Abstract
Background
Doubly Uniparental Inheritance (DUI) is a fascinating exception to matrilinear inheritance of mitochondrial DNA (mtDNA). Species with DUI are characterized by two distinct mtDNAs that are inherited either through females (F-mtDNA) or through males (M-mtDNA). DUI sex-linked mitochondrial genomes share several unusual features, such as additional protein coding genes and unusual gene duplications/structures, which have been related to the functionality of DUI. Recently, new evidence for DUI was found in the mytilid bivalve Musculista senhousia. This paper describes the complete sex-linked mitochondrial genomes of this species.
Results
Our analysis highlights that both M and F mtDNAs share roughly the same gene content and order, but with some remarkable differences. The Musculista sex-linked mtDNAs have differently organized putative control regions (CR), which include repeats and palindromic motifs, thought to provide sites for DNA-binding proteins involved in the transcriptional machinery. Moreover, in male mtDNA, two cox2 genes were found, one (M-cox2b) 123bp longer.
Conclusions
The complete mtDNA genome characterization of DUI bivalves is the first step to unravel the complex genetic signals allowing Doubly Uniparental Inheritance, and the evolutionary implications of such an unusual transmission route in mitochondrial genome evolution in Bivalvia. The observed redundancy of the palindromic motifs in Musculista M-mtDNA may have a role on the process by which sperm mtDNA becomes dominant or exclusive of the male germline of DUI species. Moreover, the duplicated M-COX2b gene may have a different, still unknown, function related to DUI, in accordance to what has been already proposed for other DUI species in which a similar cox2 extension has been hypothesized to be a tag for male mitochondria.
Background
Metazoan mitochondrial DNA (mtDNA) is generally a small molecule (15-20 kb), and although much larger mitochondrial genomes have occasionally been found, they are often products of duplications of mtDNA portions, rather than variations in gene content [1,2]. The typical mitochondrial gene complement encodes 13 protein subunits of the oxidative phosphorylation enzymes, 2 rRNAs and 22 tRNAs. However, the coding sequences (CDS) can be up to 16, the tRNAs up to 27 (source MitoZoa: http://mi.caspur.it/mitozoa see [3]), and the rRNAs can be duplicated and/or fragmented in discontinuous genes, as in oysters [4]. Generally, there is also a single large non-coding region that is known to contain regulatory elements for replication and transcription (i.e. 'Control Region', CR), but it is unclear whether it is homologous among distantly related animals or, alternatively, it independently arose from various non-coding sequences. This difficulty in establishing homology is because CRs share sequence similarity only among closely related taxa. Finally, the mtDNA is almost always a circular molecule: only the cnidarian classes Cubozoa, Scyphozoa and Hydrozoa have been found to have linear mtDNA chromosomes [5]. All metazoan mitochondrial genes have homologs in plants, fungi and/or protists [6-9].
The Mollusca is the second largest animal Phylum and currently 99 complete mitochondrial genomes are available in Genbank; among those, only 38 are from Bivalvia, the second class in terms of species richness among mollusks. So far, bivalve mtDNA displays an extraordinary amount of variation in gene arrangement, i.e. very few shared gene boundaries are detectable, and gene translocations are common across all gene classes (protein-coding genes, tRNAs and rRNAs). For this reason, bivalve mitochondrial genome may provide an excellent experimental system to review and test models of mt gene rearrangement evolution, which were mainly developed in groups with stable genomes, such as vertebrates or arthropods. In addition, gene duplications and/or losses are present in almost every bivalve taxon in which a complete mitochondrial genome is available (see [10]). It is therefore evident that efforts should be made to improve the knowledge of bivalve mitochondrial genomes.
Another interesting feature of bivalve mtDNA is its unusual transmission route, which is found in some species: while in Metazoa mtDNA is known to be usually transmitted by Strict Maternal Inheritance (SMI; [11,12]), some bivalve mollusks show a deviation from this rule, named Doubly Uniparental Inheritance (DUI; [13,14]). DUI was found in species belonging to seven different bivalve families: Donacidae, Hyriidae, Margaritiferidae, Mytilidae, Solenidae, Unionidae, and Veneridae ([15,16]). Species with DUI are characterized by the presence of two distinct gender-associated mtDNAs: one transmitted through eggs (F) and one transmitted through sperm (M). The F and M genomes show up to 52% nucleotide divergence [17]. DUI seems at first to violate the universal rule of uniparental inheritance of organelles, because males receive their mtDNA from both parents and their tissues are heteroplasmic. However the two mtDNAs segregate independently: the F-type is transmitted to the next generation only through females, while the M-type is only transmitted from father to sons, therefore both genomes are actually transmitted uniparentally.
Because of its unique features, DUI should be a choice model to address many aspects of a wide range of biological sub-fields such as mitochondria inheritance, mtDNA evolution and recombination, genomic conflicts, evolution of sex and developmental biology (see [18] for a review).
Recently, evidence for a new example of DUI was found in the mytilid Musculista senhousia [19]. In this work we characterized the two sex-linked mitochondrial genomes of M. senhousia, a step forward to the complete genetic characterization of DUI related sex-linked mitochondrial genomes. In fact, several unusual features are coming to light when analyzing mtDNAs in DUI systems, such as additional protein coding genes ([20], and references therein) and gene duplications/features [21,22]. Functional explanations for these features will require much additional work, but are needed to understand the evolution and maintenance of DUI.
Results
Mitochondrial genome features in M. senhousia
The obtained M. senhousia mtDNAs are 21,557 bp long in female (F-type) and 20,612 bp in male (M-type) (see Tables 1 and 2). Sequences are available in GenBank (Acc. No. GU001953-GU001954). The size of both F and M mitochondrial genomes are within the size range of mollusk mtDNAs sequenced to date, i.e. from 7808 bp in Batilaria cumingi to 32,115 bp in Placopecten magellanicus (source MitoZoa: http://mi.caspur.it/mitozoa; [3]).
Table 1.
Organization of female Musculista senhousia mitochondrial genome.
| Type | Name | Starts | Stops | Length | Strand | Anticodon | Start Codon | Stop Codon |
|---|---|---|---|---|---|---|---|---|
| GENE | nad3 | 1 | 390 | 390 | H | ATG | TAA | |
| UR | UR-1 | 391 | 625 | 235 | ||||
| tRNA | trnY | 626 | 691 | 66 | H | GTA | ||
| UR | UR-2 | 692 | 1234 | 543 | ||||
| tRNA | trnH | 1235 | 1299 | 65 | H | GTG | ||
| UR | UR-3 | 1300 | 1315 | 16 | ||||
| tRNA | trnI | 1316 | 1381 | 66 | H | GAT | ||
| UR | UR-4 | 1382 | 1391 | 10 | ||||
| tRNA | trnN | 1392 | 1457 | 66 | H | GTT | ||
| UR | UR-5 | 1458 | 1564 | 107 | ||||
| tRNA | trnE | 1565 | 1631 | 67 | H | TTC | ||
| LUR | LUR | 1632 | 6152 | 4521 | ||||
| GENE | cox1 | 6153 | 7736 | 1584 | H | ATG | TAA | |
| UR | UR-6 | 7737 | 8114 | 378 | ||||
| GENE | cox2 | 8115 | 8774 | 660 | H | ATA | TAA | |
| UR | UR-7 | 8775 | 8832 | 58 | ||||
| GENE | atp8 | 8833 | 8967 | 135 | H | ATG | TAA | |
| UR | UR-8 | 8968 | 9051 | 84 | H | |||
| GENE | atp6 | 9052 | 9765 | 714 | H | ATG | TAG | |
| UR | UR-9 | 9766 | 9791 | 26 | ||||
| tRNA | trnT | 9792 | 9858 | 67 | H | TGT | ||
| GENE | cob | 9835 | 11031 | 1197 | H | ATA | TAA | |
| UR | UR-10 | 11032 | 11049 | 18 | ||||
| tRNA | trnD | 11050 | 11114 | 65 | H | GTC | ||
| UR | UR-11 | 11115 | 11123 | 9 | ||||
| tRNA | trnR | 11124 | 11189 | 66 | H | TCG | ||
| tRNA | trnS(AGN) | 11191 | 11248 | 58 | H | TCT | ||
| UR | UR-12 | 11249 | 11268 | 20 | ||||
| tRNA | trnG | 11269 | 11336 | 68 | H | TCC | ||
| rRNA | rrnS | 11337 | 12154 | 818 | H | |||
| GENE | nad6 | 12155 | 12778 | 624 | H | ATG | TAA | |
| UR | UR-13 | 12779 | 12828 | 50 | ||||
| GENE | nad2 | 12829 | 13773 | 945 | H | ATA | TAA | |
| UR | UR-14 | 13774 | 13855 | 82 | ||||
| GENE | cox3 | 13856 | 14710 | 855 | H | ATG | TAA | |
| UR | UR-15 | 14711 | 14721 | 11 | ||||
| tRNA | trnK | 14722 | 14792 | 71 | H | TTT | ||
| UR | UR-16 | 14793 | 14797 | 5 | ||||
| tRNA | trnF | 14798 | 14865 | 68 | H | GAA | ||
| UR | UR-17 | 14866 | 14878 | 13 | ||||
| tRNA | trnP | 14879 | 14945 | 67 | H | TGG | ||
| UR | UR-18 | 14946 | 14977 | 32 | ||||
| tRNA | trnL(CUN) | 14978 | 15042 | 65 | H | TAG | ||
| UR | UR-19 | 15043 | 15047 | 5 | ||||
| tRNA | trnC | 15048 | 15114 | 67 | H | GCA | ||
| UR | UR-20 | 15115 | 15159 | 45 | ||||
| tRNA | trnL(UUR) | 15160 | 15223 | 64 | H | TAA | ||
| UR | UR-21 | 15224 | 15259 | 36 | ||||
| GENE | nad1 | 15260 | 16252 | 993 | H | ATG | TAA | |
| UR | UR-22 | 16253 | 16385 | 133 | ||||
| tRNA | trnM(AUA) | 16386 | 16448 | 63 | H | TAT | ||
| UR | UR-23 | 16449 | 16486 | 38 | ||||
| tRNA | trnV | 16487 | 16550 | 64 | H | TAC | ||
| UR | UR-24 | 16551 | 16695 | 145 | ||||
| GENE | nad4L | 16696 | 16911 | 216 | H | ATA | TAA | |
| UR | UR-25 | 16912 | 16988 | 77 | ||||
| GENE | nad5 | 16989 | 18738 | 1750 | H | ATA | T-- | |
| tRNA | trnA | 18739 | 18804 | 66 | H | TGC | ||
| UR | UR-26 | 18805 | 18843 | 39 | ||||
| GENE | nad4 | 18844 | 20163 | 1320 | H | ATA | TAG | |
| UR | UR-27 | 20164 | 20213 | 50 | ||||
| tRNA | trnW | 20214 | 20280 | 67 | H | TCA | ||
| UR | UR-28 | 20281 | 20285 | 5 | ||||
| tRNA | trnQ | 20286 | 20353 | 68 | H | TTG | ||
| UR | UR-29 | 20354 | 20360 | 7 | ||||
| tRNA | trnM(AUG) | 20361 | 20427 | 67 | H | CAT | ||
| rRNA | rrnL | 20428 | 21557 | 1130 | H |
Table 2.
Organization of male Musculista senhousia mitochondrial genome.
| Type | Name | Starts | Stops | Length | Strand | Anticodon | Start Codon | Stop Codon |
|---|---|---|---|---|---|---|---|---|
| GENE | nad3 | 1 | 375 | 375 | H | ATG | TAA | |
| UR | UR-1 | 376 | 433 | 58 | ||||
| tRNA | trnY | 434 | 501 | 68 | H | GTA | ||
| UR | UR-2 | 502 | 533 | 32 | ||||
| tRNA | trnH | 534 | 599 | 66 | H | GTG | ||
| UR | UR-3 | 600 | 618 | 19 | ||||
| tRNA | trnI | 619 | 688 | 70 | H | GAT | ||
| tRNA | trnN | 687 | 753 | 67 | H | GTT | ||
| LUR | LUR | 754 | 3597 | 2844 | ||||
| tRNA | trnE | 3598 | 3668 | 71 | H | TTC | ||
| UR | UR-4 | 3669 | 3708 | 40 | ||||
| GENE | cox1 | 3709 | 5292 | 1584 | H | ATG | TAA | |
| UR | UR-5 | 5293 | 5852 | 560 | ||||
| GENE | cox2b | 5853 | 6665 | 813 | H | ATG | TAA | |
| UR | UR-6 | 6666 | 6706 | 41 | ||||
| GENE | cox2 | 6707 | 7396 | 690 | H | ATA | TAA | |
| UR | UR-7 | 7397 | 7402 | 6 | ||||
| GENE | atp8 | 7403 | 7594 | 192 | H | ATG | TAG | |
| UR | UR-8 | 7595 | 7612 | 18 | ||||
| GENE | atp6 | 7613 | 8326 | 714 | H | ATG | TAA | |
| UR | UR-9 | 8327 | 8347 | 21 | ||||
| tRNA | trnT | 8348 | 8416 | 69 | H | TGT | ||
| GENE | cob | 8392 | 9588 | 1197 | H | ATA | TAA | |
| UR | UR-10 | 9589 | 9606 | 18 | ||||
| tRNA | trnD | 9607 | 9671 | 65 | H | GTC | ||
| UR | UR-11 | 9672 | 9681 | 10 | ||||
| tRNA | trnR | 9682 | 9745 | 64 | H | TCG | ||
| tRNA | trnS(AGN) | 9747 | 9806 | 60 | H | TCT | ||
| UR | UR-12 | 9807 | 9825 | 19 | ||||
| tRNA | trnG | 9826 | 9893 | 68 | H | TCC | ||
| rRNA | rrnS | 9894 | 10793 | 900 | H | |||
| GENE | nad6 | 10794 | 11417 | 624 | H | ATG | TAA | |
| UR | UR-13 | 11418 | 11472 | 55 | ||||
| GENE | nad2 | 11473 | 12417 | 945 | H | ATA | TAG | |
| UR | UR-14 | 12418 | 12444 | 27 | ||||
| GENE | cox3 | 12445 | 13299 | 855 | H | ATG | TAG | |
| tRNA | trnK | 13299 | 13366 | 68 | H | TTT | ||
| UR | UR-15 | 13367 | 13377 | 11 | ||||
| tRNA | trnF | 13378 | 13445 | 68 | H | GAA | ||
| UR | UR-16 | 13446 | 13464 | 19 | ||||
| tRNA | trnP | 13465 | 13528 | 64 | H | TGG | ||
| UR | UR-17 | 13529 | 13554 | 26 | ||||
| tRNA | trnL(CUN) | 13555 | 13621 | 67 | H | TAG | ||
| UR | UR-18 | 13622 | 13625 | 4 | ||||
| tRNA | trnC | 13626 | 13696 | 71 | H | GCA | ||
| UR | UR-19 | 13697 | 13737 | 41 | ||||
| tRNA | trnL(UUR) | 13738 | 13804 | 67 | H | TAA | ||
| UR | UR-20 | 13805 | 13840 | 36 | ||||
| GENE | nad1 | 13841 | 14836 | 996 | H | ATG | TAG | |
| tRNA | trnM(AUA) | 14835 | 14899 | 65 | H | TAT | ||
| UR | UR-21 | 14900 | 14985 | 86 | ||||
| tRNA | trnV | 14986 | 15049 | 64 | H | TAC | ||
| UR | UR-22 | 15050 | 15183 | 134 | ||||
| GENE | nad4L | 15184 | 15399 | 216 | H | ATA | TAA | |
| UR | UR-23 | 15400 | 15464 | 65 | ||||
| GENE | nad5 | 15465 | 17229 | 1765 | H | ATA | T-- | |
| tRNA | trnA | 17230 | 17294 | 65 | H | TGC | ||
| UR | UR-24 | 17295 | 17338 | 44 | ||||
| GENE | nad4 | 17339 | 18667 | 1329 | H | ATA | TAA | |
| UR | UR-25 | 18668 | 18710 | 43 | ||||
| tRNA | trnW | 18711 | 18777 | 67 | H | TCA | ||
| UR | UR-26 | 18778 | 18781 | 4 | ||||
| tRNA | trnQ | 18782 | 18848 | 67 | H | TTG | ||
| UR | UR-27 | 18849 | 18863 | 15 | ||||
| tRNA | trnM(AUG) | 18864 | 18930 | 67 | H | CAT | ||
| rRNA | rrnL | 18931 | 20612 | 1682 | H |
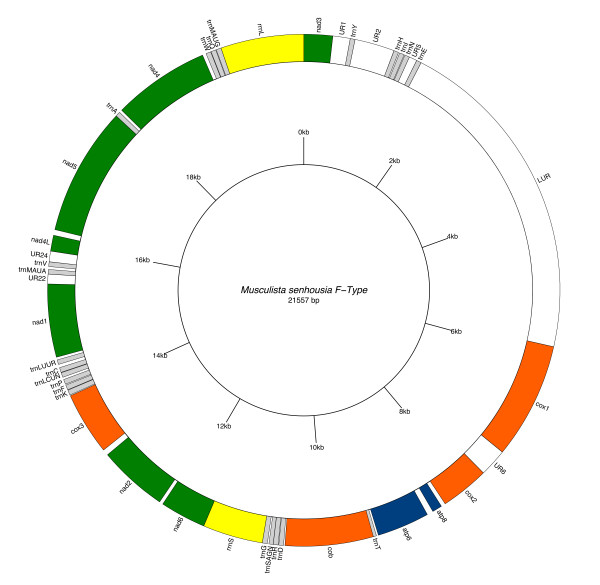
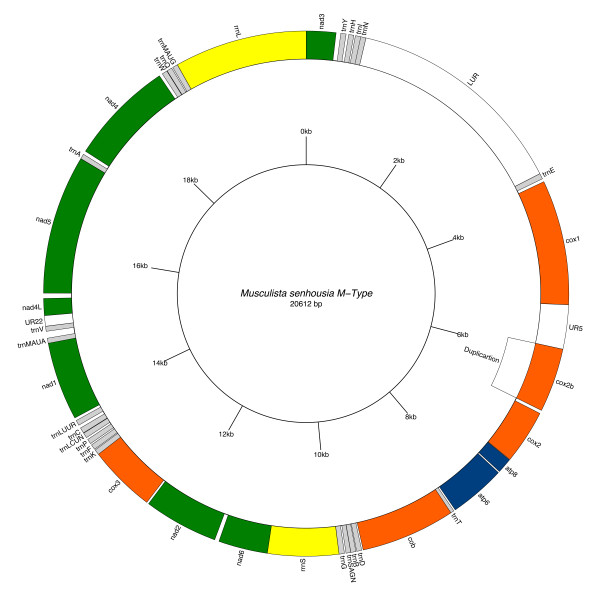
M. senhousia F and M gene arrangements are remarkably different from other fully sequenced metazoan mtDNAs (see [10] for a review). Genome annotations are reported in Figure 1 and 2, Table 1 and 2. When compared to other Mytilidae, only four gene boundaries are shared with Mytilus (tRNAs are not considered), i.e. rrnS-nad6, nad2-cox3, nad4L-nad5 and nad3-cox1, while the rest of the genome is different, thus highlighting that gene arrangement evolves rapidly within the family.
Figure 1.
Female Musculista senhousia mitochondrial genome. Gene map of the female Musculista senhousia mitochondrial genome. Shortest URs (< 100 bp) are not indicated.
Figure 2.
Male Musculista senhousia mitochondrial genome. Gene map of the male Musculista senhousia mitochondrial genome. Shortest URs (< 100 bp) are not indicated.
Comparing the two sex linked genomes, protein-coding genes may have different lengths (Table 3). Both F-type and M-type include a large number of Unassigned Regions (URs; 29 in F and 27 in M: see Tables 1, 2 and Additional File 1). Among these, the largest (4,521 and 2,844 bp in female and male respectively) are here referred as LURs (i.e. Large Unassigned Regions).
Table 3.
Length, base composition and sequence divergence of M, F genes and URs in Musculista senhousia.
| Gene/Region | F/M | Length | Base Composition (% T, C, A, G) | pD ± SE | |||
|---|---|---|---|---|---|---|---|
| UR1-27/LUR | M | 4296 | 37.8 | 11.2 | 31.4 | 19.5 | NA |
| UR1-29/LUR | F | 6798 | 37.9 | 10.4 | 30.8 | 20.9 | NA |
| rrnL | M | 1682 | 37.3 | 12.6 | 30.8 | 19.3 | 0.343 ± 0.015 |
| F | 1130 | 35.8 | 13.5 | 30.4 | 20.3 | ||
| rrnS | M | 900 | 36.0 | 11.6 | 33.1 | 19.3 | 0.093 ± 0.009 |
| F | 818 | 37.2 | 11.0 | 32.2 | 19.7 | ||
| all rRNA genes | M | 2582 | 36.3 | 12.2 | 31.6 | 19.3 | 0.209 ± 0.010 |
| F | 1948 | 36.4 | 12.4 | 31.2 | 20.0 | ||
| atp6 | M | 714 | 43.8 | 12.7 | 23.5 | 19.9 | 0.258 ± 0.016 |
| F | 714 | 42.2 | 12.9 | 23.8 | 21.1 | ||
| atp8 | M | 192 | 42.2 | 14.1 | 27.6 | 16.1 | 0.281 ± 0.037 |
| F | 135 | 43.0 | 12.6 | 25.9 | 18.5 | ||
| cox1 | M | 1584 | 38.3 | 15.9 | 24.7 | 21.1 | 0.180 ± 0.009 |
| F | 1584 | 40.0 | 14.4 | 24.4 | 21.3 | ||
| cox2 | M | 690 | 36.7 | 15.2 | 26.7 | 21.4 | 0.264 ± 0.016 |
| F | 660 | 37.4 | 14.5 | 27.3 | 20.8 | ||
| cox2b | M | 813 | 35.9 | 14.1 | 28.7 | 21.3 | 0.267 ± 0.016* |
| F | NA | NA | |||||
| cox3 | M | 855 | 42.0 | 13.1 | 23.3 | 21.6 | 0.220 ± 0.012 |
| F | 855 | 43.4 | 12.9 | 20.9 | 22.8 | ||
| cob | M | 1197 | 40.6 | 13.9 | 25.2 | 20.3 | 0.106 ± 0.009 |
| F | 1197 | 40.4 | 13.6 | 24.9 | 21.1 | ||
| nad1 | M | 996 | 39.8 | 12.2 | 26.0 | 22.0 | 0.227 ± 0.014 |
| F | 993 | 41.3 | 11.5 | 24.4 | 23.2 | ||
| nad2 | M | 945 | 44.9 | 10.8 | 24.4 | 19.9 | 0.302 ± 0.013 |
| F | 945 | 44.1 | 10.9 | 22.4 | 22.5 | ||
| nad3 | M | 375 | 44.3 | 14.1 | 21.3 | 20.3 | 0.267 ± 0.021 |
| F | 390 | 45.6 | 12.6 | 21.0 | 20.8 | ||
| nad4 | M | 1329 | 41.4 | 11.5 | 23.6 | 23.5 | 0.273 ± 0.013 |
| F | 1320 | 39.9 | 11.9 | 24.3 | 23.9 | ||
| nad4L | M | 216 | 43.5 | 8.8 | 24.5 | 23.1 | 0.199 ± 0.027 |
| F | 216 | 44.0 | 8.8 | 24.5 | 22.7 | ||
| nad5 | M | 1765 | 39.5 | 13.2 | 27.9 | 19.4 | 0.285 ± 0.011 |
| F | 1750 | 38.7 | 13.3 | 25.7 | 22.3 | ||
| nad6 | M | 624 | 43.8 | 11.4 | 25.6 | 19.2 | 0.217 ± 0.017 |
| F | 624 | 42.1 | 12.3 | 25.2 | 20.4 | ||
| all proteins | M | 12295 | 40.6 | 13.2 | 25.4 | 20.9 | 0.231 ± 0.004# |
| F | 11383 | 40.9 | 12.8 | 24.1 | 22.1 | ||
| complete | M | 20612 | 39.3 | 12.7 | 27.7 | 20.3 | NA |
| F | 21557 | 39.3 | 12.0 | 27.2 | 21.4 | ||
UR = Unassigned Regions.
NA = Not Available.
pD = p-Distance.
SE = Standard Error.
*: pD between Mcox2 and Mcox2b genes.
#: Mcox2b gene was excluded from the computation of overall pD.
Both F and M mt genomes show the same gene order and contain the full gene complement of the typical metazoan mtDNA, with two additional tRNAs: trnM and trnL (Figures 1 and 2; Tables 1 and 2). In males the cox2 gene is duplicated (Figure 2 and Table 2).
The atp8 gene was reported as missing in several bivalve mollusks, however, as recently reported [23], the lack of atp8 would rather be an annotation inaccuracy due to the extreme variability of the gene. Following [23], we found an atp8 gene in M. senhousia in both M and F genomes.
The position of the two ribosomal RNA genes, obtained through BLAST comparison, does not differ between male and female. In both sexes, rrnL is located in a region flanked by the trnM(AUG) and nad3 genes. Assuming that the first base at the 5'-end comes immediately after the trnM(AUG), and the 3'-end of the gene corresponds to the first base upstream of the start codon of nad3 gene, the length of the rrnL genes are remarkably different: the male rrnL (1,682 bp in length) is 552 bp longer than the female one (1,130 bp in length). The rrnS gene is located in a region flanked by trnS and nad6 genes and, as above, we assumed that the first base at the 5'-end comes immediately after trnG, and that the 3'-end of the gene corresponds to the first base upstream of the start codon of nad6 gene. Here, the difference in length is reduced to 82 bp: the female rrnS gene is 819 bp long while the male one is 1,087 bp.
F and M genomes of M. senhousia contain 22 tRNA genes (see Tables 1, 2 and Additional File 2). As observed in mtDNA of some other mollusks (Katharina tunicata, Cepaea nemoralis, Mytilus species complex and Argopecten irradians), two leucine tRNA genes are present in M. senhousia. These can be differentiated by their anticodons: TAA for trnL(UUR) and TAG for trnL(CUN), which are 2-fold and 4-fold redundant respectively. Consequently, tnrL is 6-fold redundant. An additional trnM was also detected, as in V. philippinarum, Mytilus species complex, Crassostrea gigas, C. hongkongensis and C. virginica. The additional tRNA coding for methionine, trnM(AUA), has the TAT anticodon.
In both male and female mtDNAs, trnS(AGN) have a shortened DHU (See Additional File 2) that is not atypical, as this arm is unpaired in many metazoan taxa [24-27]. Moreover, mispairing between bases in stems is consistent across several taxa. For example, the second base pair in the anticodon stem of trnW has a T-T mispairing in Lampsilis ornata, Mytilus, and K. tunicata and a T-G pairing in several gastropods [25].
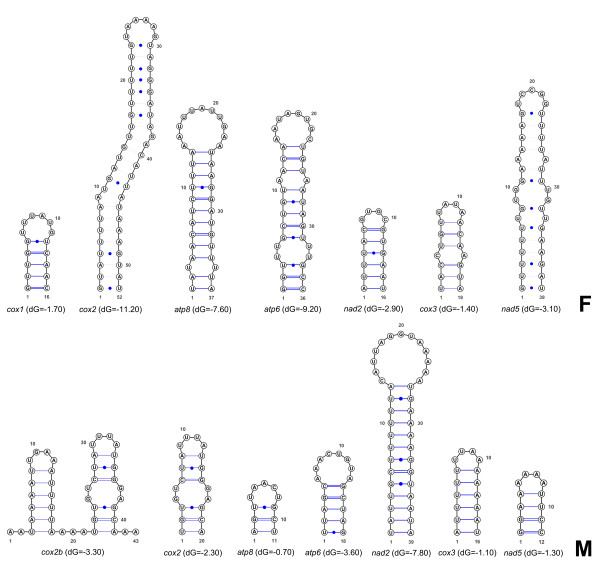
In the F mitochondrial genome of Musculista, 20 out of 22 tRNA genes are clustered in five groups of two to six (see Figure 1 and Table 1). Of the remaining two, trnT lies between atp6 and the 5'-end of cob genes (with 24 bp overlapping each other) while trnA lies between nad5 and nad4 genes. Thus, 4 of the 13 protein-coding genes (cob, nad1, nad4L and nad4) have a tRNA preceding their 5'-end. In contrast, 7 other genes (cox1, cox2, atp8, atp6, nad2, cox3 and nad5) have a non-coding sequence at their 5'-end that is capable of forming a stem and loop structure (see Figure 3).
Figure 3.
Intergenic palindromes. Putative secondary structures preceding the 5'-end of some protein-coding genes. (F) Female Musculista senhousia mitochondrial genome. (M) Male Musculista senhousia mitochondrial genome.
In male mitochondrial DNA, 19 of the 22 tRNA genes are clustered in five groups ranging from two to six (see Figure 2 and Table 2). Of the remaining three, trnT lies between atp6 and the 5'-end of cob genes (with 25 bp overlapping each other), trnA lies between nad5 and nad4 genes and trnE lies between the large unassigned region (LUR) and the 5'-end of cox1 gene. Thus, 5 of the 14 protein-coding genes (cox1, cob, nad1, nad4L and nad4) have a tRNA preceding their 5'-end, while 7 other genes (cox2b, cox2, atp8, atp6, nad2, cox3 and nad5) have a non-coding sequence preceding their 5'-end that is capable of forming a stem and loop structure (see Figure 3). In a few cases those structures contain the translation initiation codon (cox1 and cox2 in females, nad2 in males).
The nucleotide compositions of the two genomes are summarized in Table 3. Given the G content of the F and M coding strand (see Table 3), this can be considered as the heavy (H) strand of the molecule. The A+T content of the H strand is also high (66.5%, F; 67.0%, M). Variable values of A+T content are common in mollusks, and they have been reported in L. ornata (62%, [28]), Pupa strigosa (61.1%, [29]), and C. nemoralis (59.8%, [25]). In other mollusks, the A+T content is much higher (Albinaria coerulea, 70.7%, [30]; K. tunicata, 69.0%, [6]; Graptame eborea, 74.1%, [31]). Musculista values in A+T content are among the highest observed in the Phylum, and reflect the high heterogeneity of molluscan mtDNA [2]. Moreover, there is a marked bias in favor of T against C, which is not restricted to any particular class of genes and does not differ between the two genomes.
The GC and AT asymmetry between the two mitochondrial DNA strands can be expressed in terms of GC skew and AT skew calculated according to [32]: GC skew = (G-C)/(G+C) and AT skew = (A-T)/(A+T), where G, C, A, and T are the occurrences of the four bases in the H strand. In M. senhousia F and M mitochondrial genomes, the GC skew and the AT skew are F: +0.28 and -0.18, and M: +0.23 and -0.17, respectively.
In the M. senhousia male mtDNA 6 out of 14 protein genes start with the ATA codon and 8 with ATG, while in the female 7 out of 13 start with ATG and 6 with ATA (Tables 1 and 2). This pattern differs from that observed for Mytilus galloprovincialis, where 9 out of 13 protein genes start with the ATG codon, 2 with the ATA and 2 with GTG [23,33]. In all known metazoan mtDNAs, the most common start codon is ATG, and it is a general opinion that the methionine tRNA with the CAT anticodon represents the ancestral form. Moreover [24] suggested that the second methionine tRNA arose by duplication. The F and M genomes of the venerid Venerupis philippinarum also have two tRNA genes for methionine, but both have the ancestral CAT anticodon. TAA is the termination codon ten times in F and nine times in M mtDNA, while TAG is a stop codon two times in F, and four times in M. In both M and F genomes, nad5 gene is terminated by an incomplete termination codon T-- (Tables 1 and 2), with their likely completion occurring by polyadenylation after transcript processing [34].
A total of 4,098 and 3,794 amino acids residues are encoded by male and female M. senhousia mitochondrial genome respectively (Table 4). All codons do occur in both Musculista mitochondrial genomes (Table 5). UUU (phenylalanine) is the most frequent codon, followed by UUA (leucine). UUU is also the most frequent codon in M. galloprovincialis [33], in L. ornata [28] and in C. nemoralis [35], whereas UUA (leucine) is most common in A. coerulea [30], P. strigosa [29], Roboastra europaea [36], G. eborea [31], and K. tunicata [6]. These two codons are also the most frequently used in other invertebrate mtDNAs [37-42]. UUU is also very frequent in basal chordates (e.g. amphioxus, Branchiostoma lanceolatum, [43]), but not in most vertebrates, where CUA (e.g., Cyprinus, [44]; Homo sapiens, [45]) or AUU (e.g., Xenopus laevis, [46]; Danio rerio, [47]) are the most frequent.
Table 4.
Genes, gene lengths and divergences in male and female Musculista senhousia protein coding genes.
| Protein gene | Maa | Faa | pD ± SE | Ks | Ka | Ka/Ks |
|---|---|---|---|---|---|---|
| atp6 | 238 | 238 | 0.228 ± 0.026 | 0.894 | 0.156 | 0.17 |
| atp8 | 64 | 45 | 0.302 ± 0.070 | 0.581 | 0.233 | 0.40 |
| cox1 | 528 | 528 | 0.053 ± 0.009 | 0.838 | 0.042 | 0.05 |
| cox2 | 230 | 220 | 0.251 ± 0.027 | 0.877 | 0.178 | 0.20 |
| cox2b* | 271 | NA | 0.279 ± 0.029* | 0.653* | 0.223* | 0.35* |
| cox3 | 285 | 285 | 0.155 ± 0.022 | 0.811 | 0.107 | 0.13 |
| cob | 399 | 399 | 0.058 ± 0.012 | 0.346 | 0.034 | 0.10 |
| nad1 | 332 | 331 | 0.218 ± 0.022 | 0.670 | 0.145 | 0.22 |
| nad2 | 315 | 315 | 0.306 ± 0.026 | 0.843 | 0.244 | 0.29 |
| nad3 | 125 | 130 | 0.218 ± 0.034 | 0.964 | 0.162 | 0.17 |
| nad4 | 443 | 440 | 0.243 ± 0.020 | 0.931 | 0.175 | 0.19 |
| nad4L | 72 | 72 | 0.183 ± 0.045 | 0.626 | 0.107 | 0.17 |
| nad5 | 588 | 583 | 0.274 ± 0.018 | 0.862 | 0.208 | 0.24 |
| nad6 | 208 | 208 | 0.324 ± 0.031 | 0.619 | 0.268 | 0.43 |
| all proteins | 4,098 | 3,794 | 0.716 | 0.143 | 0.20 |
Maa and Faa = number of amino acids in male and female respectively.
pD = p-Distances at the amino acidic level.
Ks and Ka = divergence of protein genes in synonymous (Ks) and non synonymous (Ka) sites respectively.
SE = Standard Error.
Ka/Ks = ratio values between Ka and Ks.
*: comparisons between Mcox2 and Mcox2b genes.
Table 5.
Codon usage in male and female Musculista senhousia mitochondrial genomes.
| FEMALE | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| aa | Codon | Count | % | aa | Codon | Count | % | aa | Codon | Count | % | aa | Codon | Count | % | Codon | Count | % |
| Phe (F) | UUU | 303 | 8,0 | Ser (S) | UCU | 107 | 2,8 | Tyr (Y) | UAU | 125 | 3,3 | Cys (C) | UGU | 80 | 2,1 | UNU | 615 | 16,2 |
| UUC | 36 | 0,9 | UCC | 8 | 0,2 | UAC | 39 | 1,0 | UGC | 14 | 0,4 | UNC | 97 | 2,6 | ||||
| Leu (L) | UUA | 254 | 6,7 | UCA | 36 | 0,9 | s.c. (*) | UAA | 14 | 0,4 | Trp (W) | UGA | 53 | 1,4 | UNA | 357 | 9,4 | |
| UUG | 105 | 2,8 | UCG | 14 | 0,4 | UAG | 7 | 0,2 | UGG | 50 | 1,3 | UNG | 176 | 4,6 | ||||
| CUU | 89 | 2,3 | Pro (P) | CCU | 95 | 2,5 | His (H) | CAU | 58 | 1,5 | Arg (R) | CGU | 35 | 0,9 | CNU | 277 | 7,3 | |
| CUC | 20 | 0,5 | CCC | 13 | 0,3 | CAC | 15 | 0,4 | CGC | 7 | 0,2 | CNC | 55 | 1,5 | ||||
| CUA | 62 | 1,6 | CCA | 11 | 0,3 | Gln (Q) | CAA | 32 | 0,8 | CGA | 14 | 0,4 | CNA | 119 | 3,1 | |||
| CUG | 41 | 1,1 | CCG | 4 | 0,1 | CAG | 26 | 0,7 | CGG | 13 | 0,3 | CNG | 84 | 2,2 | ||||
| Ile (I) | AUU | 147 | 3,9 | Thr (T) | ACU | 54 | 1,4 | Asn (N) | AAU | 82 | 2,2 | Ser (S) | AGU | 71 | 1,9 | ANU | 354 | 9,3 |
| AUC | 41 | 1,1 | ACC | 9 | 0,2 | AAC | 27 | 0,7 | AGC | 30 | 0,8 | ANC | 107 | 2,8 | ||||
| Met (M) | AUA | 139 | 3,7 | ACA | 29 | 0,8 | Lys (K) | AAA | 81 | 2,1 | AGA | 90 | 2,4 | ANA | 339 | 8,9 | ||
| AUG | 62 | 1,6 | ACG | 17 | 0,4 | AAG | 33 | 0,9 | AGG | 68 | 1,8 | ANG | 180 | 4,7 | ||||
| Val (V) | GUU | 200 | 5,3 | Ala (A) | GCU | 88 | 2,3 | Asp (D) | GAU | 59 | 1,6 | Gly (G) | GGU | 102 | 2,7 | GNU | 449 | 11,8 |
| GUC | 24 | 0,6 | GCC | 17 | 0,4 | GAC | 15 | 0,4 | GGC | 39 | 1,0 | GNC | 95 | 2,5 | ||||
| GUA | 113 | 3,0 | GCA | 44 | 1,2 | Glu (E) | GAA | 44 | 1,2 | GGA | 43 | 1,1 | GNA | 244 | 6,4 | |||
| GUG | 84 | 2,2 | GCG | 22 | 0,6 | GAG | 49 | 1,3 | GGG | 89 | 2,3 | GNG | 244 | 6,4 | ||||
| NUN | 1720 | 45,4 | NCN | 568 | 15,0 | NAN | 706 | 18,6 | NGN | 798 | 21,0 | Total | 3792 | |||||
| MALE | ||||||||||||||||||
| aa | Codon | Count | % | aa | Codon | Count | % | aa | Codon | Count | % | aa | Codon | Count | % | Codon | Count | % |
| Phe (F) | UUU | 333 | 8,1 | Ser (S) | UCU | 131 | 3,2 | Tyr (Y) | UAU | 133 | 3,2 | Cys (C) | UGU | 90 | 2,2 | UNU | 687 | 16,8 |
| UUC | 57 | 1,4 | UCC | 22 | 0,5 | UAC | 36 | 0,9 | UGC | 15 | 0,4 | UNC | 130 | 3,2 | ||||
| Leu (L) | UUA | 274 | 6,7 | UCA | 36 | 0,9 | s.c. (*) | UAA | 18 | 0,4 | Trp (W) | UGA | 69 | 1,7 | UNA | 397 | 9,7 | |
| UUG | 104 | 2,5 | UCG | 6 | 0,1 | UAG | 10 | 0,2 | UGG | 46 | 1,1 | UNG | 166 | 4,1 | ||||
| CUU | 86 | 2,1 | Pro (P) | CCU | 91 | 2,2 | His (H) | CAU | 51 | 1,2 | Arg (R) | CGU | 42 | 1,0 | CNU | 270 | 6,6 | |
| CUC | 16 | 0,4 | CCC | 14 | 0,3 | CAC | 30 | 0,7 | CGC | 11 | 0,3 | CNC | 71 | 1,7 | ||||
| CUA | 55 | 1,3 | CCA | 20 | 0,5 | Gln (Q) | CAA | 40 | 1,0 | CGA | 12 | 0,3 | CNA | 127 | 3,1 | |||
| CUG | 28 | 0,7 | CCG | 8 | 0,2 | CAG | 22 | 0,5 | CGG | 8 | 0,2 | CNG | 66 | 1,6 | ||||
| Ile (I) | AUU | 178 | 4,3 | Thr (T) | ACU | 61 | 1,5 | Asn (N) | AAU | 81 | 2,0 | Ser (S) | AGU | 78 | 1,9 | ANU | 398 | 9,7 |
| AUC | 43 | 1,0 | ACC | 22 | 0,5 | AAC | 52 | 1,3 | AGC | 43 | 1,0 | ANC | 160 | 3,9 | ||||
| Met (M) | AUA | 148 | 3,6 | ACA | 35 | 0,9 | Lys (K) | AAA | 104 | 2,5 | AGA | 97 | 2,4 | ANA | 384 | 9,4 | ||
| AUG | 79 | 1,9 | ACG | 12 | 0,3 | AAG | 38 | 0,9 | AGG | 75 | 1,8 | ANG | 204 | 5,0 | ||||
| Val (V) | GUU | 193 | 4,7 | Ala (A) | GCU | 81 | 2,0 | Asp (D) | GAU | 65 | 1,6 | Gly (G) | GGU | 103 | 2,5 | GNU | 442 | 10,8 |
| GUC | 30 | 0,7 | GCC | 22 | 0,5 | GAC | 22 | 0,5 | GGC | 28 | 0,7 | GNC | 102 | 2,5 | ||||
| GUA | 106 | 2,6 | GCA | 44 | 1,1 | Glu (E) | GAA | 59 | 1,4 | GGA | 53 | 1,3 | GNA | 262 | 6,4 | |||
| GUG | 83 | 2,0 | GCG | 19 | 0,5 | GAG | 42 | 1,0 | GGG | 88 | 2,1 | GNG | 232 | 5,7 | ||||
| NUN | 1813 | 44,2 | NCN | 624 | 15,2 | NAN | 803 | 19,6 | NGN | 858 | 20,9 | Total | 4098 | |||||
Codons that match the corresponding tRNA anticodon are bold and underlined.
aa: coded amminoacid.
s.c.: stop codon.
The least used codons in males are UCG (6), CCG (8) and CGG (8), while in females they are CCG (4), CGC (7) and UAG (7). Of these, CGC is also among the least common in the mtDNA of other mollusks. Synonymous codons, whether four-fold (4FD) or two-fold (2FD) degenerate, are recognized by the same tRNA, with the exception of the methionine codons, which are recognized by different tRNAs (Table 5).
Moreover, 2,754 F and 2,967 M Musculista codons (72.6% and 72.4% in female and in male respectively) end with an A or T, a more pronounced phenomenon than what observed for a typical invertebrate codon bias. There is a strong bias against the use of C (9.3% and 11.3% in female and in male respectively) at the third position nucleotide in all codons: in detail, for residues with a fourfold degenerate third position, codon families ending with T are the most frequently used (46.7% and 46.6% in female and male respectively). This is also the case for two-fold degenerate codons. In other words, in every case an amino acid residue can be specified by any NNY codon, both female and male M. senhousia mitochondrial genomes have a much higher proportion of NNT:NNC. In fact, female showed 44.7% of T and 9.3% of C, with NNT:NNC ratio of 4.8:1; while in male the ratio's value is slightly lower: 3.9:1 (43.8% of T and 11.2% of C). At the second position, there is even a stronger bias in favor of the use of T usage (45.4% and 44.2% in female and male respectively)(see Table 6), like in M. edulis (43.5%), C. hongkongensis (42.5%), C. gigas (42.3%) and C. virginica (43.0%).
Table 6.
p-Distance (± Standard Error) of LURs repeats, subregions and motifs.
| pD | SE | ||
|---|---|---|---|
| Rep1 | Rep2 | 0,004 | 0,001 |
| A1 | A2 | 0,000 | 0,000 |
| A1/2 | A'' | 0,362 | 0,032 |
| A1/2 | A' | 0,449 | 0,035 |
| A'' | A' | 0,505 | 0,033 |
| B1 | B2 | 0,002 | 0,001 |
| B2 | B | 0,096 | 0,007 |
| B1 | B | 0,098 | 0,007 |
| C1 | C2 | 0,010 | 0,005 |
| γC1 | γC2 | 0,008 | 0,005 |
| γ2 | γ3 | 0,012 | 0,006 |
| γ2 | γ1 | 0,015 | 0,007 |
| γ3 | γ1 | 0,019 | 0,009 |
| γC1/C2 | γ3 | 0,346 | 0,027 |
| γC1/C2 | γ1/2 | 0,350 | 0,027 |
Finally, in eight 2FD and seven 4FD codon families in females and in seven 2FD and seven 4FD codon families in males, the most frequently used codon does not match the tRNA anticodon. This has been observed in other metazoan mtDNA as well [46-50] and it suggests that strict codon-anticodon complementarity does not affect the codon composition of the genome. Deviations from equal frequency of the four nucleotides in 4FD sites are common in the animal mtDNA and have been attributed to several factors, such as unequal presence of the four nucleotides in the nucleotide pool, preference of the mitochondrial gamma DNA polymerase for specific nucleotides, or asymmetrical mutation rate owing to different duration of exposure of the lagging strand during replication [40,51-54].
Comparing the two M. senhousia sex linked genomes, the most conserved protein-coding genes are cox1 and cob, and the least conserved are nad6 and atp8 (Table 4). Synonymous (Ks) and non-synonymous (Ka) substitution values between the two genomes do vary (Table 4). Ka is particularly low for cox1 (0.042), whereas Ks is not (0.838), suggesting that this gene is under some selective constraint (Ka/Ks = 0.05). The conservation of cox1 is common in animal mtDNA [55,56]. In cob gene, both K values are lower than average (Table 4) with a Ka/Ks ratio's value (0.10) which is close to that of cox1 gene.
The Large Unassigned Region (LUR)
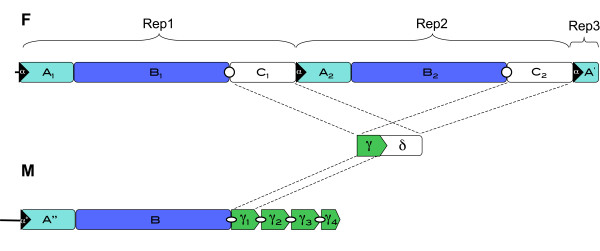
As mentioned, in the female genome the LUR (F-LUR) is 4,521 bp long and it is included between trnE and the 5'-end of cox1 gene (Figure 1 and 4, Table 1), while in the male it (M-LUR) is 2,844 bp long, and included between trnN and trnE genes (Figure 2 and 4, Table 2). Both start with a dissimilar sequence/spacer 20 and 237 bp long, respectively.
Figure 4.
Large Unassigned Regions (LURs). Schematic structure of female (F) and male (M) LURs in Musculista senhousia.
The F-LUR contains two large repeats (Figure 4: Rep1 and Rep2) about 2,150 bp long (2,149 Rep1; 2,151 Rep2), both subdividable in three regions: A, B and C (named A1, A2, B1, B2, C1 and C2; see Figure 4 and Additional File 3). Between Rep1 and Rep2, the A subregion is the most conserved (pD = 0.000, see Table 6) while C is the most variable, although with a low pD (0.010 ± 0.005). Overall, Rep1 and Rep2 have a pD of 0.004 ± 0.001. The region including the last 202 bp of the F-LUR shows some similarity (pD = 0.449 ± 0.035) to the A subregions (A1 and A2), for this reason it is indicated here as subregion A'.
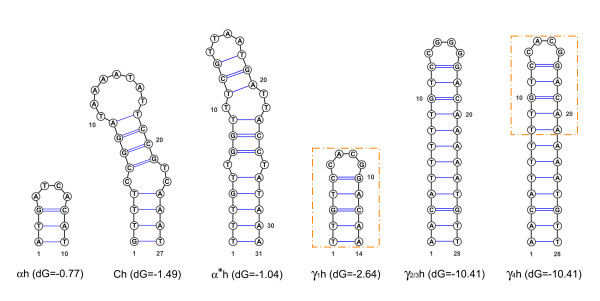
All the A-type subregions (A1, A2 and A') start with a 46 bp conserved motif, named here α, that contains a 10 bp hairpin (αh; see Figure 5). Both the subunits C (C1 and C2) begin with a hairpin 27 bp long (Ch; Figure 5). The M-LUR contains an A-like subregion showing a pD of 0.362 ± 0.032 from A1 and A2 (Table 6), indicated as A'' (Figure 4). A'' starts with a 37 bp motif, here named α*, similar to α, but 9 bp shorter and with three mutations that allow the formation of a longer hairpin, here named α*h (31 bp; Figure 5), in comparison to the female hairpin αh. The M-LUR continues with the subunit B that is the most conserved region compared to the F-LUR showing a pD from B1 and B2 of 0.098 ± 0.007 and 0.096 ± 0.007 respectively (Table 6). At the 3' end of B there is a motif, indicated as γ (Figure 4) that is similar to the first part of the subunits C. γ is repeated four times in tandem. The length of γ1, γ2 and γ3 ranges from 268 and 265 bp while the last repeat, γ4, is truncated and measures 17 bp (Additional File 3; Figure 4). The pD among the γ motifs is low and ranges from 0.008 ± 0.005 in the female (between γc1 and γc2) and 0.019 ± 0.009 between γ1 and γ3 (Table 6). The pD of the γ motifs between male and female varies from 0.346 and 0.350 ± 0.027 (Table 6). At the 5' end of each γ motif a secondary structure is present (γ1h, γ2h, γ3h and γ4h respectively; Figure 5): γ1h is 14 bp long, while the other three are 28 bp long. γ2h and γ3h are identical, γ4h has a two bases mutation at the center of the loop and γ1h is identical to the upper portion of γ4h (see Figure 5).
Figure 5.
LUR palindromes. Sequences and structures of palindromic motifs located in the Musculista senhousia LURs.
Furthermore, in line with what has been found in other DUI bivalves, including Mytilus, an ORF coding for 121 amminoacids has been found in the F-LUR of M. senhousia. This protein was proposed to have a functional role in DUI. Detailed analyses on this novel DUI related putative protein have been published in a more comparative way (see [20]).
The cox2 duplication in the male mtDNA
The male mtDNA contains an extra copy of the cox2 gene. This is not new for DUI animals, since the female mt genome of the marine clam V. philippinarum has a cox2 duplication as well (GenBank Acc. No. AB065375: Okazaki and Ueshima, unpublished).
In the female Musculista, the cox2 gene (Fcox2) is 660 bp long and is flanked by the "cox1/UR-6" and "UR-7/atp8" regions at the 5'- and 3'-end respectively (see Figure 1 and Table 1). In male mitochondrial genome, the two copies of cox2 are close to each other and linked by a little non coding region 41 bp long (UR-6). The two cox2 copies are located between "cox1/UR-5" and "UR-7/atp8" regions, and the first is 813 bp long, while the second is 690 bp long (Figure 2 and Table 2).
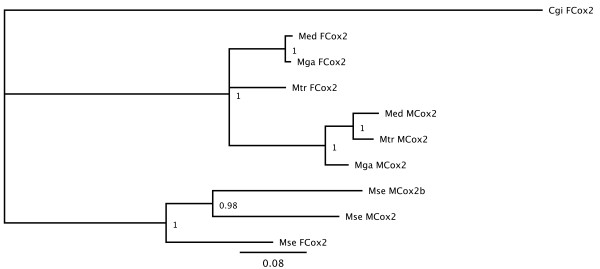
Bayesian phylogenetic analyses on Fcox2, Mcox2(690 bp), Mcox2(813 bp) genes and their homologous in Mytilus species, demonstrated that Fcox2 is more closely related to the shorter Mcox2 (690 bp), rather than to the longer one (Figure 6). For this reason, the 813 bp long Mcox2 seems to be an extra copy of the gene, and thus it is referred here as Mcox2b.
Figure 6.
Bayesian tree for the cox2 genes. Cgi: Crassostrea gigas; Med: Mytilus edulis; Mga: Mytilus galloprovincialis; Mtr: Mytilus trossulus; Mse: Musculista senhousia.
Discussion
Gene content and order of F and M Mitochondrial genomes in M. senhousia
In M. senhousia both M and F mtDNAs share the same gene content and order, except for a duplicated cox2 gene in males, and include the typical gene content of bivalve mtDNA. It has to be noted, however, that a common feature of bivalves is the apparent lack of the atp8 gene. For instance, [2] mentioned that a lack of the atp8 gene is one of several unusual features of the Mytilus mt sequence. The atp8 gene was considered missing for almost all bivalve species studied so far, including Crassostrea hongkongensis, C. gigas, C. virginica, Placopecten magellanicus, Argopecten irradians, Mizuhopecten yessoensis and Acanthocardia tuberculata. On the contrary, the apt8 gene was found in Hiatella arctica, as well as in the female mitochondrial genome of the unionid bivalve L. ornata [28]. A remarkable observation is that V. philippinarum, another species with DUI [57], was recently found to contain a putative atp8 gene [58], which was not found in the first analyses; nonetheless, this gene apparently encodes 37 amino acids only and therefore has a questionable gene function. Finally, [23] examined ORFs from several bivalve mitochondrial genomes and found two novel ORFs (F-orf-ur4 and M-orf-ur4) in the largest unassigned region of F and M mytilid ones (UR-4: see [33]). BLASTN searches against EST_others (all ESTs except human and mouse) showed that both are transcribed in Mytilus spp. BLASTX and PSI-BLAST searches using inferred aminoacid sequences of F-orf-ur4 and M-orf-ur4 failed to detect any significant sequence similarity with known proteins, so the identity of those putative proteins is still unclear. Further analyses on structure and evolution patterns suggested that the novel ORFs "represent good candidates for the previously 'missing' atp8 in mytilid mtDNAs" [23]. Therefore, following [23], we also found atp8 putative genes in both sex-linked mitochondrial genomes of M. senhousia. Our atp8 genes share the same characteristics of the above mentioned proteins, so we are confident to annotate them as Musculista atp8 genes.
Generally speaking, most mtDNAs are characterized by strand asymmetry in term of gene distribution. In both M. senhousia mt genomes, all genes are transcribed from the same strand, i.e. the asymmetry is at its highest among Metazoa. Most marine bivalves also share this feature (Mytilus species-complex, C. gigas, C. virginica, C. hongkongensis and V. philippinarum). In contrast, this is not true for the two freshwater species L. ornata [28] and Inversidens japanensis (Acc. No. AB055625 and AB055624) (see also [59]). In other mollusks, a relatively small number of mitochondrial genes are transcribed from the second strand. The scaphopods G. eborea and S. lobatum are an exception, with about an equal number of genes encoded by each strand [31,58]. The occurrence of all genes in the same strand is a relatively rare phenomenon in metazoans and, in addition to bivalves, it has been reported in some annelids (Lumbricus terrestris, [60]; Platynereis dumerilii, [61]) and brachiopods (Terebratulina retusa, [62]; Terebratalia transversa, [42]; Laqueus rubellus, [63]). Actually, almost 10% of the mitochondrial genomes examined to date do have all genes encoded in the same strand [10]. Moreover, most of the above mentioned groups, including Bivalvia, are also characterized by strong differences in gene content and/or gene order. This allowed [10] to suggest a possible correlation between these two features.
The trnS(AGN) could not be located with tRNAscan-SE [64] because of the absence of the DHU arm and therefore of a normal cloverleaf structure (see [27] for a detailed discussion), so we used the ARWEN software [65] to identify it. This unconventional tRNA was found also in several other animal groups ([27] and references therein), and it evolved very early in Metazoa [66]. In vitro analyses confirmed its functionality [67].
In Table 7, the distribution of trnS(UCN) and trnS(AGN) among bivalves is reported (only complete mitochondrial genomes included; source: http://mi.caspur.it/mitozoa see [3]). Most of the species (22) have both the tRNAs, 7 only trnS(UCN) and 3 (including M. senhousia) only trnS(AGN). Placopecten magellanicus have two copies of trnS(UCN), while Mizuhopecten yessoensis seems to lack a Serine tRNA. [68] suggested that the secondary structure of a tRNA gene between a pair of protein genes is responsible for the precise cleavage of the polycistronic primary transcript. In the absence of a tRNA, this role can be played by a stem-loop structure, the 5'-end part of the gene itself, or a combination of the two. Potential hairpin structures at protein-protein gene junctions with no intervening tRNA have been reported in several studies (e.g., [6,33,39,69,70]). Our analysis demonstrated that putative hairpins are present in all the gene junctions in which a tRNA lacks, suggesting a functional role of such intergenic sequences (Figure 3).
Table 7.
Serine tRNA [trnS(UCN) and trnS(AGN)] in bivalves.
| Taxonomy | Species (GenBank Acc. No.) | Missing | UCN | AGN | UCN+AGN |
|---|---|---|---|---|---|
| Pteriomorphia | |||||
| Mytiloida; Mytiloidea; Mytilidae | |||||
| Crenellinae | Musculista senhousia (GU001953) | x | |||
| Mytilinae | Mytilus edulis (AY823623) | x | |||
| Mytilus galloprovincialis (AY363687) | x | ||||
| Mytilus trossulus (DQ198225) | x | ||||
| Ostreoida; Ostreoidea; Ostreidae | |||||
| Saccostrea mordax (FJ841968) | x | ||||
| Crassostrea angulata (FJ841965) | x | ||||
| Crassostrea ariakensis (FJ841964) | x | ||||
| Crassostrea gigas (NC_001276) | x | ||||
| Crassostrea hongkongensis (EU266073) | x | ||||
| Crassostrea iredalei (FJ841967) | x | ||||
| Crassostrea sikamea (FJ841966) | x | ||||
| Pectinoida; Pectinoidea; Pectinidae | |||||
| Mizuhopecten yessoensis (FJ595959) | x | ||||
| Chlamys farreri (EU715252) | x | ||||
| Mimachlamys nobilis (FJ595958) | x | ||||
| Placopecten magellanicus (NC_007234)* | xx | ||||
| Argopecten irradians (NC_009687) | x | ||||
| Argopecten irradians irradians (DQ665851) | x | ||||
| Heteroconchia | |||||
| Myoida; Hiatelloidea; Hiatellidae | |||||
| Hiatella arctica (NC_008451) | x | ||||
| Veneroida; Cardioidea; Cardiidae | |||||
| Acanthocardia tuberculata (NC_008452) | x | ||||
| Veneroida; Lucinoidea; Lucinidae | |||||
| Loripes lacteus (EF043341) | x | ||||
| Lucinella divaricata (EF043342) | x | ||||
| Veneroida; Tellinoidea; Solecurtidae | |||||
| Sinonovacula constricta (EU880278) | x | ||||
| Veneroida; Veneroidea; Veneridae | |||||
| Meretrix meretrix (GQ463598) | x | ||||
| Meretrix petechialis (EU145977) | x | ||||
| Venerupis philippinarum (AB065374) | x | ||||
| Paphia euglypta (GU269271) | x | ||||
| Palaeoheterodonta | |||||
| Unionoida; Unionoidea; Unionidae | |||||
| Venustaconcha ellipsiformis (FJ809752) | x | ||||
| Ambleminae | Quadrula quadrula (FJ809750) | x | |||
| Anodontinae | Cristaria plicata (FJ986302) | x | |||
| Anodontinae | Pyganodon grandis (FJ809754) | x | |||
| Unioninae | Hyriopsis cumingii (FJ529186) | x | |||
| Inversidens japanensis (AB055624) | x | ||||
| Unio pictorium (HM014131) | x | ||||
*: Placopecten magellanicus has two copies of trnS(UCN)
Note: only species with complete mitochondrial genomes available included.
The Large Unassigned Region (LUR) and the sex-linked mt-DNA transmission
The structure of the F and M LUR palindromes found are reported on Figure 4 and 5. The presence of palindromes within a mtDNA CR is not new; in fact, the local fold symmetry created by the palindrome is thought to provide the site for DNA-binding proteins involved in the trascriptional machinery [71]. In more detail, palindromic motifs (and in general inverted repeats) have the potential to form single-stranded stem-loop cruciform structures which have been reported to be essential for replication of circular genomes in many prokaryotic and eukaryotic systems [72]. The redundancy of palindromic elements in the Musculista male LUR, when compared to that of the female, may be possibly related to an increased duplication ratio of the M mtDNA; we can also speculate that this feature may have some role in the process by which sperm mitochondrial DNA becomes dominant or exclusive of the male germline, although we know that this is also achieved through a differential segregation during early embryo development, and likely through a second, more strict, selection during primordial germ cells establishment (see [73]). Nevertheless, the question of how sperm mitochondrial DNA becomes dominant or the exclusive component of the male germline in DUI species still remains open, and may be the outcome of various coordinated processes.
The duplication of the cox2 gene
One noteworthy finding of this analysis is the cox2 gene duplication in the male mtDNA, with the duplicated gene being longer than the original one, a feature that might be somehow related to DUI. In fact, an interesting analogy is evident with unionid bivalves, in which the male cox2 gene show a 200-codon extension, which is absent in the female mtDNA. Such a feature is found in all analyzed unionids so far, and it has been related to DUI functioning [21,22,74-76]. Actually, [21,22] proposed several hypotheses for the role the cox2 extension may have for DUI, but all are dependent upon identifying a specific function for it, which is not a trivial task. Moreover, they detected in the male gonad a poly-adenylated mRNA transcript of the cox2 gene that includes the extension, and they concluded that the extension is protein-coding and functional.
[21,22] also hypothesized that the COX2 protein extension might be involved in intracellular interactions determining the survival of the male mitochondrion. In other organisms, it has been shown that upon fertilization the sperm-derived mitochondria are targeted for elimination: a key process in sperm mitochondrial degradation is ubiquitination [77], in which mitochondria of paternal derivation are tagged with Ubiquitin and then degraded. In Mytilus, in which an Ubiquitin-like process has been proposed, this degradation would be sex-specific: the sperm-derived mitochondria survive in male embryos, whereas they are eliminated in females. All that considered, [21] proposed that the COX2 extension could be involved in blocking such elimination to ensure survival of the male mitochondrion, or, alternatively, the extension could play a role in the segregation of male mitochondria to the gonad. In either case, it should be possible to detect the protein product of the extension outside of the inner mitochondrial membrane. An in situ hybridization seemed to demonstrate that the unionid male COX2 is present on both inner and outer membranes of the sperm mitochondria (see Figure 4 in [74]).
According to the above mentioned rationales, we hypothesize that the duplicated cox2b gene in male M. senhousia may represent a variant of what found in unionoidean bivalves, with proper signals for DUI mitochondrial tagging lying in the COX2 protein extension of unionid bivalves, as well as in the duplicated COX2b protein of Musculista. A support to this view comes from the observation that an additional putative Trans Membrane Helix (TMH) is found in the 41 residue long tail of the Musculista COX2b, although this tail is considerably shorter that the unionid one (200 amminoacids). Actually, five putative TMHs were found in the unionid extended C-terminus of the male COX2, which led the Authors to hypothesize that it may have a functional significance for male unionoidean bivalve reproductive success [75,76].
In analogy, we suggest that COX2b might have some function related to mitochondrial tagging, like the COX2b and the Unionid COX2 extension. Further studies are needed to gain a more clear role of such proteins in the unusual DUI system of mitochondrial inheritance. Actually, a duplication similar to the Musculista one was also found in V. philippinarum, but quite surprisingly in the female mtDNA (see unpublished GenBank annotation). This suggests that cox2 duplication may be uncoupled with maleness. Moreover, no Mytilus genomes show a similar situation for cox2 or any other gene, so either duplicated genes or a cox2 tail may not be strictly necessary to sustain DUI.
Conclusions
The characteristics of the Musculista sex-linked mtDNAs evidently add to the knowledge of DUI systems, and highlight some unexpected features, shared among distantly related DUI species. Since it is commonly accepted that DUI is rather a variation of Strict Maternal Inheritance, than a completely different mechanism, we think that DUI is a good experimental model to better understand the general rules, as well as the molecular features of Metazoan mitochondrial inheritance (see [18], for a detailed discussion). For the above mentioned reasons, the complete mtDNA genome characterization of DUI bivalves is not only a mere descriptive exercise, but rather a first step to unravel the complex genetic signals allowing Doubly Uniparental Inheritance of mitochondrial DNA, and the evolutionary implications of such unusual transmission route in mitochondrial genome evolution in Bivalvia.
Methods
Sample Collection
Alive M. senhousia specimens from Venice Lagoon (Italy) were used for this analysis. Males and females were stimulated to spawn gametes in seawater supplemented with hydrogen peroxide, according to [78]. Each emission was analyzed with a light microscope to sex specimens. A total of 10 sperm and 10 egg samples were then collected after a gentle centrifugation (3,000 g). Seawater was removed, and ethanol added before storing samples at -20°C.
PCR analyses
Total genomic DNA was extracted using the DNeasy Tissue Kit (Qiagen), and partial sequences of cytochrome b (cob) and mitochondrial ribosomal large subunit RNA (rrnL) were amplified and directly sequenced (primers reported in Table 8), as described in [79]. Sequencing reactions were performed on both strands with BigDye Terminator Cycle Sequencing Kit according to supplier's instructions (Applied Biosystem) in a 310 Genetic Analyzer (ABI) automatic sequencer.
Table 8.
Primer sequences.
| Primer name | Sequence |
|---|---|
| cobR1 | 5'-GCRTAWGCRAAWARRAARTAYCAYTCWGG-3' |
| cobF1 16Sbr2 |
5'-GGWTAYGTWYTWCCWTGRGGWCARAT-3' 5'-CCGGTCTGAACTCAGATCACGT-3' |
| 16Sar2 | 5'-CGCCTGTTTATCAAAAACAT-3' |
| F-cob383R F-16S142F |
5'-TAGGAGTTTTTATAGGGTCTGC-3' 5'-ACCTGAAGTTGTCTCATTTACC-3' |
| M-cob386R M-16S103F |
5'-GGATAGGAGTTTTTATAGGGTCTGC-3' 5'-GTGAATTTCTTAGAGTGACGATTA-3' |
1 J.L. Boore, personal communication; 2 [88]
The 20 sequences obtained for both F and M genomes were aligned (not shown), and, after checking for variable sites, used to design sex-specific primers to amplify the entire mitochondrial genome in two overlapping fragments by long PCR reactions. LongPCR was performed on one Musculista specimen per sex. To obtain the F genome, F-cob383R and F-16S142F primers were used. The M genome was amplified with M-cob386R and M-16S103F. Both pairs of primers amplified a fragment of 10-11 kb respectively. Long PCR primer sequences are reported in Table 1. LongPCR amplifications were performed on a Gene Amp® PCR System 2720 (Applied Biosystem) in 50 μl reaction volume composed of 31.5 μl of sterilized distilled water, 10 μl of 5 × Herculase II Fusion Reaction Buffer, 0.5 μl of dNTPs mix (25 mM each dNTP), 1.25 μl of each primer (10 μM), 5 μl of DNA template (25-50 ng) and 0.5 μl of Herculase II Fusion DNA Polymerase. Reaction conditions were according to supplier's recommendations: initial denaturation at 95°C for 5 min and then incubated at 95°C for 20 s, 50°C for 20 s, and 68°C for 10 min for 30 cycles and 68°C for 8 min for a final extension. Long-PCR fragments were then purified using Wizard® SV Gel and PCR Clean-Up System (Promega).
Shotgun cloning
Sequencing of the two LongPCR fragments was done using shotgun cloning: amplicons were randomly sheared to 1.2-1.5 kb DNA segments using a HydroShear device (GeneMachines). Sheared DNA was blunt end repaired at room temperature for 60 min using 6 U of T4 DNA Polymerase (Roche), 30 U of DNA Polymerase I Klenow (NEB), 10 μl of dNTPs mix, 13 μl of 10 × NEB buffer 2 in a 115 μl total volume, and then gel purified using the Wizard® SV Gel and PCR Clean-Up System (Promega). The resulting fragments were ligated into the SmaI site of a pUC18 cloning vector using the Fast-Link DNA ligation Kit (Epicentre) and electroporated into One Shot® TOP10 Electrocomp™ Escherichia coli cells (Invitrogen) using standard protocols. Clones were screened by PCR using M13 universal primers and recombinants were purified using Multiscreen (Millipore) according to the manufacturer's instructions. Clones were sequenced using M13 universal primers by Macrogen Inc. (Korea).
Raw sequences were manually corrected, and then assembled into contigs with Sequencher v.4.6 (Gene Codes). Hence, the final assemblies were based on a minimum sequence coverage of 3×.
Secondary structures and annotation
The tRNA genes were identified by their secondary structure using ARWEN [65], with invertebrate mitochondrial codon predictors. Analysis of Open Reading Frames (ORFs) was performed with the ORF Finder program of NCBI http://www.ncbi.nlm.nih.gov/projects/gorf/ using the invertebrate mitochondrial genetic code. Sequences were identified using BLASTX, PSI-BLAST [80] and BLASTN [81] as implemented by the NCBI website http://www.ncbi.nlm.nih.gov/.
For all protein coding genes, alignments were computed with ClustalW [82].
When analyzing sequence variability, pairwise p-Distances (pD), their mean values and standard errors (by the bootstrap procedure) were computed with MEGA v.5.03 [83]. In order to avoid any model of DNA substitution that can affect statistics (see [79]), the use of a pD was preferred.
The divergence of protein genes in synonymous (Ks) and non-synonymous (Ka) sites was calculated by the modified Nei-Gojobori method with the Jukes-Cantor correction; the pD at the residue level was also calculated within the MEGA v.5.03 environment [83].
Two-fold, and four-fold degenerated positions were identified using DnaSP v.5 [84]. The Sequence Manipulation Suite (http://www.bioinformatics.org/sms2; [85]) was used to estimate codon usage. Potential DNA secondary structures near or at the 5'-end of protein genes were predicted using the UNAFold software package [86] available on the DINAMelt web server (http://mfold.rna.albany.edu/?q=DINAMelt; [86]).
Bayesian analyses on cox2 genes was performed with the MrBayes 3.1 (5,000,000 generations; [87]).
Authors' contributions
MP conceived the study, participated in its design and coordination and drafted the manuscript. AR carried out the lab work and performed part of the analysis. LM and FG performed part of the analysis and drafted the manuscript. All authors read and approved the final manuscript.
Supplementary Material
The Unassigned Regions (URs) in the female and male mtDNAs of Musculista senhousia. Annotation and length of Unassigned Regions (URs) in the female (Mse_URs_F) and male (Mse_URs_M) mtDNAs of Musculista senhousia.
tRNAs in the female and male mtDNAs of Musculista senhousia. Annotation, length and structures of tRNAs in the female (Mse_trn_F) and male (Mse_trn_M) mtDNAs of Musculista senhousia.
Structure of the female (F-LUR) and male (M-LUR) Large Unassigned Regions of Musculista senhousia mtDNA. Schematic table of repeats and hairpin structures in the Large Unassigned Regions (LURs) of the female and male Musculista senhousia mtDNAs (F-LUR and M-LUR).
Contributor Information
Marco Passamonti, Email: marco.passamonti@unibo.it.
Andrea Ricci, Email: and.ricci@gmail.com.
Liliana Milani, Email: liliana.milani@unibo.it.
Fabrizio Ghiselli, Email: fabrizio.ghiselli@unibo.it.
Acknowledgements
We want to thank Edoardo Turolla (C.Ri.M., Goro, Italy) for providing us Musculista samples. This work was supported by the University and Research Italian Ministry (MIUR PRIN07, grant number 2007NSHJL8_002 to MP) and the "Canziani Bequest" fund (University of Bologna, grant number A.31.CANZELSEW to MP).
References
- Boyce TM, Zwick ME, Aquadro CF. Mitochondrial DNA in the bark weevils: size, structure and heteroplasmy. Genetics. 1989;123:825–836. doi: 10.1093/genetics/123.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore JL. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27:1767–1780. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupi R, de Meo PD, Picardi E, D'Antonio M, Paoletti D, Castrignanò T, Pesole G, Gissi C. MitoZoa: a curated mitochondrial genome database of metazoans for comparative genomics studies. Mitochondrion. 2010;10(2):192–199. doi: 10.1016/j.mito.2010.01.004. [DOI] [PubMed] [Google Scholar]
- Milbury CA, Lee JC, Cannone JJ, Gaffney PM, Gutell RR. Fragmentation of the large subunit ribosomal RNA gene in oyster mitochondrial genomes. BMC Genomics. 2010;11:485. doi: 10.1186/1471-2164-11-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridge D, Cunningham CW, Schierwater B, DeSalle R, Buss LW. Class-level relationships in the phylum Cnidaria: evidence from mitochondrial genome structure. Proc Natl Acad Sci USA. 1992;89:8750–8753. doi: 10.1073/pnas.89.18.8750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore JL, Brown WM. Complete DNA sequence of the mitochondrial genome of the black chiton Katharina tunicata. Genetics. 1994;138:423–443. doi: 10.1093/genetics/138.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levings CS, III, Vasil IK. The Molecular Biology of Plant Mitochondria. Boston, MA: Kluwer Academic Publishers; 1995. [Google Scholar]
- Gray MW, Lang BF, Cedergren R, Golding GB, Lemieux C, Sankoff D, Turmel M, Brossard N, Delage E, Littlejohn TG, Plante I, Rioux P, Saint-Louis D, Zhu Y, Burger G. Genome structure and gene content in protist mitochondrial DNAs. Nucleic Acids Res. 1998;26:865–878. doi: 10.1093/nar/26.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquin B, Laforest MJ, Forget L, Roewer I, Wang Z, Longcore J, Lang BF. The fungal mitochondrial genome project: evolution of fungal mitochondrial genomes and their gene expression. Curr Genet. 1997;31:380–395. doi: 10.1007/s002940050220. [DOI] [PubMed] [Google Scholar]
- Gissi C, Iannelli F, Pesole G. Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity. 2008;101:301–320. doi: 10.1038/hdy.2008.62. [DOI] [PubMed] [Google Scholar]
- Gillham N. Organelle Genes And Genomes. New York, Oxford: Oxford University Press; 1994. [Google Scholar]
- Birky CW Jr. The inheritance of genes in mitochondria and chloroplasts: laws, mechanisms, and models. Annu Rev Genet. 2001;35:125–148. doi: 10.1146/annurev.genet.35.102401.090231. [DOI] [PubMed] [Google Scholar]
- Zouros E, Ball AO, Saavedra C, Freeman KR. Mitochondrial DNA inheritance. Nature. 1994;368:818. doi: 10.1038/368818a0. [DOI] [PubMed] [Google Scholar]
- Skibinski DO, Gallagher C, Beynon CM. Mitochondrial DNA inheritance. Nature. 1994;368:817–818. doi: 10.1038/368817b0. [DOI] [PubMed] [Google Scholar]
- Breton S, Doucet-Beaupré HD, Stewart DT, Hoeh WR, Blier PU. The unusual system of doubly uniparental inheritance of mtDNA: isn't one enough? Trends Genet. 2007;23:465–474. doi: 10.1016/j.tig.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Theologidis I, Fodelianakis S, Gaspar MB, Zouros E. Doubly uniparental inheritance (DUI) of mitochondrial DNA in Donax trunculus (Bivalvia: Donacidae) and the problem of its sporadic detection in Bivalvia. Evolution Int J Org Evolution. 2008;62:959–970. doi: 10.1111/j.1558-5646.2008.00329.x. [DOI] [PubMed] [Google Scholar]
- Doucet-Beaupré H, Breton S, Chapman EG, Blier PU, Bogan AE, Stewart DT, Hoeh WR. Mitochondrial phylogenomics of the Bivalvia (Mollusca): searching for the origin and mitogenomic correlates of doubly uniparental inheritance of mtDNA. BMC Evol Biol. 2010;10:50. doi: 10.1186/1471-2148-10-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passamonti M, Ghiselli F. Doubly Uniparental Inheritance: Two Mitochondrial Genomes, One Precious Model for Organelle DNA Inheritance and Evolution. DNA Cell Biol. 2009;28:79–89. doi: 10.1089/dna.2008.0807. [DOI] [PubMed] [Google Scholar]
- Passamonti M. An unusual case of gender-associated mitochondrial DNA heteroplasmy: the mytilid Musculista senhousia (Mollusca Bivalvia) BMC Evol Biol. 2007. p. S7. [DOI] [PMC free article] [PubMed]
- Breton S, Ghiselli F, Passamonti M, Milani L, Stewart DT, Hoeh WR. Evidence for a Fourteenth mtDNA-Encoded Protein in the Female-Transmitted mtDNA of Marine Mussels (Bivalvia: Mytilidae) PLoS One. 2011;6(4):e19365. doi: 10.1371/journal.pone.0019365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curole JP, Kocher TD. Ancient sex-specific extension of the cytochrome c oxidase II gene in bivalves and the fidelity of doubly-uniparental inheritance. Mol Biol Evol. 2002;19:1323–1328. doi: 10.1093/oxfordjournals.molbev.a004193. [DOI] [PubMed] [Google Scholar]
- Curole JP, Kocher TD. Evolution of a unique mitotype-specific protein-coding extension of the cytochrome c oxidase II gene in freshwater mussels (Bivalvia: Unionoida) J Mol Evol. 2005;61:381–389. doi: 10.1007/s00239-004-0192-7. [DOI] [PubMed] [Google Scholar]
- Breton S, Stewart DT, Hoeh WR. Characterization of a mitochondrial ORF from the gender-associated mtDNAs of Mytilus spp. (Bivalvia: Mytilidae): Identification of the "missing" ATPase 8 gene. Mar Genomics. 2010;3:11–18. doi: 10.1016/j.margen.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Hoffmann RJ, Boore JL, Brown WM. A novel mitochondrial genome organization for the blue mussel, Mytilus edulis. Genetics. 1992;131:397–412. doi: 10.1093/genetics/131.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki N, Ueshima R, Terrett JA. Evolution of pulmonate gastropod mitochondrial genomes: comparisons of gene organizations of Euhadra, Cepaea and Albinaria and implications of unusual tRNA secondary structures. Genetics. 1997;145:749–758. doi: 10.1093/genetics/145.3.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K, Yokobori S, Oshima T, Ueda T, Watanabe K. The cephalopod Loligo bleekeri mitochondrial genome: multiplied noncoding regions and transposition of tRNA genes. J Mol Evol. 2002;54:486–500. doi: 10.1007/s00239-001-0039-4. [DOI] [PubMed] [Google Scholar]
- Sheffield NC, Song H, Cameron SL, Whiting MF. A comparative analysis of mitochondrial genomes in Coleoptera (Arthropoda: Insecta) and genome descriptions of six new beetles. Mol Biol Evol. 2008;25:2499–2509. doi: 10.1093/molbev/msn198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serb JM, Lydeard C. Complete mtDNA sequence of the North American freshwater mussel, Lampsilis ornata (Unionidae): an examination of the evolution and phylogenetic utility of mitochondrial genome organization in Bivalvia (Mollusca) Mol Biol Evol. 2003;20:1854–1866. doi: 10.1093/molbev/msg218. [DOI] [PubMed] [Google Scholar]
- Kurabayashi A, Ueshima R. Complete sequence of the mitochondrial DNA of the primitive opisthobranch gastropod Pupa strigosa: systematic implication of the genome organization. Mol Biol Evol. 2000;17:266–277. doi: 10.1093/oxfordjournals.molbev.a026306. [DOI] [PubMed] [Google Scholar]
- Hatzoglou E, Rodakis GC, Lecanidou R. Complete sequence and gene organization of the mitochondrial genome of the land snail Albinaria coerulea. Genetics. 1995;140:1353–1366. doi: 10.1093/genetics/140.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore JL, Medina M, Rosenberg LA. Complete sequences of the highly rearranged molluscan mitochondrial genomes of the scaphopod Graptacme eborea and the bivalve Mytilus edulis. Mol Biol Evol. 2004;21:1492–1503. doi: 10.1093/molbev/msh090. [DOI] [PubMed] [Google Scholar]
- Perna NT, Kocher TD. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J Mol Evol. 1995;41:353–358. doi: 10.1007/BF01215182. [DOI] [PubMed] [Google Scholar]
- Mizi A, Zouros E, Moschonas N, Rodakis GC. The complete maternal and paternal mitochondrial genomes of the Mediterranean mussel Mytilus galloprovincialis: implications for the doubly uniparental inheritance mode of mtDNA. Mol Biol Evol. 2005;22:952–967. doi: 10.1093/molbev/msi079. [DOI] [PubMed] [Google Scholar]
- Fernandez-Silva P, Enriquez JA, Montoya J. Replication and transcription of mammalian mitochondrial DNA. Exp Physiol. 2003;88:4156. doi: 10.1113/eph8802514. [DOI] [PubMed] [Google Scholar]
- Terrett JA, Miles S, Thomas RH. Complete DNA sequence of the mitochondrial genome of Cepaea nemoralis (Gastropoda: Pulmonata) J Mol Evol. 1996;42:160–168. doi: 10.1007/BF02198842. [DOI] [PubMed] [Google Scholar]
- Grande C, Templado J, Cervera JL, Zardoya R. The complete mitochondrial genome of the nudibranch Roboastra europaea (Mollusca: Gastropoda) supports the monophyly of opisthobranchs. Mol Biol Evol. 2002;10:1672–1685. doi: 10.1093/oxfordjournals.molbev.a003990. [DOI] [PubMed] [Google Scholar]
- Garesse R. Drosophila melanogaster mitochondrial DNA: gene organization and evolutionary considerations. Genetics. 1988;118:649–663. doi: 10.1093/genetics/118.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantatore P, Roberti M, Rainaldi G, Gadaleta MN, Saccone C. The complete nucleotide sequence, gene organization, and genetic code of the mitochondrial menome of Paracentrotus lividus. J Biol Chem. 1989;264:10965–10975. [PubMed] [Google Scholar]
- Okimoto R, Macfarlane JL, Clary DO, Wolstenholme DR. The mitochondrial genomes of two nematodes, Caenorhabditis elegans and Ascaris suum. Genetics. 1992;130:471–498. doi: 10.1093/genetics/130.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakawa S, Kumazawa Y, Araki T, Himeno H, Miura K, Watanabe K. Strand-specific nucleotide composition bias in echinoderm and vertebrate mitochondrial genomes. J Mol Evol. 1991;32:511–520. doi: 10.1007/BF02102653. [DOI] [PubMed] [Google Scholar]
- De Giorgi C, Martiradonna A, Lanave C, Saccone C. Complete sequence of the mitochondrial DNA in the sea urchin Arbacia lixula: conserved features of the echinoid mitochondrial genome. Mol Phyl Evol. 1996;5:323–332. doi: 10.1006/mpev.1996.0027. [DOI] [PubMed] [Google Scholar]
- Helfenbein KG, Brown WM, Boore JL. The complete mitochondrial genome of the articulate brachiopod Terebratalia transversa. Mol Biol Evol. 2001;18:1734–1744. doi: 10.1093/oxfordjournals.molbev.a003961. [DOI] [PubMed] [Google Scholar]
- Spruyt N, Delarbre C, Gachelin G, Laudet V. Complete sequence of the amphioxus (Branchiostoma lanceolatum) mitochondrial genome: relations to vertebrates. Nucleic Acids Res. 1998;26:3279–3285. doi: 10.1093/nar/26.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YS, Huang FL, Lo TB. The complete nucleotide sequence and gene organization of carp (Cyprinus carpio) mitochondrial genome. J Mol Evol. 1994;38:138–155. doi: 10.1007/BF00166161. [DOI] [PubMed] [Google Scholar]
- Ingman M, Kaessmann H, Paabo S, Gyllensten U. Mitochondrial genome variation and the origin of modern humans. Nature. 2000;408:708–713. doi: 10.1038/35047064. [DOI] [PubMed] [Google Scholar]
- Roe BA, Ma DP, Wilson RK, Wong JF. The complete nucleotide sequence of the Xenopus laevis mitochondrial genome. J Biol Chem. 1985;260:9759–9774. [PubMed] [Google Scholar]
- Broughton RE, Milam JE, Roe BA. The complete sequence of the zebrafish (Danio rerio) mitochondrial genome and evolutionary patterns in vertebrate mitochondrial DNA. Genome Res. 2001;11:1958–1967. doi: 10.1101/gr.156801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolstenholme DR. Animal mitochondrial DNA: structure and evolution. Int Rev Cytol. 1992;141:173–216. doi: 10.1016/s0074-7696(08)62066-5. [DOI] [PubMed] [Google Scholar]
- Rand DM, Kann LM. Mutation and selection at silent and replacement sites in the evolution of animal mitochondrial DNA. Genetica. 1998;102/103:393–407. [PubMed] [Google Scholar]
- Crease TJ. The complete sequence of the mitochondrial genome of Daphnia pulex (Cladocera: Crustacea) Gene. 1999;233:89–99. doi: 10.1016/S0378-1119(99)00151-1. [DOI] [PubMed] [Google Scholar]
- Sueoka N. On the genetic basis of variation and heterogeneity of DNA base composition. Proc Natl Acad Sci USA. 1962;48:582–592. doi: 10.1073/pnas.48.4.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jermiin LS, Crozier RH. The cytochrome b region in the mitochondrial DNA of the ant Tetraponera rufoniger: sequence divergence in Hymenoptera may be associated with nucleotide content. J Mol Evol. 1994;38:282–294. doi: 10.1007/BF00176090. [DOI] [PubMed] [Google Scholar]
- Jermiin LS, Foster PG, Graur D, Lowe RM, Crozier RH. Unbiased estimation of symmetrical directional mutation pressure from protein-coding DNA. J Mol Evol. 1996;42:476–480. doi: 10.1007/BF02498643. [DOI] [PubMed] [Google Scholar]
- Jermiin LS, Graur D, Lowe RM, Crozier RH. Analysis of directional mutation pressure and nucleotide content in mitochondrial cytochrome b genes. J Mol Evol. 1994;39:160–173. doi: 10.1007/BF00163805. [DOI] [PubMed] [Google Scholar]
- Pesole G, Gissi C, De Chirico A, Saccone C. Nucleotide substitution rate of mammalian mitochondrial genomes. J Mol Evol. 1999;48:427–434. doi: 10.1007/PL00006487. [DOI] [PubMed] [Google Scholar]
- Saccone C, De Giorgi C, Gissi C, Pesole G, Reyes A. Evolutionary genomics in Metazoa: the mitochondrial DNA as a model system. Gene. 1999;238:195–209. doi: 10.1016/S0378-1119(99)00270-X. [DOI] [PubMed] [Google Scholar]
- Passamonti M, Scali V. Gender-associated mitochondrial DNA heteroplasmy in the venerid clam Tapes philippinarum (Mollusca Bivalvia) Curr Genet. 2001;39:117–124. doi: 10.1007/s002940100188. [DOI] [PubMed] [Google Scholar]
- Dreyer H, Steiner G. The complete sequences and gene organisation of the mitochondrial genomes of the heterodont bivalves Acanthocardia tuberculata and Hiatella arctica and the first record for a putative Atpase subunit 8 gene in marine bivalves. Front Zool. 2006;3:13. doi: 10.1186/1742-9994-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton S, Stewart DT, Blier PU. Role-reversal of gender-associated mitochondrial DNA affects mitochondrial function in Mytilus edulis (Bivalvia: Mytilidae) J Exp Zool B Mol Dev Evol. 2009;312:108–117. doi: 10.1002/jez.b.20251. [DOI] [PubMed] [Google Scholar]
- Boore JL, Brown WM. Complete sequence of the mitochondrial DNA of the annelid worm Lumbricus terrestris. Genetics. 1995;141:305–319. doi: 10.1093/genetics/141.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore JL, Brown WM. Mitochondrial genomes of Galathealinum, Helobdella, and Platynereis: sequence and gene arrangement comparisons indicate that Pogonophora is not a phylum and Annelida and Arthropoda are not sister taxa. Mol Biol Evol. 2000;17:87–106. doi: 10.1093/oxfordjournals.molbev.a026241. [DOI] [PubMed] [Google Scholar]
- Stechmann A, Schlegel M. Analysis of the complete mitochondrial DNA sequence of the brachiopod Terebratulina retusa places Brachiopoda within the protostomes. Proc R Soc Lond B Biol Sci. 1999;266:2043–2052. doi: 10.1098/rspb.1999.0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi Y, Endo K, Tajima F, Ueshima R. The mitochondrial genome of the brachiopod Laqueus rubellus. Genetics. 2000;155:245–259. doi: 10.1093/genetics/155.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laslett D, Canbäck B. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 2008;24(2):172–175. doi: 10.1093/bioinformatics/btm573. [DOI] [PubMed] [Google Scholar]
- Garey JR, Wolstenholme DR. Platyhelminth mitochondrial DNA: evidence for early evolutionary origin of a tRNA(serAGN) that contains a dihydrouridine arm replacement loop, and of serine-specifying AGA and AGG codons. J Mol Evol. 1989;28(5):374–387. doi: 10.1007/BF02603072. [DOI] [PubMed] [Google Scholar]
- Hanada T, Suzuki T, Yokogawa T, Takemoto-Hori C, Sprinzl M, Watanabe K. Translation ability of mitochondrial tRNAsSer with unusual secondary structures in an in vitro translation system of bovine mitochondria. Genes Cells. 2001;6(12):1019–1030. doi: 10.1046/j.1365-2443.2001.00491.x. [DOI] [PubMed] [Google Scholar]
- Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- Bibb MJ, Van Etten RA, Wright CT, Walberg MW, Clayton DA. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981;26:167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- Clary DO, Wolstenholme DR. The mitochondrial DNA molecule of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol. 1985;22:252–271. doi: 10.1007/BF02099755. [DOI] [PubMed] [Google Scholar]
- Arunkumar KP, Nagaraju J. Unusually long palindromes are abundant in mitochondrial control regions of insects and nematodes. PLoS ONE. 2006;1:e110. doi: 10.1371/journal.pone.0000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AK. Palindrome regeneration by template strand-switching mechanism at the origin of DNA replication of porcine circovirus via the rolling-circle melting-pot replication model. J Virol. 2004;78:9016–9029. doi: 10.1128/JVI.78.17.9016-9029.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiselli F, Milani L, Passamonti M. Strict sex-specific mtDNA segregation in the germ line of the DUI species Venerupis philippinarum (Bivalvia: Veneridae) Mol Biol Evol. 2011;28(2):949–961. doi: 10.1093/molbev/msq271. [DOI] [PubMed] [Google Scholar]
- Chakrabarti R, Walker JM, Chapman EG, Shepardson SP, Trdan RJ, Curole JP, Watters GT, Stewart DT, Vijayaraghavan S, Hoeh WR. Reproductive function for a C-terminus extended, male-transmitted cytochrome c oxidase subunit II protein expressed in both spermatozoa and eggs. FEBS Lett. 2007;581:5213–5219. doi: 10.1016/j.febslet.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti R, Walker JM, Stewart DT, Trdan RJ, Vijayaraghavan S, Curole JP, Hoeh WR. Presence of a unique male-specific extension of C-terminus to the cytochrome c oxidase subunit II protein coded by the male-transmitted mitochondrial genome of Venustaconcha ellipsiformis (Bivalvia: Unionoidea) FEBS Lett. 2006;580:862–866. doi: 10.1016/j.febslet.2005.12.104. [DOI] [PubMed] [Google Scholar]
- Chapman EG, Piontkivska H, Walker JM, Stewart DT, Curole JP, Hoeh WR. Extreme primary and secondary protein structure variability in the chimeric male-transmitted cytochrome c oxidase subunit II protein in freshwater mussels: evidence for an elevated amino acid substitution rate in the face of domain-specific purifying selection. BMC Evol Biol. 2008;8:165. doi: 10.1186/1471-2148-8-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutovsky P, Moreno RD, Ramalho-Santos J, Dominko T, Simerly C, Schatten G. Ubiquitin tag for sperm mitochondria. Nature. 1999;402:371–372. doi: 10.1038/46466. [DOI] [PubMed] [Google Scholar]
- Morse DE, Duncan H, Hooker N, Morse A. Hydrogen peroxide induces spawning in molluscs, with activation of prostaglandin endoperoxide synthtase. Science. 1977;196:298–300. doi: 10.1126/science.403609. [DOI] [PubMed] [Google Scholar]
- Passamonti M, Boore JL, Scali V. Molecular evolution and recombination in gender-associated mitochondrial DNAs of the Manila clam Tapes philippinarum. Genetics. 2003;164:603–611. doi: 10.1093/genetics/164.2.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank: update. Nucleic Acids Res. 2004;32:D23–26. doi: 10.1093/nar/gkh045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ, Clustal W. improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specifc gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol. 2011. in press . [DOI] [PMC free article] [PubMed]
- Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Stothard P. The Sequence Manipulation Suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques. 2000;28:1102–1104. doi: 10.2144/00286ir01. [DOI] [PubMed] [Google Scholar]
- Markham NR, Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005;33:W577–W581. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Palumbi S, Martin A, Romano S, McMillan WO, Stice L, Grabowski G. The simple fools guide to PCR. Honolulu, Special publication Department of Zoology and Kewalo Marine Laboratory, University of Hawaii; 1991. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The Unassigned Regions (URs) in the female and male mtDNAs of Musculista senhousia. Annotation and length of Unassigned Regions (URs) in the female (Mse_URs_F) and male (Mse_URs_M) mtDNAs of Musculista senhousia.
tRNAs in the female and male mtDNAs of Musculista senhousia. Annotation, length and structures of tRNAs in the female (Mse_trn_F) and male (Mse_trn_M) mtDNAs of Musculista senhousia.
Structure of the female (F-LUR) and male (M-LUR) Large Unassigned Regions of Musculista senhousia mtDNA. Schematic table of repeats and hairpin structures in the Large Unassigned Regions (LURs) of the female and male Musculista senhousia mtDNAs (F-LUR and M-LUR).