressive (tA); and Gaussian, Matérn (GM).
| Neighbor correlation | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| 0.61 | 0.83 | 0.94 | |||||||
| Error model | |||||||||
| Method | GA | tA | GM | GA | tA | GM | GA | tA | GM |
| Lik | 0.96 | 0.96 | 0.60 | 0.90 | 0.92 | 0.61 | 0.96 | 0.95 | 0.58 |
| EES | 0.92 | 0.90 | 0.74 | 0.76 | 0.81 | 0.75 | 0.89 | 0.90 | 0.72 |
| EE | 0.90 | 0.90 | 0.70 | 0.75 | 0.80 | 0.71 | 0.86 | 0.88 | 0.69 |
Shown is the proportion of times the P-values under the alternative were smaller than the 5th percentile of the null. The parametric approach (Lik) has the highest power when the covariance structure of the error is correctly specified. This is true for both Gaussian and t-distributed errors. The estimating equations approaches have slightly less power than the likelihood method with correctly specified correlation. When the covariance structure is incorrectly specified, the estimating equations approaches have greater power. Estimating equations with shrinkage (EES) appears to have slightly higher power than estimating equations without shrinkage (EES).
Simulation studies: Comparison with parametric functional method with unstructured covariance
Yap et al. (2009) proposed using regularized covariance estimation within the framework of parametric likelihood-based functional mapping. The regularization parameter was selected by 10-fold cross-validation and assumed constant throughout a marker interval.
We performed simulations using the same scenario as in Yap et al. (2009). We simulated a 100-cM linkage group with six equally spaced markers with a QTL at 32 cM. The associated phenotypes were sampled from a multivariate Gaussian distribution with logistic function describing the mean function conditional on the QTL genotypes. There are three genotypes with three mean curves following logistic functions as in Equation 7: the first genotype has θ0 = 30, θ1 = 5, θ2 = 0.5; the second genotype has θ0 = 28.5, θ1 = 5, θ2 = 0.5; and the third genotype has θ0 = 27.5, θ1 = 5, θ2 = 0.5. Each individual is observed at 10 time points. The residual error was assumed to be multivariate normal, with three different covariance structures:
Σ1 is autoregressive with σ2 = 3, ρ = 0.6.
Σ2 is equicorrelated with σ2 = 3, ρ = 0.5.
Σ3 is an “unstructured” covariance matrix, as given in Yap et al. (2009) (reproduced in File S1). It does not have an easily described structure.
All parameter values including the covariance matrices are taken from Yap et al. (2009). We ran 10,000 simulation replicates to obtain stable estimates; Yap et al. (2009) had 100 simulation replicates.
The LOD score (or linkage test statistic) was calculated every 4 cM, resulting in 26 loci. As in the simulations reported by Yap et al. (2009), for each simulation, we estimated the location of the QTL as the location of the maximum LOD score. The average, standard deviation, and root mean squared error of QTL location estimates were compared. Lower root mean squared error indicates a better method. (Note that we are unable to compare the power of the methods as we do not have access to the software used in the Yap et al. 2009 article and are restricted to what was reported in the article.) The results are in Table 3.
Table 3 . Mean, standard deviation, and root mean squared error of the genome scan peak in simulations using our method and that of Yap et al. (2009).
| Yap (nonpara) | Yap (autoreg) | EE (Wald) | EE (residual) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Covariance | n | Mean | SD | rmse | Mean | SD | rmse | Mean | SD | rmse | Mean | SD | rmse |
| Σ1 | 100 | 32.8 | 9.9 | 9.9 | 33.2 | 7.7 | 7.8 | 33.0 | 10.4 | 10.5 | 32.8 | 9.09 | 9.1 |
| 400 | 31.5 | 2.8 | 2.8 | 31.8 | 3.2 | 3.2 | 32.1 | 2.9 | 2.9 | 32.1 | 2.9 | 2.9 | |
| Σ2 | 100 | 32.6 | 7.6 | 7.6 | 35.3 | 15.7 | 16.0 | 32.7 | 7.7 | 7.7 | 33.1 | 12.0 | 12.0 |
| 400 | 31.7 | 2.6 | 2.6 | 32.0 | 5.4 | 5.4 | 32.1 | 2.4 | 2.4 | 32.2 | 3.7 | 3.7 | |
| Σ3 | 100 | 33.2 | 22.2 | 22.2 | 46.5 | 27.4 | 31.0 | 40.6 | 26.5 | 27.9 | 39.3 | 25.3 | 26.3 |
| 400 | 32.3 | 11.9 | 11.9 | 43.6 | 26.4 | 28.9 | 33.1 | 10.8 | 10.9 | 33.1 | 10.7 | 10.8 | |
The true QTL is at 32 cM. The columns labeled “Yap (nonpara)” and “Yap (autoreg)” are derived from Tables 1 and 2 of Yap et al. (2009). They refer to the results of using the likeihood-based methods of Yap et al. (2009) with an estimated regularized covariance and autocorrelated covariance, respectively. The columns labeled “EE (Wald)” and “EE (residual)” refer to our estimating equations approach with the Wald statistic and the residual error statistic, respectively. Yap et al. (2009) performed 100 simulation replicates, whereas we used 10,000 replicates, which gave stable estimates. For each method we report the mean position of the genome scan maximum (“mean”) over simulation replicates, the standard deviation (“SD”), and the root mean squared error (“rmse”). Note that Yap et al. (2009) reported standard error, which we converted to standard deviation as the latter is independent of the number of simulation replicates. The simulations were performed with three error structures corresponding to an autocorrelated covariance (Σ1), an equicorrelated covariance (Σ2), and an unstructured covariance (Σ3).
We found that our method performed comparably to Yap et al.’s (2009) method for the most part, except for Σ3 with a small sample size of 100, where Yap et al.’s (2009) method had better mean and slightly smaller standard deviation. But looking at the large values of standard deviations for all methods, it is clear that no method performed satisfactorily with this small sample size. For Σ3 with sample size 400, the performance of both methods was again comparable. Note, however, that our method is considerably simpler to implement and has lower computational complexity.
Data analysis
Please refer to the Mouse Behavioral Data section for the details of the mapping population, markers, and phenotypic data collection. Our analytic goal was to detect genetic loci that contribute to individual variation in the shape of the curve describing how ASP changes with time of day. We applied our method using the Wald statistic and the residual error statistic. We also applied a nonfunctional approach informed by prior knowledge and a visual exploration of the data. The approach tries to mimic what we might do if we did not have a functional method at our disposal. It parallels the approach taken recently by Nishi et al. (2010) for similar homecage movement data in consomic strains. Note that a credible parametric model for mouse behavior cannot be easily specified, and thus we cannot use a parametric functional method.
Nonfunctional method
We constructed six measures on the basis of the marginal distribution of the ASP curves as shown in Figure 1. Note that the change in active state probability with time of day exhibited by the mice appears to be divided into five phases. During the dark cycle, there is an onset peak (7–9 pm) in active state probability that then decreases somewhat, exhibiting a broad peak from 9 pm to 4 am followed by a dark cycle nadir from 4 to 6 am. An offset peak (6–8 am) in the active state probability then occurs near the end of the dark cycle. Finally, the active state probability is low throughout the majority of the light cycle. On the basis of these observations we segmented the day into the following phases to measure the probability that a mouse was in the active state:
Daily: This is the mean over all time points.
Dark cycle onset peak: This is the mean over 7–9 pm.
Mid-dark cycle: This is the mean over 9 pm–4 Am.
Dark cycle nadir: This is the mean over 4–6 Am.
Dark cycle offset peak: This is the mean over 6–8 Am.
Light cycle: This is the mean over 8 Am-12 noon and 2–7 pm.
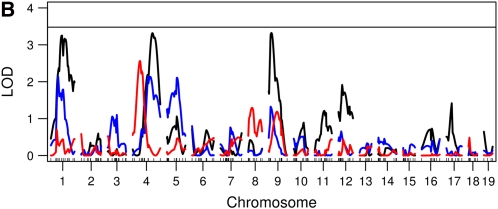
We performed genome scans using the Haley–Knott method (Haley and Knott 1992) for each of these measures. To correct for the fact that we used six correlated genome scans, we calculated a genome-wide threshold for the maximum of the six genome scans, using 1000 permutations. Using the 5% multiple-scan corrected threshold we found only one locus on chromosome 1 for daily ASP (Figure 3).
Figure 3 .
Genome scans using a nonfunctional approach. (A) Genome scans for total daily ASP (black), dark cycle onset peak (blue), and mid-dark cycle (red). (B) Genome scans for dark cycle nadir (black), dark cycle offset peak (blue), and light cycle activity (red). The solid horizontal lines are the 5% permutation threshold for the maximum of the six genome scans, which adjusts for the fact that we are examining six correlated genome scans.
Functional method
We used B-splines as our basis functions. We applied 10-fold cross-validation to the behavioral data, ignoring the genotype data, to select the number of basis functions to use for smoothing. We selected the smallest number of basis functions that gave a residual sum of squares within one standard deviation of the least value. This led us to select B-splines with 16 d.f. with equally spaced knots. These basis functions were used for all genome scans.
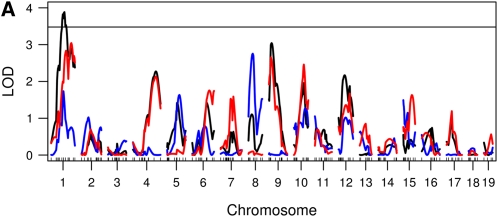
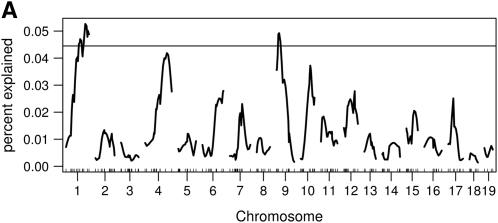
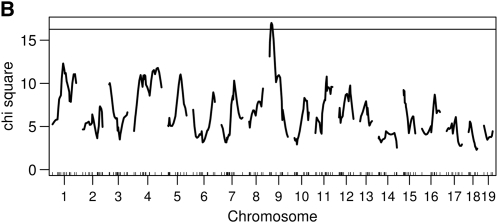
We performed genome scans using the Haley–Knott method with both the Wald statistic and the residual error statistics. We used 1000 permutations to establish genome-wide statistical significance. Using the functional approach and the 5% genome-wide threshold, we found two loci (on chromosomes 1 and 9) using the residual error statistic (Figure 4A) and one locus on chromosome 9 using the Wald statistic (Figure 4B). The estimated genetic effects of the two loci are shown in Figure 5. Thus, in this example, the Wald statistic appears to be less sensitive than the residual error statistic.
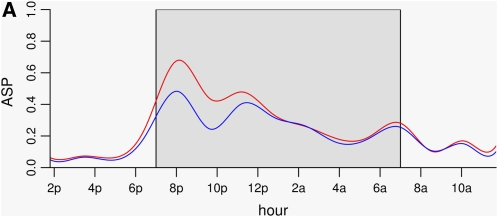
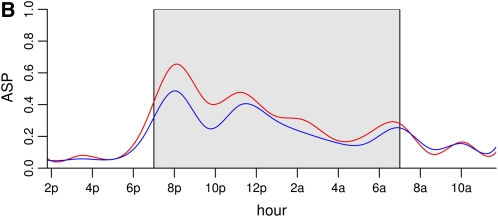
Figure 5 .
Functional effect of two genetic loci on chromosome 1 ((A)) and chromosome 9 ((B)). The dark period is shaded gray. Both loci appear to have the greatest effect in the high activity periods; there is little difference between the two genotypes in the low activity period concentrated between 8 am and 5 am. The chromosome 1 locus effect is largely between 6 pm and 1 am. The chromosome 9 locus affects activity between 6 pm and 7 am. For both loci there is evidence of the dark cycle nadir shifting a little.
Figure 4 .
Genome scans using the functional approach. (A) A genome scan using the residual error statistics expressed as a proportion of the integrated error explained by a locus. The horizontal line is the 5% permutation threshold. (B) A genome scan using the Wald statistic and the corresponding 5% permutation threshold.
Discussion
We have presented a new approach based on estimating equations for mapping function-valued traits. It is an attractive alternative to likelihood-based methods, especially when we have incomplete knowledge of the covariance structure or when the number of time points exceeds the number of samples. While information about the covariance can be incorporated to improve efficiency, our method is robust to its misspecification. Relative to a correctly specified likelihood model, the estimating equations approach may be slightly less efficient. However, such loss was modest in our simulations and other studies (Chandler and Bate 2007). We thus believe that our method would be a good choice in most settings where the analyst is uncertain about the error distribution. The mouse behavioral data we presented are such an example. Likelihood methods may be preferred, say, for traits such as growth curves, which have a long history of study, lending confidence to the data model.
Misplaced confidence in the covariance structure may have a price, as our simulations indicate. Likelihood-based methods with a misspecified covariance may not have the target type-I error and may have lower power than the estimation equations method. Although the anticonservative behavior of a misspecified likelihood observed in our simulations may be rectified by a permutation test, that has an additional computational cost. This cost is manageable for QTL mapping in experimental crosses, but less so in GWAS where a correctly calibrated P-value is desired at each locus. Thus, the estimating equations Wald statistic has an edge in GWAS.
There are several ways to deal with more time points than the sample size, which engenders a singular covariance matrix. Zhao et al. (2007) used a wavelet transform for time series dimension reduction as a precursor to parametric functional mapping. Our method, on the other hand, has dimension reduction built in via the basis functions, which in fact could be wavelets. While Zhao et al. (2007) used wavelets for dimension reduction as a separate step, we perform dimension reduction and functional mapping in one step.
Computational efficiency, as opposed to statistical efficiency, plays a bigger role for high-dimensional data. If the number of time points is very large or if model selection, permutation testing, or multiple imputation is needed, computational time increases severalfold. In such settings, our method has a clear advantage, as it is based on two low-dimensional least-squares operations: it can be computed quickly and does not involve any complex optimization procedures. The computational advantage is particularly pronounced when covariance structure defies easy specification. While Yap et al.’s (2009) method does not rely on specification of covariance structure, it comes at the expense of a generalized EM algorithm that must numerically solve a nonlinear optimization problem as part of the M-step. For large-scale studies with dense genotyping, computational efficiency is essential.
The estimating equation approach for mapping function-valued traits can be adapted for different situations. Our formulation assumes that the error variance is independent of the mean, which might not hold for traits whose point-wise marginal distribution is markedly non-Gaussian such as Poisson-distributed count data. A simple change to the sandwich estimator in Equation 5 can be used to obtain a robust variance estimate in this scenario. This involves replacing the estimate by a sum of n terms with the same form as in Equation 5, with Σ replaced by the empirical multivariate residual for each individual. Another extension is when trait data on all individuals are not observed on the same set of time points. While small amounts of missing trait data can be dealt with by smoothing or imputation, the more general problem of uneven time points is more challenging. The approach of Yao et al. (2005) may offer a resolution under Gaussian modeling assumptions.
In summary, our estimating equation method based on a general functional linear model makes few distributional assumptions. It has broad applicability to genetic studies and has opened an attractive alternative avenue to functional mapping distinct from likelihood methods. The possible improvements and adaptations listed above suggest the method’s promise and flexibility. Data and software used in this paper are available in File S2. The mouse behavior data also is available at http://www.qtlarchive.org; updated versions of the software will be posted at http://www.biostat.ucsf.edu/sen/software/.
Acknowledgments
Genotyping was carried out in the DNA Technologies Core at the University of California, Davis, Genome Center. We thank Ethelyn Layco for animal care, sample preparation for Illumina genotyping, and assistance with behavioral experiments. We thank Rongling Wu for providing Matlab code for the likelihood method that we adapted for use in R, Karl Broman and Tom Juenger for helpful comments, and Sandra Erickson for suggesting we work on mapping function-valued traits. Some computations were performed using the University of California, San Francisco, Biostatistics High Performance Computing System. This research was supported by grants from the National Institutes of Health (GM078338, GM074244, and DK072187).
Literature Cited
- Broman K. W., 2001. Review of statistical methods for QTL mapping in experimental crosses. Lab Anim. (NY) 30(7): 44–52 [PubMed] [Google Scholar]
- Chandler R. E., Bate S., 2007. Inference for clustered data using the independence loglikelihood. Biometrika 94(1): 167–183 [Google Scholar]
- Churchill G. A., Doerge R. W., 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggle P., Liang K., Heagerty P., Zeger S., 2002. Analysis of Longitudinal Data, Ed. 2 Oxford University Press, London/New York/Oxford [Google Scholar]
- Godambe V., 1960. An optimum property of regular maximum likelihood estimation. Ann. Math. Stat. 31: 1208–1211 [Google Scholar]
- Golub G., Van Loan C., 1996. Matrix Computations. Johns Hopkins University Press, Baltimore [Google Scholar]
- Goulding E., Schenk A., Juneja P., MacKay A., Wade J., et al. , 2008. A robust automated system elucidates mouse home cage behavioral structure. Proc. Natl. Acad. Sci. USA 105(52): 20575–20582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley C., Knott S., 1992. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity 69: 315–324 [DOI] [PubMed] [Google Scholar]
- Kao C. H., 2000. On the difference between maximum likelihood and regression interval mapping in the analysis of quantitative trait loci. Genetics 156: 855–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauermann G., Carroll R., 2001. A note on the efficiency of sandwich covariance matrix estimation. J. Am. Stat. Assoc. 96(456): 1387–1396 [Google Scholar]
- Kingsolver J., Gomulkiewicz R., Carter P., 2001. Variation, selection and evolution of function-valued traits. Genetica 112: 87–104 [PubMed] [Google Scholar]
- Kirkpatrick M., Heckman N., 1989. A quantitative genetic model for growth, shape, reaction norms, and other infinite-dimensional characters. J. Math. Biol. 27(4): 429–450 [DOI] [PubMed] [Google Scholar]
- Kramer M., Vaughn T., Pletscher L., King-Ellison K., Adams E., et al. , 1998. Genetic variation in body weight gain and composition in the intercross of Large (LG/J) and Small (SM/J) inbred strains of mice. Genet. Mol. Biol. 21(2): 211–218 [Google Scholar]
- Lander E. S., Botstein D., 1989. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange C., Whittaker J., 2001. Mapping quantitative trait loci using generalized estimating equations. Genetics 159: 1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leamy L. J., Klingenberg C. P., Sherratt E., Wolf J. B., Cheverud J. M., 2008. A search for quantitative trait loci exhibiting imprinting effects on mouse mandible size and shape. Heredity 101(6): 518–526 [DOI] [PubMed] [Google Scholar]
- Ledoit O., Wolf M., 2004. A well-conditioned estimator for large-dimensional covariance matrices. J. Multivariate Anal. 88(2): 365–411 [Google Scholar]
- Liang K., Zeger S., 1986. Longitudinal data analysis using generalized linear models. Biometrika 73: 13–22 [Google Scholar]
- Lin M., Wu R., 2006. A joint model for nonparametric functional mapping of longitudinal trajectory and time-to-event. BMC Bioinformatics 7(1): 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Wu R., 2009. A Bayesian algorithm for functional mapping of dynamic traits. Algorithms 2: 667–691 [Google Scholar]
- Ma C., Casella G., Wu R., 2002. Functional mapping of quantitative trait loci underlying the character process: a theoretical framework. Genetics 161: 1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch C. E., Searle S. R., Neuhaus J. M., 2008. Generalized, Linear, and Mixed Models, Ed. 2 Wiley, New York [Google Scholar]
- Meyer K., Kirkpatrick M., 2010. Better estimates of genetic covariance matrices by “bending” using penalized maximum likelihood. Genetics 185: 1097–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezey J., Houle D., 2005. The dimensionality of genetic variation for wing shape in Drosophila melanogaster. Evolution 59(5): 1027–1038 [PubMed] [Google Scholar]
- Nishi A., Ishiii A., Takahashi A., Shiroishi T., Koide T., 2010. QTL analysis of measures of mouse home-cage activity using B6/MSM consomic strains. Mamm. Genome 21: 477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletcher S., Geyer C., 1999. The genetic analysis of age-dependent traits: modeling the character process. Genetics 153: 825–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay J., Silverman B. W., 2005. Functional Data Analysis, Ed. 2 Springer-Verlag, Berlin/Heidelberg, Germany/New York [Google Scholar]
- Rapp J. P., 2000. Genetic analysis of inherited hypertension in the rat. Physiol. Rev. 80: 131–172 [DOI] [PubMed] [Google Scholar]
- Rotnitzky A., Jewell N., 1990. Hypothesis-testing of regression parameters in semiparametric generalized linear-models for cluster correlated data. Biometrika 77(3): 485–497 [Google Scholar]
- Schafer J., Strimmer K., 2005. A shrinkage approach to large-scale covariance matrix estimation and implications for functional genomics. Stat. Appl. Genet. Mol. Biol. 4(1): 32. [DOI] [PubMed] [Google Scholar]
- Sen S., Churchill G. A., 2001. A statistical framework for quantitative trait mapping. Genetics 159: 371–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen S., Satagopan J. M., Churchill G. A., 2005. Quantitative trait loci study design from an information perspective. Genetics 170: 447–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q., Faraway J., 2004. An F test for linear models with functional responses. Stat. Sin. 14(4): 1239–1257 [Google Scholar]
- Shimomura K., Low-Zeddies S., King D., Steeves T., Whiteley A., et al. , 2001. Genome-wide epistatic interaction analysis reveals complex genetic determinants of circadian behavior in mice. Genome Res. 11(6): 959–980 [DOI] [PubMed] [Google Scholar]
- Wu R., Lin M., 2006. Functional mapping—how to map and study the genetic architecture of dynamic complex traits. Nat. Rev. Genet. 7(3): 229–237 [DOI] [PubMed] [Google Scholar]
- Wu R., Ma C., Lin M., Casella G., 2004. A general framework for analyzing the genetic architecture of developmental characteristics. Genetics 166: 1541–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Wu R., Casella G., 2009. Nonparametric functional mapping of quantitative trait loci. Biometrics 65: 30–39 [DOI] [PubMed] [Google Scholar]
- Yang R., Xu S., 2007. Bayesian shrinkage analysis of quantitative trait loci for dynamic traits. Genetics 176: 1169–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R., Tian Q., Xu S., 2006. Mapping quantitative trait loci for longitudinal traits in line crosses. Genetics 173: 2339–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao F., Müller H., Wang J., 2005. Functional data analysis for sparse longitudinal data. J. Am. Stat. Assoc. 100(470): 577–590 [Google Scholar]
- Yap J. S., Fan J., Wu R., 2009. Nonparametric modeling of longitudinal covariance structure in functional mapping of quantitative trait loci. Biometrics 65(4): 1068–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Li H., Hou W., Wu R., 2007. Wavelet-based parametric functional mapping of developmental trajectories with high-dimensional data. Genetics 176: 1879–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]