SUMMARY
Clinical and genomic evidence suggests that the metastatic potential of a primary tumor may be dictated by pro-metastatic events that have additional oncogenic capability. To test this deterministic hypothesis, we adopted a comparative oncogenomics-guided function-based strategy involving (i) comparison of global transcriptomes of two genetically engineered mouse models with contrasting metastatic potential, (ii) genomic and transcriptomic profiles of human melanoma, (iii) functional genetic screen for enhancers of cell invasion and (iv) evidence of expression selection in human melanoma tissues. This integrated effort identified 6 genes that are potently pro-invasive and oncogenic. Further, we show that one such gene, ACP5, confers spontaneous metastasis in vivo, engages a key pathway governing metastasis and is prognostic in human primary melanomas.
INTRODUCTION
Cancers are highly heterogeneous on both the genomic and cellular levels such that similarly staged early disease can exhibit radically different clinical outcomes – from cure following surgical removal of the primary tumor to death within months of diagnosis due to widespread metastasis. Metastasis is responsible for the majority of cancer-related mortality and involves multiple interrelated steps by which primary tumor cells spread to establish cancerous lesions at distant sites (Gupta and Massague, 2006). To become metastatic, tumor cells acquire a number of biological capabilities to overcome barriers of dissemination and distant growth such as invasion, anoikis resistance, extravasation, colonization and growth in new microenvironments. Each of these biological attributes can be conferred by genetic or epigenetic events observed in tumors (Hanahan and Weinberg, 2011), supporting the thesis that biological heterogeneity of cancers, including metastatic potential, is dictated by underlying genomic alterations.
While significant data exists in support of a classical model of stepwise accumulation of genetic events which endow increasing malignant potential, the identification of extensive genome rearrangements in early stage cancers (driven in part by telomere crisis) (Rudolph et al., 2001; Chin et al., 2004) raise the possibility that some tumors may acquire genomic alterations with significant metastatic potential early in their evolution. Such tumors would inherently carry higher risk of metastasis despite early diagnoses. This deterministic model is consistent with the finding that transcriptomic profiles of primary tumors share striking resemblance with their metastatic lesions (Perou et al., 2000), and gene expression patterns of the primary bulk tumor can predict the likelihood of recurrence or metastatic spread, e.g. MammaPrint® and OncotypeDx® (van’t Veer et al., 2002; Paik et al., 2004). Furthermore, the prognostic significance of these gene expression signatures supports the view that information on metastatic propensity is encoded in the bulk of the primary tumor (van’t Veer et al., 2002; van de Vijver et al., 2002; Ramaswamy et al., 2003).
Together, these observations lead one to posit that pro-metastatic genetic alterations acquired early at primary tumor stage might themselves be classical oncogenes and tumor suppressor genes which can confer a selective growth advantage during tumorigenesis, and if so, such genes would be subject to recurrent genomic alterations in cancer (i.e., amplification and loss). The existence and identification of such pro-metastasis oncogenes could therefore provide both prognostic markers as well as therapeutic targets for inherently aggressive early stage cancers. In this study, using melanoma as a disease model given its cardinal feature of high metastatic propensity, we sought to validate the concept of oncogenic driver of metastasis or metastasis oncogenes through systematic identification of putative metastasis driving genes which also confer transforming oncogenic activity in early stage cancers.
RESULTS
Evolutionarily-conserved, differentially expressed genes with metastatic potential
The enormous genomic complexity of human melanoma and the less than complete certainty surrounding occult metastatic disease in any given human patient prompted us to compare two extensively characterized genetically engineered mouse (GEM) models of human melanoma with distinct metastatic profiles. The selected melanoma models are (i) the HRASV12G-driven mouse melanoma model (Tyr-rtTA;Tet-HRASV12G;Ink4a/Arf−/−, hereafter “iHRAS*”), which develops aggressive cutaneous melanomas that do not metastasize (Chin et al., 1997; Chin et al., 1999), and (ii) a Met-driven GEM model (Tyr-rtTA;Tet-Met;Ink4a/Arf−/−, hereafter “iMet”), which develops metastatic melanomas. The iMet model expresses an inducible Met transgene (Tet-Met) and is constructed following a similar engineering strategy used for the iHRAS model (Ganss et al., 1994; Chin et al., 1997; Chin et al., 1999) (see Supplemental Information for details). Tet-Met transgenic animals were bred with transgenic mice carrying the reverse tetracycline transactivator under the control of tyrosinase gene promoter–enhancer elements (designated Tyr–rtTA) (Gossen et al., 1995). Given the frequency and demonstrated relevance of INK4a/Arf deletions in melanoma (Hussussian et al., 1994; Kamb et al., 1994), these compound transgenic alleles were further intercrossed onto an INK4a/Arf null background to generate cohorts of single and double transgenic mice (designated iMet) deficient for INK4a/ARF whose melanocytes express Met upon induction with doxycycline (Figure 1A).
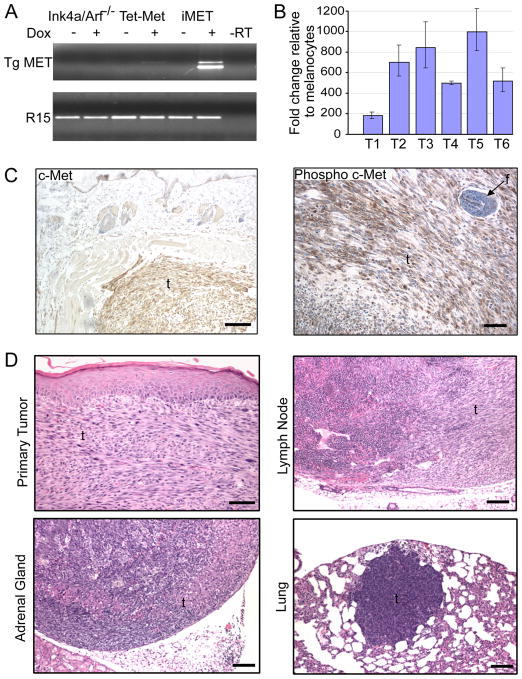
Figure 1. Melanocyte-specific MET expression promotes formation of cutaneous metastatic melanoma.
(A) Melanocytes were harvested from the indicated animals (Ink4a/Arf−/−, Tet-Met and iMet) and adapted to culture for total RNA extraction following treatment with or without doxycycline (Dox). Expression of MET (Tg MET) was assayed by RT-PCR using transgene-specific primers. R15, ribosomal protein R15 internal control; -RT, no reverse transcriptase PCR control.
(B) RT-qPCR was performed to analyze HGF expression in MET-induced primary melanomas (T1–T6). Tumor expression data is normalized to expression in two Ink4a/Arf−/− melanocyte cell lines. Error bars indicate +/− SD
(C) Immunohistochemical staining of total c-Met and phosphorylated c-Met in a MET-induced primary melanoma. Scale bar = 100 μm (left) and 50 μm (right)
(D) H&E stained sections of a primary cutaneous spindle cell melanoma in the dorsal skin of an iMet transgenic mouse induced with doxycycline and distal metastases residing in lymph node, adrenal gland and lung. Scale bar = 50 μm (primary tumor) and 100 μm (metastases). f = follicle; t = tumor.
iMet mice develop melanomas at sites of skin wounding with an average latency of 12 weeks (Table S1). These lesions are positive for prototypical melanocyte markers and express phospho-Met receptor and its ligand hepatocyte growth factor (HGF) (Figure 1B–C and Figure S1A–B). These iMet melanomas uniformly metastasize to lymph nodes and show occasional dissemination to the adrenal glands and lung parenchyma, which are common sites for metastases in human melanoma (Figure 1D). In sharp contrast, the iHRAS* melanoma model develops aggressive cutaneous melanomas which do not metastasize (Chin et al., 1997; Chin et al., 1999). Consistent with the contrasting metastatic potential of iMet and iHRAS* primary tumors, only iMet melanoma-derived cell lines were able to seed and grow to large macroscopic lesions in tail-vein experimental metastasis assays (Figure S1C).
Using these two GEM models as “extreme cases”, we compared the transcriptomic profiles of primary cutaneous melanomas from iHRAS* and iMet models to define 1597 gene probe sets with ≥2-fold differential expression at a false discovery rate <0.05. This list of differentially expressed genes was next intersected with genes residing in recurrent copy number aberrations (CNAs) in human metastatic melanoma (GEO accession # GSE7606) and/or genes exhibiting significant differential expression between primary and metastatic melanomas in human (Kabbarah et al., 2010). This comparative oncogenomics analysis led to a list of 360-genes comprised of 295 up-regulated/amplified and 65 down-regulated/deleted candidates (Figure 2A; Table S2), representing differentially expressed genes in primary melanoma that are correlated with metastatic potential. Compared with the 1597 probe set, this cross-species intersected list of 360 genes was significantly more enriched for cancer-relevant functional networks based on Ingenuity Pathway Analysis (IPA; Figure S2A).
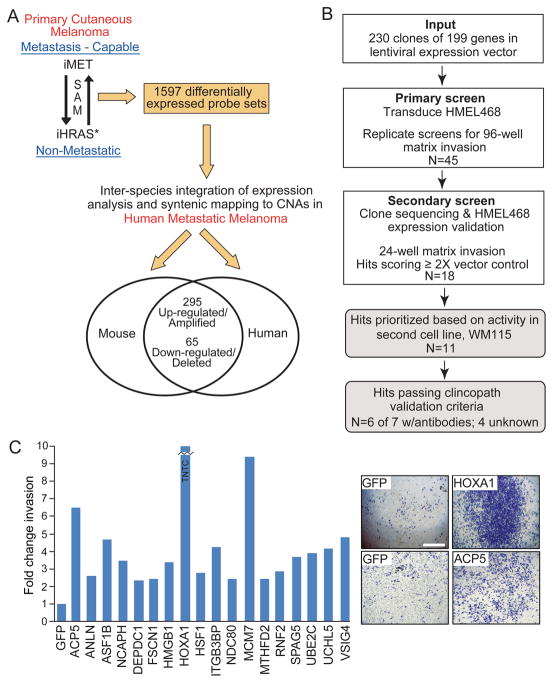
Figure 2. Multi-dimensional genomic analyses and low-complexity functional genetic screen for cell invasion.
(A) Schematic illustrating the integrative cross-species oncogenomics comparison.
(B) Flowchart depicting the low-complexity genetic screen for invasion and validation processes.
(C) Histogram of 18 pro-invasion genes satisfying sequencing, expression and secondary screen verification efforts. Shown are representative invasion chamber images for HMEL468 cells stably expressing HOXA1 and ACP5. GFP = negative control; TNTC = Too numerous to count. Scale bar = 1.6 mm.
Identification of pro-invasion oncogenes
From the above cross-species triangulated gene list for metastatic potential, we set out to identify functionally active metastasis drivers in primary melanomas following the experimental outline in Figure 2B. In particular, we designed a genetic screen for invasion based on the rationale that the ability of primary melanoma cells to invade downward into the dermis and subcutis is significantly correlated with metastasis, and a primary melanoma with pro-invasive genetic events is more likely to metastasize early, hence, we postulated that metastasis drivers in such primary melanoma would harbor pro-invasive activity. Here, we elected to focus on the 295 up-regulated genes using a gain-of-function screening design given their possible therapeutic potential. The human ORFeome collection (http://horfdb.dfci.harvard.edu/) contained 230 open reading frame (ORF) cDNAs corresponding to 199 of the 295 unique up-regulated/amplified candidates (Table S3), which were then transferred to a lentiviral expression system for transduction into HMEL468 (PMEL/hTERT/CDK4(R24C)/p53DD), a TERT-immortalized primary human melanocyte line engineered to express BRAFV600E (Garraway et al., 2005). For the primary screen, we utilized a 96-well transwell invasion assay with fluorometric readout to measure the ability of candidate genes to enhance migration and invasion of HMEL468 through Matrigel, which simulates extracellular matrix. Lentiviral expression vectors encoding GFP and NEDD9 (Kim et al., 2006; O’Neill et al., 2007; Sanz-Moreno et al., 2008; Izumchenko et al., 2009) were used as negative and positive controls, respectively. The primary screen was performed in duplicate, and 45 candidates that reproducibly scored two standard deviations from the GFP control were considered as primary screen hits (Figure S2B; Table S3). Secondary validation of these 45 candidate genes was performed by assaying their invasive ability in standard 24-well Matrigel invasion chambers, togehter with parallel sequencing and expression verification (see Supplemental Information), yielding 18 genes (Table S4) possessing >2-fold enhancement of invasion compared to the GFP control (Figure 2C and Table 1). As a frame of reference, the positive control pro-metastasis gene NEDD9, which has been shown to be required for cell movement (Sanz-Moreno et al., 2008) and in vivo metastasis of breast cancers (Izumchenko et al., 2009), enhanced invasion by 1.5-fold in this system (data not shown).
Table 1.
Invasion validation and progression-correlated expression analysis.
| Metastasis Oncogene Candidates | Gene ID | Gene Name | HMEL468 Invasion Screen | Invasion Activity in WM115 | Clinicopathological Validation# |
Oncogenic In Vivo | ||
|---|---|---|---|---|---|---|---|---|
| Transformationa | Progressionb | Passed? | ||||||
| ACP5 | 54 | acid phosphatase 5, tartrate resistant | 6.5X | 2.1X | p=0.001 | p=0.026 | yes | Yes |
| FSCN1 | 6624 | fascin homolog 1, actin-bundling protein | 2.4X | 2.2X | n.s. | p=0.026 | yes | Yes |
| HOXA1 | 3198 | homeobox A1 | TNTC | 6.1X | p<0.001 | n.s. | yes | Yes |
| HSF1 | 3297 | heat shock transcription factor 1 | 2.8X | 4.4X | p=0.003 | n.s. | yes | Yes |
| NDC80 | 10403 | NDC80 homolog, kinetochore component | 2.4X | 2.2X | n.s. | p=0.034 | yes | Yes |
| VSIG4 | 11326 | V-set & immunoglobulin domain cont. 4 | 4.8X | 2.1X | p<0.001 | n.s. | yes | Yes |
| NCAPH | 23397 | non-SMC condensin I complex, subunit H | 3.5X | 2.1X | n.s. | n.s. | no | n.t. |
|
| ||||||||
| ASF1B | 55723 | ASF1 anti-silencing function 1 homolog B | 4.7X | 2.0X | No antibody | n.f. | ||
| MTHFD2 | 10797 | methylenetetrahydrofolate dehydrogenase | 2.4X | 2.5X | No antibody | n.f. | ||
| RNF2 | 6045 | ring finger protein 2 | 2.9X | 3.4X | No antibody | n.f. | ||
| SPAG5 | 10615 | sperm associated antigen 5 | 3.2X | 2.5X | No antibody | n.f. | ||
|
| ||||||||
| ANLN | 54443 | anillin, actin binding protein | 2.6X | n.s. | ||||
| DEPDC1 | 55635 | DEP domain containing 1 | 2.3X | n.s. | ||||
| HMGB1 | 3146 | high-mobility group box 1 | 3.4X | n.s. | ||||
| ITGB3BP | 23421 | integrin beta 3 binding protein | 4.2X | n.s. | ||||
| MCM7 | 4176 | minichromosome maintenance complex 7 | 9.4X | n.s. | ||||
| UBE2C | 11065 | ubiquitin-conjugating enzyme E2C | 3.9X | n.s. | ||||
| UCHL5 | 51377 | ubiquitin carboxyl-terminal hydrolase L5 | 4.1X | n.s. | ||||
n.t. = not tested; n.s. = not significant; n.f. = not followed-up; TNTC = too numerous to count
Fisher’s test for significance between means
Assoc with transformation means Primary or Mets > than Nevi
Assoc with Progression means Mets > than Primary
To prioritize downstream validation efforts, we next assayed the 18 candidates for ability to confer a 2-fold increase of invasion in a second melanoma cell system, WM115. This identified 11 robust pro-invasion genes (Table 1). Mindful of the artificial nature of in vitro invasion screen and limitation of an over-expression system, we then interrogated the expression patterns of these pro-invasion genes in human melanocytic lesions for evidence of human relevance, specifically increasing expression from benign to malignant and/or from primary to metastasis lesions as criteria for clinicopathological validation. To this end, we rigorously screened commercially available antibodies and successfully qualified and optimized conditions of 7 antibodies for proteins encoded by 7 of the 11 genes for quantitative immunofluorescence staining on formalin-fixed paraffin-embedded tissue. Using the AQUA® platform (Camp et al., 2002), we quantitated protein expression levels on the Yale Melanoma Progression Tissue Microarray (YTMA98) containing 20 specimens each of benign nevi, primary melanoma and melanoma metastases. As summarized in Table 1, proteins encoded by 6 of 7 (ACP5, FSCN1, HOXA1, HSF1, NDC80, VSIG4) pro-invasion genes showed significantly higher expression across the benign-to-malignant and/or primary-to-metastasis transitions in human (Table 1 and Figure S3), qualifying them as validated pro-invasion genes in human melanomas.
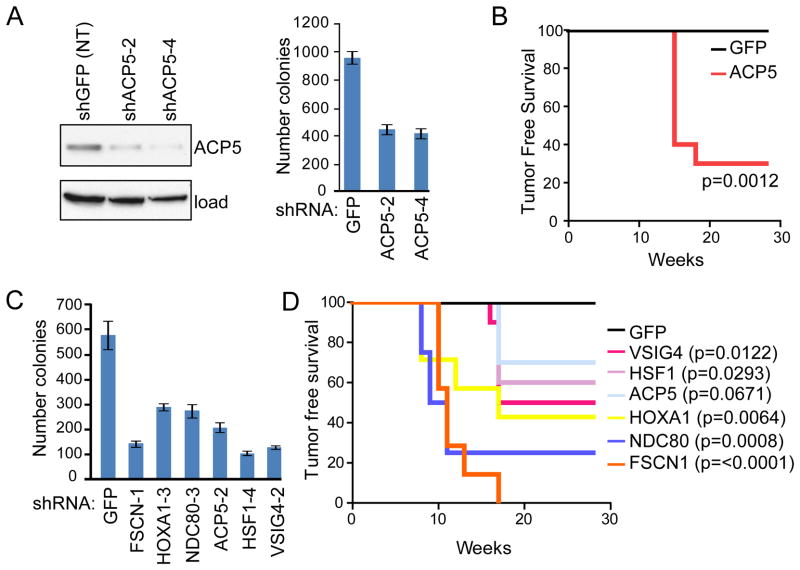
The acquisition of metastasis drivers in some early stage tumors might reflect their roles as bona fide oncogenes that could provide a proliferative advantage to the emergent primary tumors as speculated by Bernards and Weinberg (Bernards and Weinberg, 2002). To test this hypothesis, we examined the oncogenic potential of the 6 validated pro-invasion genes by assaying their requirement in maintaining the tumorigenic phenotype of established human melanoma cells in vitro and their ability to transform immortalized human melanocytes in vivo. For example, using anchorage independent growth as a surrogate for tumorigenic phenotype, depletion of ACP5 using two independent shRNAs in the human melanoma cell line 1205Lu resulted in a 56% reduction in soft agar colony formation (p = 0.0001; Figure 3A). Conversely, HMEL468 melanocytes (1×106 cells/injection) stably expressing ACP5 became robustly tumorigenic when subcutaneously implanted into the right flank of athymic nude mice (p=0.0012, Figure 3B). Importantly, extending these assays to the remaining 5 pro-invasion genes, we found that knockdown of all 6 in M619 and C918 human melanoma cells significantly decreased colony formation when compared with non-targeting (shGFP) shRNA (Figure 3C and Figure S4). Similarly, mice injected with HMEL468 cells over-expressing each of the 6 genes developed tumors, compared to none of the animals injected with GFP control HMEL468 cells after 30 weeks of observation (Figure 3D). Together, these complementary loss- and gain-of-function studies demonstrated unequivocally that all 6 of these pro-invasion genes are oncogenic. These results are striking given that transforming activity of these genes was not screened for in the course of their identification.
Figure 3. Assessment of oncogenic activity by pro-invasion genes.
(A) 1205Lu melanoma cells expressing non-targeting control (shGFP; NT) or individual shRNA hairpins against ACP5 (shACP5-2 and -4) were assayed for effects on anchorage-independent growth in soft agar. Immunoblot depicts ACP5 protein knockdown with indicated hairpins.
(B) Kaplan-Meier tumor-free survival analysis for xenograft assays in Ncr-Nude mice using non-tumorigenic HMEL468 cells (1×106 cells/injection site) stably expressing GFP or ACP5 (n=10 each). P-value calculated by log-rank test.
(C) M619 melanoma cells expressing non-targeting control (GFP) or individual shRNA hairpins against the indicated candidates were assayed for effects on anchorage-independent growth in soft agar as in (A). See Figure S4 for additional data using C918 melanoma cells and complementary knockdown verification data.
(D) Kaplan-Meier tumor-free survival analysis for xenograft assays in Ncr-Nude mice using non-tumorigenic HMEL468 cells stably expressing the indicated genes as in (B). Log-rank calculated P-values for individual candidates indicated at right of plot. Error bars indicate +/− SD
In summary, from the initial cross-species differentially-expressed list of 199 genes enlisted into the functional screen for cell invasion, 18 candidate metastasis oncogenes were identified. Of these, 7 candidates were prioritized for multi-level functional and clinicopathological validation, 6 were confirmed as potent pro-invasion oncogenes, capable of robust transforming and invasive activities in immortalized non-transformed human melanocytes and whose expressions positively correlated with human melanoma transformation or progression.
Functional and clinical validation of ACP5
Our integrated functional genomics screen and validation above have identified 6 pro-invasion oncogenes that are posited to confer enhanced metastasis risk in vivo and therefore carry prognostic significance in patients diagnosed with primary melanomas. To seek evidence in support of this, we next focused on ACP5 as a proof-of-concept example based on the observations that (i) ACP5 was the only gene exhibiting significant expression correlation with transformation as well as progression (Table 1) and (ii) ACP5 has been used as a histochemical marker of osteoclastic activity, which is increased in conditions of bone diseases including bone metastases (Halleen et al., 2001; Capeller et al., 2003; Lyubimova et al., 2004).
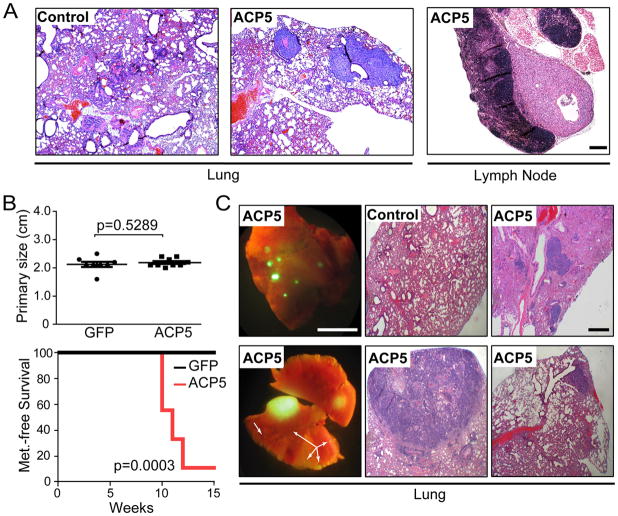
To investigate ACP5’s ability to drive distal metastasis in vivo, we over-expressed ACP5 or GFP control in the human melanoma cell line 1205Lu, which shows minimal to no distal metastasis from skin tumor sites. One million cells were then implanted into a subcutaneous site in the skin on one flank of athymic nude mice (n=5) and followed for primary tumor growth. When tumor size reached 2cm2, animals were sacrificed and examined for macro and micro metastasis in lymph nodes and distal organ systems. Consistent with its invasive activity, animals bearing ACP5-expressing melanomas in the subcutaneous sites developed spontaneous metastasis to the lung and lymph nodes (n=2; Figure 4A) while none in the control cohort harbored any metastatic lesion despite similar tumor penetrance in both cohorts (n=5 each). Additionally, based on the prognostic significance of these genes in human breast cancers (see below), we also utilized NB008 (mTerc−/−, p53+/−; mTerc), a well-characterized, non-metastatic cell line originating from a spontaneous murine mammary adenocarcinoma (mTerc−/−, p53+/−) engineered to re-express mTerc. Specifically, GFP-labeled NB008 cells stably expressing ACP5 or vector control were orthotopically implanted into the right inguinal mammary fat pad of athymic nude mice. Macroscopic GFP-positive lesions in the lungs were scored at necropsy when primary mammary tumors reached 2cm, the maximum size allowed by our experimental protocol (Figure 4B). As shown by Kaplan-Meier metastasis-free survival analysis, GFP-positive macro-metastasis was detected in the lungs of 89% (8/9) of mice bearing ACP5-expressing tumors, whereas none (0/8) of the animals injected with control tumor cells presented with lung nodules (p=0.0003; Figure 4B). Histopathological examination confirmed presence of macro- and micro-metastases (Figure 4C). Together, these results show that ACP5 is a bona fide metastasis driver in vivo.
Figure 4. In vivo metastasis studies.
(A) Representative H&E staining of lung and lymph node metastases in athymic mice (2/5) harboring subcutaneous tumors generated from 1205Lu melanoma cells expressing ACP5. No metastases (0/5 animals) were detected in the GFP-expressing control cohort. Scale bar = 200 μm.
(B) Mammary fat pad metastasis assay using GFP-positive non-metastatic murine breast adenocarcinoma cells (NB008; 2×104 cells/injection site) stably expressing vector control or ACP5. Shown are endpoint primary tumor size (top) and Kaplan-Meier metastasis-free survival analysis (bottom). P-values calculated by two-tailed t-test (top) and log-rank (bottom).
(C) Representative images of GFP-positive lung metastases and H&E stained sections of infiltrated lung from the ACP5 cohort. Arrows denote micro metastases. Scale bar = 5.5 mm (left 2 panels) 300 μm (right 4 H&E panels).
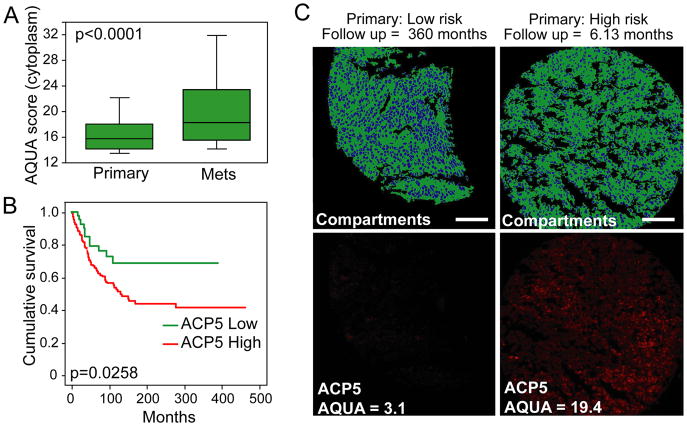
Next, to investigate the prognostic significance of ACP5 expression in human primary melanomas, we again employed the quantitative immunofluorescence measurement of ACP5 protein expression on a tissue microarray (YTMA59) containing 196 cases of primary melanomas and 299 cases of metastatic melanomas annotated for survival outcome (Berger et al., 2005; Gould Rothberg et al., 2009). As observed in the clinicopathological validation study (Figure S3), ACP5 staining was primarily cytoplasmic, and the differential distributions of staining intensity were significantly higher in the metastatic lesions compared to primary specimens (Figure 5A; ANOVA P<0.0001). Importantly, the ACP5 protein level in primary melanomas correlates with survival of patients, for which a significantly shorter melanoma-specific survival was observed in cases with higher cytoplasmic ACP5 levels (log rank p=0.0258; Figure 5B–C and Figure S5). Collectively, our data therefore show that ACP5 is not only a pro-invasion oncogene but also a prognostic biomarker in human primary melanomas.
Figure 5. ACP5 expression on melanoma tissue microarrays.
(A) Box plot demonstrating the distribution of ACP5 cytoplasmic scores for primary (n=182) and metastatic (n=325) lesions on the Yale Melanoma Outcome Annotated TMA (YTMA59). P-value calculated by mixed model ANOVA. Error bars indicate data within 1.5 interquartile range of the mean.
(B) Primary tumors from (A) were divided into quartiles based on cytoplasmic expression of ACP5. Shown is comparing melanoma-specific survival of the the lowest quartile (green) with the other 3 quartiles (red). P-value calculated by log-rank test.
(C) Representative staining of ACP5 (red) across histospot tumor specimens on YTMA59. S100/GP100 (green) defines tumor and nonnuclear compartments, and DAPI (blue) defines the nuclear compartments. Scale bar = 100 μm.
See also Figure S5.
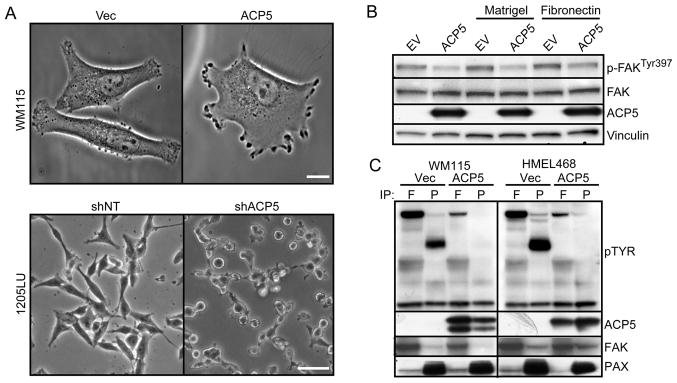
On the cell biological level, over-expression and RNAi-knockdown of ACP5 resulted in striking morphological changes such as cell spreading and cell rounding, respectively (Figure 6A), prompting us to consider the possibility that ACP5 could modulate phosphorylation of focal adhesion complexes that are integral to cell attachment and motility. Indeed, over-expression of ACP5 in melanoma cells led to a reproducible decrease in FAK auto-phosphorylation at Tyr397 (Figure 6B) and global FAK tyrosine phosphorylation beyond its autophosphorylation site (Figure 6C). Similar analysis uncovered a more significant effect of ACP5 over-expression on tyrosine phosphorylation of Paxillin (PAX; Figure 6C), including Tyr118 (Figure S6), which is thought to serve as a critical docking site for other signaling molecules. Live-cell imaging of ACP5 over-expressing cells translated these biochemical changes to increased cell movement (Movies S1 and S2), consistent with our data on ACP5’s activity on cell invasion. Given the literature implicating the FAK complex activity in metastasis (Zheng and Lu, 2009), this mechanistic link thus further substantiates the functional role of ACP5 in invasion and points to the FAK complex as a possible point of therapeutic intervention in high-risk primary melanoma with high ACP5 expression.
Figure 6. ACP5 expression modulates phosphorylation status of adhesion molecules.
A) Morphology of WM115 (top) cells without (Vec) or with ACP5 over-expression or 1205Lu cells (bottom) treated with a control shRNA (shNT) or an shRNA targeting ACP5 (shACP5). Scale bar = 10 μm (top) and 5 μm (bottom).
(B) Immunoblot analysis of indicated proteins of WM115 cells expressing empty vector (EV) or ACP5 and grown on plates without coating or coated with Matrigel or Fibronectin as indicated.
(C) Protein lysates extracted from WM115 and HMEL468 cells were immunoprecipitated (IP) with antibodies against focal adhesion kinase (FAK or F) and paxillin (PAX or P) for immunoblotting with the indicating antibodies. Tyrosine phosphorylation (pTyr) is determined by anti-pTyr immunoblot analysis.
Metastasis oncogenes are not lineage-specific
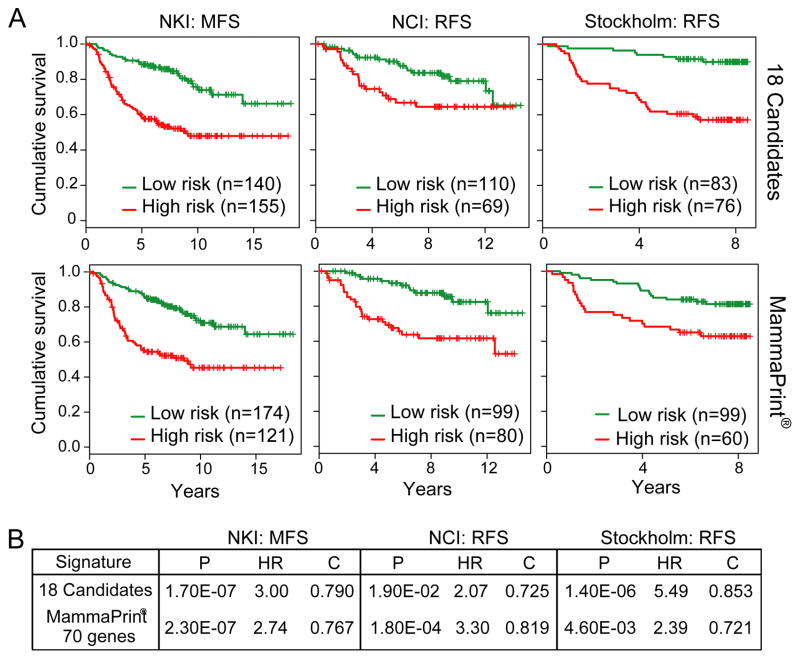
While the majority of pro-invasion genes identified from our integrated functional genetic screen have not been linked to metastasis (see Discussion), the actin-bundling protein FSCN1 is reported to be prognostic in numerous cancer types (Hashimoto et al., 2005), and recently shown to be required for metastasis (Chen et al., 2010). This led us to explore the prognostic relevance of these pro-invasion genes in other tumor types using RNA expression, given the limitation of antibody reagent for quantitative protein-based assays. We focused specifically on breast cancer based on the availability of 3 independent cohorts of transcriptome datasets on Stage I/II breast adenocarcinomas with outcome (recurrence or metastasis-free survival) annotation (van de Vijver et al., 2002; Pawitan et al., 2005; Sotiriou et al., 2006). As summarized in Figure 7A, expression levels of the 18 pro-invasion genes were able to stratify patients by K-mean clustering into two subgroups with significant differences in metastasis-free or recurrence-free survival by Kaplan-Meier survival analysis in all 3 independent datasets. Moreover, by C-statistics, these 18 genes were comparable to the 70-genes in the FDA-approved Mammaprint® in their ability to prognosticate recurrence or metastasis (Figure 7B). These data are remarkable in light of the fact that these genes were discovered in melanoma. Such cross-tumor prognostic significance reinforces the human relevance and highlights the power of this integrative functional genomics approach for discovery of metastasis oncogenes that can function across different tumor types.
Figure 7. Kaplan-Meier survival curves in breast cancer cohorts.
(A) K-means clustering analysis based on the 18-gene pro-invasion oncogene (top) and Mammaprint® (bottom) signature using three independent cohorts of early-staged breast cancers: NKI metastasis-free survival (MFS)(van de Vijver et al., 2002); NCI recurrence-free survival (RFS)(Sotiriou et al., 2006); and Stockholm RFS(Pawitan et al., 2005). P-values calculated by log-rank test.
(B) Comparison of the 18-gene signature performance with the Mammaprint® (Agendia, Huntington Beach, CA) prognostic signature using the patient cohorts specified in (A). HR = Hazard ratio; C = C statistics.
DISCUSSION
In this study, we employed well-defined GEM models, comparative oncogenomics, and functional genomics to identify genes capable of driving invasion and transformation in early-staged melanomas. The genomic and biological homogeneity of GEM tumors and filtering power of cross-species comparisons proved highly effective in generating a shorter, more biologically significant list of genes enriched for cancer- and metastasis-relevant networks than either human or mouse datasets alone. Subsequent functional screen and stringent validation efforts identified high priority drivers of invasion – the key biological process that correlates with metastatic potential in melanoma. Finally, although oncogenic activity was not screened for, it is remarkable that every one of the 6 pro-invasion genes is robustly transforming in vivo, a finding that supports the hypothesis that drivers of metastasis in early-staged primary tumors also serve as professional oncogenes promoting tumorigenesis.
Of the 6 validated metastasis oncogenes, most are not known or implicated in metastasis although some have been linked to cancer. For example, HSF1 (Heat Shock Factor 1) is a regulator of cell transformation and in vivo tumorigenesis (Dai et al., 2007), and HSF1-deficient cells exhibit markedly impaired migration and MAP kinase signaling (O’Callaghan-Sunol and Sherman, 2006). In a transgenic mouse model with over-expression of NDC80, a component of the spindle checkpoint, tumor development was reported in multiple organs (Diaz-Rodriguez et al., 2008), and depletion of NDC80 impairs tumor growth (Gurzov and Izquierdo, 2006). HOXA1 (Homeobox Transcription factor 1) has oncogenic activity in mammary tumor models (Zhang et al., 2003) and is up-regulated in multiple human cancers including breast, squamous cell carcinoma and melanoma (Chariot and Castronovo, 1996; Maeda et al., 2005; Abe et al., 2006). VSIG4 (V-set and immunoglobulin domain containing 4) is a cell surface protein whose expression is mainly restricted to macrophages where it functions as a potent T-cell inhibitor (Vogt et al., 2006; Xu et al., 2010). Based on its significantly higher expression in aggressive breast and ovarian tissues compared to benign tissues, ACP5 expression has been suggested to represent a progression marker (Honig et al., 2006; Adams et al., 2007), consistent with our data in melanoma. Although we have shown in model systems that ACP5 over-expression alone was sufficient in conferring distal metastasis in vivo, frank metastasis in patients most certainly requires cooperation of a multitude of genetic alterations, each driving one or more steps in the metastatic cascade. Therefore, one may speculate that there would be metastasis oncogenes that are drivers of other biological processes (besides invasion) that are required for metastasis thus a similar integrated functional genomics approach could be powerful in aiding their discovery.
The majority of cancer-related deaths result from metastases. With the improvement of early detection capability by serum biomarkers and imaging advances, an increasing number of cancer cases will be diagnosed and surgically resected prior to apparent metastatic spread, leading to better overall survival relative to high-stage disease. At the same time, it is long-recognized that equivalent low-stage cancers are clinically heterogeneous with a subset exhibiting high-risk behavior, recurring with metastatic spread in the years ahead. The precise identification of such high-risk cases would enable more aggressive management in adjuvant setting, while avoiding unnecessary treatment in those patients cured by surgical intervention alone. Therefore, there is a growing need for the development of molecular-based prognostic biomarkers that can stratify risk for metastasis in the early-stage cancer population which constitutes an increasing proportion of cancer diagnoses each year. Transcriptomic and genomic characterization of human cancers supports the presence of molecular signals resident in primary tumors that can predict risk for metastasis. The development of MammaPrint® and OncotypeDx® has provided a strong measure of clinical proof of this concept. In comparison to the predominantly statistical correlative analyses from which these signatures were derived, the approach reported here focuses on discovery of functional drivers of the metastatic process that are also oncogenic in early-stage cancers. Given their functional nature, we believe that the mechanism-of-action through which these pro-invasion oncogenes drive metastasis would inform evidence-based therapeutic decisions in the adjuvant setting, in addition to themselves being rational points for therapeutic intervention. In this regard, the convergence of emerging targeted therapeutics for melanoma (such as the selective BRAF inhibitor) and identification of pro-invasion oncogenes as prognostic biomarkers (such as ACP5) will offer a real opportunity to stratify a molecularly high-risked subpopulation among early-stage primary melanoma patients for clinical investigation aimed to explore the efficacy of these new therapies in the prevention of recurrence and metastasis.
EXPERIMENTAL PROCEDURES
GEM Mouse Models for Melanoma, Comparative Data Analyses and In Vivo Tumor Assays
All mice were bred and maintained under defined conditions at the Dana-Farber Cancer Institute (DFCI), and all procedures were approved by the Animal Care and Use Committee of DFCI and conformed to the legal mandates and national guidelines for the care and maintenance of laboratory animals. The tetracycline-inducible MET-driven mouse (iMet) model (Tyr-rtTA;Tet-Met;Ink4a/Arf−/−) was constructed similar to the previously described iHRAS* model (Tyr-rtTA;Tet-HRASV12G;Ink4a/Arf−/−)(Chin et al., 1999). Mice were sacrificed according to institute guidelines and organs were fixed in 10% buffered formalin and paraffin embedded. Tissue sections were stained with H&E to enable classification of the lesions and detection of tumor metastasis. For detection of c-Met protein, tumor sections were immunostained with total c-Met and phospho c-Met (Tyr1349) antibodies (Cell Signaling Technology). iMet tumors were additionally immunostained with S100 antibody (Sigma). RNA from cutaneous melanomas derived from iMet or iHRAS* models were profiled on Affymetrix Gene Chips and resultant transcriptomes were compared using Significance Analysis of Microarray (SAM 2.0) to generate a phenotype-based (metastatic capable or not) differentially expressed gene list. Cross-species triangulation to human gene expression and copy number aberrations was based on ortholog mapping. See Supplemental Information for more details.
For xenograft tumorigenicity studies, HMEL468 cells were transduced with pLenti6/V5 DEST-generated virus for stable expression of GFP (control) or the indicated genes. Following selection with blasticidin (Invitrogen; 5 μg/ml) for 5–7 days, 1.0×106 cells [prepared in Hanks Balanced Salts (HBS) at 1:1 with Matrigel] were injected subcutaneously into the right flank of NCr-Nude (Taconic) mice. Two-tailed t-test calculations were performed using Prism 4 (Graphpad). In vivo metastasis assays were performed by 1) subcutaneous skin tumor assays using 1205Lu cells stably-expressing GFP (control) or ACP5 and 2) orthotopic mammary fad pad assays using non-metastatic NB008 adenocarcinoma cells stably-expressing vector (control) or ACP5 as described in Supplemental Information
Cell Culture
HMEL468 primed melanocytes were a subclone of PMEL/hTERT/CDK4(R24C)/p53DD/BRAFV600E cells as described (Garraway et al., 2005). The non-metastatic NB008 cell line was established from a spontaneous tumor isolated from the breast of a G4 52-week old female mTerc−/−, p53+/− mouse. GFP-mTerc was re-introduced into the resulting cell line by lentiviral transduction prior to use in these studies. The WM115 melanoma cell line was obtained from the Wistar Institute, and the 1205Lu melanoma cell line was obtained from the American Type Culture Collection. M619 and C918 melanoma lines have been described previously (Maniotis et al., 1999). All cell lines were propagated at 37°C and 5% CO2 in humidified atmosphere in RPMI 1640 medium supplemented with 10% FBS.
Invasion Screen and Transwell Invasion Assays
The low complexity genetic screen for cell invasion was performed using Tert-immortalized melanocytes HMEL468 in 96-well modified Boyden chambers coated with Matrigel (96-well tumor invasion plates; BD Bioscience) following the manufacture’s recommendations. Invaded cells were detected with labeling using 4 uM Calcein AM (BD Bioscience) and measured by fluorescence at 494/517 nm (Abs/Em) after 20 hrs incubation at 37°C and 5% CO2. Positive-scoring candidates were identified as those scoring 2x standard deviations from the vector control. See Supplemental Information for details on the screen and individual clones. Validation assays for cell invasion were performed in standard 24-well invasion chambers containing Matrigel (BD Bioscience) following the manufacture’s recommendations. Following 18–20 hrs incubation at 37°C and 5% CO2, chambers were fixed in 10% formalin, stained with crystal violet for manual counting or by pixel quantitation with Adobe Photoshop (Adobe). Data was normalized to input cells to control for differences in cell number (loading control).
Automated Quantitative Analysis (AQUA®)
Uses of human tissues in this study are approved by the Yale institutional IRB, HIC protocol number 9500008219 including consent and waived consent. AQUA® analysis and the Yale Melanoma Arrays and tissue microarray construction have been described previously (Camp et al., 2002; Gould Rothberg et al., 2009). Arrays were stained with the following antibodies: monoclonal anti-Fascin1 diluted 1:500 (clone 55K2, Santa Cruz Biotechnology, Inc.), polyclonal anti-HOXA1 diluted 1:50 (BO1P, Abnova), polyclonal anti-HSF1 diluted 1:2500 (AO1, Abnova), monoclonal anti-NDC80 diluted 1:50 (clone 1A10, Abnova), monoclonal anti-ACP5 diluted 1:100 (clone 26E5, Abcam), polyclonal anti-NCAPH diluted 1:750 (Bethyl Laboratories, Inc.), and polyclonal anti-VSIG4 diluted 1:1000 (ab56037, Abcam). See Supplemental Information for full details.
Anchorage Independent Growth Assays
Soft-agar colony formation assays were performed on 6-well plates in triplicate for cells transduced with pLKO-shGFP (Open Biosystems) or shRNA (Bill Hahn, DFCI/Broad Institute; available via Open Biosystems) hairpins targeting the indicated genes (see Supplemental Information for additional clone information). Cells were selected for 5 days with 2.5 μg/ul puromycin, and 1×104 cells were mixed thoroughly in cell growth medium containing 0.4% SeaKem LE agarose (Fisher) in RPMI + 10% FBS, followed by plating onto bottom agarose prepared with 0.65 % agarose in RPMI + 10% FBS. Each well was allowed to solidify and subsequently covered in 1 ml RPMI + 10% FBS + P/S, which was refreshed every 4 days. Colonies were stained with 0.05% (wt/vol) iodonitrotetrazolium chloride (Sigma) and scanned at 1200 dpi using a flatbed scanner, followed by counting and two-tailed t-test calculation using Prism 4 (Graphpad). Verification of knockdown was achieved by qRT-PCR (described in Supplemental Information) using gene-specific primer sets (SABiosciences).
Co-immunoprecipitation and immunoblotting
For immunoprecipitation studies, lysates were prepared in NP-40 buffer (20 mMTris-HCl, pH 8.0, 150 mMNaCl, 2 mM EDTA, 1% NP40) containing 1 mM PMSF, 1x Protease Inhibitor Cocktail (Roche) and 1x Phosphatase inhibitor (Calbiochem) for immunoprecipitation. Anti-Paxillin (Abcam) or anti-FAK (Santa Cruz) antibody was added to cell lysates for 2 hr at 4°C with rocking, followed by incubation overnight with protein G sepharose (Roche) at 4°C with rocking. Immunoprecipitates were washed 3x for 10 min with lysis buffer, eluted by the addition of SDS loading buffer after centrifugation and resolved on NuPAGE 4–12% Bis-Tris gels (Invitrogen) for immunoblotting on PVDF (Millipore). The following antibodies were used for immunoblotting following the manufacture’s recommendations: anti-FAK (Santa Cruz); anti-FAK (Tyr397; Cell Signaling); anti-Paxillin (Abcam); anti-Paxillin (Tyr118; Cell Signaling); anti-Vinculin (Santa Cruz); anti-V5 (for ACP5 detection; Invitrogen) and anti-phospho-tyrosine (Millipore).
Cell Imaging
Single-plane phase image was collected on a Nikon Ti with a 40x Plan-Apochromatic phase objective NA 0.95 and a Clara camera using Andor iQ software (Andor Technology). Time lapse phase images were collected on a Nikon TE2000-E with a 10x phase objective and an OrcaER camera (Hamamatsu) at the Dana-Farber Cancer Institute Confocal and Light Microscopy Core. Shutters, stage position, and camera were controlled by NIS-Elements software (Nikon, Melville, NY). Images were collected every 2 minutes at 6–12 stage positions for 20 hours. A representative time lapse movie for vector and ACP5 over-expressing cells are shown. For the Quicktime® movie, every 10th frame was used (20 min time points) and playback is 15 frames per second.
Breast cancer prognostic studies
Expression patterns of the 18 candidate pre-invasion oncogenes and MammaPrint® 70-gene signature (Agendia, Huntington Beach, CA) were used for Kaplan-Meier survival analyses of the indicated breast cancer datasets by K-means clustering using the survival package in R.
Supplementary Material
Article Highlights.
Integrates melanoma mouse models and human genomics data to derive cancer gene list.
Uses a function-based screening approach to identify oncogenic metastasis drivers.
Shows that ACP5 engages the FAK signaling complex and is prognostic in melanoma.
Metastatic events present in early tumors can reflect their oncogenic capability.
SIGNIFICANCE.
Early-stage melanoma is often cured by surgical excision, yet some cases without clinical evidence of dissemination recur with lethal metastatic disease despite successful surgical removal of the primary tumor. Elucidation of the molecular basis underlying such aggressive biology has been a longstanding focus, with the goal of identifying prognostic biomarkers and rational therapeutics for high-risk patients diagnosed with early-stage disease who are in need of further treatment in adjuvant setting. This study illustrates how one can exploit and integrate genetically engineered mouse models, cross-species cancer genomics knowledge, and functional screens to identify robust pro-invasion drivers of metastasis that are also bona fide oncogenes.
Acknowledgments
Authors wish to thank members of the Chin laboratory for helpful discussion, particularly Shan Jiang for mouse colony work and Bob Xiong for computational assistance. Authors acknowledge generous gifts Drs. Marc Vidal and David Hill of the human ORFeome in Center for Cancer System Biology for ORF clones, and Dr. David Fisher for the HMEL468 primed melanocytes (PMEL/hTERT/CDK4(R24C)/p53DD/BRAFV600E). Wide-field microscopy images for this study were acquired in the Confocal and Light Microscopy Core Facility at the Dana Farber Cancer Institute. K.L.S was supported by a postdoctoral fellowship from the American Cancer Society and was previously supported by a National Institute of Health Training Grant appointment in the Department of Dermatology at Brigham and Women’s Hospital, Boston, MA. C.N. was supported by a fellowship from FCT (Praxis XXI/BD/21794/99). R.v.D. was supported by a KWF fellowship for medical specialists. This work is supported by grants from the NIH (RO1 CA93947, U01 CA84313 and P50 CA93683) to L. Chin.
Footnotes
ACCESSION NUMBERS
Expression array data for the iMet and iRas tumors generated by these studies have been deposited into the GEO database with accession GSE29074.
Supplemental Information includes Extended Experimental Procedures, 6 figures, and 4 tables and can be found with this article online at ???.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abe M, Hamada J, Takahashi O, Takahashi Y, Tada M, Miyamoto M, Morikawa T, Kondo S, Moriuchi T. Disordered expression of HOX genes in human non-small cell lung cancer. Oncol Rep. 2006;15:797–802. [PubMed] [Google Scholar]
- Adams LM, Warburton MJ, Hayman AR. Human breast cancer cell lines and tissues express tartrate-resistant acid phosphatase (TRAP) Cell Biol Int. 2007;31:191–195. doi: 10.1016/j.cellbi.2006.09.022. [DOI] [PubMed] [Google Scholar]
- Berger AJ, Davis DW, Tellez C, Prieto VG, Gershenwald JE, Johnson MM, Rimm DL, Bar-Eli M. Automated quantitative analysis of activator protein-2alpha subcellular expression in melanoma tissue microarrays correlates with survival prediction. Cancer Res. 2005;65:11185–11192. doi: 10.1158/0008-5472.CAN-05-2300. [DOI] [PubMed] [Google Scholar]
- Bernards R, Weinberg RA. A progression puzzle. Nature. 2002;418:823. doi: 10.1038/418823a. [DOI] [PubMed] [Google Scholar]
- Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8:1323–1327. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- Capeller B, Caffier H, Sutterlin MW, Dietl J. Evaluation of tartrate-resistant acid phosphatase (TRAP) 5b as serum marker of bone metastases in human breast cancer. Anticancer Res. 2003;23:1011–1015. [PubMed] [Google Scholar]
- Chariot A, Castronovo V. Detection of HOXA1 expression in human breast cancer. Biochem Biophys Res Commun. 1996;222:292–297. doi: 10.1006/bbrc.1996.0737. [DOI] [PubMed] [Google Scholar]
- Chen L, Yang S, Jakoncic J, Zhang JJ, Huang XY. Migrastatin analogues target fascin to block tumour metastasis. Nature. 2010;464:1062–1066. doi: 10.1038/nature08978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K, de Solorzano CO, Knowles D, Jones A, Chou W, Rodriguez EG, Kuo WL, Ljung BM, Chew K, Myambo K, et al. In situ analyses of genome instability in breast cancer. Nat Genet. 2004;36:984–988. doi: 10.1038/ng1409. [DOI] [PubMed] [Google Scholar]
- Chin L, Pomerantz J, Polsky D, Jacobson M, Cohen C, Cordon-Cardo C, Horner JW, 2nd, DePinho RA. Cooperative effects of INK4a and ras in melanoma susceptibility in vivo. Genes Dev. 1997;11:2822–2834. doi: 10.1101/gad.11.21.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L, Tam A, Pomerantz J, Wong M, Holash J, Bardeesy N, Shen Q, O’Hagan R, Pantginis J, Zhou H, et al. Essential role for oncogenic Ras in tumour maintenance. Nature. 1999;400:468–472. doi: 10.1038/22788. [DOI] [PubMed] [Google Scholar]
- Dai C, Whitesell L, Rogers AB, Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130:1005–1018. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Rodriguez E, Sotillo R, Schvartzman JM, Benezra R. Hec1 overexpression hyperactivates the mitotic checkpoint and induces tumor formation in vivo. Proc Natl Acad Sci U S A. 2008;105:16719–16724. doi: 10.1073/pnas.0803504105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganss R, Montoliu L, Monaghan AP, Schutz G. A cell-specific enhancer far upstream of the mouse tyrosinase gene confers high level and copy number-related expression in transgenic mice. EMBO J. 1994;13:3083–3093. doi: 10.1002/j.1460-2075.1994.tb06607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, Beroukhim R, Milner DA, Granter SR, Du J, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- Gould Rothberg BE, Berger AJ, Molinaro AM, Subtil A, Krauthammer MO, Camp RL, Bradley WR, Ariyan S, Kluger HM, Rimm DL. Melanoma prognostic model using tissue microarrays and genetic algorithms. J Clin Oncol. 2009;27:5772–5780. doi: 10.1200/JCO.2009.22.8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Gurzov EN, Izquierdo M. RNA interference against Hec1 inhibits tumor growth in vivo. Gene Ther. 2006;13:1–7. doi: 10.1038/sj.gt.3302595. [DOI] [PubMed] [Google Scholar]
- Halleen JM, Alatalo SL, Janckila AJ, Woitge HW, Seibel MJ, Vaananen HK. Serum tartrate-resistant acid phosphatase 5b is a specific and sensitive marker of bone resorption. Clin Chem. 2001;47:597–600. [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y, Skacel M, Adams JC. Roles of fascin in human carcinoma motility and signaling: prospects for a novel biomarker? Int J Biochem Cell Biol. 2005;37:1787–1804. doi: 10.1016/j.biocel.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Honig A, Rieger L, Kapp M, Krockenberger M, Eck M, Dietl J, Kammerer U. Increased tartrate-resistant acid phosphatase (TRAP) expression in malignant breast, ovarian and melanoma tissue: an investigational study. BMC Cancer. 2006;6:199. doi: 10.1186/1471-2407-6-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussussian CJ, Struewing JP, Goldstein AM, Higgins PA, Ally DS, Sheahan MD, Clark WH, Jr, Tucker MA, Dracopoli NC. Germline p16 mutations in familial melanoma. Nat Genet. 1994;8:15–21. doi: 10.1038/ng0994-15. [DOI] [PubMed] [Google Scholar]
- Izumchenko E, Singh MK, Plotnikova OV, Tikhmyanova N, Little JL, Serebriiskii IG, Seo S, Kurokawa M, Egleston BL, Klein-Szanto A, et al. NEDD9 promotes oncogenic signaling in mammary tumor development. Cancer Res. 2009;69:7198–7206. doi: 10.1158/0008-5472.CAN-09-0795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabbarah O, Nogueira C, Feng B, Nazarian RM, Bosenberg M, Wu M, Scott KL, Kwong LN, Xiao Y, Cordon-Cardo C, et al. Integrative genome comparison of primary and metastatic melanomas. PLoS One. 2010;5:e10770. doi: 10.1371/journal.pone.0010770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamb A, Gruis NA, Weaver-Feldhaus J, Liu Q, Harshman K, Tavtigian SV, Stockert E, Day RS, 3rd, Johnson BE, Skolnick MH. A cell cycle regulator potentially involved in genesis of many tumor types. Science. 1994;264:436–440. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- Kim M, Gans JD, Nogueira C, Wang A, Paik JH, Feng B, Brennan C, Hahn WC, Cordon-Cardo C, Wagner SN, et al. Comparative oncogenomics identifies NEDD9 as a melanoma metastasis gene. Cell. 2006;125:1269–1281. doi: 10.1016/j.cell.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Lyubimova NV, Pashkov MV, Tyulyandin SA, Gol’dberg VE, Kushlinskii NE. Tartrate-resistant acid phosphatase as a marker of bone metastases in patients with breast cancer and prostate cancer. Bull Exp Biol Med. 2004;138:77–79. doi: 10.1023/b:bebm.0000046945.95479.d6. [DOI] [PubMed] [Google Scholar]
- Maeda K, Hamada J, Takahashi Y, Tada M, Yamamoto Y, Sugihara T, Moriuchi T. Altered expressions of HOX genes in human cutaneous malignant melanoma. Int J Cancer. 2005;114:436–441. doi: 10.1002/ijc.20706. [DOI] [PubMed] [Google Scholar]
- Maniotis AJ, Folberg R, Hess A, Seftor EA, Gardner LM, Pe’er J, Trent JM, Meltzer PS, Hendrix MJ. Vascular channel formation by human melanoma cells in vivo and in vitro: vasculogenic mimicry. Am J Pathol. 1999;155:739–752. doi: 10.1016/S0002-9440(10)65173-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Callaghan-Sunol C, Sherman MY. Heat shock transcription factor (HSF1) plays a critical role in cell migration via maintaining MAP kinase signaling. Cell Cycle. 2006;5:1431–1437. doi: 10.4161/cc.5.13.2915. [DOI] [PubMed] [Google Scholar]
- O’Neill GM, Seo S, Serebriiskii IG, Lessin SR, Golemis EA. A new central scaffold for metastasis: parsing HEF1/Cas-L/NEDD9. Cancer Res. 2007;67:8975–8979. doi: 10.1158/0008-5472.CAN-07-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- Pawitan Y, Bjohle J, Amler L, Borg AL, Egyhazi S, Hall P, Han X, Holmberg L, Huang F, Klaar S, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7:R953–964. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- Rudolph KL, Millard M, Bosenberg MW, DePinho RA. Telomere dysfunction and evolution of intestinal carcinoma in mice and humans. Nat Genet. 2001;28:155–159. doi: 10.1038/88871. [DOI] [PubMed] [Google Scholar]
- Sanz-Moreno V, Gadea G, Ahn J, Paterson H, Marra P, Pinner S, Sahai E, Marshall CJ. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 2008;135:510–523. doi: 10.1016/j.cell.2008.09.043. [DOI] [PubMed] [Google Scholar]
- Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, Smeds J, Nordgren H, Farmer P, Praz V, Haibe-Kains B, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- van ‘t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- Vogt L, Schmitz N, Kurrer MO, Bauer M, Hinton HI, Behnke S, Gatto D, Sebbel P, Beerli RR, Sonderegger I, et al. VSIG4, a B7 family-related protein, is a negative regulator of T cell activation. J Clin Invest. 2006;116:2817–2826. doi: 10.1172/JCI25673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Sun Z, Li L, Liu J, He J, Song D, Shan G, Liu H, Wu X. Induction of T cells suppression by dendritic cells transfected with VSIG4 recombinant adenovirus. Immunol Lett. 2010;128:46–50. doi: 10.1016/j.imlet.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhu T, Chen Y, Mertani HC, Lee KO, Lobie PE. Human growth hormone-regulated HOXA1 is a human mammary epithelial oncogene. J Biol Chem. 2003;278:7580–7590. doi: 10.1074/jbc.M212050200. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Lu Z. Paradoxical roles of FAK in tumor cell migration and metastasis. Cell Cycle. 2009;8:3474–3479. doi: 10.4161/cc.8.21.9846. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.