Abstract
During protein synthesis, the ribosome controls the movement of transfer RNA (tRNA) and messenger RNA (mRNA) by means of large-scale structural rearrangements. We describe structures of the intact bacterial ribosome from Escherichia coli that reveal how the ribosome binds tRNA in two functionally distinct states, determined to a resolution of ~3.2 Å by x-ray crystallography. One state positions tRNA in the peptidyl-tRNA binding site. The second, a fully rotated state, is stabilized by ribosome recycling factor (RRF) and binds tRNA in a highly bent conformation in a hybrid peptidyl/exit (P/E) site. The structures help to explain how the ratchet-like motion of the two ribosomal subunits contributes to the mechanisms of translocation, termination, and ribosome recycling.
Protein biosynthesis by the ribosome proceeds in defined phases of initiation, protein elongation, termination, and ribosome recycling (1). Understanding the molecular mechanism of translation requires high-resolution descriptions of the motions in the ribosome that enable key translational events (1-3). A ratchet-like rotation of the small ribosomal subunit relative to the large ribosomal subunit (4) is crucial to the positioning of tRNAs in intermediate – or hybrid – binding sites, in which the 3’-CCA termini and acceptor stems of tRNA advance by one site on the large subunit while the anticodon elements of tRNA remain fixed on the small subunit (5). Binding of tRNAs in hybrid sites is central to mRNA and tRNA movements on the ribosome when they are translocated after each peptide bond is formed, during termination, and during ribosome recycling (6, 7). However, the molecular basis for ribosome positioning of tRNAs in hybrid sites has been unclear.
Atomic resolution x-ray crystal structures of the bacterial ribosome with ligands bound have revealed molecular details of conformational rearrangements taking place in the unratcheted ribosome (1). The first molecular descriptions of intermediate states of ribosome ratchet-like rotation at atomic resolution were provided by x-ray crystal structures of the E.coli 70S ribosome (8), with additional sub-steps proposed based on cryo-EM reconstructions (9). A post-translocation rotated state of the ribosome was recently identified by cryo-EM (10), in a conformation similar to that of the Saccharomyces cerevisiae 80S ribosome in the absence of bound substrates (11, 12).
After the termination of protein synthesis, ribosome recycling is required to free ribosomes from the mRNA transcript to enable further rounds of translation. In bacteria and organelles, ribosome recycling factor (RRF) binds in the tRNA binding cleft of the 70S ribosome at the interface of the large (50S) and small (30S) subunits and interacts with the 50S subunit peptidyl transferase center (PTC) (13, 14). In so doing, RRF sterically occludes deacylated tRNA binding in the peptidyl-tRNA site (P site, P/P configuration) to favor tRNA positioning in the hybrid peptidyl/exit tRNA binding site (P/E configuration) (Fig. 1A) (15, 16). In the P/E configuration, tRNA is bound simultaneously to the P site of the small (30S) subunit and to the E site of the large (50S) subunit (5). Binding of the GTPase elongation factor-G (EF-G) to the RRF-ribosome complex and subsequent GTP hydrolysis lead to the dissociation of ribosomal subunits (17).
Fig. 1.
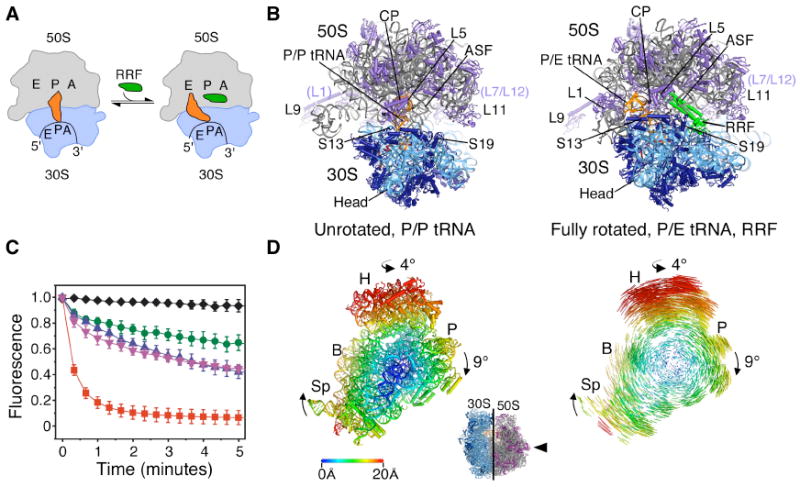
Ribosome recycling in bacteria and organelles. (A) Steps of ribosome recycling. After termination, ribosomes with deacylated tRNA in the P site undergo a structural rearrangement to a fully rotated state in which tRNA adopts a P/E hybrid state of binding and RRF is bound in the 50S P site. EF-G then catalyzes subunit dissociation (not shown). (B) Global views of the ribosome in a post-termination state (L) and intermediate state of recycling (R). The small subunit rRNA and proteins are colored light and dark blue, respectively, with the large subunit rRNA and proteins colored grey and magenta, respectively. Bound tRNA (orange), mRNA (dark red), and RRF (green) are also shown. (C) The dependence of subunit release on RRF, EF-G and GTP under crystallographic buffer conditions. Release was monitored by the loss of Cy5-labeled L1 fluorescence in 50S subunits from surface-immobilized ribosome complexes carrying Cy3-labeled tRNAPhe in the P site. Complexes imaged in the absence of factors (black diamonds) or in the presence of 10μM RRF (green circles), 20 μM EF-G/2mM GTP (pink inverted triangles), 10 μM RRF/20 μM EF-G/2mM GDPNP (blue triangles) or 10 μM RRF/20 μM EF-G/2mM GTP (red squares). Data reflect the mean +/- SD of normalized Cy5 fluorescence intensity as a function of time from three experimental replicates. (D) Conformational changes in the 70S ribosome during ratcheting. View of the 30S subunit from the perspective of the 50S subunit (inset). Shifts between equivalent RNA phosphorus atoms and protein Cα atoms in the unrotated (R0) and fully rotated (RF) states are color coded as indicated by the scale. Ribosomes were superimposed using the 50S subunit as the frame of reference (37). Difference vectors between equivalent phosphorus or Cα atoms of the 30S subunits in the unrotated and fully rotated ribosome structures are shown on the right.
Here, we determined structures of the intact Escherichia coli 70S ribosome at a resolution of ~3.2 Å (Table S1, S2) (12), based on crystals that contain two independent copies of the ribosome per asymmetric unit in a “top-top” polysome configuration (18). One ribosome adopts an unrotated state, with tRNAPhe bound in the peptidyl-tRNA (P/P) binding site (Fig. 1B) (19) that mimics a post-termination state of the translation cycle. The second ribosome adopts a fully rotated conformation that contains tRNAPhe bound in the hybrid P/E binding site and RRF bound at the ribosomal subunit interface (Fig. 1B). This structure is thought to represent an early intermediate in bacterial ribosome recycling (Fig. 1A) (15). Under these buffer conditions, single-molecule Fluorescence Resonance Energy Transfer (smFRET) measurements showed RRF achieved approximately 50% maximal stabilization of the fully rotated, P/E hybrid configuration of the post-termination complex (Fig. S1) and supported EF-G- and GTP hydrolysis-dependent ribosome recycling (Fig. 1C, Fig. S1) (12).
When compared to the post-termination ribosome complex, the 30S subunit of the RRF-bound ribosome is rotated ~9° relative to the 50S subunit. An approximately orthogonal rotation of the head domain of the 30S subunit of ~4° swivels the head domain in the direction of the ribosomal E site on the 50S subunit. These motions of the 30S subunit into the rotated state result in shifts at the periphery of the ribosome of more than 20 Å (Fig. 1D) that direct deacylated P-site tRNA into the P/E hybrid site. The tRNA anticodon stem-loop (ASL) and mRNA move laterally by ~6 Å relative to the 50S subunit, coupled to the motion of the 30S subunit platform domain (Fig. 1D, Fig. 2A). When tRNA moves into the P/E site from the P/P site, ASL of the tRNA remains in contact with the 30S subunit head and platform domains (Fig. 2B, Fig. S2A) (19), but breaks its interactions with 23S ribosomal RNA (rRNA) helix H69 in the large subunit (19) (Fig. 2B, Movie S1).
Fig. 2.
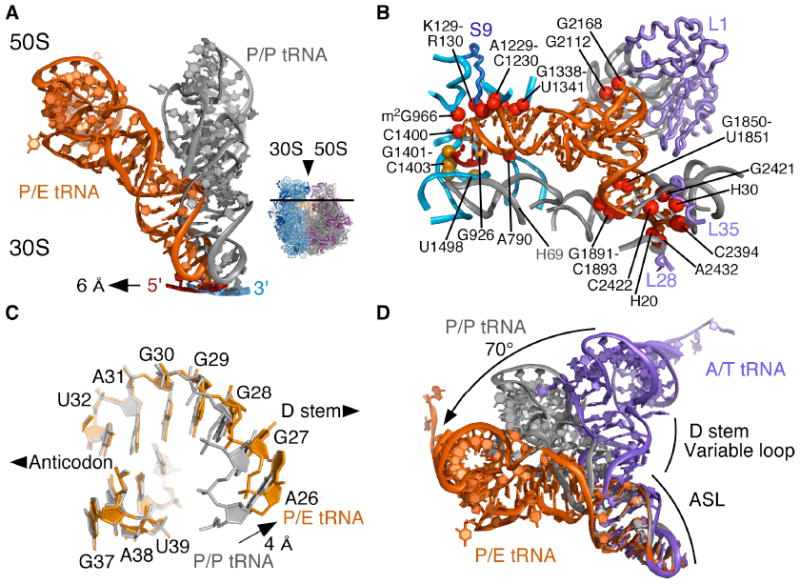
Conformation of tRNA in the P/E hybrid state. (A) Movement of P/E tRNA and mRNA towards the E site when compared to P/P tRNA and mRNA. The direction of view is shown to the right. (B) View of mRNA and P/E tRNA interactions with the 30S subunit P site and 50S subunit E site. Residues that contact mRNA (gold) and P/E tRNA (red) are shown. Colors for the ribosome, mRNA and tRNA as in Fig. 1. (C) View of the P/E tRNA ASL/D stem junction (orange). P/P tRNA (grey) is shown for comparison, with an arrow indicating the widening of the helix major groove. (D) Comparison of ASL/D stem junctions between P/E tRNA (orange), P/P tRNA (grey), and A/T tRNA (purple). A/T tRNA structure is a homology model adapted from (12, 21). The bending angle for the A/T to P/E conformational change (70°) is shown.
Bound in the hybrid P/E site, tRNAPhe is severely kinked at the junction between the ASL and D stem when compared to tRNAPhe bound in the P/P site. Although the conformation of the anticodon and two closing base pairs of the ASL region remain essentially unchanged, the major groove widens by ~4 Å at the junction of the ASL and D stem (Fig. 2C, Fig. S2). The kink between the ASL and D stems allows the acceptor stem of P/E tRNA to swing by ~37° into the 50S E site (Fig. 2D) (12). This abrupt kink contrasts with the more distributed bend that occurs in mRNA decoding complexes bound to elongation factor EF-Tu (A/T state, (20, 21)), in which tRNA bends in the opposite direction. Comparing P/E tRNA to A/T tRNA, the total extent of tRNA bending at the ASL/D-stem junction amounts to ~70° (Fig. 2D, Movie S1).
In the large subunit E site, P/E tRNA contacts the ribosome in a similar manner to tRNA bound in the E/E site (19)(Fig. 2B). Nucleotides G2112 and G2168 in 23S rRNA, part of the protein L1-containing arm of the 50S subunit, stack on the D-loop and T-loop of P/E tRNA (Fig. 1C, Fig. 2B). Consistent with biochemical studies of the mechanism of translocation (22), nucleotide A76 at the acceptor end of P/E tRNA stacks between nucleotides in helix H88 of 23S rRNA (Fig. 2B, Fig. S3), where the terminal ribose engages the Watson-Crick face of nucleotide C2394 (19, 23). In contrast to the positioning of C75 in E-site tRNA in the bacterium T. thermophilus (19) and in the archaeal large subunit (23), in E. coli, nucleotide C75 in P/E tRNA stacks on nucleotide A2432 in 23S rRNA, away from the tRNA acceptor stem (Fig. S3). The striking divergence of the 50S E site contacts contrasts with the high level of conservation in the peptidyl transferase center, supporting the notion that the ribosomal E site evolved relatively late, and has continued to diverge (19, 24).
The molecular contacts between the two ribosomal subunits are composed of both rRNA and ribosomal proteins, with the central contacts, or bridges, conserved across kingdoms (11, 25). In the fully-rotated state, the pivot point for inter-subunit ratcheting occurs at bridge B3 (Fig. 3A, Fig. S4), which maintains the same conformation and contacts when compared to the unrotated ribosome (25). Bridge B3 is composed of a cross-strand adenosine stacking motif (26) in which residues A1418 and A1483 within helix 44 (h44) of 16S rRNA in the 30S subunit dock into the minor groove of helix 71 (H71) in 23S rRNA of the 50S subunit. Residues A1418 and A1483 lie within adjacent sheared G-A base pairs that coordinate an inner-sphere magnesium ion that possibly contributes to subunit association in all organisms (25, 27) (Fig. 3B).
Fig. 3.
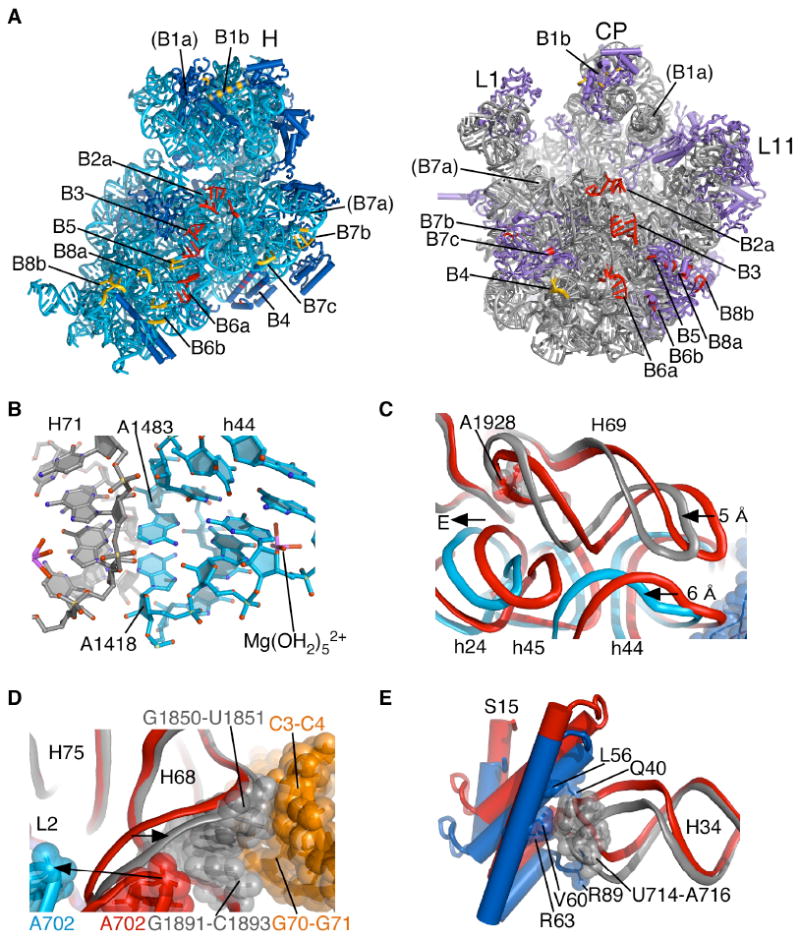
Inter-subunit contacts in the fully rotated state. (A) Global view of inter-subunit contacts of the fully rotated state. Elements in each ribosomal subunit that contact rRNA in opposite subunit are color-coded red, while elements in each subunit that contact ribosomal proteins in the opposite subunit are color-coded gold. Ribosomal RNAs and proteins are colored as in Figure 1. Bridge numbering is adapted from (11, 25). The tip of helix H38 in bridge B1a is disordered in the present structures. (B) Bridge B3 serves as the pivot of inter-subunit rotation. The Mg2+ ion involved in inner-sphere coordination to the tandem sheared GA pairs in 16S rRNA and a fully hydrated Mg2+ ion in 23S rRNA are also shown. Ribosomal RNA colored as in Fig. 1. (C) Compression of helix H69 in 23S rRNA due to inter-subunit rotation. The direction of view is similar to Fig. 1. Color coding of the fully rotated ribosome (R) as in Fig. 1, with unrotated ribosome (U) in red. Nucleotide A1928 in 23S rRNA, nearly invariant in position, is shown for reference. (D) Movement of H68 due to disruption of A702 interactions and packing with P/E tRNA. Nucleotides involved in H68 packing with P/E tRNA are indicated. Elements of the fully rotated ribosome are colored as in Fig. 1. Elements of the unrotated ribosome are shown in red. Arrows indicate movement from the unrotated to fully rotated state. (E) Bridge B4 in the fully rotated state compared to that in state R0 (red). Residues involved in direct contact in the fully rotated state are shown. Coloring for the fully rotated state as in Fig. 1.
In the aminoacyl-tRNA (A) and P sites, bridge B2a involves contacts between 23S rRNA helix H69 in the 50S subunit and 16S rRNA residues at the end of helix h44 in the 30S subunit, and is preserved in both the unrotated and fully-rotated states of the ribosome (Fig. 3A). In both states, residue A1913 of H69 penetrates the minor groove of the h44 mRNA decoding site. However, in going from the unrotated to fully-rotated state, the P-site tRNA anticodon and mRNA (Fig. 2A) and the end of helix h44 move laterally by ~6 Å towards the E site (Fig. 3C). Remarkably, the interactions between H69 and h44 are maintained during this movement due to a ~5 Å compression of H69 (Fig. 3C, Movie S2). In part, this compression is enabled by disruption of the terminal base pair (C1925-G1929) of H69 and extrusion of the nearly universally conserved uridine U1926 (28) from the tight U-turn motif at the base of H69 (25) (Fig. S5, Movie S2).
The observed conformational rearrangements in bridge B2a may help explain how antibiotics such as viomycin that target translocation stabilize the fully rotated state of the ribosome (29, 30). Viomycin and the related antibiotic capreomycin bind to the unrotated state of the ribosome in the vicinity of nt A1913 in 23S rRNA (31), the only nucleotide whose contacts with h44 change appreciably during inter-subunit rotation. Aminoglycosides such as neomycin, which bind to two sites in bridge B2a (32, 33), may favor the fully rotated state of the ribosome by stabilizing the compressed conformation of helix H69.
On the opposite end of the tRNA binding cleft, bridge B7a is disrupted due to the rotation of the 30S platform domain (Fig. 1D). In the unrotated state, nucleotide A702 in 16S rRNA stacks on an A-A dinucleotide platform near the end of helix H68 of 23S rRNA (12, 34). This interaction involves a hydrogen bond between N1 of A702 and G1846 in 23S rRNA (25). Consistent with chemical probing data used to identify hybrid tRNA binding sites (5), rotation of the 30S platform domain into the fully rotated position results in a ~13 Å displacement of A702 away from H68 that exposes the base pairing face of A702 to solvent (Fig. 3D). Consistent with biochemical observations (35), H68 moves in the opposite direction by 2-3 Å to pack in the minor groove of the acceptor stem of P/E tRNA (Fig. 3D) likely helping to stabilize tRNA in the P/E hybrid site.
The absence of bridge B7a in the fully rotated state appears to be partially compensated for by new contacts between protein L2 in the large subunit and helices h23 and h24 in 16S rRNA (Bridges B7b, B7c; Fig. 3A). However, the most significant stabilizing contact to the 30S platform region in both the unrotated and fully rotated ribosome configurations remains bridge B4, which in bacteria involves intimate contacts between the hairpin loop at the end of helix H34 in 23S rRNA of the large subunit and protein S15 in the small subunit. Helix H34 bends by ~7 Å, or 12°, due to inter-subunit rotation and slightly adjusts how nucleotide A715 packs on the hydrophobic surface of protein S15 (25)(Fig. 3E). Compensation for the loss of bridge B7a in the fully rotated state may also result from the formation of more extensive interactions between the 30S subunit body domain and the 50S subunit near bridge B8. In bridge B8, large subunit proteins L14 and L19 interact more strongly with helices h8 and h14 in the 30S subunit (Fig. 3A).
In the fully rotated state, the head domain of the 30S subunit swivels as a rigid body in the direction of tRNA movement, rearranging bridge B1b to place the central alpha helix of protein S13 directly across from protein L5 in the 50S subunit (Fig. 1C) (36). This lateral change in protein S13 position correlates with tRNA binding in the hybrid P/E site and may help control the position of tRNAs on the ribosome (37). Thus, the contacts between protein S13 and protein L5 probably play an important role in the ribosome ratcheting mechanism. Consistent with this view, deletions in protein S13 result in more rapid and lower fidelity translocation of mRNA and tRNA (38). Mutations in the other major contact between the 30S subunit head domain and helix H38 in the 50S subunit, bridge B1a, have a similar effect (39).
In the fully rotated ribosome, RRF binds in the P-site and A-site cleft of the 50S subunit, precluding tRNA binding in either site. Its 3-helix bundle domain (domain I) runs nearly parallel to the subunit interface, with alpha helix 3 packed tightly against helix H71 in 23S rRNA (Fig. 4A). Mutations in this region result in lethal or temperature-sensitive phenotypes (40). In addition, conserved amino acids within the tip of RRF domain I (41) interact with rRNA nucleotides of the universally conserved P loop element of the peptidyl transferase center (Fig. 4A). These sets of interactions appear to be the same in both the unrotated and fully rotated states of the ribosome (32, 42), suggesting that they are necessary but not sufficient for the recycling mechanism.
Fig. 4.
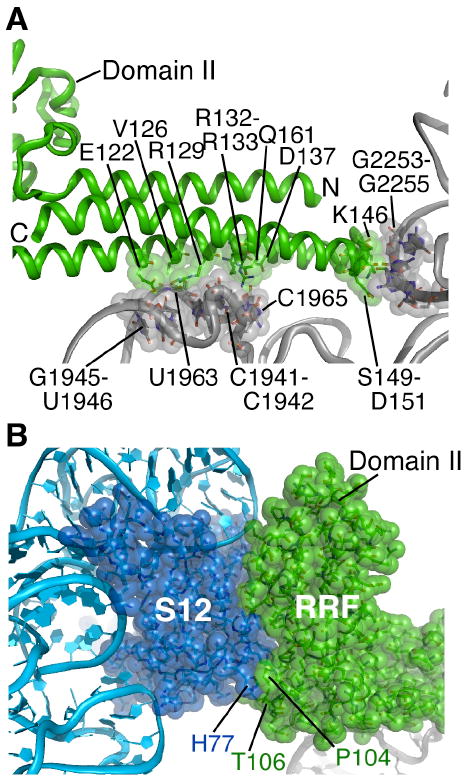
RRF interactions with the ribosome in the fully rotated state. (A) Contacts between RRF domain I and the P and A sites of the 50S subunit. Amino acids in RRF (green) and nucleotides in 23S rRNA (grey) in direct contact are shown. Helix H69 and the 30S subunit are behind the view shown. (B) Contacts between RRF and protein S12 in the 30S subunit. Amino acids at the junction of RRF domains I and II that interact closely with S12 are indicated. RRF, S12 and rRNAs colored as in Fig. 1.
Additional points of contact between RRF and the fully rotated ribosome occur between conserved amino acids near the junction of domains I and II in RRF and ribosomal protein S12 of the small subunit (Fig. 4B)(41). Domain II of RRF is more constrained in its position in the ratcheted state compared to its location in the unratcheted ribosome (32, 42). As suggested by cryo-EM reconstructions of the ribosome in complexes with RRF (14, 15), RRF domain II likely serves a steric function in ribosome recycling. Docking of EF-G from a cryo-EM reconstruction of the ribosome in a rotated conformation related to translocation (10) onto the ratcheted 70S ribosome structure determined here shows significant overlap between domain II of RRF and domains IV and V of EF-G (Fig. S6). Thus, EF-G binding to the RRF-bound ribosome likely entails large-scale rearrangements in RRF, EF-G, and the ribosome (15).
When compared to other structures of the ribosome, the structure of the fully rotated state of the ribosome provides critical insights into the molecular description of the ratcheting mechanism in translation. As simple mRNAs can be translated in the absence of exogenous factors like EF-G (43), the ribosome itself serves as a Brownian ratchet (2, 44), with tRNA substrates likely serving as the “teeth”. A notable feature of the ratcheting mechanism is the use of RNA secondary structural elements to control large-scale conformational rearrangements in the ribosome. These include RNA stem-loops in bridges B2a and B4 that adjust as the 30S subunit rotates relative to the 50S subunit (8, 11, 15, 45, 46), helix H68 in 23S rRNA adjacent to bridge B7a and P/E tRNA, RNA helices H76 and H42 in the L1 and L11 arms of the large subunit, respectively (1, 2, 11, 36, 47), and helix h28 in 16S rRNA which directs swiveling of the 30S subunit head domain (25). Helix h28 likely serves as the “spring” in the ratcheting process, helping to position the “pawl” between the small subunit P and E sites (3, 10, 25, 44). The hinge-like motion in P/E tRNA observed here, when compared to P/P tRNA, suggests that the conserved tertiary structure of tRNA is required not only for mRNA decoding (20, 21, 48), but also for translocation, termination, and ribosome recycling (10, 49). Intact P-site tRNA is required for translocation (50), a requirement that may in part be due to need for a large distortion of tRNA in the P/E binding site. This distortion may be used to tune the energetics of the transition between the pre-translocation state and post-translocation state of the ribosome. Future structural studies of ribosome complexes with EF-G will be required to explain how this factor controls the conformational events described here to accelerate translocation and ribosome recycling.
Supplementary Material
Acknowledgments
We thank K. Hamadani for purified RRF, K. Nierhaus for tRNAPhe overexpression plasmids, K. Frankel, S. Classen, and G. Meigs for help with data measurement at the SIBYLS and 8.3.1 beamlines at the Advanced Light Source (ALS), R. Kanagalaghatta, D. Neau, F. Murphy, and I. Kourinov for help with data measurement at ID-24 at the Advanced Photon Source (APS), J. Headd for help with Phenix refinement, and J. Holton for useful crystallographic discussions. We also thank J. Doudna and H. Noller for helpful comments on the manuscript. Atomic coordinates and structure factors are deposited in the Protein Data Bank (accession codes 3R8N and 3R8S for the fully rotated state RF, 3R8O and 3R8T for the unrotated state R0). This work was funded by the National Institutes of Health (GM65050 to J.H.D.C., GM079238 to S.C.B, and GM074127-04S1, GM088674 and P01-GM63210 project IV to J.S.R.), NCI grant CA92584 for the SIBYLS and 8.3.1 beam lines at the ALS, and NRCC grant RR-15301 for the Northeastern Collaborative Access Team beam lines at 24-ID at the APS, and by the Department of Energy (DE-AC03 76SF00098 for the SIBYLS and 8.3.1 beamlines at the ALS, and DE-AC02-06CH11357 for the APS). M.B.F. is a trainee in the Weill Cornell/Rockefeller University/Sloan-Kettering Tri-Institutional MD-PhD Program supported by US National Institutes of Health Medical Scientist Training Program grant GM07739. J.N. is supported by a Human Frontiers in Science Program Postdoctoral Fellowship.
References and Notes
- 1.Schmeing TM, Ramakrishnan V. Nature. 2009;461:1234. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- 2.Munro JB, Sanbonmatsu KY, Spahn CM, Blanchard SC. Trends Biochem Sci. 2009;34:390. doi: 10.1016/j.tibs.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunkle JA, Cate JH. Annu Rev Biophys. 2010;39:227. doi: 10.1146/annurev.biophys.37.032807.125954. [DOI] [PubMed] [Google Scholar]
- 4.Frank J, Agrawal RK. Nature. 2000;406:318. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 5.Moazed D, Noller HF. Nature. 1989;342:142. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 6.Semenkov YP, Rodnina MV, Wintermeyer W. Nat Struct Biol. 2000;7:1027. doi: 10.1038/80938. [DOI] [PubMed] [Google Scholar]
- 7.Zavialov AV, Ehrenberg M. Cell. 2003;114:113. doi: 10.1016/s0092-8674(03)00478-1. [DOI] [PubMed] [Google Scholar]
- 8.Zhang W, Dunkle JA, Cate JH. Science. 2009;325:1014. doi: 10.1126/science.1175275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, Stark H. Nature. 2010;466:329. doi: 10.1038/nature09206. [DOI] [PubMed] [Google Scholar]
- 10.Ratje AH, et al. Nature. 2010;468:713. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Science. 2010;330:1203. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- 12.Materials and methods are available as supporting material on Science online.
- 13.Lancaster L, Kiel MC, Kaji A, Noller HF. Cell. 2002;111:129. doi: 10.1016/s0092-8674(02)00938-8. [DOI] [PubMed] [Google Scholar]
- 14.Agrawal RK, et al. Proc Natl Acad Sci U S A. 2004;101:8900. doi: 10.1073/pnas.0401904101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao N, et al. Mol Cell. 2005;18:663. doi: 10.1016/j.molcel.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 16.Sternberg SH, Fei J, Prywes N, McGrath KA, Gonzalez RL., Jr Nat Struct Mol Biol. 2009;16:861. doi: 10.1038/nsmb.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Savelsbergh A, Rodnina MV, Wintermeyer W. RNA. 2009;15:772. doi: 10.1261/rna.1592509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brandt F, et al. Cell. 2009;136:261. doi: 10.1016/j.cell.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 19.Selmer M, et al. Science. 2006;313:1935. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 20.Schmeing TM, et al. Science. 2009;326:688. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. Science. 2010;330:835. doi: 10.1126/science.1194460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lill R, Robertson JM, Wintermeyer W. EMBO J. 1989;8:3933. doi: 10.1002/j.1460-2075.1989.tb08574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmeing TM, Moore PB, Steitz TA. RNA. 2003;9:1345. doi: 10.1261/rna.5120503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bokov K, Steinberg SV. Nature. 2009;457:977. doi: 10.1038/nature07749. [DOI] [PubMed] [Google Scholar]
- 25.Schuwirth BS, et al. Science. 2005;310:827. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 26.Cate JH, et al. Science. 1996;273:1678. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 27.Shenvi CL, Dong KC, Friedman EM, Hanson JA, Cate JH. RNA. 2005;11:1898. doi: 10.1261/rna.2192805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cannone JJ, et al. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ermolenko DN, et al. Nat Struct Mol Biol. 2007;14:493. doi: 10.1038/nsmb1243. [DOI] [PubMed] [Google Scholar]
- 30.Cornish PV, Ermolenko DN, Noller HF, Ha T. Mol Cell. 2008;30:578. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stanley RE, Blaha G, Grodzicki RL, Strickler MD, Steitz TA. Nat Struct Mol Biol. 2010;17:289. doi: 10.1038/nsmb.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borovinskaya MA, et al. Nat Struct Mol Biol. 2007;14:727. doi: 10.1038/nsmb1271. [DOI] [PubMed] [Google Scholar]
- 33.Feldman MB, Terry DS, Altman RB, Blanchard SC. Nat Chem Biol. 2010;6:54. doi: 10.1038/nchembio.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cate JH, et al. Science. 1996;273:1696. doi: 10.1126/science.273.5282.1696. [DOI] [PubMed] [Google Scholar]
- 35.Feinberg JS, Joseph S. Proc Natl Acad Sci U S A. 2001;98:11120. doi: 10.1073/pnas.211184098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valle M, et al. Cell. 2003;114:123. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 37.Frank J, Gao H, Sengupta J, Gao N, Taylor DJ. Proc Natl Acad Sci U S A. 2007;104:19671. doi: 10.1073/pnas.0708517104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cukras AR, Green R. J Mol Biol. 2005;349:47. doi: 10.1016/j.jmb.2005.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Komoda T, et al. J Biol Chem. 2006;281:32303. doi: 10.1074/jbc.M607058200. [DOI] [PubMed] [Google Scholar]
- 40.Janosi L, et al. J Mol Biol. 2000;295:815. doi: 10.1006/jmbi.1999.3401. [DOI] [PubMed] [Google Scholar]
- 41.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. Nucleic Acids Res. 2010;38:W529. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weixlbaumer A, et al. Nat Struct Mol Biol. 2007;14:733. doi: 10.1038/nsmb1282. [DOI] [PubMed] [Google Scholar]
- 43.Gavrilova LP, Kostiashkina OE, Koteliansky VE, Rutkevitch NM, Spirin AS. J Mol Biol. 1976;101:537. doi: 10.1016/0022-2836(76)90243-6. [DOI] [PubMed] [Google Scholar]
- 44.Spirin AS. J Biol Chem. 2009;284:21103. doi: 10.1074/jbc.X109.001552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Connell SR, et al. Mol Cell. 2007;25:751. doi: 10.1016/j.molcel.2007.01.027. [DOI] [PubMed] [Google Scholar]
- 46.Spahn CM, et al. EMBO J. 2004;23:1008. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Valle M, et al. Nat Struct Biol. 2003;10:899. doi: 10.1038/nsb1003. [DOI] [PubMed] [Google Scholar]
- 48.Valle M, et al. EMBO J. 2002;21:3557. doi: 10.1093/emboj/cdf326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li W, Frank J. Proc Natl Acad Sci U S A. 2007;104:16540. doi: 10.1073/pnas.0708094104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Joseph S, Noller HF. EMBO J. 1998;17:3478. doi: 10.1093/emboj/17.12.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


