Abstract
Certain residues have no known function yet are co-conserved across distantly related protein families and diverse organisms, suggesting that they perform critical roles associated with as-yet-unidentified molecular properties and mechanisms. This raises the question of how to obtain additional clues regarding these mysterious biochemical phenomena with a view to formulating experimentally testable hypotheses. One approach is to access the implicit biochemical information encoded within the vast amount of genomic sequence data now becoming available. Here, a new Gibbs sampling strategy is formulated and implemented that can partition hundreds of thousands of sequences within a major protein class into multiple, functionally-divergent categories based on those pattern residues that best discriminate between categories. The sampler precisely defines the partition and pattern for each category by explicitly modeling unrelated, non-functional and related-yet-divergent proteins that would otherwise obscure the analysis. To aid biological interpretation, auxiliary routines can characterize pattern residues within available crystal structures and identify those structures most likely to shed light on the roles of pattern residues. This approach can be used to define and annotate automatically subgroup-specific conserved domain profiles based on statistically-rigorous empirical criteria rather than on the subjective and labor-intensive process of manual curation. Incorporating such profiles into domain database search sites (such as the NCBI BLAST site) will provide biologists with previously inaccessible molecular information useful for hypothesis generation and experimental design. Analyses of P-loop GTPases and of AAA+ ATPases illustrate the sampler's ability to obtain such information.
Keywords: protein sequence/structural analysis, Markov chain Monte Carlo sampling, Bayesian partitioning with pattern selection
1. INTRODUCTION
A major goal of modern biology is to understand how protein molecular machines work at the atomic level in the context of the living cell (Alberts, 1998). Although the catalytic mechanisms of certain proteins are relatively well understood, much less is known regarding mechanisms mediating other aspects of protein function, such as energy-dependent coordinated conformational changes. Moreover, the existence of mysterious, as yet unidentified molecular mechanisms is suggested by the fact that related proteins from diverse organisms often strikingly conserve specific residues whose functions, thus far, are completely unknown. For instance, an invariant tyrosine and a nearly invariant isoleucine that is never replaced by a leucine occur far from the active site within Ran GTPases (based on 276 sequences from 9 metazoan, 4 fungal, 2 plant, and 10 protozoan phyla). This suggests that relatively minor side-chain modifications at these sites, such as removal of an oxygen or rearrangement of a methyl group, are consistently eliminated by natural selection and that such residues thus establish interactions with precise geometric or chemical constraints required for some critical unknown function. Often such residues are conserved across families that are associated with distinct multi-component complexes and that vary widely in their protein-protein interactions, localization, turnover, kinetics of binding, et cetera—indicating that the roles of these residues transcend functions or properties specific to individual families and instead involve general mechanisms shared by otherwise functionally-divergent proteins.
Delving deeper into the biochemical roles of such residues through experimentation requires that we first obtain sufficient preliminary information to formulate plausible hypotheses. An important source of such information is, of course, protein crystal structure analysis. Nevertheless, even with a complete set of functionally-relevant structural conformations, identifying the underlying biochemical mechanisms of a protein is non-trivial. One reason for this is that, at the atomic level, matter and energy obey the laws of quantum electrodynamics (Feynman, 1985) and thus do not behave like particles or waves or anything else that we have ever seen. This counterintuitive behavior of quantum phenomena hinders our ability to conceptualize molecular processes correctly.
To help work around these inherent limitations, we can utilize another source of implicit information regarding molecular mechanisms, genomic sequence data—the cell’s own library for encoding those mechanisms. As such, sequence data may provide valuable clues regarding protein biochemical properties and mechanisms that, thus far, have escaped our attention. One way to access this information is to use a statistical approach: Just as a statistical analysis of patterns of inherited traits can provide clues regarding underlying genetic mechanisms, a statistical analysis of patterns in protein sequences can provide clues regarding underlying biochemical mechanisms. Residue patterns that have been conserved for a billion years or more presumably reflect strong selective pressures maintaining structural and mechanistic similarities. Divergent patterns that are conserved in descendent proteins maintaining a particular divergent function likewise reflect structural and mechanistic differences. Thus such patterns presumably correspond to conservation and divergence of underlying molecular mechanisms that are responsible for critical biochemical properties, which I define broadly here to include all properties that are required for a protein’s function. When examined in the light of available structural data and of published biochemical analyses, patterns of co-conserved residues can suggest plausible hypotheses regarding underlying biological processes. These hypotheses can be tested by experimentally determining, for example, whether an engineered protein harboring those patterns acquires a particular biochemical property. Hence, without postulating a specific mechanism, we can still associate certain residue patterns with certain biochemical properties.
The identification of such patterns is complicated by the probabilistic nature of protein sequence data: Sequences possessing certain biochemical properties are characterized by inherent variability, such that each protein family is best viewed as a distribution of (similar) sequences corresponding to a peak within a high dimensional ‘sequence space’ or, when viewed from an evolutionary perspective, within a ‘fitness’ landscape (Romero and Arnold, 2009). This landscape may be quite irregular insofar as small peaks and valleys can form on top of larger peaks whenever a protein subgroup either gains additional biochemical properties or loses certain aspects of ancestral properties. Moreover, protein sequences may encode multiple biochemical properties in various combinations and to varying degrees. As a result, we should not expect characteristic patterns to be clearly defined across related sequences (i.e., to be associated with isolated, sharply-defined peaks in sequence space), but rather to be obscured by various degrees of evolutionary and combinatorial noise. To make biological sense of sequence data, it is helpful to extract from this noise those canonical patterns that are most typical of (albeit not fully conserved within) specific categories of functionally-divergent proteins. Such canonical patterns are helpful for the same reason that simple, idealized models help make sense of complex phenomena in physics.
In order to identify canonical patterns in this way, we previously developed a procedure, termed Bayesian Partitioning with Pattern Selection (BPPS) (Neuwald, 2007a; Neuwald et al., 2003). BPPS relies on a Gibbs sampling strategy (Liu, 2008) to identify a residue pattern that most distinguishes one set of protein sequences (termed the ‘foreground’) from another, functionally divergent sequence set (termed the ‘background’). Gibbs sampling is a Bayesian Markov chain Monte Carlo (MCMC) procedure that relies, as does the scientific method itself, on iterating between empirical testing and model refinement until convergence on the most probable models given the input data. For BPPS the input data consists of a (typically very large) set of related, multiply-aligned protein sequences. A Gibbs sampling strategy is required because, a priori, we know neither which sequences to assign to the foreground, nor which positions are pattern positions, nor exactly which residues are conserved at each pattern position. More specifically, the BPPS procedure samples over two random variables: a sequence pattern and a set of indicators for assigning each sequence in the alignment to either the foreground or the background partition. While doing so, it explores possible pattern-partition pairs, searching for one where the pattern maximally distinguishes the foreground sequences from the background sequences (Fig. 1). To ensure that pattern residues are conserved due to strong positive selective pressure (rather than merely to recent common descent), we require conservation of patterns across distinct phyla or kingdoms.
Figure 1.
Input and output for the BPPS sampler. The main alignment is partitioned into a ‘foreground’ (colored horizontal bars) and a ‘background’ (gray horizontal bars) based on conserved foreground pattern positions (dark blue vertical bars) that diverge from (or contrast with) the background residues at those positions (white vertical bars). For this reason, the output is termed a “contrast alignment”. The heights of the bars above the alignment quantify the selective constraints imposed on divergent residue positions.
This ‘single category’ (sc)BPPS sampler serves as a starting point for the ‘multiple category’ (mc)BPPS sampler described here. The mcBPPS sampler is substantially more powerful in the following respects: (i) By representing multiple pattern-partition pairs, it can optimally model the sequence space of an entire protein class. (In contrast, the scBPPS sampler merely identifies one globally or locally optimal pattern-partition.) (ii) It uses an enhanced statistical formulation that avoids over-fitting by minimizing the number of free parameters. (iii) It sets up a stringent competition between functional-divergent categories for pattern residues, thereby defining each pattern-partition pair with greater clarity. (iv) It models protein subgroups more precisely by explicitly modeling (and eliminating) problematic sequences that tend to obscure an analysis. Such problematic sequences are likely to include, for example, pseudogene products and other nonfunctional proteins, related proteins that have undergone additional functional divergence, and unrelated and erroneous sequences. (v) Unlike the scBPPS sampler, it includes an option that does not require assignment of ‘gold standard’ sequences to a foreground partition in order to help define that partition, though—to address specific questions—it still allows specification of such gold standard sequences. (vi) Perhaps most importantly, it can be used to generate (automatically) protein domain profiles, in which multiple categories of pattern residues are annotated. And (vii), when merged into the National Center for Biotechnology Information (NCBI) conserved domain database, such annotated profiles can aid experimental design by suggesting (via web-based BLAST searches linked to the NCBI Cn3D viewer) (Wang et al., 2000) plausible hypotheses regarding mysterious, as-yet-unidentified biochemical properties and mechanisms.
2. RESULTS
The mcBPPS sampler addresses the following problem: We are given a (typically large) set of aligned input sequences corresponding to a major protein class. Each of these sequences belongs to one out of N subgroups, though we know not which one. The sampler’s task is to infer the most likely subgroup to which each sequence belongs based on M functionally-divergent categories, each of which is associated with a canonical pattern that likewise needs to be inferred. These are termed ‘differentiating patterns’ because they correspond to residues that are co-conserved within one or more subgroups (that together constitute the foreground) but that diverge within one or more other subgroups (constituting the background) and that thus differentiate between the foreground and the background.
As most commonly applied, the user selects the number of subgroups and assigns one or more ‘seed sequences’ to each of the subgroups. Typically, less than a dozen seed sequences are selected from distinct phyla for each subgroup to ensure that pattern residues are conserved due to functional constraints rather than merely to recent common descent. These seed sequences serve as Bayesian priors or—if viewed as a ‘missing data’ problem (Little and Rubin, 2002)—as ‘labeled’ sequences that are required to remain in their pre-assigned subgroups during sampling and that thus help define each subgroup. The remaining (unlabeled) sequences are assigned to subgroups through Bayesian inference. This strategy focuses the sampler on those properties of the sequence data of primary interest to the user.
The user chooses which comparisons are to be made by providing a N × M table (termed a hyperpartition) that specifies which of the N subgroups (one per row) to place in the foreground and background partitions (as well as which subgroups to leave out) for each of M divergent categories (i.e., one category per column). Thus each column in a hyperpartition corresponds to a single contrast alignment (Fig. 2) where those subgroups assigned to the foreground are indicated by a ‘+’ in the table, those assigned to the background by a ‘−’ and the ‘non-participating’ subgroups by a ‘o’. (Further information is given in the supplementary file.) The phylogenetic relationships between subgroups are useful when creating a hyperpartition, and indeed a phylogenetic tree can be directly converted into a hyperpartition (Fig. 2). However, as discussed below (and as illustrated in the supplementary file), subgroups from distinct clades sometimes share certain co-conserved residues that would be unexpected based solely on their phylogeny. For this and other reasons, the hyperpartition is not required to conform strictly to a phylogenetic tree.
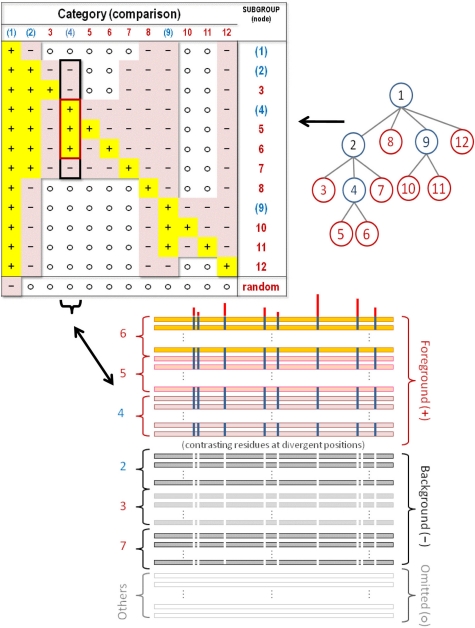
Figure 2.
A hyperpartition table (top left), which corresponds to the multiple category functional divergence model optimized by the mcBPPS sampler. Each row in the table corresponds to a distinct divergent subgroup and each column corresponds to a distinct contrast alignment. A column is converted into the corresponding contrast alignment—such as the one shown for column 4 (bottom)—as follows: Each subgroup corresponding to a row with a ‘+’ symbol in that column is assigned to the foreground, each subgroup with a ‘−’ symbol in that column is assigned to the background, and each subgroup with an ‘o’ symbol in that column is assigned to the non-participating partition. A tree (top right) can also represent the hierarchical relationships between functionally divergent subgroups in this table, where, for example, the subtrees rooted at nodes 5, 4, 2 and 1 could correspond to a subfamily, a family, a superfamily and the entire protein class, respectively. Such a tree can be converted into a hyperpartition, as follows: Each node of the tree corresponds to both a column and a row in the hyperpartition table. The subtree rooted at a particular node corresponds to the foreground for that column in the table, whereas the rest of the subtree rooted at the parent of that node corresponds to the background. Hence sequences corresponding to the subtree of a node share a pattern (albeit often imperfectly) that corresponds to that node’s column (i.e., contrast alignment) in the table. Note that internal nodes in the tree corresponds to miscellaneous subgroups—that is to sequences sharing a common pattern with, but lacking patterns specific to subgroups corresponding to that node’s descendent nodes. For the root node a set of random sequences serves as the background; the sampler will favor assignment of aberrant input sequences to this random subgroup. The program tree2hpt (which is distributed with the mcBPPS program) may be used to convert a tree in Newick format into the corresponding hyperpartition. Although any tree can be converted into a hyperpartition, some hyperpartitions (such as the one shown in Table 2 below) cannot be converted into a tree.
2.1. The scBPPS model
The starting point for the mcBPPS sampler is the scBPPS procedure (Neuwald et al., 2003), which samples over the joint distribution that is defined (logarithmically) by:
| (1) |
where X is an n × k matrix representing a multiple alignment of n sequences and k columns, xi,j is a 20-dimensional vector of all 0's except for a lone ‘1’ indicating the observed residue type, R is a vector indicating which rows (i.e., sequences) belong to the foreground (Ri = 1) or background (Ri = 0) partitions, C is a vector indicating which columns do (Cj = 1) or do not (Cj = 0) differentiate the foreground from the background, Θ is an array of vectors representing the amino acid compositions at each column position for each partition, 〈·,·〉 denotes the inner product of two vectors, and models the foreground composition at pattern positions where θj ≡ (θj,1,…,θj, 20)T is the background amino acid frequency vector for column j, the parameter α specifies the expected background ‘contamination’ at pattern positions in the foreground, and δAj is a vector that specifies the pattern residues at position j. At the positions defined by C, we require that the pattern match a consensus residue that is derived from the seed sequences and that, in this way, helps define the foreground partition. Note that, at non-pattern positions, the vector θj corresponds to the overall (foreground and background) composition. The first two terms on the right hand side of Equation (1) correspond to the logarithm of the likelihood. The third through sixth terms correspond to the logarithm of the product of the prior probabilities (which are defined as for Equation 2 below).
2.2. A Function-based Alphabet
Presumably the most important feature discriminating between two functionally divergent categories of proteins is whether the foreground residues at pattern positions can perform a critical function that the corresponding background residues cannot. I capture this feature by collapsing the 20 letter amino acid alphabet A at each position j down to a two-letter alphabet consisting of a functional residue set Aj and a complementary non-functional residue set . The vector A defines an array of k such functional residue sets: one Aj for each position in the alignment. In keeping with the minimum description length principle (Grunwald, 2007), this allows us to focus on the relevant properties of the sequence data, namely, whether or not residues at position j can perform an implicit function. Moreover, this has the added benefits of increasing the statistical power by decreasing the number of parameters and of speeding up the sampler.
In order to sample pattern residue sets in this way, for each amino acid residue y, I define a set of permissible functional residue sets as those subsets of the alphabet that contain y along with zero or more additional residues with positive (non-integer) BLOSUM62 (Henikoff and Henikoff, 1992) log-odds scores both with y and with each other; that is, as:
(This provides a statistical basis for defining biochemically similar residues because, within related proteins, such residue pairs occur more often than expected by chance.) Hence, for differentiating columns I require that Aj ∈ A(yj) where yj is the seed alignment consensus residue for column j. For example, if yj = T then A(yj) = {{T}, {S,T}, {T,N}, {S,T,N}}). For non-differentiating columns Aj = Ø. Moreover, the column indicator variable is redefined as
This allows us to define independent priors for A (and indirectly for C at the same time) as (product categorical distributions), where:
where 0 < q < 1; κ is a normalizing constant; and ‖Aj‖ denotes the number of residue sets in A(yj) with cardinality |Aj|. For example, if yj = V,
and Aj = {V,L}, then ‖Aj‖ =3 because there are three sets in A(yj) with a cardinality of 2. Formulated in this way, ρAf is geometrically down-weighted based on |Aj| and linearly down-weighted based on ‖Aj‖. The former disfavors the inclusion of additional residues to the pattern set whenever those residues are only marginally elevated in the foreground relative to the background. The latter spreads out the prior probability equally over all residue sets having the same cardinality. By default q = 0.5 ; lowering or raising the value of q increases or decreases, respectively, the prior probability associated with larger residue sets.
Thus the logarithm of the joint distribution of all the variables for this modified scBPPS model is log P(X, R, Θ, α, A) =
| (2) |
where the variable C depends entirely on the variable A and where the priors for α, Θ and R are defined as:
2.3. The mcBPPS model
The modified scBPPS sampler is generalized into the mcBPPS sampler by introducing the notion of a hyperpartition, as follows. Consider an N-fold partitioning of the input sequences into disjoint sets, which we denote as the N-dimensional vector S. Thus each Sz corresponds to a distinct, functionally-specialized subgroup (within the protein class) that is ‘labeled’ through association with a (user-defined) seed alignment, from which is derived a consensus sequence as required for the Bayesian statistical formulation. Based on these sets, a hyperpartition H is defined as a length M array, each element of which (1≤h≤M) corresponds to a 3-tuple denoting a tripartitioning of the set indices 1≤z≤N such that the foreground, background and “non-participating” sequence sets correspond to
The multiple categories are modeled by adding an extra dimension to the variables R, C, Θ, α and A and by redefining R (based on S and H) such that otherwise Rh,i = 0, and by redefining the prior for R based on the priors for S, which are modeled as independent categorical distributions:
where, by default, σSz = N−1 for all z (uniform distribution).
The statistical model. To formulate the mcBPPS statistical model, we first define a “global background model” (GBM), for which the joint distribution of the sequence data and of an array of 20-dimensional (i.e., non-collapsed alphabet) residue frequency vectors Θ′ is defined (logarithmically) by . Note that this GBM independently models the residue frequencies observed in each column of the alignment, but not how those residues are distributed among the N sequence sets—which, instead, is captured by the following pattern-partition models (PPMs). These PPMs are based on a conditional version of Equation (2) that uses a collapsed alphabet and which is denoted as P(Rh, Θh, αh, Ah, H, S| Uh) where Uh ≡ R¬h, Θ¬h, α¬h, A¬h, Θ′ denotes the conditional universe of discourse and where the ¬h subscripts indicate that the corresponding vector includes every element except the h-th element. Note, however, that we cannot compute a complete data log-likelihood by summing the data log-likelihoods for the GBM and the PPM inasmuch as this doubly models the residue frequencies at each position. Instead, we add the data log-likelihood ratio (LLR) of the PPM versus the same PPM except where Θh is defined as for non-differentiating columns; that is, we add: LLRh(X|Rh, Θh, αh, Ah, Uh, H, S) =
| (3) |
where the function
indicates the participating sequences for the h-th PPM. For differentiating columns denotes an alternative residue frequency vector for column j that is computed using the same collapsed two-residue alphabet, but with all the foreground sequences merged in with the background partition. For non-differentiating columns (i.e., where Ch,j = 0) Ah,j = Ø, so that the residue frequency vectors are 1-dimensional with α = 1, and therefore, using the convention (based on continuity) that 0log 0 = 0, non-differentiating columns contribute nothing to Equation 3. As a result, this LLR adjusts the GBM log-likelihood upward or downward proportional to how likely or unlikely it is that the sequence data harbors each pattern-partition pair.
The preceding discussion focuses on the relationship between the GBM and a specific PPM, but we also need to ensure that there is no overlap between different PPMs—that is, that we can justify treating all the differentiating columns as statistically independent. This is the case for PPMs whose pattern positions are distinct or whose participating sequence sets are disjoint. In addition, given a collapsed alphabet, two or more models of a shared differentiating column can be treated as approximately independent under the following conditions: (i) the PPMs’ foreground or background partitions are disjoint; (ii) their pattern residue sets are disjoint; or (iii) for each pair of PPMs, both the foreground partition and the pattern residue set of one PPM are proper subsets of those of the other PPM. Satisfying the column independence conditions in this way leads to a dependency between PPMs, which is modeled by enumerating, for each column, every permissible combination of categories and of collapsed alphabets satisfying these conditions and then defining (uniform) priors for each (within p(A)) and setting the priors for prohibited combinations to zero. The resulting number of combinations is large, but manageable (due to consensus-imposed pattern restrictions); however, the formulation is too complicated to be helpful here. To avoid overtraining and to speed up the sampler, a maximum number of pattern positions for each category h can be specified; this is straightforward to model using appropriately-modified indicator variables.
Given these restrictions, we estimate the multiple-category log-likelihood as: log P(X, R, Θ, α, A, H, S) =
| (4) |
where the logs of the PPM prior probabilities (the last 5 terms) ensure that the adjusted distribution sums to 1. We can also compute a multiple-category (mc)-LLR by subtracting from Equation (4) the value obtained when all of the columns are non-differentiating. Due to the conservative nature of the Bayesian formulation, this mc-LLR provides a measure of significance: if the aligned input sequences are randomly shuffled within each column, then this value is expected to be negative—unless, that is, all of the PPMs likewise lack differentiating columns, in which case it is zero.
Hyperpartition restrictions. When creating a hyperpartition several restrictions are imposed. First, we require inclusion of an ‘aberrant sequence set’ (denoted as S0) that contains a large number of random prior pseudocounts and that serves as the background for the largest foreground partition(s) (e.g., the root in Fig. 2). This allows the sampler to eliminate unrelated sequences, pseudogene products and other non-functional or erroneous sequences from the analysis. Second, the remaining partition assignments must be either derived from a phylogenetic tree (as in Fig. 2) or else arbitrarily defined based on two conditions: (i) Each of the sets (except S0) must be assigned to at least one foreground partition. And (ii) no two sets can share identical foreground assignments (as this would prohibit the sampler from discriminating between sets). An analysis based on an arbitrarily-defined hyperpartition (Table 2) is illustrated below.
Table 2.
Hyperpartition for AAA+ domains.
| Category | Subgroup: | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| + | − | − | − | − | o | + | + | + | − | o | − | − | + | + | o | o | RfcA | Clamp Loaders |
| + | − | − | − | − | o | + | + | + | − | o | − | + | − | + | o | o | Ctf18 | |
| + | − | − | − | − | + | + | + | + | + | − | + | o | o | − | − | o | RfcBCD | |
| + | − | − | − | − | + | + | + | − | + | + | − | o | o | o | − | o | RfcE | |
| + | − | − | − | − | o | + | o | o | o | o | o | o | o | o | o | o | MiscRfc | |
| + | − | − | − | − | + | o | − | o | o | o | o | o | o | o | + | + | γ | |
| o | − | − | − | − | + | o | o | o | o | o | o | o | o | o | + | − | δ′ | |
| + | − | − | − | + | o | o | o | o | o | o | o | o | o | o | o | o | RuvBlike | |
| + | − | − | + | − | − | − | o | o | o | o | o | o | o | o | o | o | ClpLon | |
| + | − | + | − | − | − | − | o | o | o | o | o | o | o | o | o | o | NtrClike | |
| + | + | − | − | − | − | − | o | o | o | o | o | o | o | o | o | o | AAA | |
| + | − | − | − | − | − | − | o | o | o | o | o | o | o | o | o | o | MiscAAA+ | |
| − | o | o | o | o | o | o | o | o | o | o | o | o | o | o | o | o | Random | |
| 1 | 3 | 5 | 7 | 9 | 11 | 13 | 15 | 17 | ||||||||||
2.4. Sampling and optimization strategies
Instead of directly sampling sequences between the foreground and background partitions, as for the scBPPS sampler, the mcBPPS sampler moves sequences between sets (i.e., the Sz) and, as a result, indirectly changes the foreground and background partitions for each category as specified by the hyperpartition. With minor modifications to account for sampling between partitions indirectly in this way and for multiple categories, the mcBPPS sampling strategies are as described for the scBPPS sampler (Neuwald et al., 2003)—except, that is, for the following optimization strategy for functional residue sets.
Optimization of Aj. Because the functional residue set Aj at each position in each PPM is unknown to us a priori, these need to be inferred from the data. Conditional on all the other variables we can determine the relative probability of the observed residues in an aligned column. Since we are working on one column and one category at a time, we ignore the indicators j and h for notational clarity. Hence, the probability associated with a column residue composition vector θ = (θf, θn) (where and θn = 1 − θf) is given by:
where n1,f,n1,n and n0,f,n0,n are the numbers of functional and non-function residues in the foreground and background, respectively. Based on this formulation, we can conduct the following Metropolis algorithm:
Randomly select an alternative functional residue set A′ ∈ (A(y) ∪ {Ø}) ∧ A′ ≠ A (where y is the consensus residue at that position) and propose to change A to A′;
- Accept the proposal with probability:
Note that for non-differentiating columns A = Ø and n1,f = n1,n = 0.
Sampling versus optimization. The mcBPPS sampler has two applications. The first is to sample a set of random variables from the posterior probability distribution, where each variable is a length M array of pattern-partition pairs. Such a sample can be used to estimate both the predictive probability that a specific protein belongs to a specific subgroup (with implicit shared functional and/or mechanistic features) and the predictive probability that specific pattern residues are distinguishing (i.e., functionally-critical) features of these subgroups. The focus here, however, is on a second application, namely to optimally define the functionally-divergent categories (i.e., the partition assignments) and the corresponding pattern residues. The sampler applies three heuristics to converge on such an optimum more rapidly. First it initializes each of the Sz sets to contain those input sequences with the best pair-wise similarity score against the consensus sequence for that set. Second, based on these initialized sets and the corresponding partitions, it computes, for each category, a ‘seed pattern’ based on those residues that are both prevalent in the foreground but rare in the background; this is done by identifying functional residue sets that maximize the relative entropy—an information theoretical measure of the ‘distance’ between foreground and background distributions. Finally, simulated annealing (Kirkpatrick et al., 1983) is applied during convergence to ‘drop into’ a nearby (ideally global) optimum (see Fig. S14 in the supplementary file).
2.5. Creating a hyperpartition with and without user-supplied information
To create a hyperpartition for a major protein class, several strategies can be used. One strategy is to rely on web resources, such as the NCBI conserved domain database (CDD) (Marchler-Bauer et al., 2002), which provides both a curated multiple alignment of representative subgroups and a corresponding tree (in Newick format) reflecting the (presumed) evolutionary relationships between subgroups. An NCBI CDD curated alignment can be used as input to the MAPGAPS program (Neuwald, 2009c) in order to obtain an input alignment for the mcBPPS sampler. Note, however, that one or more of the relationships represented in the corresponding phylogenetic tree may fail to correspond to significant co-conserved patterns; in such cases, the mcBPPS sampler will fail to converge on a contrast alignment with a positive log-likelihood score, which it is programmed to reject. An alternative strategy is to run the scBPPS sampler to identify prominently co-conserved pattern-partition pairs prior to constructing a mcBPPS hyperpartition. Multilevel hierarchical relationships can be identified through recursive application of the scBPPS sampler to the foreground or background alignment from a previous scBPPS analysis. As a third strategy, the mcBPPS sampler includes an option where it will create a starting hyperpartition automatically—that is, in the absence of user-supplied seed sequences and a hyperpartition. (For an example of such automatically generated output for Rossmann fold proteins, see the supplementary file.) In this case, the mcBPPS sampler first searches for small sets of seed sequences in the input alignment; within each seed set the sequences both are selected from distinct phyla and are closely related to each other. (For this, the input alignment needs to include the phylum and kingdom for each sequence.) It then creates a simple tree consisting of a root node corresponding to all of the proteins in the class and one leaf node for each of the seed alignments. From this tree it creates a starting hyperpartition. This automated procedure can be applied recursively in order to construct more complex trees and corresponding hyperpartitions, which can then be edited by the user. Again, the sampler will reject nodes in the tree (and the corresponding contrast alignments) with LLR’s (Equation 3) below a specified conservative cutoff (100 nats by default). (The information is in nats because LLRs are computed as natural logarithms.) The example presented in section 3.2 below, illustrates how to design a hyperpartition based on a set of biochemical questions.
3. APPLICATION
The mcBPPS sampler’s biological relevance and performance is illustrated here through analyses of three major protein classes, P-loop GTPases (Leipe et al., 2002), AAA+ domains (Neuwald et al., 1999) and helicases (Abdelhaleem, 2010). In order to facilitate comparison with the scBPPS sampler and to avoid burying novel biological findings within this (statistically-oriented) paper’s supplemental files, two of these analyses focus on proteins that we have studied in considerable detail previously: Ras-like GTPases (Neuwald, 2007b, 2009a, 2009b; Neuwald et al., 2003) and DNA clamp loader subunits (Neuwald, 2005, 2006a, 2006b, 2007a; Neuwald et al., 1999). For detailed descriptions and biological interpretations of these mcBPPS analyses, see the supplementary file and these previous publications. The mcBPPS sampler was also applied to an entirely new analysis of Arf and Arf-like GTPases resulting in novel biological findings. To bring these findings to the attention of the biological community, this analysis was recently published in Biology Direct, a rapid publication online journal that features an open review process. Readers are referred to this publication (Neuwald, 2010) for evaluations of the mcBPPS sampler’s biological relevance by three expert referees. Finally, the supplementary file reports another new (albeit preliminary) mcBPPS analysis of superfamily 1 and 2 helicases (Abdelhaleem, 2010); an in-depth analysis will be published separately (Neuwald, in preparation). For an evaluation of the sampler’s convergence behavior, its robustness when confronted with variable input and the choice of tuning parameters in prior distributions, see the supplementary file.
For such analyses, an accurate input alignment can be obtained in about an hour by performing a MAPGAPS (Neuwald, 2009c) search of the protein sequence databases using, as the query, a curated alignment of representative sequences within the protein class. Such curated alignments can be obtained from the NCBI CD database (ftp.ncbi.nih.gov/pub/mmdb/cdd/) or from other protein domain databases. For the analyses here, however, the curated alignments (except for the independently-generated PSI-BLAST GTPase alignment in Fig. S15 of the supplementary file) were obtained as described in Methods. These analyses took about one to two hours to run on a 64-bit Linux workstation. Unlike the helicase hyperpartition, which was derived from the corresponding phylogenetic tree, the hyperpartitions for P-loop GTPases and AAA+ domains were designed to address specific questions regarding the proteins of interest. An auxiliary program (see Methods) was applied to the sampler’s output to generate 3D-visualization scripts for structural analysis of pattern residues within available structures. The Ras-like GTPase and DNA clamp loader analyses are summarized as follows.
3.1. P-loop GTPases
Given an input alignment of 66,386 unique P-loop GTPase sequences, the mcBPPS sampler searched for optimal pattern-partition pairs based on the hyperpartition in Table 1. The pattern-partition pair in column 1 distinguishes well-defined P-loop GTPases from related sequences corresponding to pseudogene products, non-functional proteins, or otherwise aberrant sequences. The background partition includes 30,000 random sequences (as prior pseudocounts) for this category—which thus serve as a ‘sink’ for eliminating sequences that would otherwise obscure the overall analysis. Columns 2 and 3 correspond to two major divergent subclasses, the SIMIBI and TRAFAC GTPases (Leipe et al., 2002), respectively. Columns 4–6 correspond to three divergent groups within the TRAFAC subclass (Leipe et al., 2002), namely Era/Obg, translation elongation factor-related (EF-like) GTPases and Ras-like GTPases. Columns 7–15 correspond to divergent subgroups of Ras-like GTPases. The hyperpartition in Table 1 also includes two miscellaneous subgroups: MiscRasLike and MiscTRAFAC; these accommodate sequences that generally conserve the canonical features of that particular “supergroup”, but that fail to conserve features of the explicitly-modeled subgroups within that supergroup.
Table 1.
Hyperpartition for P-loop GTPases.
| Category1 | Subgroup2: | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| + | - | + | − | − | + | − | − | − | − | − | − | + | + | Galpha | Ras-Like GTPases | TRAFAC subclass |
| + | − | + | − | − | + | − | − | − | − | − | + | − | + | Arl | ||
| + | − | + | − | − | + | − | − | − | − | + | − | − | + | Sar | ||
| + | − | + | − | − | + | − | − | − | + | − | − | − | − | Rab | ||
| + | − | + | − | − | + | − | − | + | + | − | − | − | − | Ran | ||
| + | − | + | − | − | + | − | + | − | + | − | − | − | − | RhoLike | ||
| + | − | + | − | − | + | + | − | − | o | − | − | − | − | Ras | ||
| + | − | + | − | − | + | − | − | − | o | − | − | − | − | MiscRasLike | ||
| + | − | + | − | + | − | o | o | o | o | o | o | o | o | EF-like | ||
| + | − | + | + | − | − | o | o | o | o | o | o | o | o | Era/Obg | ||
| + | − | + | − | − | − | o | o | o | o | o | o | o | o | MiscTRAFAC | ||
| + | + | o | − | − | o | o | o | o | o | o | o | o | o | SIMIBI | ||
| − | − | − | − | − | o | o | o | o | o | o | o | o | o | Random | ||
| 5 | ||||||||||||||||
Each column corresponds to a functionally-divergent category. The symbols ‘+’, ‘−’ and ‘o’ indicates that the subgroup in that row is assigned to that column’s foreground, background, and non-participating partitions, respectively.
Each row corresponds to a protein subgroup; miscellaneous subgroups are italicized.
For each non-miscellaneous subgroup the sampler outputs a multi-level “hierarchical” alignment consisting of one contrast alignment for each column in which that subgroup has been assigned to the foreground partition. Each of these contrast alignments highlights, within that subgroup’s seed alignment, those conserved residues that most distinguish the foreground from the corresponding background partition. Fig, 3 shows a 3-tier hierarchical alignment for Rab GTPases corresponding to columns 3, 6 and 10 of Table 1. The patterns associated with these three categories correspond to three structural features (Fig. 4): (i) the guanine nucleotide-binding pocket (Vetter and Wittinghofer, 2001); (ii) a charge-dipole pocket, formation of which is highly correlated with formation of an unusual, outward-directed switch II helix (Neuwald, 2009a); and (iii) a ‘glycine brace’ (Neuwald, 2009b) proposed to stabilize guanine-nucleotide binding loop hinge points. These alignments corroborate earlier analyses (Neuwald, 2007b, 2009a, 2009b) with two exceptions: due to correction of previous misaligned switch I regions, the middle alignment in Fig. 3 reclassifies—from the Ras-like to the TRAFAC categories—a switch I threonine that coordinates with the GTP-bound Mg++ ion (Thr61 in Fig. 3) and highlights one additional residue, a glycine (Gly63 in Fig. 3) that appears to serve as a switch I hinge point. Thus this mcBPPS analysis identifies these (and other) Ras-like GTPase distinguishing features more rapidly, precisely and directly than our previous approach, which required multiple scBPPS analyses in conjunction with various ‘intervention strategies’.
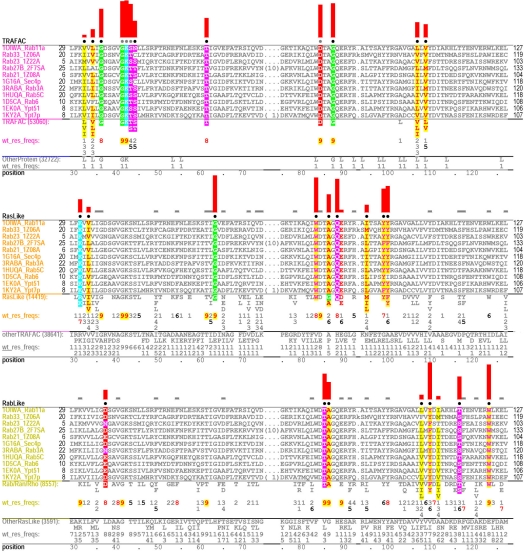
Figure 3.
Contrast alignments for Rab GTPases. The seed alignment for the Rab subgroup (Table 1, row 4) is highlighted in three ways to reveal patterns characteristic of three functionally-divergent categories for which Rab is in the foreground: top, patterns distinguishing TRAFAC class from other proteins (column 3 in Table 1); middle, patterns distinguishing Ras-like from other TRAFAC GTPases (column 6); and bottom, patterns distinguishing Rab, Ran and Rho from Arf/Arl/Gα GTPases (column 10). Characteristic foreground and background residues at each position are shown below each alignment and, directly below these, corresponding frequencies are given in integer tenths. (The actual alignments are too large to show every sequence, of course.) A ‘7’, for example, indicates that the corresponding residue occurs in 70–80% of the sequences. The histograms above highlighted columns quantify selective pressures imposed on pattern residues. See the supplementary file for the full size version of the alignment and for detailed descriptions and interpretations.
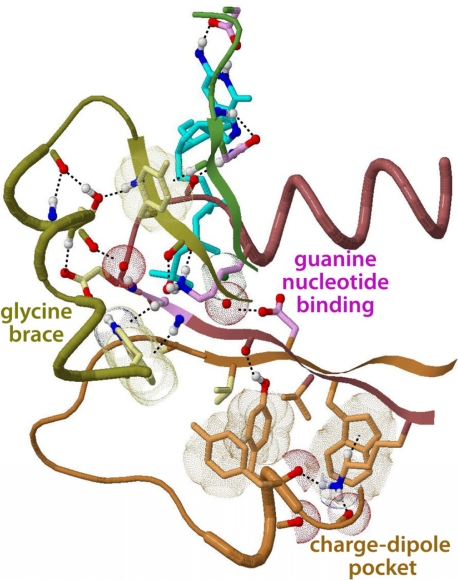
Figure 4.
Structural locations of pattern residues within Rab11 bound to a GTP analog (Pasqualato et al., 2004). Color scheme: residues with magenta-, orange- and yellow-colored side-chains correspond to the top, middle and bottom alignments in Fig. 3, respectively. In this structure, the Ras-like residues (orange side-chains) form a ‘charge-dipole pocket’ (Neuwald, 2009a) whereas the Rab/Rho/Ran-associated residues (yellow side-chains) form a ‘glycine brace’ (Neuwald, 2009b).
3.2. DNA clamp loaders
Choosing a specific hyperpartition configuration also depends on the various subgroups’ biochemical properties, which do not necessarily follow their phylogeny. A phylogenetic tree is typically determined based on overall sequence similarity, whereas sub-regions within each of the corresponding proteins may evolve in different ways. As a result, subsets of proteins from different clades can, in principle, share certain biochemical properties, presumably inherited from an ancestor of both clades, but lost in other members of both clades due to a relaxation of selective constraints upon those other members. This is illustrated by an analysis of AAA+ domains with a focus on eukaryotic Replication Factor C (RFC) DNA clamp loaders (see Fig. 5, the hyperpartition in Table 2 and the supplementary file).
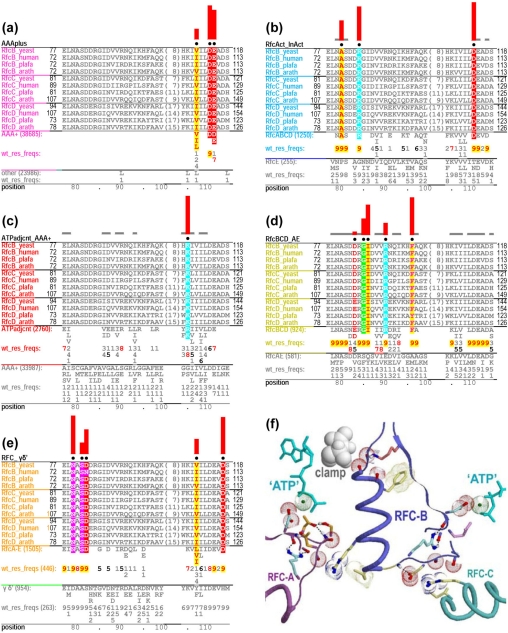
Figure 5.
Contrast alignments and corresponding structural features for RFC-B, -C and -D subunits. (a)–(e) Contrast alignments corresponding to columns 1, 9, 6, 12, and 8 of Table 2, respectively. The RFC-BCD seed alignment used throughout spans the region from the ATP binding site to just beyond a helix whose C-terminus binds to the DNA clamp. (f) The structure of the corresponding region within the RFC-complex bound to the clamp and to an ATP analog (Bowman et al., 2004). Residue side-chains are colored to match the color of the corresponding alignment’s leftmost column (which lists sequence identifiers). For clarity, some pattern residues are not shown (e.g., those highlighted in (e) for RFC-B). See the supplementary file for the full length alignments and detailed structural and functional interpretations.
RFC DNA clamp loaders (O’Donnell and Kuriyan, 2006) consist of five evolutionarily-related, functionally-specialized subunits (denoted A, B, C, D and E). Biochemically, RFC-ABCD are all active ATPases whereas RFC-E is not, and RFC-BCDE all interact with the ATP-binding site of an adjacent RFC subunit, whereas RFC-A does not. Such sequence similarities and differences reflect functional similarities and differences that, presumably, are associated with each subunit’s specialized role within the clamp loader complex. Based on this information, we construct a hyperpartition that addresses the following questions: Which RFC residues: (i) Catalyze ATP hydrolysis? (Compare all AAA+ ATPases with other proteins; column 1 in Table 2.) (ii) Directly couple ATP hydrolysis to clamp loading? (Compare RFC-ABCD, which are active ATPases, with RFC-E, which is not; column 9.) (iii) Trans-activate ATP hydrolysis? (Compare RFC-BCDE, which contact an adjacent ATPase subunit, with RFC-A, which does not; column 10.) (iv) Differentiate RFC subunits from active bacterial clamp loader subunits? (Compare RFCs with γ; column 8.) (v) Differentiate all trans-acting clamp loader subunits from all other AAA+ subunits? (Compare trans-acting clamp loader subunits with other AAA+ subunits; column 6.) Table 2 addresses similar questions regarding bacterial clamp loader γ and δ’ subunits, which were analyzed along with the RFC subunits (see the supplementary file). Because RuvB-like AAA+ subunits are more closely related to clamp loader subunits than are other AAA+ subunits, these are modeled specifically (column 5) and in some cases are assigned to non-participating partitions so that they would not obscure important, co-conserved residues. Other major AAA+ subgroups (columns 2–4) are also modeled specifically so that the sampler can more readily distinguish these from clamp loader subunits.
Some of the sampler’s output is shown in Fig. 5. This reveals that RFC-BCD shares co-conserved residues both with RFC-A (because they all hydrolyze ATP) but not with RFC-E (Figs 5b and S6D), and with RFC-E (because they all trans-activate ATP hydrolysis) but not with RFC-A (Fig. S6E). Likewise, RFC-BCD share certain co-conserved residues with both RFC-A and RFC-E (Figs 5e and S6C), and with neither RFC-A nor RFC-B (Figs 5d and S6F). Moreover, all five RFC subunits conserve residues both in common with (Figs 5c and S6B) and distinct from (Figs 5e and S6C) the corresponding bacterial clamp loader subunits. Note that these diverse relationships cannot be represented by a tree. This single mcBPPS analysis rapidly, precisely and directly characterizes clamp loader AAA+ domains that were previously characterized only after much more involved scBPPS analyses (Neuwald, 2005, 2006a, 2006b, 2007a).
4. DISCUSSION
The mcBPPS sampler addresses the problem of obtaining statistical clues from vast amounts of sequence data regarding unknown biochemical phenomena as a starting point for biological discovery. Note that this is a fundamentally different problem from that addressed by “functional subtype” prediction (FSP) methods (Carro et al., 2006; Chakrabarti et al., 2007; Feenstra et al., 2007; Gu and Vander Velden, 2002; Hannenhalli and Russell, 2000; Kalinina et al., 2004; Lichtarge et al., 1996; Livingstone and Barton, 1996; Mihalek et al., 2004; Mirny and Gelfand, 2002; Pirovano et al., 2006; Ye et al., 2008). FSP methods aim to predict residue functions that are sufficiently well-understood to allow benchmarking (Capra and Singh, 2008; Chakrabarti and Panchenko, 2009); in particular, they typically are designed to detect residues directly involved in substrate specificity. Validation against benchmark data sets is, of course, important for such tools. However, some of the most interesting phenomena in Nature are those that initially defy prevailing paradigms. For this reason, the mcBPPS sampler lets the data itself reveal its most statistically striking properties without making assumptions about the types of residues to be identified. It is further distinguished from each of the FSP methods in at least several of the following respects: (i) It does not require that the input alignment be partitioned into divergent subsets beforehand; this is unlike many (Chakrabarti et al., 2007; Feenstra et al., 2007; Hannenhalli and Russell, 2000; Kalinina et al., 2004; Livingstone and Barton, 1996; Mirny and Gelfand, 2002; Pirovano et al., 2006; Ye et al., 2008), though not all (Carro et al., 2006; Gu and Vander Velden, 2002; Lichtarge et al., 1996; Mihalek et al., 2004) such methods. (ii) It has a rigorous statistical basis. One FSP method (Marttinen et al., 2006) is Bayesian-based, though it lacks a MCMC sampling component. (iii) It is designed for very large input alignments (of up to a million or more sequences). (iv) It can be used to estimate predictive probabilities for membership within each subgroup. (v) It automatically separates out unrelated and aberrant sequences. And (vi) it identifies multiple categories of patterns within individual proteins of interest and (vii) thereby introduces Bayesian multilevel modeling (Snijders and Bosker, 1999) to protein sequence analysis.
Although the scBPPS and mcBPPS samplers address similar problems, the mcBPPS sampler identifies functionally critical residues more precisely, is much faster, can address questions that neither the scBPPS sampler nor other programs are able to address and (in conjunction with auxiliary routines) can define and generate structurally-annotated protein domain profiles automatically. By setting up a stringent competition between categories for both pattern positions and sequences, both the conserved residues most distinctive of and the sequences within each category are more precisely defined. Protein evolution often leads to three or more functionally divergent groups: one group harboring certain ‘canonical’ features, another lacking them, and one or more groups that—due to further divergence—harbor some but not all of the canonical features. For example, the RFC clamp loader complex performs three distinct functions: one associated with DNA replication (Tsurimoto and Stillman, 1989), another with sister chromatid cohesion (Bermudez et al., 2003) and a third with DNA damage checkpoints (Ellison and Stillman, 2003; Majka and Burgers, 2003). As a result, the ancestral RFC-A subunit has further diverged functionally (and in sequence) into at least three distinct subunits, one for each of these cellular functions (Majka and Burgers, 2004); one of these is CTF-18 in Table 2. The mcBPPS sampler prevents such divergent features from obscuring an analysis (without having to identify and remove such sequences beforehand) by explicitly modeling divergent and miscellaneous RFC-A subgroups. Likewise, an input alignment is nearly certain to harbor a significant number of pseudogene products and other related, non-functional sequences. For example, among fifty naturally-expressed protein tyrosine phosphatase-related transcripts identified in humans, twelve (or 24%) were determined experimentally to be pseudogene products (Andersen et al., 2004). The mcBPPS sampler addresses this problem by explicitly modeling aberrant sequences.
The mcBPPS sampler generates multilevel contrast alignments that facilitate comparisons between various functionally-divergent categories. This allows the mcBPPS sampler to search for a single global optimum, whereas the scBPPS sampler could only identify more than one functionally-divergent category by searching for local optima in the posterior probability distribution. Likewise, by explicitly modeling all 20 amino acid residues the scBPPS sampler inadvertently disfavors certain biologically-relevant patterns. For example, if the pattern residues are acidic and the foreground is ‘contaminated’ with serine, whereas the background lacks serine, then the scBPPS statistical model imposes a penalty—even though this situation implies further (not less) divergence of the foreground from the background. The mcBPPS sampler’s improved statistical model avoids this problem.
The mcBPPS sampler’s speed is illustrated by the Ras-like GTPase and AAA+ clamp loader analyses described here. Our earlier scBPPS analyses of these proteins were performed over a period of several years and required: (i) Considerable data manipulation and processing (such as, for example, elimination of spurious sequences and pulling out specific sequence sets for subgroup-to-subgroup comparisons). (ii) Intervention strategies—such as guiding the sampler into suboptimal solutions (corresponding, for example, to specific subfamilies) rather than letting it converge on an optimal solution (corresponding, for example, to a superfamily). And (iii) time-consuming interactive structural analyses to interpret the biological relevance of pattern residues. By contrast, the mcBPPS sampler (with auxiliary structural routines) can quickly perform such analyses in parallel, obtain more clear-cut results and dramatically speed up the biological interpretation of those results. And, although not emphasized here, the mcBPPS sampler can provide predictive probabilities for alternative subgroup assignments and, in a similar manner, probabilistically assign functions based on co-classification with functionally-verified proteins.
The mcBPPS sampler constitutes a substantial step toward fully automating protein classification, structural/functional annotation, and the construction of subgroup-specific domain profiles. It can also annotate likely functionally-critical residues—and (in conjunction with other routines) their corresponding structural interactions—automatically, based on empirical, statistically-based criteria. For speed and sensitivity it will be increasingly important to search a query sequence against domain profiles rather than against millions (and soon billions) of individual protein sequences. Thus a principle application of the mcBPPS sampler is automated generation of annotated profiles for incorporation into the Pfam (Finn et al., 2008) and NCBI conserved domain (Marchler-Bauer et al., 2009) databases. Most importantly, when linked into web-based BLAST searches and into the NCBI Cn3D viewer, such annotated profiles will provide biologists with previously inaccessible molecular information regarding mysterious, as-yet-unidentified biochemical phenomena.
5. METHODS
5.1. Input alignments
The mcBPPS sampler requires an (ideally very high quality) multiple sequence alignment as input. This is typically accomplished using the MAPGAPS program (Neuwald, 2009c) (http://mapgaps.igs.umaryland.edu). MAPGAPS identifies and aligns database sequences that share significant similarity to at least one sequence within a manually-curated alignment, which serves as the query. (To ensure that seed sequences are selected from distinct phyla, taxonomic information can be added to a sequence database using another auxiliary program.) Using the query alignment as a template, it then multiply aligns the detected sequences with accuracy comparable to that of the query alignment (assuming that the query alignment adequately represents the various subgroups within the associated protein class). A good source for such template alignments is the NCBI conserved domain database (Marchler-Bauer et al., 2009). For the two analyses described above, however, we used our own, template alignments, which were curated with the help of our Bayesian and MAPGAPS multiple alignment procedures (Neuwald, 2009c; Neuwald and Liu, 2004; Neuwald et al., 1997). Together, these approaches facilitate the construction of accurate alignments containing vast numbers of sequences.
5.2. Other input files
The sequences within a curated alignment can also serve as seed sequences for the mcBPPS analysis. The seed sequence phylogenetic tree can be used to generate the corresponding hyperpartition (using an auxiliary program called ‘tree2hpt’). Alternatively, the user may curate both a set of seed sequences for each subgroup and a hyperpartition designed to ask specific questions regarding functionally divergent subgroups (as is illustrated by the GTPase and AAA+ analyses here).
5.3. Down weighting for sequence redundancy
The sampler deals with overrepresentation of certain sequences using the weighting scheme of (Henikoff and Henikoff, 1994), as implemented within the PSI-BLAST program (Altschul et al., 1997); to avoid rounding errors this was implemented using integer sequence weights, where the integers 1 to 100 correspond to weights of .01 to 1.0, respectively.
5.4. Structural analysis
The mcBPPS package includes an unpublished auxiliary routine termed the Structural Analysis of Residue Patterns (SARP) program. The SARP program takes as input several files generated by the mcBPPS program and a file that lists the locations of the corresponding structural coordinate files. (A MAPGAPS search of the NCBI pdbaa sequence database can be performed to detect related proteins of known structure.) The SARP program searches the structural coordinate files (up to ten thousand or more) for interactions involving pattern residues. The interactions include both classical (Baker and Hubbard, 1984) and weak (Toth et al., 2001; Wahl and Sundaralingam, 1997; Weiss et al., 2001) hydrogen bonds, aromatic-aromatic interactions (Burley and Petsko, 1985), and van der Waals contacts (determined based on a standard distance of 4.5 Å). The REDUCE program (Word et al., 1999) was used to attach hydrogen atoms prior to hydrogen bond determination. Residue interactions between subunits are assessed based on contact surface area (Lee and Richards, 1971). SARP uses heuristics to order the structures based on how likely they are to shed light on the roles of pattern residues; this ordering allows the user to examine the most informative structures first. The heuristic used to determine this ordering is based primarily on the number of hydrogen bonds between pattern residues weighted by the strength of the interaction. For example, strong hydrogen bonds have greater weight than weak hydrogen bonds. However this current heuristic is tentative, as I am actively exploring ways to improve this scoring scheme. SARP also outputs a list of pattern residue interactions for each structure; various auxiliary routines are used to convert these lists into scripts for various structural visualization programs; for the examples described here RasMol (Sayle and Milner-White, 1995) scripts were generated. Future versions of SARP will include an option to generate PyMOL (DeLano, 2002) scripts and 3D visualizations using Cn3D (Wang et al., 2000).
5.5. Annotated domain profiles
The mcBPPS program outputs an alignment for each of the subgroups modeled by the hyperpartition along with the corresponding canonical patterns. The SARP program reports the structurally most conserved 3D interactions between pattern residues across all of the structures searched; this flags the most informative pattern residues and interactions for structural annotation. Together these output files can be used to generate a structurally annotated conserved domain profile for each subgroup.
5.6. Program availability
C++ implementations of the mcBPPS and tree2hpt programs are freely available at http://chain.igs.umaryland.edu.
Supplementary File
6. REFERENCES
- Abdelhaleem M. Helicases: an overview. Methods Mol Biol. 2010;587:1–12. doi: 10.1007/978-1-60327-355-8_1. [DOI] [PubMed] [Google Scholar]
- Alberts B. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell. 1998;92(3):291–4. doi: 10.1016/S0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JN, Jansen PG, Echwald SM, Mortensen OH, Fukada T, Del Vecchio R, Tonks NK, Moller NP. A genomic perspective on protein tyrosine phosphatases: gene structure, pseudogenes, and genetic disease linkage. FASEB J. 2004;18(1):8–30. doi: 10.1096/fj.02-1212rev. [DOI] [PubMed] [Google Scholar]
- Baker EN, Hubbard RE. Hydrogen bonding in globular proteins. Prog Biophys Mol Biol. 1984;44(2):97–179. doi: 10.1016/0079-6107(84)90007-5. [DOI] [PubMed] [Google Scholar]
- Bermudez VP, Maniwa Y, Tappin I, Ozato K, Yokomori K, Hurwitz J. The alternative Ctf18-Dcc1-Ctf8-replication factor C complex required for sister chromatid cohesion loads proliferating cell nuclear antigen onto DNA. Proc Natl Acad Sci U S A. 2003;100(18):10237–42. doi: 10.1073/pnas.1434308100. Epub 2003 Aug 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman GD, O’Donnell M, Kuriyan J. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature. 2004;429(6993):724–30. doi: 10.1038/nature02585. [DOI] [PubMed] [Google Scholar]
- Burley SK, Petsko GA. Aromatic-aromatic interaction: a mechanism of protein structure stabilization. Science. 1985;229(4708):23–8. doi: 10.1126/science.3892686. [DOI] [PubMed] [Google Scholar]
- Capra JA, Singh M. Characterization and prediction of residues determining protein functional specificity. Bioinformatics. 2008;24(13):1473–80. doi: 10.1093/bioinformatics/btn214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carro A, Tress M, de Juan D, Pazos F, Lopez-Romero P, del Sol A, Valencia A, Rojas AM. TreeDet: a web server to explore sequence space. Nucleic Acids Res. 2006;34(Web Server issue):W110–5. doi: 10.1093/nar/gkl203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S, Bryant SH, Panchenko AR. Functional specificity lies within the properties and evolutionary changes of amino acids. J Mol Biol. 2007;373(3):801–10. doi: 10.1016/j.jmb.2007.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S, Panchenko AR. Ensemble approach to predict specificity determinants: benchmarking and validation. BMC Bioinformatics. 2009;10:207. doi: 10.1186/1471-2105-10-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. “The PyMOL Molecular Graphics System”. 2002. http://www.pymol.org.
- Ellison V, Stillman B. Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5′ recessed DNA. PLoS Biol. 2003;1(2):E33. doi: 10.1371/journal.pbio.0000033. Epub 2003 Nov 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feenstra KA, Pirovano W, Krab K, Heringa J. Sequence harmony: detecting functional specificity from alignments. Nucleic Acids Res. 2007;35(Web Server issue):W495–8. doi: 10.1093/nar/gkm406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feynman R. QED: The Strange Theory of Light and Matter. Princeton, NJ: Princeton University Press; 1985. [Google Scholar]
- Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2008;36(Database issue):D281–8. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunwald PD. The minimum description length principle. Boston: MIT Press; 2007. [Google Scholar]
- Gu X, Vander Velden K. DIVERGE: phylogeny-based analysis for functional-structural divergence of a protein family. Bioinformatics. 2002;18(3):500–1. doi: 10.1093/bioinformatics/18.3.500. [DOI] [PubMed] [Google Scholar]
- Hannenhalli SS, Russell RB. Analysis and prediction of functional sub-types from protein sequence alignments. J Mol Biol. 2000;303(1):61–76. doi: 10.1006/jmbi.2000.4036. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A. 1992;89:10915–9. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG. Position-based sequence weights. J Mol Biol. 1994;243(4):574–8. doi: 10.1016/0022-2836(94)90032-9. [DOI] [PubMed] [Google Scholar]
- Kalinina OV, Mironov AA, Gelfand MS, Rakhmaninova AB. Automated selection of positions determining functional specificity of proteins by comparative analysis of orthologous groups in protein families. Protein Sci. 2004;13(2):443–56. doi: 10.1110/ps.03191704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick S, Gelatt CD, Vecchi MP. Optimization by simulated annealing. Science. 1983;220:671–680. doi: 10.1126/science.220.4598.671. [DOI] [PubMed] [Google Scholar]
- Lee B, Richards FM. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971;55(3):379–400. doi: 10.1016/0022-2836(71)90324-X. [DOI] [PubMed] [Google Scholar]
- Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317(1):41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- Lichtarge O, Bourne HR, Cohen FE. An evolutionary trace method defines binding surfaces common to protein families. J Mol Biol. 1996;257(2):342–58. doi: 10.1006/jmbi.1996.0167. [DOI] [PubMed] [Google Scholar]
- Little RJA, Rubin DB. Statistical Analysis with Missing Data. 2nd ed. New York: Wiley-Interscience; 2002. [Google Scholar]
- Liu JS. Monte Carlo Strategies in Scientific Computing of Springer Series in Statistics. New York: Springer-Verlag; 2008. [Google Scholar]
- Livingstone CD, Barton GJ. Identification of functional residues and secondary structure from protein multiple sequence alignment. Methods Enzymol. 1996;266:497–512. doi: 10.1016/S0076-6879(96)66031-5. [DOI] [PubMed] [Google Scholar]
- Majka J, Burgers PM. Yeast Rad17/Mec3/Ddc1: a sliding clamp for the DNA damage checkpoint. Proc Natl Acad Sci U S A. 2003;100(5):2249–54. doi: 10.1073/pnas.0437148100. Epub 2003 Feb 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majka J, Burgers PM. The PCNA-RFC families of DNA clamps and clamp loaders. Prog Nucleic Acid Res Mol Biol. 2004;78:227–60. doi: 10.1016/S0079-6603(04)78006-X. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Liebert CA, Liu C, Lu F, Lu S, Marchler GH, Mullokandov M, Song JS, Tasneem A, Thanki N, Yamashita RA, Zhang D, Zhang N, Bryant SH. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37(Database issue):D205–10. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Panchenko AR, Shoemaker BA, Thiessen PA, Geer LY, Bryant SH. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002;30(1):281–3. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marttinen P, Corander J, Toronen P, Holm L. Bayesian search of functionally divergent protein subgroups and their function specific residues. Bioinformatics. 2006;22(20):2466–74. doi: 10.1093/bioinformatics/btl411. [DOI] [PubMed] [Google Scholar]
- Mihalek I, Res I, Lichtarge O. A family of evolution-entropy hybrid methods for ranking protein residues by importance. J Mol Biol. 2004;336(5):1265–82. doi: 10.1016/j.jmb.2003.12.078. [DOI] [PubMed] [Google Scholar]
- Mirny LA, Gelfand MS. Using orthologous and paralogous proteins to identify specificity-determining residues in bacterial transcription factors. J Mol Biol. 2002;321(1):7–20. doi: 10.1016/S0022-2836(02)00587-9. [DOI] [PubMed] [Google Scholar]
- Neuwald AF. Evolutionary clues to eukaryotic DNA clamp-loading mechanisms: analysis of the functional constraints imposed on replication factor C AAA+ ATPases. Nucleic Acids Res. 2005;33(11):3614–28. doi: 10.1093/nar/gki674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald AF. Bayesian shadows of molecular mechanisms cast in the light of evolution. Trends Biochem Sciences. 2006a;31(7):374–382. doi: 10.1016/j.tibs.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Neuwald AF. Hypothesis: bacterial clamp loader ATPase activation through DNA-dependent repositioning of the catalytic base and of a transacting catalytic threonine. Nucleic Acids Res. 2006b;34(18):5280–90. doi: 10.1093/nar/gkl519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald AF. The CHAIN program: forging evolutionary links to underlying mechanisms. Trends Biochem Sciences. 2007a;32(00):487–493. doi: 10.1016/j.tibs.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Neuwald AF. Gα-Gβγ dissociation may be due to retraction of a buried lysine and disruption of an aromatic cluster by a GTP-sensing Arg-Trp pair. Protein Science. 2007b;16(11):2570–2577. doi: 10.1110/ps.073098107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald AF. The charge-dipole pocket: a defining feature of signaling pathway GTPase on/off switches. J Mol Biol. 2009a;390(1):142–53. doi: 10.1016/j.jmb.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald AF. The glycine brace: a component of Rab, Rho, and Ran GTPases associated with hinge regions of guanine- and phosphate-binding loops. BMC Struct Biol. 2009b;9:11. doi: 10.1186/1472-6807-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald AF. Rapid detection, classification and accurate alignment of up to a million or more related protein sequences. Bioinformatics. 2009c;25(15):1869–1875. doi: 10.1093/bioinformatics/btp342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald AF. Bayesian classification of residues associated with protein functional divergence: Arf and Arf-like GTPases. Biol Direct. 2010;5:66. doi: 10.1186/1745-6150-5-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9(1):27–43. [PubMed] [Google Scholar]
- Neuwald AF, Kannan N, Poleksic A, Hata N, Liu JS. Ran’s C-terminal, basic patch and nucleotide exchange mechanisms in light of a canonical structure for Rab, Rho, Ras and Ran GTPases. Genome Res. 2003;13(4):673–692. doi: 10.1101/gr.862303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald AF, Liu JS. Gapped alignment of protein sequence motifs through Monte Carlo optimization of a hidden Markov model. BMC Bioinformatics. 2004;5(1):157. doi: 10.1186/1471-2105-5-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald AF, Liu JS, Lipman DJ, Lawrence CE. Extracting protein alignment models from the sequence database. Nucleic Acids Research. 1997;25(9):1665–1677. doi: 10.1093/nar/25.9.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell M, Kuriyan J. Clamp loaders and replication initiation. Curr Opin Struct Biol. 2006;16(1):35–41. doi: 10.1016/j.sbi.2005.12.004. Epub 2006 Jan 11. [DOI] [PubMed] [Google Scholar]
- Pasqualato S, Senic-Matuglia F, Renault L, Goud B, Salamero J, Cherfils J. The structural GDP/GTP cycle of Rab11 reveals a novel interface involved in the dynamics of recycling endosomes. J Biol Chem. 2004;279(12):11480–8. doi: 10.1074/jbc.M310558200. [DOI] [PubMed] [Google Scholar]
- Pirovano W, Feenstra KA, Heringa J. Sequence comparison by sequence harmony identifies subtype-specific functional sites. Nucleic Acids Res. 2006;34(22):6540–8. doi: 10.1093/nar/gkl901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero PA, Arnold FH. Exploring protein fitness landscapes by directed evolution. Nat Rev Mol Cell Biol. 2009;10(12):866–76. doi: 10.1038/nrm2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayle RA, Milner-White EJ. RASMOL: biomolecular graphics for all. Trends Biochem Sci. 1995;20(9):374. doi: 10.1016/S0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- Snijders TAB, Bosker RJ. Multilevel analysis : an introduction to basic and advanced multilevel modeling. London; Thousand Oaks, Calif: Sage Publications; 1999. [Google Scholar]
- Toth G, Watts CR, Murphy RF, Lovas S. Significance of aromatic-backbone amide interactions in protein structure. Proteins. 2001;43(4):373–81. doi: 10.1002/prot.1050. [DOI] [PubMed] [Google Scholar]
- Tsurimoto T, Stillman B. Purification of a cellular replication factor, RF-C, that is required for coordinated synthesis of leading and lagging strands during simian virus 40 DNA replication in vitro. Mol Cell Biol. 1989;9(2):609–19. doi: 10.1128/mcb.9.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294(5545):1299–304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- Wahl MC, Sundaralingam M. C-H...O hydrogen bonding in biology. Trends Biochem Sci. 1997;22(3):97–102. doi: 10.1016/S0968-0004(97)01004-9. [DOI] [PubMed] [Google Scholar]
- Wang Y, Geer LY, Chappey C, Kans JA, Bryant SH. Cn3D: sequence and structure views for Entrez. Trends Biochem Sci. 2000;25(6):300–2. doi: 10.1016/S0968-0004(00)01561-9. [DOI] [PubMed] [Google Scholar]
- Weiss MS, Brandl M, Suhnel J, Pal D, Hilgenfeld R. More hydrogen bonds for the (structural) biologist. Trends Biochem Sci. 2001;26(9):521–3. doi: 10.1016/S0968-0004(01)01935-1. [DOI] [PubMed] [Google Scholar]
- Word JM, Lovell SC, Richardson JS, Richardson DC. Asparagine and glutamine: using hydrogen atom contacts in the choice of side-chain amide orientation. J Mol Biol. 1999;285(4):1735–47. doi: 10.1006/jmbi.1998.2401. [DOI] [PubMed] [Google Scholar]
- Ye K, Feenstra KA, Heringa J, Ijzerman AP, Marchiori E. Multi-RELIEF: a method to recognize specificity determining residues from multiple sequence alignments using a Machine-Learning approach for feature weighting. Bioinformatics. 2008;24(1):18–25. doi: 10.1093/bioinformatics/btm537. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.