Abstract
Background
Allergic contact dermatitis is an inflammatory skin disease that affects a significant proportion of the population. This disease is caused by an adverse immune response towards chemical haptens, and leads to a substantial economic burden for society. Current test of sensitizing chemicals rely on animal experimentation. New legislations on the registration and use of chemicals within pharmaceutical and cosmetic industries have stimulated significant research efforts to develop alternative, human cell-based assays for the prediction of sensitization. The aim is to replace animal experiments with in vitro tests displaying a higher predictive power.
Results
We have developed a novel cell-based assay for the prediction of sensitizing chemicals. By analyzing the transcriptome of the human cell line MUTZ-3 after 24 h stimulation, using 20 different sensitizing chemicals, 20 non-sensitizing chemicals and vehicle controls, we have identified a biomarker signature of 200 genes with potent discriminatory ability. Using a Support Vector Machine for supervised classification, the prediction performance of the assay revealed an area under the ROC curve of 0.98. In addition, categorizing the chemicals according to the LLNA assay, this gene signature could also predict sensitizing potency. The identified markers are involved in biological pathways with immunological relevant functions, which can shed light on the process of human sensitization.
Conclusions
A gene signature predicting sensitization, using a human cell line in vitro, has been identified. This simple and robust cell-based assay has the potential to completely replace or drastically reduce the utilization of test systems based on experimental animals. Being based on human biology, the assay is proposed to be more accurate for predicting sensitization in humans, than the traditional animal-based tests.
Background
Allergic contact dermatitis (ACD) is a common inflammatory skin disease characterized by eczema and recurrent episodes of itching [1]. The disease affects a significant proportion of the population, with prevalence rates of 7.2% to 18.6% in Europe [2,3], and the incidence is increasing due to repeated exposure to sensitizing chemicals. ACD is a type IV delayed-type hypersensitivity response caused mainly by reactive T helper 1 (Th1) and interferon (IFN)γ producing CD8+ T cells, at site of contact with small chemical haptens in previously exposed, and immunologically sensitized, individuals [4]. Dendritic cells (DC) in the epidermis initiate the immune reactions by responding to haptens bound to self-molecules subsequently activating T cell-mediated immunity.
The REACH (Registration, Evaluation, and Authorization of Chemicals) regulation requires that all new and existing chemicals within the European Union, involving approximately 30.000 chemicals, should be tested for hazardous effects [5]. As the identification of potential sensitizers currently requires animal testing, the REACH legislation will have a huge impact on the number of animals needed for testing. Further, the 7th Amendment to the Cosmetics Directive posed a ban on animal tests for the majority of cosmetic ingredients for human use, to be in effect by 2009, with the exceptions of some tests by 2013. Thus, development of reliable in vitro alternatives to experimental animals for the assessment of sensitizing capacity of chemicals is urgent. To date, no non-animal replacements are available for identification of skin sensitizing chemicals, instead the preferred assay is the mouse Local Lymph Node Assay (LLNA) [6], followed by the Guinea pig maximization test (GPMT) [7]. An in vitro alternative to these animal models should exhibit improved reliability, accuracy and importantly correlate to human reactivity.
DCs play key roles in the immune response by bridging the essential connections between innate and adaptive immunity. Upon stimulation, they can rapidly produce large amounts of mediators that affect chemotaxis and activation of other cells at the site of inflammation, and can selectively respond to various pathogens and environmental factors, by fine-tuning the cellular response through antigen-presentation. Thus, exploring and utilizing the immunological decision-making by DCs during stimulation with sensitizers, could serve as a potent test strategy for the prediction of sensitization.
Factors that complicate and impede the use of primary DCs as a test platform include adaptable phenotypes and specialized functions of different DC subpopulations, in addition to their wide and sparse distribution. Thus, the development of assays based on the predictability of DC function must rely on alternative cell types or mimics of in vivo DCs. For this purpose, a cell line with DC characteristics would be advantageous, as it constitutes a stable, reproducible and unlimited supply of cells. MUTZ-3 is an unlimited source of CD34+ DC progenitors. Upon differentiation, MUTZ-3 can acquire phenotypes comparable to immature DCs or Langerhans-like DCs [8], present antigens through CD1d, MHC class I and II and induce specific T-cell proliferation [9]. Differentiated MUTZ-3 can also display a mature transcriptional and phenotypic profile upon stimulation with inflammatory cytokines [10].
In this report, we present a novel test principle for the prediction of skin sensitizers. To simplify the assay procedures and increase reproducibility, we employed progenitor MUTZ-3 cells, without further differentiation, and subjected the cells to stimulation with a large panel of sensitizing chemicals, non-sensitizing chemicals, and controls. The transcriptional response to chemical stimulation was assessed by genome-wide profiling. From data analysis, a biomarker signature of 200 transcripts was identified, which completely separated the response induced by sensitizing chemicals vs. non-sensitizing chemicals and the predictive power of the signature was illustrated, using ROC curves. The biomarker signature includes transcripts involved in relevant biological pathways, such as oxidative stress, DC maturation and cytokine responses, which further could shed light on molecular interactions involved in the process of sensitization. In conclusion, we have identified a biomarker signature with potent predictive power, which we propose as an in vitro assay for the identification of human sensitizing chemicals.
Results
The cellular rationale of the in vitro cell culture system
DCs are essential immunoregulatory cells of the immune system demonstrated by their unique property to recognize antigen for the initiating of T cell responses, and their potent regulatory function in skewing immune responses. This makes them obvious targets for assay development. However, primary DCs constitute a heterogeneous and minor population of cells not suited for screening and the choice would be a human DC-like cell line, with characteristics compared to primary DCs. Since no leukemic cell line with DC-like properties has been reported [11], the generation of human DC-like cell lines relies on available myeloid leukemia cell lines. MUTZ-3 is a human acute myelomonocytic leukemia cell line with a potent ability to mimic primary human DCs [11]. Similar to immature primary DCs, MUTZ-3 progenitors express CD1a, HLA-DR and CD54, as well as low levels of CD80 and CD86 (Figure 1). The MUTZ-3 population also contains three subpopulations of CD14+, CD34+ and double negative cells, previously reported to be transitional differentiation steps from a proliferative CD34+ progenitor into a non-proliferative CD14+ DC precursor [8]. Consequently, constitutively differentiating progenitor MUTZ-3 cells were used as the basis for a test system.
Figure 1.
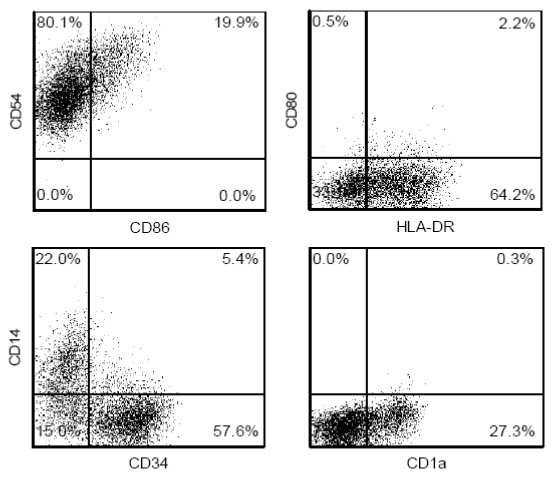
Phenotype of MUTZ-3 cells prior to stimulation with sensitizing and non-sensitizing chemicals. Cell surface expression levels of CD14, CD1a, CD34, CD54, CD80, CD86 and HLA-DR were assessed with flow cytometry. Gates were set to exclude debris and dead cells, and quadrants were established by comparing with relevant isotype controls. Results are shown from one representative experiment out of six.
CD86 surface expression in response to sensitizer stimulation
CD86 is the most extensively studied biomarker for sensitization to date, using e.g. monocyte derived dendritic cells (MoDCs) or human cell lines and their progenitors, such as THP-1, U-937 and KG-1. Thus, as a reference, cell surface expression of CD86 was measured with flow cytometry after 24 h stimulation, using 20 sensitizers and 20 non-sensitizers, as well as vehicle controls (Figure 2). CD86 was significantly up-regulated on cells stimulated with 2-aminophenol, kathon CG, 2-nitro-1,4-phenylendiamine, 2,4-dinitrochlorobenzene, 2-hydroxyethyl acrylate, cinnamic aldehyde, p-phenylendiamine, resorcinol, potassium dichromate, and 2-mercaptobenzothiazole. Hence, an assay based on measurement of a single biomarker, such as CD86, would give a sensitivity of 50% and a specificity of 100%. Consequently, CD86 cannot classify skin sensitizers, using a system based on MUTZ-3 cells.
Figure 2.
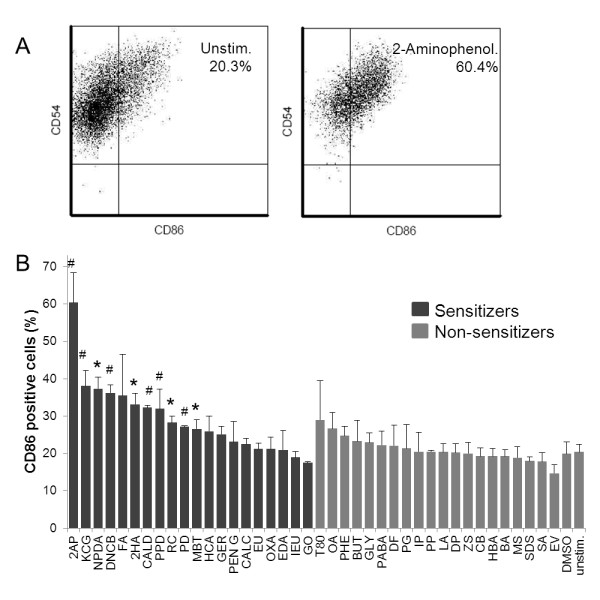
Changes in CD86 expression following stimulation with sensitizing and non-sensitizing chemicals. Cell surface expression levels of CD86 were monitored after stimulation with chemicals for 24 h. A). Chemical-induced up regulation of CD86, in terms of changes in frequency of positive cells, were determined by flow cytometry, as exemplified by the comparison of 2-aminophenol-stimulated cells (right dotplot) and unstimulated controls (left dot plot). Results are shown from one representative experiment out of three. Gates were set to exclude debris and dead cells, and quadrants were established by comparing with relevant isotype controls. B) Compilation of frequencies of CD86-positive cells after 24 h of stimulation. Statistical analysis was performed using Student's t test. *p < 0.05, # p < 0.01.
Analysis of the transcriptional profiles in chemically stimulated MUTZ-3 cells
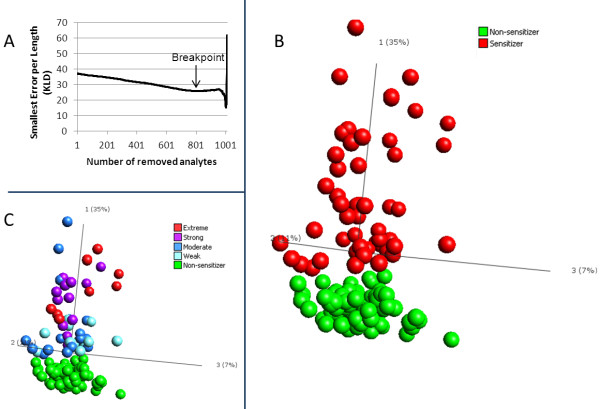
The genomic expression arrays were then used to test the same 20 sensitizers and 20 non-sensitizers, in triplicates. The vehicle controls, such as DMSO and distilled water, were included in twelve replicates. In total, a data set was generated based on 144 samples. RMA normalization and quality controls of the samples revealed that the oxazolone and cinnamic aldehyde samples were significant outliers and had to be removed, or they would have dominated the data set prohibiting biomarker identification (data not shown). In addition, one of the replicates of potassium permanganate had to be removed due to a faulty array. This left a data set consisting of 137 samples, each with data from measurements of 29,141 transcripts. In order to mine the data set for information specific for sensitizers vs. non-sensitizers, the software Qlucore Omics Explorer 2.1 was used, which enable real time principal component analysis (PCA) analysis. The input genes were at the same time sorted after desired criteria, i.e. sensitizers and non-sensitizers, based on ANOVA p-value selection. Two different ANOVA analyses were performed (Figure 3). First, Figure 3A and 3B show PCA plots based on 1010 transcripts with a p-value of ≤ 2.0 × 10-6, from a one-way ANOVA analysis, comparing sensitizing vs. non-sensitizing chemicals. As can be seen in Figure 3A, a clear discrimination can be made between the two groups, with non-sensitizers forming a condensed cloud in the lower part of the figure (green), while sensitizers stretch upwards in various directions (red). However, a complete separation is not achieved between the two groups at this level of significance. From Figure 3B, now colored according to stimulating agent, it is evident that one or more replicate of glyoxal, eugenol, hexylcinnamic aldehyde, isoeugenol, resorcinol, penicillin G and ethylendiamine grouped together with the control group. In addition, one replicate or more of the non-sensitizers tween 80, octanoic acid and phenol grouped closely with the sensitizers. Secondly, Figure 3C and 3D show PCA plots based on 1137 genes, with p-values ≤ 7.0 × 10-21, from a multi-group ANOVA analysis, comparing each individual stimulation. Identifying this large number of genes at this level of significance provided strong indications of the power in the data set. In Figure 3D, it is clear that the replicates group together, indicating high quality data. The triplicate samples of potassium dichromate have a discrete profile, which demonstrate a substantial impact of the cells compared to non-sensitizers. Furthermore, 2-hydroxyethyl acrylate, 2-aminophenol, kathon CG, formaldehyde, 2-nitro-1,4-phenylendiamine, 2,4-dinitrochlorobenzoic acid, p-phenylendiamine, 2-mercaptobenzothiazole, cinnamic alcohol and resorcinol have replicates that group together, separate from the negative group. Still, as can be seen in Figure 3C as well as in 3A, complete separation is not achieved with neither of the gene signatures of 1010 and 1137 genes both selected on p-values.
Figure 3.
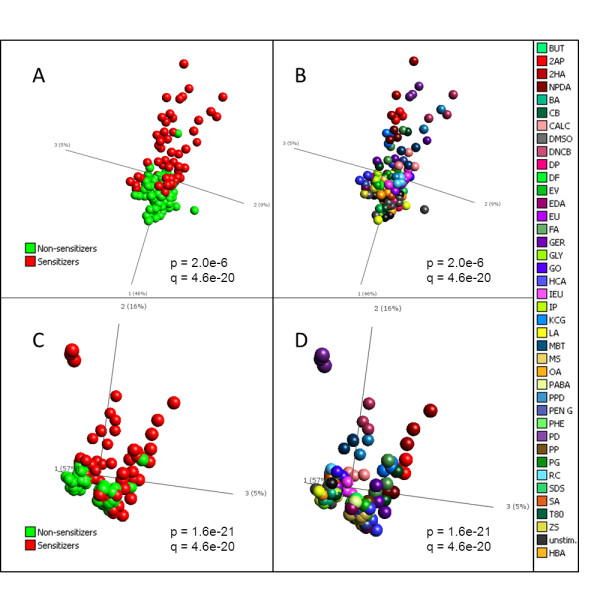
Principal component analysis of transcripts differentially expressed after chemical stimulation. mRNA levels in MUTZ-3 cells stimulated for 24 h with 20 sensitizing and 20 non-sensitizing chemicals were assessed with transcriptomics, using Affymetrix Human Gene 1.0 ST arrays. Structures and similarities in the gene expression data set were investigated, using principal component analysis (PCA) in the software Qlucore. A) PCA of genes differentially expressed in cells stimulated with sensitizing (red) versus non-sensitizing (green) chemicals (1010 genes identified with one-way ANOVA). B) PCA of genes differentially expressed in cells stimulated with sensitizing versus non-sensitizing chemicals (1010 genes), but now samples are colored by the compound used for stimulation. C) PCA of genes differentially expressed when comparing the different stimulations with 2-way ANOVA (1137 genes). Samples are colored according to sensitizing (red) and non-sensitizing (green) chemicals. D) PCA of genes differentially expressed when comparing the different stimulations with 2-way ANOVA (1137 genes), but now samples are colored by the compound used for stimulation. P, p-value from ANOVA. Q, p-value corrected for multiple hypothesis testing.
Backward elimination identifies genes with the most discriminatory power
Even though the data set contains genes with p-values down to 1 × 10-17, lowering the p-value cutoff did not achieve complete separation between sensitizers and non-sensitizers. Gene signatures entirely selected on p-values does not provide the best possible predictive power, since the information is per se not orthogonal. To further reduce the number of transcripts for a predictive biomarker signature, we employed an algorithm for backward elimination (Figure 4A). The algorithm removes genes one by one while taking into account not only the impact of genes individually, but how they perform collectively with the entire selected gene signature. For each gene eliminated, the Kullback-Leibler divergence (KLD) value is lowered, until a breakpoint is reached, at which point 200 genes remained. Continuing eliminating genes at this point causes the KLD to rise again, indicating that information is being lost (Figure 4A). Therefore, the 200 genes with lowest KLD value were selected for further analysis. PCA of the 200 analytes now revealed that they have the ability to completely separate sensitizers from non-sensitizers, indicating that these transcripts can be used as predictors for sensitizing properties of unknown samples (Figure 4B). Importantly, by coloring the samples in the PCA by their potency, according to LLNA, it is clear that potency can also be predicted (Figure 4C), as extreme and strong sensitizers tend to group further from the non-sensitizers, while moderate and extreme sensitizers group closer to non-sensitizers. The 200 genes are termed the "Prediction Signature" and their identities are listed in Table 1. In addition, the transcriptional profiles of the differentially expressed genes are presented in a heatmap (Figure 5).
Figure 4.
Identification and PCA analysis of Prediction Signature. A) The number of differentially expressed significant genes in cells stimulated with sensitizing versus non-sensitizing chemicals (1010 genes) was reduced, using Backward Elimination. The lowest KLD is observed after elimination of 810 analytes, referred to as the Breakpoint. The remaining 200 genes are considered to be the top predictors in the data set, and are termed Prediction Signature. B) Complete separation between sensitizers (red) and non-sensitizers (green) is observed with PCA of the Prediction Signature. C) Same PCA as in B, now with samples colored according to their potency in LLNA.
Table 1.
Prediction Signature
| Gene Title | Gene Symbol | Entrez Gene ID | Affymetrix HuGene 1.0 ST ID | Validation Call frequency (%) |
|---|---|---|---|---|
| 4-aminobutyrate aminotransferase | ABAT | 18 | 7993126 | 30 |
| abhydrolase domain containing 5 | ABHD5 | 51099 | 8079153 | 85 |
| alkaline ceramidase 2 | ACER2 | 340485 | 8154563 | 95 |
| ATP citrate lyase | ACLY | 47 | 8015460 | 85 |
| actin-related protein 10 homolog (S. cerevisiae) | ACTR10 | 55860 | 7974587 | 75 |
| ADAM metallopeptidase domain 20 | ADAM20 | 8748 | 7979927 | 35 |
| aldehyde dehydrogenase 18 fam., member A1 | ALDH18A1 | 5832 | 7935230 | 75 |
| aldehyde dehydrogenase 1 fam., member B1 | ALDH1B1 | 219 | 8155327 | 70 |
| anaphase promoting complex subunit 1 | ANAPC1 | 64682 | 8043349 | 55 |
| anaphase promoting complex subunit 5 | ANAPC5 | 51433 | 7967149 | 25 |
| ankyrin repeat, fam. A (RFXANK-like), 2 | ANKRA2 | 57763 | 8112596 | 100 |
| ADP-ribosylation factor GTPase activating protein 3 | ARFGAP3 | 26286 | 8076515 | 55 |
| Rho GTPase activating protein 9 | ARHGAP9 | 64333 | 7964436 | 75 |
| ankyrin repeat and SOCS box-containing 7 | ASB7 | 140460 | 7986433 | 65 |
| ATPase, H+ transporting, lysosomal 38 kDa, V0 subunit d1//ATPase, H+ transporting, lysosomal 38 kDa, V0 subunit d1 | ATP6V0D1//ATP6V0D1 | 9114//9114 | 8002041 | 10 |
| ATPase, H+ transporting, lysosomal 9 kDa, V0 subunit e1 | ATP6V0E1 | 8992 | 8110022 | 75 |
| ATPase, H+ transporting, lysosomal 50/57 kDa, V1 subunit H | ATP6V1H | 51606 | 8150797 | 100 |
| B-cell CLL/lymphoma 7A | BCL7A | 605 | 7959354 | 85 |
| bridging integrator 2 | BIN2 | 51411 | 7963289 | 80 |
| bleomycin hydrolase | BLMH | 642 | 8014008 | 15 |
| brix domain containing 1//ribosome production factor 2 homolog (S. cerevisiae) | BXDC1//RPF2 | 84154//84154 | 8062211 | 40 |
| chromosome 11 open reading frame 61 | C11orf61 | 79684 | 7952445 | 55 |
| chromosome 11 open reading frame 67//integrator complex subunit 4 | C11orf67//INTS4 | 28971//92105 | 7942783 | 50 |
| chromosome 12 open reading frame 57 | C12orf57 | 113246 | 7953564 | 40 |
| chromosome 13 open reading frame 18 | C13orf18 | 80183 | 7971486 | 50 |
| chromosome 15 open reading frame 24 | C15orf24 | 56851 | 7987172 | 50 |
| chromosome 19 open reading frame 46//alkB, alkylation repair homolog 6 (E. coli) | C19orf46//ALKBH6 | 163183//84964 | 8036242 | 30 |
| chromosome 19 open reading frame 54 | C19orf54 | 284325 | 8036956 | 95 |
| chromosome 1 open reading frame 174 | C1orf174 | 339448 | 7911897 | 40 |
| chromosome 1 open reading frame 183 | C1orf183 | 55924 | 7918552 | 85 |
| chromosome 20 open reading frame 111 | C20orf111 | 51526 | 8066402 | 65 |
| chromosome 20 open reading frame 24 | C20orf24 | 55969 | 8062326 | 20 |
| chromosome 3 open reading frame 62//ubiquitin specific peptidase 4 (proto-oncogene) | C3orf62//USP4 | 375341//7375 | 8087374 | 40 |
| chromosome 9 open reading frame 89 | C9orf89 | 84270 | 8156404 | 100 |
| coactivator-associated arginine methyltransferase 1 | CARM1 | 10498 | 8025766 | 60 |
| CD33 molecule | CD33 | 945 | 8030804 | 45 |
| CD86 molecule | CD86 | 942 | 8082035 | 45 |
| CD93 molecule | CD93 | 22918 | 8065359 | 50 |
| cytochrome c oxidase subunit VIIa polypeptide 2 like | COX7A2L | 9167 | 8051777 | 45 |
| corticotropin releasing hormone binding protein | CRHBP | 1393 | 8106418 | 45 |
| chondroitin sulfate N-acetylgalactosaminyltransferase 2 | CSGALNACT2 | 55454 | 7927146 | 90 |
| cytochrome P450, fam. 51, subfam. A, polypeptide 1 | CYP51A1 | 1595 | 8140864 | 85 |
| DDRGK domain containing 1 | DDRGK1 | 65992 | 8064601 | 60 |
| DEAD (Asp-Glu-Ala-As) box polypeptide 19A | DDX19A | 55308 | 7997059 | 95 |
| DEAD (Asp-Glu-Ala-Asp) box polypeptide 21 | DDX21 | 9188 | 7927936 | 60 |
| 24-dehydrocholesterol reductase | DHCR24 | 1718 | 7916432 | 100 |
| 7-dehydrocholesterol reductase | DHCR7 | 1717 | 7950067 | 80 |
| DEAH (Asp-Glu-Ala-His) box polypeptide 33 | DHX33 | 56919 | 8011861 | 100 |
| DnaJ (Hsp40) homolog, subfam. B, member 4 | DNAJB4 | 11080 | 7902512 | 100 |
| DnaJ (Hsp40) homolog, subfam. B, member 9 | DNAJB9 | 4189 | 8135480 | 25 |
| DnaJ (Hsp40) homolog, subfam. C, member 5 | DNAJC5 | 80331 | 8064208 | 10 |
| DnaJ (Hsp40) homolog, subfam. C, member 9 | DNAJC9 | 23234 | 7934320 | 55 |
| DNA-damage regulated autophagy modulator 2//choline/ethanolamine phosphotransferase 1 | DRAM2//CEPT1 | 128338//10390 | 7918474 | 100 |
| D-tyrosyl-tRNA deacylase 1 homolog (S. cerevisiae) | DTD1 | 92675 | 8061211 | 45 |
| ER degradation enhancer, mannosidase alpha-like 2 | EDEM2 | 55741 | 8065855 | 80 |
| ecotropic viral integration site 2B | EVI2B | 2124 | 8014063 | 60 |
| fam. with sequence similarity 36, member A//non-protein coding RNA 201 | FAM36A//NCRNA00201 | 116228//284702 | 7911085 | 15 |
| fam. with sequence similarity 86, member A | FAM86A | 196483 | 7999304 | 25 |
| Fas (TNF receptor superfam., member 6) | FAS | 355 | 7929032 | 70 |
| fatty acid synthase | FASN | 2194 | 8019392 | 100 |
| F-box protein 10//translocase of outer mitochondrial membrane 5 homolog (yeast) | FBXO10//TOMM5 | 26267//401505 | 8161229 | 40 |
| MGC44478 | FDPSL2A | 619190 | 8140443 | 55 |
| ferredoxin reductase | FDXR | 2232 | 8018236 | 40 |
| forkhead box O4 | FOXO4 | 4303 | 8168205 | 80 |
| ferritin, heavy polypeptide-like 5 | FTHL5 | 2509 | 8126948 | 95 |
| fucosidase, alpha-L- 2, plasma | FUCA2 | 2519 | 8129974 | 20 |
| growth arrest-specific 2 like 3 | GAS2L3 | 283431 | 7957850 | 70 |
| ganglioside induced differentiation associated protein 2 | GDAP2 | 54834 | 7918955 | 80 |
| growth differentiation factor 11 | GDF11 | 10220 | 7956026 | 65 |
| glutaredoxin (thioltransferase) | GLRX | 2745 | 8113214 | 90 |
| guanine nucleotide binding protein-like 3 (nucleolar)-like | GNL3L | 54552 | 8167797 | 85 |
| glucosamine-phosphate N-acetyltransferase 1 | GNPNAT1 | 64841 | 7979196 | 90 |
| glutathione reductase | GSR | 2936 | 8150112 | 40 |
| GTF2I repeat domain containing 2//GTF2I repeat domain containing 2B | GTF2IRD2//GTF2IRD2B | 84163//389524 | 8133549 and 8140170 | 50 and 30 |
| general transcription factor IIIC, polypeptide 2, beta 110 kDa | GTF3C2 | 2976 | 8051075 | 55 |
| HMG-box transcription factor 1//component of oligomeric golgi complex 5 | HBP1//COG5 | 26959//10466 | 8135392 | 65 |
| histone cluster 1, H1c | HIST1H1C | 3006 | 8124397 | 45 |
| histone cluster 1, H1e | HIST1H1E | 3008 | 8117377 | 95 |
| histone cluster 1, H2ae | HIST1H2AE | 3012 | 8117408 | 45 |
| histone cluster 1, H2be | HIST1H2BE | 8344 | 8117389 | 15 |
| histone cluster 1, H3g | HIST1H3G | 8355 | 8124440 | 35 |
| histone cluster 1, H3j | HIST1H3J | 8356 | 8124537 | 60 |
| histone cluster 1, H4a | HIST1H4A | 8359 | 8117334 | 10 |
| histone cluster 2, H2ac//histone cluster 2, H2aa3//histone cluster 2, H2aa4 | HIST-2H2AC//2H2AA3//2H2AA4 | 8338//8337//723790 | 7905079 and 7919619 | 75 and 75 |
| histone cluster 2, H2bf//histone cluster 2, H2be//histone cluster 2, H2ba | HIST-2H2BF//2H2BE//2H2BA | 440689//8349//337875 | 7919606 | 50 |
| high-mobility group box 3 | HMGB3 | 3149 | 8170468 | 5 |
| 3-hydroxy-3-methylglutaryl-Coenzyme A reductase//3-hydroxy-3-methylglutaryl-CoA reductase | HMGCR//HMGCR | 3156//3156 | 8106280 | 90 |
| 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 (soluble)//3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) | HMGCS1//HMGCS1 | 3157//3157 | 8111941 | 80 |
| heme oxygenase (decycling) 1 | HMOX1 | 3162 | 8072678 | 10 |
| heterogeneous nuclear ribonucleoprotein L | HNRNPL | 3191 | 8036613 | 30 |
| insulin receptor substrate 2 | IRS2 | 8660 | 7972745 | 35 |
| iron-sulfur cluster scaffold homolog (E. coli) | ISCU | 23479 | 7958414 | 100 |
| interferon stimulated exonuclease gene 20 kDa-like 2 | ISG20L2 | 81875 | 7921110 | 45 |
| potassium voltage-gated channel, Isk-related fam., member 3 | KCNE3 | 10008 | 7950409 | 25 |
| keratinocyte growth factor-like protein 1//fibroblast growth factor 7 (keratinocyte growth factor)//keratinocyte growth factor-like protein 2//hypothetical protein FLJ20444 | KGFLP1//FGF7//KGFLP2//FLJ20444 | 387628//2252//654466//403323 | 8155530 | 70 |
| lysophosphatidic acid receptor 1 | LPAR1 | 1902 | 8163257 | 10 |
| leucine-rich PPR-motif containing | LRPPRC | 10128 | 8051882 | 65 |
| lymphocyte antigen 96 | LY96 | 23643 | 8146934 | 35 |
| mitogen-activated protein kinase kinase 1//small nuclear RNA activating complex, polypeptide 5, 19 kDa | MAP2K1//SNAPC5 | 5604//10302 | 7984319 | 30 |
| mitogen-activated protein kinase 13 | MAPK13 | 5603 | 8119016 | 60 |
| methyltransferase like 2A | METTL2A | 339175 | 8009008 | 45 |
| microsomal glutathione S-transferase 3 | MGST3 | 4259 | 7906978 | 70 |
| mitochondrial ribosomal protein L30 | MRPL30 | 51263 | 8043848 | 30 |
| mitochondrial ribosomal protein L4 | MRPL4 | 51073 | 8025586 | 40 |
| mitochondrial ribosomal protein S17//glioblastoma amplified sequence//zinc finger protein 713 | MRPS17//GBAS//ZNF713 | 51373//2631//349075 | 8132922 | 60 |
| mitochondrial poly(A) polymerase//golgi autoantigen, golgin subfam. a, 6 pseudogene | MTPAP//LOC729668 | 55149//729668 | 7932834 | 45 |
| 5-methyltetrahydrofolate-homocysteine methyltransferase | MTR | 4548 | 7910752 | 15 |
| neighbor of BRCA1 gene 1 | NBR1 | 4077 | 8007471 | 20 |
| nuclear import 7 homolog (S. cerevisiae) | NIP7 | 51388 | 7996934 | 75 |
| NLR fam., pyrin domain containing 12 | NLRP12 | 91662 | 8039096 | 35 |
| nucleolar protein fam. 6 (RNA-associated) | NOL6 | 65083 | 8160682 | 95 |
| NAD(P)H dehydrogenase, quinone 1 | NQO1 | 1728 | 8002303 | 45 |
| nuclear receptor binding protein 1 | NRBP1 | 29959 | 8040927 | 20 |
| nucleotide binding protein-like | NUBPL | 80224 | 7973826 | 10 |
| nudix (nucleoside diphosphate linked moiety X)-type motif 14 | NUDT14 | 256281 | 7981566 | 35 |
| nuclear fragile × mental retardation protein interacting protein 1 | NUFIP1 | 26747 | 7971361 | 60 |
| nucleoporin 153 kDa | NUP153 | 9972 | 8124059 | 25 |
| olfactory receptor, fam. 5, subfam. B, member 21 | OR5B21 | 219968 | 7948330 | 50 |
| PAS domain containing serine/threonine kinase | PASK | 23178 | 8060205 | 55 |
| PRKC, apoptosis, WT1, regulator | PAWR | 5074 | 7965112 | 30 |
| PDGFA associated protein 1 | PDAP1 | 11333 | 8141273 | 35 |
| phosphodiesterase 1B, calmodulin-dependent | PDE1B | 5153 | 7955943 | 85 |
| phosphoribosylformylglycinamidine synthase | PFAS | 5198 | 8004804 | 60 |
| pleckstrin homology-like domain, fam. A, member 3 | PHLDA3 | 23612 | 7923372 | 75 |
| phosphoinositide-3-kinase adaptor protein 1 | PIK3AP1 | 118788 | 7935337 | 20 |
| PTEN induced putative kinase 1 | PINK1 | 65018 | 7898663 | 70 |
| phosphomannomutase 2 | PMM2 | 5373 | 7993148 | 65 |
| partner of NOB1 homolog (S. cerevisiae) | PNO1 | 56902 | 8042381 | 40 |
| polymerase (RNA) II (DNA directed) polypeptide E, 25 kDa | POLR2E | 5434 | 8032149 | 80 |
| polymerase (RNA) III (DNA directed) polypeptide E (80 kD) | POLR3E | 55718 | 7993973 | 30 |
| protein phosphatase 1D magnesium-dependent, delta isoform//protein phosphatase, Mg2+/Mn2+ dependent, 1D | PPM1D//PPM1D | 8493//8493 | 8008922 | 80 |
| phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 | PREX1 | 57580 | 8066848 | 100 |
| proline-serine-threonine phosphatase interacting protein 1 | PSTPIP1 | 9051 | 7985099 | 95 |
| prothymosin, alpha | PTMA | 5757 | 7954006 and 7961022 | 20 and 15 |
| RAB33B, member RAS oncogene fam. | RAB33B | 83452 | 8097507 | 40 |
| renin binding protein | RENBP | 5973 | 8175933 | 65 |
| replication factor C (activator 1) 2, 40 kDa | RFC2 | 5982 | 8140151 | 30 |
| ribonuclease H1 | RNASEH1 | 246243 | 8050079 | 90 |
| ring finger protein 146 | RNF146 | 81847 | 8121927 | 50 |
| ring finger protein 24 | RNF24 | 11237 | 8064766 | 100 |
| ring finger protein 26 | RNF26 | 79102 | 7944510 | 95 |
| ribosomal protein SA//small nucleolar RNA, H/ACA box 62 | RPSA//SNORA62 | 3921//6044 | 8078918 | 75 |
| RNA pseudouridylate synthase domain containing 2 | RPUSD2 | 27079 | 7982753 | 45 |
| ribosomal RNA processing 12 homolog (S. cerevisiae) | RRP12 | 23223 | 7935425 | 75 |
| retinoid × receptor, alpha | RXRA | 6256 | 8159127 | 5 |
| scavenger receptor class B, member 2 | SCARB2 | 950 | 8101158 | 70 |
| SERPINE1 mRNA binding protein 1 | SERBP1 | 26135 | 7916836 | 95 |
| splicing factor proline/glutamine-rich (polypyrimidine tract binding protein associated)//splicing factor proline/glutamine-rich | SFPQ//SFPQ | 6421//6421 | 7914791 | 40 |
| solute carrier fam. 25, member 32//DDB1 and CUL4 associated factor 13 | SLC25A32//DCAF13 | 81034//25879 | 8152255 | 100 |
| solute carrier fam. 35, member B3 | SLC35B3 | 51000 | 8123825 | 40 |
| solute carrier fam. 37 (glucose-6-phosphate transporter), member 4 | SLC37A4 | 2542 | 7952132 | 55 |
| solute carrier fam. 5 (sodium-dependent vitamin transporter), member 6 | SLC5A6 | 8884 | 8051030 | 95 |
| sphingomyelin phosphodiesterase 4, neutral membrane (neutral sphingomyelinase-3) | SMPD4 | 55627 | 8055183 | 40 |
| small nucleolar RNA host gene 1 (non-protein coding)//small nucleolar RNA, C/D box 26 | SNHG1//SNORD26 | 23642//9302 | 7948908 | 20 |
| small nucleolar RNA host gene 12 (non-protein coding) | SNHG12 | 85028 | 7914202 | 10 |
| small nucleolar RNA, H/ACA box 45 | SNORA45 | 677826 | 7938293 | 25 |
| sorting nexin fam. member 27 | SNX27 | 81609 | 7905444 | 35 |
| spinster homolog 2 (Drosophila)//MYB binding protein (P160) 1a | SPNS2//MYBBP1A | 124976//10514 | 8011640 | 45 |
| sprouty homolog 2 (Drosophila) | SPRY2 | 10253 | 7972217 | 75 |
| squalene epoxidase | SQLE | 6713 | 8148280 | 95 |
| sterol regulatory element binding transcription factor 2 | SREBF2 | 6721 | 8073522 | 45 |
| ST3 beta-galactoside alpha-2,3-sialyltransferase 6 | ST3GAL6 | 10402 | 8081219 | 100 |
| serine/threonine kinase 17b | STK17B | 9262 | 8057887 | 90 |
| transmembrane anterior posterior transformation 1 | TAPT1 | 202018 | 8099506 | 65 |
| taste receptor, type 2, member 5 | TAS2R5 | 54429 | 8136647 | 40 |
| tubulin folding cofactor E-like | TBCEL | 219899 | 7944623 | 55 |
| tectonic fam. member 2 | TCTN2 | 79867 | 7959638 | 40 |
| toll-like receptor 6 | TLR6 | 10333 | 8099841 | 30 |
| transmembrane protein 150B | TMEM150B | 284417 | 8039453 | 25 |
| transmembrane protein 55A | TMEM55A | 55529 | 8151756 | 90 |
| transmembrane protein 59 | TMEM59 | 9528 | 7916372 | 90 |
| transmembrane protein 97 | TMEM97 | 27346 | 8005839 | 95 |
| tumor necrosis factor receptor superfam., member 10c, decoy without an intracellular domain | TNFRSF10C | 8794 | 8145244 | 75 |
| translocase of outer mitochondrial membrane 34 | TOMM34 | 10953 | 8066461 | 35 |
| translocase of outer mitochondrial membrane 40 homolog (yeast) | TOMM40 | 10452 | 8029521 | 40 |
| tumor protein p53 inducible protein 3 | TP53I3 | 9540 | 8050702 | 30 |
| tumor protein p53 inducible nuclear protein 1 | TP53INP1 | 94241 | 8151890 | 100 |
| twinfilin, actin-binding protein, homolog 2 (Drosophila)//toll-like receptor 9 | TWF2//TLR9 | 11344//54106 | 8087860 | 65 |
| thioredoxin reductase 1 | TXNRD1 | 7296 | 7958174 | 55 |
| ubiquitin-fold modifier conjugating enzyme 1 | UFC1 | 51506 | 7906662 | 95 |
| ubiquitin specific peptidase 10 | USP10 | 9100 | 7997633 | 30 |
| vesicle-associated membrane protein 3 (cellubrevin) | VAMP3 | 9341 | 7897370 | 40 |
| valyl-tRNA synthetase | VARS | 7407 | 8125091 and 8178609 | 10 and 10 |
| vacuolar protein sorting 37 homolog A (S. cerevisiae) | VPS37A | 137492 | 8144774 | 60 |
| zinc finger protein 211 | ZNF211 | 10520 | 8031792 | 45 |
| zinc finger protein 223 | ZNF223 | 7766 | 8029360 | 65 |
| zinc finger protein 561 | ZNF561 | 93134 | 8033795 | 60 |
| zinc finger protein 79 | ZNF79 | 7633 | 8158022 | 100 |
| --- | --- | --- | 7910385 | 40 |
| --- | --- | --- | 7946567 | 15 |
| --- | --- | --- | 7966223 | 45 |
| --- | --- | --- | 7979694 | 40 |
| --- | --- | --- | 8130495 | 30 |
| --- | --- | --- | 8180237 | 60 |
| --- | --- | --- | 8180268 | 85 |
| --- | --- | --- | 8180417 | 85 |
The table shows the biomarker genes found by t-test and Backward Elimination. Genes were annotated, using the NetAffx database from Affymetrix (http://www.affymetrix.com, Santa Clara USA). When found, the Entrez Gene ID http://www.ncbi.nlm.nih.gov/gene was chosen as the gene identifier. The validation call frequency (%) is the occurrence of each gene in the 20 Test Gene Signatures obtained in the validation step.
Figure 5.
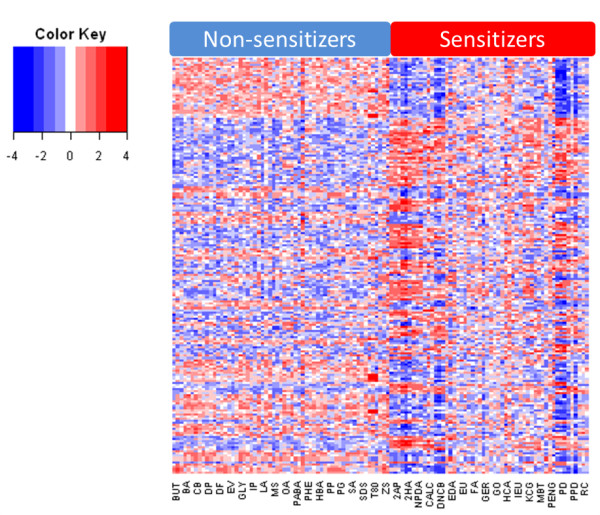
Transcriptional profiles of sensitizers and non-sensitizers. Hierarchical clustering of the genes in the Prediction Signature. Samples are grouped as sensitizer or non-sensitizer, and all replicates are included. Each row represents one gene, which is scaled to have a mean of zero and standard deviation of one, with colors representing the number of standard deviations from the mean.
Interrogation of the analysis used to identify the Prediction Signature
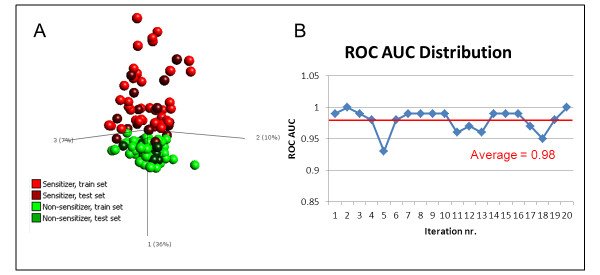
To validate the predictive power of our signature, we used a machine learning method called the Support Vector Machine (SVM) [12], which maps the data from a training set in space in order to maximize the separation of gene expression induced by sensitizing and non-sensitizing chemicals. As training set, 70% of the data set was selected randomly and the entire selection process was repeated. Starting with 29,141 transcripts, the signature was reduced to 200 transcripts, termed "Test Gene Signature", using ANOVA filtering and backward elimination, as described above. The remaining 30% of the data set was used to test each signature. The partitioning of the data set into subsets of 70% training data set and 30% test data set was done in a stratified random manner, while maintaining the relation of sensitizers and non-sensitizers. Thereafter, the Test Gene Signature was used to train an SVM model with the training set, and the predictive power of the model was assessed with the test set. This entire process was iterated 20 times. The frequency by which each gene in the Prediction Signature was included in the Test Gene Signatures is reported in table 1. Figure 6A shows a PCA plot based on the Test Gene Signature from one representative iteration. Clearly, the separation between sensitizers and non-sensitizers resembles the one observed for the Prediction Signature in Figure 4B. In Figure 6A, the samples of the sensitizing and non-sensitizing chemicals in the test set have been colored dark red and dark green respectively, indicating that they are not contributing to the principal components of the plot, but are merely plotted based on their expression values of the selected Test Gene Signature. As can be seen, sensitizers from the test set group with sensitizers from the training set, while non-sensitizers from the test set group with non-sensitizers from the training set. The final outcome of the SVM training and validation can be seen in Figure 6B, where the areas under the ROC curve are plotted for each iteration. The average area under the ROC curve of 0.98 confirms the ability to discriminate sensitizers from control samples. Based on this average, the estimated prediction performance of the assay reveals an accuracy of 99%, sensitivity of 99% and specificity of 99%. While this experiment does not validate the prediction power of the Prediction Signature per se, it does indeed validate the method by which it has been selected, supporting the claim that the Prediction Signature is capable of accurately predicting sensitizing properties of unknown samples.
Figure 6.
Validation of selection procedure of Prediction Signature. The method by which the Prediction Signature was constructed was validated by repeating the process on 70% randomly selected data (training set). The remaining 30% of data was used as a test set for signature validation. The process was repeated for 20 iterations. A) A representative PCA of one of the 20 iterations, which demonstrates that the Test Gene Signature can separate skin sensitizers from non-sensitizers. Only the samples of the 70% training set, displayed in bright colors, were used to build the space of the first three principal components. The test set samples, displayed in dark colors, were plotted into this space based on expression levels of the analytes in the Test Gene Signature. B) An SVM was trained on the 70% training set, and validated with the 30% test set. The areas under the ROC curve from 20 such randomizations are plotted, yielding an average AUC value of 0.98. This indicated that the classification of samples in the test set was correct.
Interactome, molecular functions and canonical pathways involving the Prediction Signature
Using Ingenuity Pathways Analysis (IPA, Ingenuity Systems Inc.), 184 of the 200 molecules in the signature were characterized with regard to the interactome, known functions and (canonical) pathways. The remaining 16 molecules could not be mapped to any unique IPA entries. The dominating functions identified were small molecule biochemistry (39 molecules), cell death (33), lipid metabolism (25), hematological system development (18), cell cycle (18), molecular transport (17), cellular growth and proliferation (16), and carbohydrate metabolism (15) (Table 2).
Table 2.
Dominating functions of the Prediction signature
| Function | Number of molecules from signature | Molecule names | Most prominent sub functions |
|---|---|---|---|
| small molecule biochemistry | 39 | ABHD5, ACLY, ALDH18A1, BLMH, CD86, CSGALNACT2, CYP51A1, DHCR24, DHCR7, DNAJC5, FAS, FASN, FDXR, FOXO4, GLRX, GNPNAT1, HMGCR, HMOX1, IRS2, LPAR1, LY96, MGST3, MTR, NQO1, PASK, PDE1B, PINK1, PMM2, RENBP, RXRA, SLC25A32, SLC37A4, SLC5A6, SMPD4, SQLE, SREBF2, ST3GAL6, TLR6, TMEM55A | Metabolism (24), biosynthesis (15), modification (12), synthesis (11) |
| cell death | 33 | CD33, DDX19A, DHCR24, DNAJB9, DNAJC5, FAS, FASN, FDXR, FOXO4, GLRX, GNPNAT1, GSR, HIST1H1C, HMGB3, HMOX1, IRS2, LPAR1, MAP2K1, MAPK13, NQO1, PAWR, PDE1B, PHLDA3, PINK1, PPM1D, RXRA, SERBP1, SPRY2, STK17B, TLR6, TNFRSF10C, TP53INP1, TXNRD1 | Apoptosis (30), cell death (13) |
| lipid metabolism | 25 | ABHD5, ACLY, CYP51A1, DHCR24, DHCR7, FAS, FASN, FDXR, FOXO4, HMGCR, HMOX1, IRS2, LPAR1, LY96, MGST3, PASK, RENBP, RXRA, SLC37A4, SMPD4, SQLE, SREBF2, ST3GAL6, TLR6, TMEM55A | Metabolism (18), synthesis (11), modification (11) |
| hematological system development | 18 | CARM1, CD33, CD86, FAS, FOXO4, HMGB3, HMGCR, HMOX1, IRS2, LY96, NBR1, NQO1, PAWR, PIK3AP1, PPM1D, STK17B, TP53INP1, VAMP3 | Proliferation (10), quantity (7) |
| cell cycle | 18 | ABHD5, ANAPC5, DNAJB4, DTD1, FAS, FASN, FOXO4, GDF11, HBP1, HMOX1, IRS2, MAP2K1, PAWR, PPM1D, RXRA, SFPQ, SPRY2, TP53INP1 | Cell cycle progression (13), G2 phase (5) |
| molecular transport | 17 | ABHD5, DNAJC5, FAS, FOXO4, HMOX1, LPAR1, MTR, NQO1, PASK, PINK1, RENBP, RXRA, SLC25A32, SLC37A4, SLC5A6, SREBF2, TLR6 | Accumulation (9), quantity (5) |
| cellular growth and proliferation | 16 | CD33, CD86, FAS, GNPNAT1, HMOX1, IRS2, LPAR1, LY96, MAP2K1, PAWR, PIK3AP1, PPM1D, RXRA, SPRY2, STK17B, TP53INP1 | Proliferation (16), growth (4) |
| carbohydrate metabolism | 15 | ABHD5, ACLY, CSGALNACT2, FAS, FASN, FUCA2, GNPNAT1, IRS2, LY96, NQO1, PMM2, RENBP, SLC37A4, ST3GAL6, TMEM55A | Metabolism (9), biosynthesis (5) |
Dominating functions in the molecular signature. 184 of the 200 molecules were functionally investigated, using IPA. Only functions populated by 15 or more genes were included in the present study.
Pathways possibly invoked by the molecules in the signature were also investigated using IPA. Those most highly populated involved NRF2-mediated oxidative response (10), xenobiotic metabolism signaling (8), protein ubiquitination pathway (7), LPS/IL-1 mediated inhibition of RXR function (6), aryl hydrocarbon receptor signaling (6) and protein kinase A signaling (6). These pathways are known to take part in reactions provoked by foreign substances, xenobiotics, which supports a relevant biology behind the genomic signature.
Discussion
Allergic contact dermatitis (ACD) is an inflammatory skin disease caused by an adaptive immune response to normally innocuous chemicals [13]. Small molecular weight chemicals, so-called haptens, can bind self-proteins in the skin, which enables internalization of the protein-bound allergenic chemical by skin dendritic cell (DC). DCs, under the influence of the local microenvironment, process the protein-hapten complex, migrate to the local lymph nodes and activate naïve T cells. The initiation and development of allergen-specific responses, mainly effector CD8+ T cells and Th1 cells, and production of immunoregulatory proteins, are hallmarks of the immune activation observed in ACD. ACD is also the most common manifestation of immunotoxicity observed in humans [13] and hundreds of chemicals have been shown to cause sensitization in skin [14]. The driving factors and molecular mechanisms involved in sensitization are still unknown even though intense research efforts have been carried out to characterize the immunological responses towards allergenic chemicals. The REACH legislation requires that all chemicals produced over 1 ton/year are tested for hazardous properties such as toxicity and allergenicity [5], which increase the demand for accurate assays with predictive power for hazard identification. Additionally, the 7th Amendment to the Cosmetics Directive (76/768/EEC) poses a complete ban on using animal experimentation for testing cosmetic ingredients by 2013 if a scientifically reliable method is available. Thus, there is a significant need for predictive test methods that are based on human cells. Today, the identification of potential human sensitizers relies on animal experimentation, in particular the murine local lymph node assay (LLNA) [6]. The LLNA is based upon measurements of proliferation induced in draining lymph nodes of mice after chemical exposure [15]. Chemicals are defined as sensitizers if they provoke a three-fold increase in proliferation compared to control, and the amount of chemical required for the increase is the EC3 value. Thus, the LLNA can also be used to categorize the chemicals based on sensitization potency. However, LLNA is, besides the obvious ethical implications, also time consuming and expensive. Human sensitization data often stem from human maximization tests (HMT) [16] and human patch tests (HPT). In an extensive report from the Interagency Coordinating Committee on the Validation of Alternative Methods (ICCVAM), the performance characteristics of LLNA were compared to other available animal-based methods and human sensitization data (HMT and HPT) [17]. The LLNA performance in comparison to human data (74 assessments) revealed an accuracy of 72%, a sensitivity of 72% and a specificity of 67%.
Various human cell lines and primary cells involved in sensitization have been evaluated as predictive test system, such as epithelial cells, dendritic cells and T cells, however, no validated test assay is currently available. THP-1, U937, KG-1 and MUTZ-3, naive or differentiated, are among the human myeloid cell lines most extensively evaluated as platforms for DC-based in vitro assays, as reviewed in [18]. These cells are easy to grow and enable standardization of protocols. U937 and THP-1 are currently being evaluated in pre-validation stage for prediction of skin sensitization. The Human Cell Line Activation Test (hCLAT) is based upon analysis of CD86 and/or CD54 expression on THP-1 cells after chemical stimulation [19,20]. The Myeloid U937 Skin Sensitization Test (MUSST) also involves analysis of CD86 [21]. These assays are thus very limited in readout. As CD86 is among the markers most extensively studied, we evaluated the expression level of this marker in our assay. We demonstrated its relevance but also its insufficient predictive power (Figure 2), since only 10 out of 20 sensitizing chemicals induced a significant up regulation of CD86. Various other single biomarkers have been suggested to be up regulated upon stimulation with sensitizing chemicals, such as CD40, CD80, CD54, CXCL8, IL-1β, MIP-1β, p38 MAPK, as reviewed in [18], yet single-handedly, none of them have enough predictive power to discriminate between sensitizing and non-sensitizing chemicals. The analysis of biomarker signatures, i.e. combination of biomarkers, has been shown to be superior in molecular diagnostic of cancer and superior to any single biomarker. Consequently, we therefore utilized the power of global transcriptomics and screened the gene regulation induced by a large set of well-defined chemicals and controls in search of predictive biomarker combinations.
The large number of differentially expressed genes in MUTZ-3 cells stimulated with sensitizing chemicals vs. non-sensitizing controls revealed that MUTZ-3 indeed had a capacity to differentiate between these two groups. Efforts have previously been done to create assays based on genome analysis in various cell systems, such as e.g. CD34+-progenitor cells-derived DCs [22-24]. While such assays might provide in vivo like environments, primary cells are not well suited for a high-throughput format considering both donor-dependent variations as well as ethical aspect of such cell sources. Furthermore, previous efforts within in vitro assay development for sensitization that rely on full genome analysis have used a limited set of testing compounds.
The present study utilized in all 40 compounds and efforts were made to divide these compounds into two subsets, for training and testing respectively. While these experiments have resulted in successful predictions (data not shown), it is our experience that sensitizing compounds differ greatly in their induced gene expression profile, as can be seen in Figure 3D. In this perspective, we strived to include as many training compounds as possible when identifying our Prediction Signature, and did not exclude any compounds for validation. Instead, we validated the method by which the Prediction Signature was identified, by subdividing the samples into training and test sets at random, using unseen data for validation, to avoid overfitting. At present, the Prediction Signature consists of 200 transcripts, based on Figure 4A. Continuing the elimination process beyond 200 transcripts causes loss of information, as seen by the rise of KLD. Experiments have shown that correct classifications are possible even with further reduced signatures, down to 11 genes (data not shown). A reduction of signature size could be assessed in conjunction with validation of the assay, using untested positive and negative compounds in a new test set. By reducing the signature size at this point, the risk of biasing the signature towards this data set increases, making it harder to correctly classify unknown samples. Additional test compounds will also serve to assess the frequency of extreme transcriptional profile outliers, such as Oxazolone and Cinnamic aldehyde, which had to be removed from the analysis performed in this study. A number of reasons may be attributed to the fact that these compounds were not compatible with the assay, such as solubility in the cell media or extreme toxic effects. In those cases, other in vitro alternatives may complement this assay, so that the safety assessment of chemicals for sensitization includes a battery of in vitro assays. Naturally, an additional data set with blinded compounds is essential to validate whether the assay truly performs as estimated by the random subdivisions into training and test sets.
Of note, our Prediction Signature is able to predict the potency of sensitizing compounds, as defined by the LLNA (Figure 4C). However, the potency predicted by LLNA and that of our classifier do not match for all samples. Notably, the moderate sensitizer 2-hydroxyethyl acrylate showed resemblance to strong and extreme sensitizers with respect to gene expression profile. Similarly, the moderate sensitizers ethylendiamine, hexylcinnamic aldehyde, and glyoxal grouped together with weak sensitizers. These findings support the fact that sensitizing potency, as defined, may need revising.
By studying the identity of the transcripts and their involvement in intracellular signaling pathways, we were also able to confirm the biological relevance of the Prediction Signature. Using IPA, we found that the most highly populated pathways were nuclear factor-erythroid 2-related factor 2 (NRF2) mediated oxidative response, xenobiotic metabolism signaling, protein ubiquitination pathway, LPS/IL-1 mediated inhibition of Retinoic X receptor (RXR) function, aryl hydrocarbon receptor (AHR) signaling, and protein kinase A (PKA) signaling. These pathways are all known to take part in reactions provoked by xenobiotics, and several were associated with oxidative stress. Furthermore, Toll-like receptor (TLR) signaling is among the top pathways found in IPA. Recent studies on assay development for prediction of sensitization in vitro have to a large extent focused on how danger signals are provided to antigen-presenting cells, inducing pro-inflammatory cytokines and chemokines, as well as co-stimulatory molecules needed for a specific T-cell response. We hypothesize that these signals are provided through the innate immune responses, in analogy with infections, as reviewed in [25].
The primary pathways found in this study involved NRF2 signaling. This is a pathway activated by Reactive Oxygen Species (ROS), and is a defense mechanism to xenobiotics and response to cellular stress. In the resting cell, NRF2 is bound by kelch-like ECH-associated protein 1 (KEAP1) and located in the cytosol. In the response to ROS activity, KEAP1 is targeted for ubiquitination and protesomal degradation, resulting in the translocation of NRF2 to the nucleus, where it activates transcription of genes containing anti-oxidant response elements (ARE) in their promoter region [26]. The functions of genes transcribed by NRF2 association to ARE include regulation of inflammation, migration of DC and anti-oxidant defense enzymes, such as NADPH quinone oxidoreductase 1 (NQO1) and glutathione S-transferases (GST) [27,28], genes found in the Prediction Signature. Furthermore, the NRF2/KEAP1/ARE pathway has previously been described as activated in response to skin sensitizers, inducing maturation of dendritic cells [29].
Similarly, AHR is a transcription factor in the cytosol that is activated by binding to ligands, which includes a wide range of xenobiotic chemicals, such as halogenated aromatic hydrocarbons, polyphenols and a number of pharmaceuticals [30]. In the absence of a ligand, AHR is bound by a complex of chaperon proteins, keeping it in the cytosol. Upon ligand binding, AHR is translocated to the nucleus, where it dimerizes with aryl hydrocarbon receptor nuclear translocator (ARNT) [30]. The ARNT/AHR heterodimer then binds to xenobiotic response elements (XRE) in promoter regions of target genes. The typical target genes for XRE include enzymes for drug metabolism, such as the cytochrome P450 (CYP) superfamily, as well as cytoprotective enzymes mediating defense against oxidative stress, such as NQO1 [31]. Interestingly, while NQO1 is under control of both NRF2 and AHR, with both ARE and XRE in the promoter region, it has also been shown that AHR is among the target genes for the activated NRF2 pathway and vice versa [32]. Thus, a battery of protective enzymes are induced in response to a variety of xenobiotics, possibly through a number of signaling pathways, ultimately leading to the maturation of dendritic cells, as also indicated by the present data. The protein ubiquitination pathway is involved in degradation of short-lived or regulatory proteins involved in many cellular processes, such as the cell cycle, cell proliferation, apoptosis, DNA repair, transcription regulation, cell surface receptors and ion channels regulation, and antigen presentation. Of note, both NRF2 and AHR are in the resting cell bound by proteins that are targeted for ubiquitination upon ligand binding.
RXR is a nuclear receptor, with retinoic acid as the most prominent natural ligand [33]. It has previously been described as important for xenobiotics recognition and glutathione homeostasis, with cytoprotective enzymes as target genes [34,35].
TLR signaling is known to play a major role in dendritic cell maturation, as they activate transcription of a number of pro-inflammatory cytokines, chemokine-receptors for homing to lymph nodes and co-stimulatory molecules [36-38]. While TLR6 and TLR9 are present in our Prediction Signature, others have reported TLR4 as a crucial mediator of contact allergy to nickel [39]. As these receptors all signal through nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), it is not surprising that different compounds activate different receptors, considering the chemical diversity of skin sensitizers, as discussed above.
Lastly, PKA signaling is a vastly versatile pathway activated by numerous stimuli, and, to the best of knowledge, this pathway has not previously been reported in association with skin sensitization. However, individual species of CYPs are known to be phosphorylated by PKA, in response to elevated levels of cyclic adenosine monophosphate (cAMP), triggered by xenobiotics. In addition, cAMP levels influence the nuclear translocation of AHR, connecting these two pathways and their impact on CYP activity [40].
Conclusion
In this paper, we have demonstrated the predictive power of a genomic biomarker signature, which correctly classifies sensitizers and non-sensitizers. The biomarker signature was derived from the human DC-like cell line MUTZ-3, which was challenged with a panel of 40 reference chemical compounds. The biomarker genes were shown to be biologically relevant, as demonstrated by their involvement in cytoprotective mechanisms and pathways triggered by xenobiotic substances, supporting their relevance as predictor genes for skin sensitization. The findings reported in this paper might impact the development of in vitro assays for assessment of skin sensitization, which is crucial in order to replace the animal models currently in use.
Methods
Chemicals
A panel of 40 chemical compounds, consisting of 20 sensitizers and 20 non-sensitizers were used for cell stimulations. The sensitizers were 2,4-dinitrochlorobenzene, cinnamaldehyde, resorcinol, oxazolone, glyoxal, 2-mercaptobenzothiazole, eugenol, isoeugenol, cinnamic alcohol, p-phenylendiamine, formaldehyde, ethylendiamine, 2-hydroxyethyl acrylate, hexylcinnamic aldehyde, potassium dichromate, penicillin G, kathon CG (MCI/MI), 2-aminophenol, geraniol and 2-nitro-1,4-phenylendiamine. The non-sensitizers were sodium dodecyl sulphate, salicylic acid, phenol, glycerol, lactic acid, chlorobenzene, p-hydrobenzoic acid, benzaldehyde, diethyl phtalate, octanoic acid, zinc sulphate, 4-aminobenzoic acid, methyl salicylate, ethyl vanillin, isopropanol, dimethyl formamide, 1-butanol, potassium permanganate, propylene glycol and tween 80 (Table 3). All chemicals were from Sigma-Aldrich, St. Louis, MO, USA. Compounds were dissolved in either dimethyl sulfoxide (DMSO) or distilled water. Prior to stimulations, the cytotoxicity of all compounds was monitored, using propidium iodide (PI) (BD Biosciences, San Diego, CA) using protocol provided by the manufacturer. The relative viability of stimulated cells was calculated as
Table 3.
List of reference chemicals used in assay development
| Compound | Abbreviation | Potency | LLNA | HMT1 | HPTA1 |
|---|---|---|---|---|---|
| Sensitizers | |||||
| 2,4-Dinitrochlorobenzene | DNCB | Extreme [15] | + [15] | ||
| Oxazolone | OXA | Extreme [15] | + [15] | ||
| Potassium dichromate | PD | Extreme [14] | + [14] | + | + |
| Kathon CG (MC/MCI) | KCG | Extreme [14,45] | + [14,46] | ||
| Formaldehyde | FA | Strong [15] | + [15] | + | + |
| 2-Aminophenol | 2AP | Strong [46] | + [47] | ||
| 2-nitro-1,4-Phenylendiamine | NPDA | Strong [46] | + [47] | ||
| p-Phenylendiamine | PPD | Strong [47] | + [48] | + | + |
| Hexylcinnamic aldehyde | HCA | Moderate [15] | + [15] | ||
| 2-Hydroxyethyl acrylate | 2HA | Moderate [46] | + [47] | + | |
| 2-Mercaptobenzothiazole | MBT | Moderate [46] | + [47] | + | + |
| Glyoxal | GO | Moderate [46] | + [47] | + | |
| Cinnamaldehyde | CALD | Moderate [47] | + [48] | + | + |
| Isoeugenol | IEU | Moderate [47] | + [48] | + | |
| Ethylendiamine | EDA | Moderate [14] | + [14] | ||
| Resorcinol | RC | Moderate [48] | + [49] | - | + |
| Cinnamic alcohol | CALC | Weak [46] | + [48] | ||
| Eugenol | EU | Weak [47] | + [48] | + | |
| Penicillin G | PEN G | Weak [47] | + [48] | + | |
| Geraniol | GER | Weak [14] | + [14] | - | + |
| Non-sensitizers | |||||
| 1-Butanol | BUT | - [50] | |||
| 4-Aminobenzoic acid | PABA | - [51] | - | + | |
| Benzaldehyde | BA | - [52] | |||
| Chlorobenzene | CB | - [14] | |||
| Diethyl phthalate | DP | - [48] | |||
| Dimethyl formamide | DF | - [46] | |||
| Ethyl vanillin | EV | - [52] | |||
| Glycerol | GLY | - [48] | |||
| Isopropanol | IP | - [48] | |||
| Lactic acid | LA | - [14] | |||
| Methyl salicylate | MS | - [14] | - | ||
| Octanoic acid | OA | - [53] | |||
| Propylene glycol | PG | - [51] | |||
| Phenol | PHE | - [53] | - | ||
| p-Hydroxybenzoic acid | HBA | - [54] | |||
| Potassium permanganate | PP | - | |||
| Salicylic acid | SA | - [14] | - | ||
| Sodium dodecyl sulphate | SDS | +2 [14,53] | - | ||
| Tween 80 | T80 | - [20] | + | ||
| Zinc sulphate | ZS | +2 [55] |
List of sensitizers and non-sensitizers used in assay development. 1) HMT, Human Maximization Test; HPTA, Human Patch Test Allergen. Information is derived from [17]. 2) False positives in LLNA.
For toxic compounds, the concentration yielding 90% relative viability (Rv90) was used. For non-toxic compounds, a concentration of 500 μM was used. For non-toxic compounds that were insoluble at 500 μM in medium, the highest soluble concentration was used. For compounds dissolved in DMSO, the final concentration of DMSO in each well was 0.1%. The vehicle and concentrations used for each compound are listed in Table 4.
Table 4.
Concentrations and vehicles used for each reference chemical
| Compound | Abbreviation | Vehicle | Max solubility (μM) |
Rv90 (μM) |
Concentration in culture (μM) |
|---|---|---|---|---|---|
| Sensitizers | |||||
| 2,4-Dinitrochlorobenzene | DNCB | DMSO | - | 4 | 4 |
| Oxazolone | OXA | DMSO | 250 | - | 250 |
| Potassium dichromate | PD | Water | 51.02 | 1.5 | 1.5 |
| Kathon CG (MC/MCI)1 | KCG | Water | - | 0.0035% | 0.0035% |
| Formaldehyde | FA | Water | - | 80 | 80 |
| 2-Aminophenol | 2AP | DMSO | - | 100 | 100 |
| 2-nitro-1,4-Phenylendiamine | NPDA | DMSO | - | 300 | 300 |
| p-Phenylendiamine | PPD | DMSO | 566 | 75 | 75 |
| Hexylcinnamic aldehyde | HCA | DMSO | 32.34 | - | 32.24 |
| 2-Hydroxyethyl acrylate | 2HA | Water | - | 100 | 100 |
| 2-Mercaptobenzothiazole | MBT | DMSO | 250 | - | 250 |
| Glyoxal | GO | Water | - | 300 | 300 |
| Cinnamaldehyde | CALD | Water | - | 120 | 120 |
| Isoeugenol | IEU | DMSO | 641 | 300 | 300 |
| Ethylendiamine | EDA | Water | - | - | 500 |
| Resorcinol | RC | Water | - | - | 500 |
| Cinnamic alcohol | CALC | DMSO | 500 | - | 500 |
| Eugenol | EU | DMSO | 649 | 300 | 300 |
| Penicillin G | PEN G | Water | - | - | 500 |
| Geraniol | GER | DMSO | - | - | 500 |
| Non-sensitizers | |||||
| 1-Butanol | BUT | DMSO | - | - | 500 |
| 4-Aminobenzoic acid | PABA | DMSO | - | - | 500 |
| Benzaldehyde | BA | DMSO | 250 | - | 250 |
| Chlorobenzene | CB | DMSO | 98 | - | 98 |
| Diethyl phthalate | DP | DMSO | 50 | - | 50 |
| Dimethyl formamide | DF | Water | - | - | 500 |
| Ethyl vanillin | EV | DMSO | - | - | 500 |
| Glycerol | GLY | Water | - | - | 500 |
| Isopropanol | IP | Water | - | - | 500 |
| Lactic acid | LA | Water | - | - | 500 |
| Methyl salicylate | MS | DMSO | - | - | 500 |
| Octanoic acid | OA | DMSO | 504 | - | 500 |
| Propylene glycol | PG | Water | - | - | 500 |
| Phenol | PHE | Water | - | - | 500 |
| p-Hydroxybenzoic acid | HBA | DMSO | 250 | - | 250 |
| Potassium permanganate | PP | Water | 38 | - | 38 |
| Salicylic acid | SA | DMSO | - | - | 500 |
| Sodium dodecyl sulphate | SDS | Water | - | 200 | 200 |
| Tween 80 | T80 | DMSO | - | - | 500 |
| Zinc sulphate | ZS | Water | 126 | - | 126 |
List of concentrations and vehicles used for each testing compound. 1) Kathon CG is a mixture of the compounds MC and MCI. The concentration of this mixture is given in %.
Chemical exposure of the cells
The human myeloid leukemia-derived cell line MUTZ-3 (DSMZ, Braunschweig, Germany) was maintained in α-MEM (Thermo Scientific Hyclone, Logan, UT) supplemented with 20% (volume/volume) fetal calf serum (Invitrogen, Carlsbad, CA) and 40 ng/ml rhGM-CSF (Bayer HealthCare Pharmaceuticals, Seattle, WA), as described [10]. Cultures were maintained at 200.000 cells/ml during expansion, with a media change every 3-4 days. No differentiating steps were performed. Instead, the proliferating progenitor MUTZ-3 was used for stimulations, as delivered by the supplier. Prior to each experiment, the cells were immunophenotyped using flow cytometry as a quality control. Cells were seeded in 6-well plates at 200.000 cells/ml. Stock solutions of each compound were prepared in either DMSO or distilled water, and were subsequently diluted so the in-well concentrations corresponded to the Rv90 value, and in-well concentrations of DMSO were 0.1%. Cells were incubated for 24 h at 37°C and 5% CO2. Thereafter, cells were harvested and analyzed by flow cytometry. In parallel, harvested cells were lysed in TRIzol reagent (Invitrogen) and stored at -20°C until RNA extraction. Stimulations with chemicals were performed in three individual experiments, so that triplicates samples were obtained.
Phenotypic analysis with flow cytometry
All cell surface staining and washing steps were performed in PBS containing 1% BSA (w/v). Cells were incubated with specific mouse mAbs for 15 min at 4°C. The following mAbs were used for flow cytometry: FITC-conjugated CD1a (DakoCytomation, Glostrup, Denmark), CD34, CD86, and HLA-DR (BD Biosciences), PE-conjugated CD14 (DakoCytomation), CD54 and CD80 (BD Biosciences). Mouse IgG1, conjugated to FITC or PE were used as isotype controls (BD Biosciences) and PI was used to assess cell viability. FACSDiva software was used for data acquisition with FACSCanto II instrument (BD Bioscience). 10,000 events were acquired and gates were set based on light scatter properties to exclude debris and nonviable cells. Further data analysis was performed using FCS Express V3 (De Novo Software, Los Angeles, CA).
Preparation of cRNA and gene chip hybridization
RNA isolation and gene chip hybridization was performed as described [41]. Briefly, RNA from unstimulated and chemical-stimulated MUTZ-3 cells, from triplicate experiments, were extracted and analyzed. The preparation of labeled sense DNA was performed according to Affymetrix GeneChip™ Whole Transcript (WT) Sense Target Labeling Assay (100 ng Total RNA Labeling Protocol) using the recommended kits and controls (Affymetrix, Santa Clara, CA). Hybridization, washing and scanning of the Human Gene 1.0 ST Arrays were performed according to the manufacturer's protocol (Affymetrix). The microarray data have been deposited in the Array Express database http://www.ebi.ac.uk/arrayexpress/ with accession number E-MTAB-670.
Microarray data analysis and statistical methods
The microarray data were normalized and quality checked with the RMA algorithm, using Affymetrix Expression Console (Affymetrix). Genes that were significantly regulated when comparing sensitizers with non-sensitizers were identified using one-way ANOVA, with false discovery rate (FDR) as a correction for multiple hypothesis testing. In order to reduce the large number of identified significant genes, we applied an algorithm developed in-house for Backward Elimination of analytes [42]. With this method, we train and test a Support Vector Machine (SVM) model [12] with leave-one out cross-validation, with one analyte left out. This process is iterated until each analyte has been left out once. For each iterative step, a Kullback-Leibler divergence (KLD) is recorded, yielding N KLDs, where N is the number of analytes. The analyte that was left out when the smallest KLD was observed is considered to provide the least information in the data set. Thus, this analyte is eliminated and the iterations proceed, this time with N-1 analytes. In this manner, the analytes are eliminated one by one until a panel of markers remain that have been selected based on the ability of each analyte to contribute with orthogonal information for the discrimination of skin sensitizers vs. non-sensitizers. The selected biomarker profile of 200 transcripts were designated the "Prediction Signature". The scripts for Backwards Elimination and Support Vector Machines were programmed for R [43], with the additional package e1071 [44]. ANOVA analyses and visualization of results with Principal Component Analysis were performed in Qlucore Omics Explorer 2.1 (Qlucore AB, Lund, Sweden). Hierarchical clustering for the heatmap was performed in R.
Interrogation of the method for identification of the Prediction Signature
The data set was divided into a training set and a test set, consisting of 70% and 30%, of the chemical compounds, respectively. The division was performed randomly, while maintaining the proportions of sensitizers and non-sensitizers in each subset at the same ratio as in the complete data set. A biomarker signature was identified in the training set, using ANOVA filtering and Backward Elimination, as described above. This test signature was used to train an SVM, using the training set, which was thereafter applied to predict the samples of the test set. The process was repeated 20 times and the distribution of the area under the Receiver Operating Characteristic (ROC) curve [45] was used as a measurement of the performance of the model.
Assessment of biological functions of Prediction Signature using pathway analysis
In order to investigate the biological functions the gene profile of the 200 genes derived from the Backward Elimination was analyzed, using the Ingenuity Pathway Analysis software, IPA, (Ingenuity Systems, Inc. Mountain View, USA). The gene profile was analyzed using the 'Build' and 'Path Explorer' functions to build an interactome of the core genes from the Prediction Signature together with connecting molecules, as suggested by IPA. The molecules of the signature were connected using the shortest known paths. In this process only human data from primary cells, cell lines and epidermal tissue was used. Public identifiers were used to map genes in IPA. All molecules except for endogenous and chemical drugs were allowed in the network and all kinds of connections were allowed. Known 'Functions' and 'Canonical Pathways' from IPA were mapped to the signature using the 'Overlay' function. The most densely populated pathways and functions were reported. All were significant, using the built in IPA statistical measures (p-values for functions and -log(p-values) for pathways).
Abbreviations
ACD: atopic contact dermatitis; AML: acute myeloid leukemia cell; APC: Antigen Presenting Cell; DC: Dendritic Cell; GM-CSF: Granulocyte macrophage colony-stimulating factor; GPMT: Guinea pig maximization test; HMT: Human Maximation Test; HPTA: Human Patch Test Allergen; IL: Interleukin; LLNA: Local Lymph Node Assay; PCA: Principal Component Analysis.
Competing interests
The authors have applied for a patent related to the content of this article.
Authors' contributions
ML and CB designed the study strategy. HJ and ML set up and optimized the cell-based assay. HJ performed the cellular stimulations with chemicals. HJ and ML wrote the manuscript. AA and HJ analyzed the microarray data and prepared the figures. All authors revised and approved the manuscript.
Contributor Information
Henrik Johansson, Email: henrik.johansson@immun.lth.se.
Malin Lindstedt, Email: malin.lindstedt@immun.lth.se.
Ann-Sofie Albrekt, Email: ann-sofie.albrekt@immun.lth.se.
Carl AK Borrebaeck, Email: carl.borrebaeck@immun.lth.se.
Acknowledgements
This work was supported by grants from the Swedish Fund for Research Without Animal Experiments, Faculty of Engineering (LTH), the Swedish Research Council (K2010-79X-21371-01-3) and the European Commission as part of the Integrated project 'Novel Testing Strategies for in vitro Assessment of Allergens; Sens-it-iv' (LSHB-CT-2005-018681). We would like to thank Ann-Charlott Olsson for microarray sample preparation and Dr. Anders Carlsson for the backward elimination algorithm.
References
- Akhavan A, Cohen SR. The relationship between atopic dermatitis and contact dermatitis. Clin Dermatol. 2003;21(2):158–162. doi: 10.1016/S0738-081X(02)00372-3. [DOI] [PubMed] [Google Scholar]
- Mortz CG, Lauritsen JM, Bindslev-Jensen C, Andersen KE. Prevalence of atopic dermatitis, asthma, allergic rhinitis, and hand and contact dermatitis in adolescents. The Odense Adolescence Cohort Study on Atopic Diseases and Dermatitis. Br J Dermatol. 2001;144(3):523–532. doi: 10.1046/j.1365-2133.2001.04078.x. [DOI] [PubMed] [Google Scholar]
- Nielsen NH, Linneberg A, Menne T, Madsen F, Frolund L, Dirksen A, Jorgensen T. Allergic contact sensitization in an adult Danish population: two cross-sectional surveys eight years apart (the Copenhagen Allergy Study) Acta Derm Venereol. 2001;81(1):31–34. doi: 10.1080/000155501750208155. [DOI] [PubMed] [Google Scholar]
- Fonacier LS, Dreskin SC, Leung DY. Allergic skin diseases. J Allergy Clin Immunol. pp. S138–149. [DOI] [PubMed]
- EC 1907/2006. Regulation (EC) No 1907/2006 of the European Parliament and of the Council of 18 December 2006 concerning the Registration, Evaluation, Authorisation and Restriction of Chemicals (REACH), establishing a European Chemicals Agency, amending Directive 1999/45/EC and repealing Council Regulation (EEC) No 793/93 and Commission Regulation (EC) No 1488/94 as well as Council Directive 76/769/EEC and Commission Directives 91/155/EEC, 93/67/EEC, 93/105/EC and 2000/21/EC.
- Basketter DA, Evans P, Fielder RJ, Gerberick GF, Dearman RJ, Kimber I. Local lymph node assay - validation, conduct and use in practice. Food Chem Toxicol. 2002;40(5):593–598. doi: 10.1016/S0278-6915(01)00130-2. [DOI] [PubMed] [Google Scholar]
- Magnusson B, Kligman AM. The identification of contact allergens by animal assay. The guinea pig maximization test. J Invest Dermatol. 1969;52(3):268–276. doi: 10.1038/jid.1969.42. [DOI] [PubMed] [Google Scholar]
- Santegoets SJ, Masterson AJ, van der Sluis PC, Lougheed SM, Fluitsma DM, van den Eertwegh AJ, Pinedo HM, Scheper RJ, de Gruijl TD. A CD34(+) human cell line model of myeloid dendritic cell differentiation: evidence for a CD14(+)CD11b(+) Langerhans cell precursor. J Leukoc Biol. 2006;80(6):1337–1344. doi: 10.1189/jlb.0206111. [DOI] [PubMed] [Google Scholar]
- Masterson AJ, Sombroek CC, De Gruijl TD, Graus YM, van der Vliet HJ, Lougheed SM, van den Eertwegh AJ, Pinedo HM, Scheper RJ. MUTZ-3, a human cell line model for the cytokine-induced differentiation of dendritic cells from CD34+ precursors. Blood. 2002;100(2):701–703. doi: 10.1182/blood.V100.2.701. [DOI] [PubMed] [Google Scholar]
- Larsson K, Lindstedt M, Borrebaeck CA. Functional and transcriptional profiling of MUTZ-3, a myeloid cell line acting as a model for dendritic cells. Immunology. 2006;117(2):156–166. doi: 10.1111/j.1365-2567.2005.02274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasaiyaah J, Yong K, Katz DR, Kellam P, Chain BM. Dendritic cells and myeloid leukaemias: plasticity and commitment in cell differentiation. Br J Haematol. 2007;138(3):281–290. doi: 10.1111/j.1365-2141.2007.06622.x. [DOI] [PubMed] [Google Scholar]
- Cortes C, Vapnik V. Support-Vector Networks. Machine Learning. 1995;20(3):273–297. [Google Scholar]
- Kimber I, Basketter DA, Gerberick GF, Dearman RJ. Allergic contact dermatitis. Int Immunopharmacol. 2002;2(2-3):201–211. doi: 10.1016/S1567-5769(01)00173-4. [DOI] [PubMed] [Google Scholar]
- Gerberick GF, Ryan CA, Kern PS, Schlatter H, Dearman RJ, Kimber I, Patlewicz GY, Basketter DA. Compilation of historical local lymph node data for evaluation of skin sensitization alternative methods. Dermatitis. 2005;16(4):157–202. [PubMed] [Google Scholar]
- Kimber I, Basketter DA, Butler M, Gamer A, Garrigue JL, Gerberick GF, Newsome C, Steiling W, Vohr HW. Classification of contact allergens according to potency: proposals. Food Chem Toxicol. 2003;41(12):1799–1809. doi: 10.1016/S0278-6915(03)00223-0. [DOI] [PubMed] [Google Scholar]
- Kligman AM. The identification of contact allergens by human assay. 3. The maximization test: a procedure for screening and rating contact sensitizers. J Invest Dermatol. 1966;47(5):393–409. doi: 10.1038/jid.1966.160. [DOI] [PubMed] [Google Scholar]
- Haneke KE, Tice RR, Carson BL, Margolin BH, Stokes WS. ICCVAM evaluation of the murine local lymph node assay. Data analyses completed by the National Toxicology Program Interagency Center for the Evaluation of Alternative Toxicological Methods. Regul Toxicol Pharmacol. 2001;34(3):274–286. doi: 10.1006/rtph.2001.1498. [DOI] [PubMed] [Google Scholar]
- dos Santos GG, Reinders J, Ouwehand K, Rustemeyer T, Scheper RJ, Gibbs S. Progress on the development of human in vitro dendritic cell based assays for assessment of the sensitizing potential of a compound. Toxicol Appl Pharmacol. 2009;236(3):372–382. doi: 10.1016/j.taap.2009.02.004. [DOI] [PubMed] [Google Scholar]
- Ashikaga T, Yoshida Y, Hirota M, Yoneyama K, Itagaki H, Sakaguchi H, Miyazawa M, Ito Y, Suzuki H, Toyoda H. Development of an in vitro skin sensitization test using human cell lines: the human Cell Line Activation Test (h-CLAT). I. Optimization of the h-CLAT protocol. Toxicol In Vitro. 2006;20(5):767–773. doi: 10.1016/j.tiv.2005.10.012. [DOI] [PubMed] [Google Scholar]
- Sakaguchi H, Ashikaga T, Miyazawa M, Yoshida Y, Ito Y, Yoneyama K, Hirota M, Itagaki H, Toyoda H, Suzuki H. Development of an in vitro skin sensitization test using human cell lines; human Cell Line Activation Test (h-CLAT). II. An inter-laboratory study of the h-CLAT. Toxicol In Vitro. 2006;20(5):774–784. doi: 10.1016/j.tiv.2005.10.014. [DOI] [PubMed] [Google Scholar]
- Ade N, Martinozzi-Teissier S, Pallardy M, Rousset F. Activation of U937 cells by contact sensitizers: CD86 expression is independent of apoptosis. J Immunotoxicol. 2006;3(4):189–197. doi: 10.1080/15476910600978038. [DOI] [PubMed] [Google Scholar]
- Hooyberghs J, Schoeters E, Lambrechts N, Nelissen I, Witters H, Schoeters G, Van Den Heuvel R. A cell-based in vitro alternative to identify skin sensitizers by gene expression. Toxicol Appl Pharmacol. 2008;231(1):103–111. doi: 10.1016/j.taap.2008.03.014. [DOI] [PubMed] [Google Scholar]
- Lambrechts N, Vanheel H, Hooyberghs J, De Boever P, Witters H, Van Den Heuvel R, Van Tendeloo V, Nelissen I, Schoeters G. Gene markers in dendritic cells unravel pieces of the skin sensitization puzzle. Toxicol Lett. 2010;196(2):95–103. doi: 10.1016/j.toxlet.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Lambrechts N, Vanheel H, Nelissen I, Witters H, Van Den Heuvel R, Van Tendeloo V, Schoeters G, Hooyberghs J. Assessment of chemical skin-sensitizing potency by an in vitro assay based on human dendritic cells. Toxicol Sci. 2010;116(1):122–129. doi: 10.1093/toxsci/kfq108. [DOI] [PubMed] [Google Scholar]
- Freudenberg MA, Esser PR, Jakob T, Galanos C, Martin SF. Innate and adaptive immune responses in contact dermatitis: analogy with infections. G Ital Dermatol Venereol. 2009;144(2):173–185. [PubMed] [Google Scholar]
- Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci USA. 2002;99(18):11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsch A, Emter R. Skin sensitizers induce antioxidant response element dependent genes: application to the in vitro testing of the sensitization potential of chemicals. Toxicol Sci. 2008;102(1):110–119. doi: 10.1093/toxsci/kfm259. [DOI] [PubMed] [Google Scholar]
- Ade N, Leon F, Pallardy M, Peiffer JL, Kerdine-Romer S, Tissier MH, Bonnet PA, Fabre I, Ourlin JC. HMOX1 and NQO1 genes are upregulated in response to contact sensitizers in dendritic cells and THP-1 cell line: role of the Keap1/Nrf2 pathway. Toxicol Sci. 2009;107(2):451–460. doi: 10.1093/toxsci/kfn243. [DOI] [PubMed] [Google Scholar]
- Mizuashi M, Ohtani T, Nakagawa S, Aiba S. Redox imbalance induced by contact sensitizers triggers the maturation of dendritic cells. J Invest Dermatol. 2005;124(3):579–586. doi: 10.1111/j.0022-202X.2005.23624.x. [DOI] [PubMed] [Google Scholar]
- Abel J, Haarmann-Stemmann T. An introduction to the molecular basics of aryl hydrocarbon receptor biology. Biol Chem. 2010;391(11):1235–1248. doi: 10.1515/BC.2010.128. [DOI] [PubMed] [Google Scholar]
- Jaiswal AK. Human NAD(P)H:quinone oxidoreductase (NQO1) gene structure and induction by dioxin. Biochemistry. 1991;30(44):10647–10653. doi: 10.1021/bi00108a007. [DOI] [PubMed] [Google Scholar]
- Kohle C, Bock KW. Coordinate regulation of Phase I and II xenobiotic metabolisms by the Ah receptor and Nrf2. Biochem Pharmacol. 2007;73(12):1853–1862. doi: 10.1016/j.bcp.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Allenby G, Bocquel MT, Saunders M, Kazmer S, Speck J, Rosenberger M, Lovey A, Kastner P, Grippo JF, Chambon P. et al. Retinoic acid receptors and retinoid × receptors: interactions with endogenous retinoic acids. Proc Natl Acad Sci USA. 1993;90(1):30–34. doi: 10.1073/pnas.90.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Konishi T, Han G, Campwala KH, French SW, Wan YJ. The role of hepatocyte RXR alpha in xenobiotic-sensing nuclear receptor-mediated pathways. Eur J Pharm Sci. 2002;15(1):89–96. doi: 10.1016/S0928-0987(01)00211-1. [DOI] [PubMed] [Google Scholar]
- Wu Y, Zhang X, Bardag-Gorce F, Robel RC, Aguilo J, Chen L, Zeng Y, Hwang K, French SW, Lu SC. et al. Retinoid × receptor alpha regulates glutathione homeostasis and xenobiotic detoxification processes in mouse liver. Mol Pharmacol. 2004;65(3):550–557. doi: 10.1124/mol.65.3.550. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. Toll-like receptor and RIG-I-like receptor signaling. Ann N Y Acad Sci. 2008;1143:1–20. doi: 10.1196/annals.1443.020. [DOI] [PubMed] [Google Scholar]
- Palm NW, Medzhitov R. Pattern recognition receptors and control of adaptive immunity. Immunol Rev. 2009;227(1):221–233. doi: 10.1111/j.1600-065X.2008.00731.x. [DOI] [PubMed] [Google Scholar]
- Beutler B. Microbe sensing, positive feedback loops, and the pathogenesis of inflammatory diseases. Immunol Rev. 2009;227(1):248–263. doi: 10.1111/j.1600-065X.2008.00733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Raghavan B, Muller V, Vogl T, Fejer G, Tchaptchet S, Keck S, Kalis C, Nielsen PJ, Galanos C. et al. Crucial role for human Toll-like receptor 4 in the development of contact allergy to nickel. Nat Immunol. 2010;11(9):814–819. doi: 10.1038/ni.1919. [DOI] [PubMed] [Google Scholar]
- Oesch-Bartlomowicz B, Oesch F. Phosphorylation of xenobiotic-metabolizing cytochromes P450. Anal Bioanal Chem. 2008;392(6):1085–1092. doi: 10.1007/s00216-008-2315-2. [DOI] [PubMed] [Google Scholar]
- Lindstedt M, Schiott A, Bengtsson A, Larsson K, Korsgren M, Greiff L, Borrebaeck CA. Genomic and functional delineation of dendritic cells and memory T cells derived from grass pollen-allergic patients and healthy individuals. Int Immunol. 2005;17(4):401–409. doi: 10.1093/intimm/dxh220. [DOI] [PubMed] [Google Scholar]
- Carlsson A, Wingren C, Kristensson M, Rose C, Fernö M, Olsson H, Jernström H, Ek S, Gustavsson E, Ingvar C, Ohlsson M, Peterson C, Borrebaeck CAK. Molecular serum portraits in patients with primary breast cancer predict the devlopment of distant metastases. PNAS. in press . [DOI] [PMC free article] [PubMed]
- R: A language and environment for statistical computing. R Foundation for Statistical Computing. http://www.R-project.org
- R package e1071. http://cran.r-project.org/web/packages/e1071/index.html
- Lasko TA, Bhagwat JG, Zou KH, Ohno-Machado L. The use of receiver operating characteristic curves in biomedical informatics. J Biomed Inform. 2005;38(5):404–415. doi: 10.1016/j.jbi.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Warbrick EV, Dearman RJ, Basketter DA, Kimber I. Influence of application vehicle on skin sensitization to methylchloroisothiazolinone/methylisothiazolinone: an analysis using the local lymph node assay. Contact Dermatitis. 1999;41(6):325–329. doi: 10.1111/j.1600-0536.1999.tb06181.x. [DOI] [PubMed] [Google Scholar]
- Roberts DW, Patlewicz G, Kern PS, Gerberick F, Kimber I, Dearman RJ, Ryan CA, Basketter DA, Aptula AO. Mechanistic applicability domain classification of a local lymph node assay dataset for skin sensitization. Chem Res Toxicol. 2007;20(7):1019–1030. doi: 10.1021/tx700024w. [DOI] [PubMed] [Google Scholar]
- Gerberick GF, Robinson MK, Ryan CA, Dearman RJ, Kimber I, Basketter DA, Wright Z, Marks JG. Contact allergenic potency: correlation of human and local lymph node assay data. Am J Contact Dermat. 2001;12(3):156–161. doi: 10.1053/ajcd.2001.23926. [DOI] [PubMed] [Google Scholar]
- Basketter DA, Sanders D, Jowsey IR. The skin sensitization potential of resorcinol: experience with the local lymph node assay. Contact Dermatitis. 2007;56(4):196–200. doi: 10.1111/j.1600-0536.2006.01008.x. [DOI] [PubMed] [Google Scholar]
- Ryan CA, Gerberick GF, Cruse LW, Basketter DA, Lea L, Blaikie L, Dearman RJ, Warbrick EV, Kimber I. Activity of human contact allergens in the murine local lymph node assay. Contact Dermatitis. 2000;43(2):95–102. doi: 10.1034/j.1600-0536.2000.043002095.x. [DOI] [PubMed] [Google Scholar]
- APPENDIX C; Comparative LLNA: BrdU-FC, Traditional LLNA, Guinea Pig Skin Sensitization, and Human Data. http://iccvam.niehs.nih.gov/methods/immunotox/fcLLNA/Appx/AppendixC_LLNA_FC07Jan08FD.pdf
- Patlewicz G, Basketter DA, Smith CK, Hotchkiss SA, Roberts DW. Skin-sensitization structure-activity relationships for aldehydes. Contact Dermatitis. 2001;44(6):331–336. doi: 10.1034/j.1600-0536.2001.044006331.x. [DOI] [PubMed] [Google Scholar]
- Basketter DA, Gerberick GF, Kimber I. Strategies for identifying false positive responses in predictive skin sensitization tests. Food Chem Toxicol. 1998;36(4):327–333. doi: 10.1016/S0278-6915(97)00158-0. [DOI] [PubMed] [Google Scholar]
- Ashby J, Basketter DA, Paton D, Kimber I. Structure activity relationships in skin sensitization using the murine local lymph node assay. Toxicology. 1995;103(3):177–194. doi: 10.1016/0300-483X(95)03132-Y. [DOI] [PubMed] [Google Scholar]
- The Murine Local Lymph Node Assay: A Test Method for Assessing the Allergic Contact Dermatitis Potential of Chemicals/Compounds. http://iccvam.niehs.nih.gov/docs/immunotox_docs/llna/llnarep.pdf