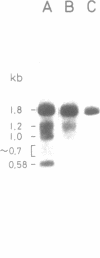
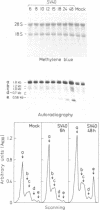
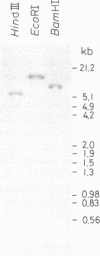
Abstract
We have isolated and sequenced a mouse replacement variant histone H3.3 cDNA. It corresponds to the most abundant mRNA expressed from a unique gene by the use of one out of three polyadenylation sites. The 3' non coding region of H3.3 is very long (approximately 1100 nt) and highly conserved throughout evolution since it is about 95% homologous to the 3' non coding region of the chicken H3.3B gene. We studied the expression of the H3.3 gene during SV40- and polyoma-induced mitotic host reaction in confluent, Go-arrested primary mouse kidney cell cultures. H3.3 replacement variant mRNA steady state levels increased during the Go to S-phase transition, apparently as the result of two mechanisms: one related to cell growth, whereas the other was linked to cellular DNA synthesis. The latter mechanism was however far less pronounced than with replication histone variant mRNAs. The biological implications of these results are discussed.
Full text
PDF
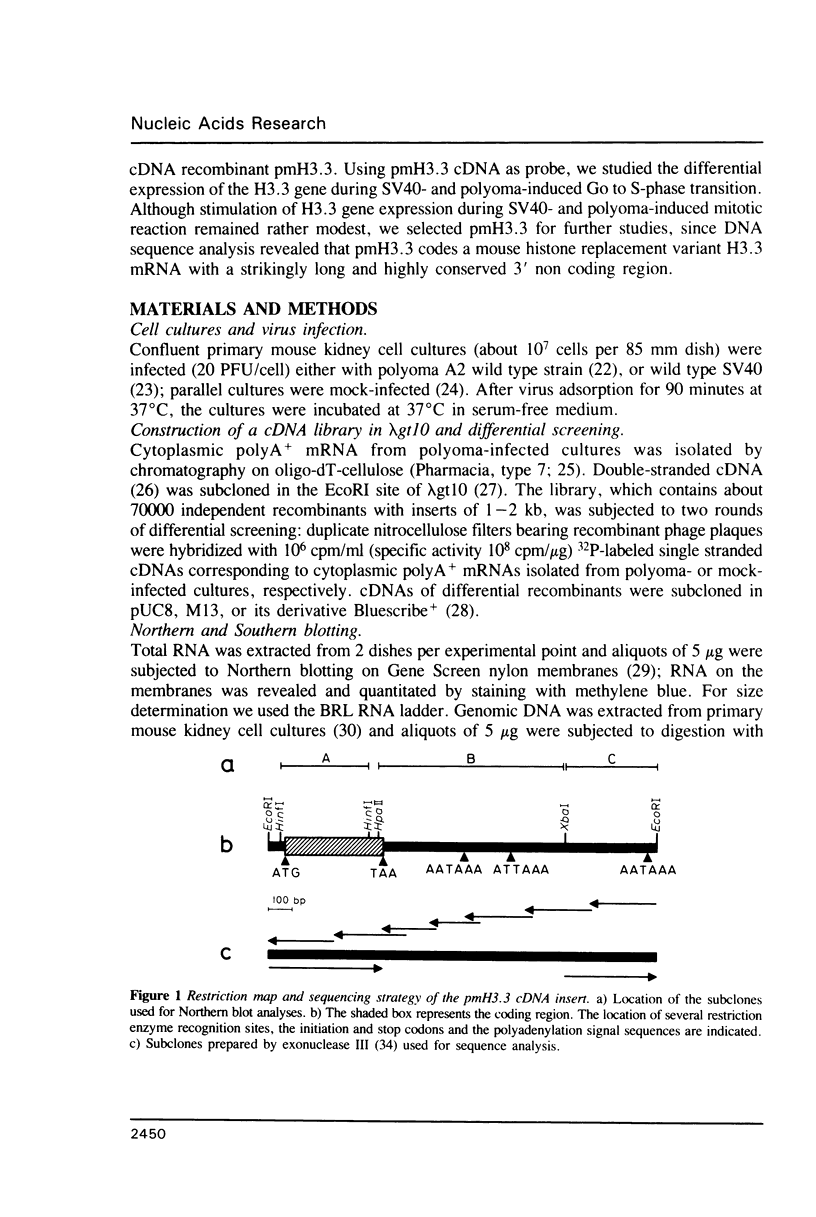










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allis C. D., Glover C. V., Bowen J. K., Gorovsky M. A. Histone variants specific to the transcriptionally active, amitotically dividing macronucleus of the unicellular eucaryote, Tetrahymena thermophila. Cell. 1980 Jul;20(3):609–617. doi: 10.1016/0092-8674(80)90307-4. [DOI] [PubMed] [Google Scholar]
- Allis C. D., Richman R., Gorovsky M. A., Ziegler Y. S., Touchstone B., Bradley W. A., Cook R. G. hv1 is an evolutionarily conserved H2A variant that is preferentially associated with active genes. J Biol Chem. 1986 Feb 5;261(4):1941–1948. [PubMed] [Google Scholar]
- Almendral J. M., Sommer D., Macdonald-Bravo H., Burckhardt J., Perera J., Bravo R. Complexity of the early genetic response to growth factors in mouse fibroblasts. Mol Cell Biol. 1988 May;8(5):2140–2148. doi: 10.1128/mcb.8.5.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso A., Breuer B., Bouterfa H., Doenecke D. Early increase in histone H1(0) mRNA during differentiation of F9 cells to parietal endoderm. EMBO J. 1988 Oct;7(10):3003–3008. doi: 10.1002/j.1460-2075.1988.tb03163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Bonner W. M., West M. H., Stedman J. D. Two-dimensional gel analysis of histones in acid extracts of nuclei, cells, and tissues. Eur J Biochem. 1980 Aug;109(1):17–23. doi: 10.1111/j.1432-1033.1980.tb04762.x. [DOI] [PubMed] [Google Scholar]
- Brickell P. M., Latchman D. S., Murphy D., Willison K., Rigby P. W. The class I major histocompatibility antigen gene activated in a line of SV40-transformed mouse cells is H-2Dd, not Qa/Tla. Nature. 1985 Jul 11;316(6024):162–163. doi: 10.1038/316162a0. [DOI] [PubMed] [Google Scholar]
- Brush D., Dodgson J. B., Choi O. R., Stevens P. W., Engel J. D. Replacement variant histone genes contain intervening sequences. Mol Cell Biol. 1985 Jun;5(6):1307–1317. doi: 10.1128/mcb.5.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran B. H., Reffel A. C., Stiles C. D. Molecular cloning of gene sequences regulated by platelet-derived growth factor. Cell. 1983 Jul;33(3):939–947. doi: 10.1016/0092-8674(83)90037-5. [DOI] [PubMed] [Google Scholar]
- Dodgson J. B., Yamamoto M., Engel J. D. Chicken histone H3.3B cDNA sequence confirms unusual 3' UTR structure. Nucleic Acids Res. 1987 Aug 11;15(15):6294–6294. doi: 10.1093/nar/15.15.6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel J. D., Sugarman B. J., Dodgson J. B. A chicken histone H3 gene contains intervening sequences. Nature. 1982 Jun 3;297(5865):434–436. doi: 10.1038/297434a0. [DOI] [PubMed] [Google Scholar]
- Gauchat J. F., Weil R. On the functional roles of simian virus 40 large and small T-antigen in the induction of a mitotic host response. Nucleic Acids Res. 1986 Dec 9;14(23):9339–9351. doi: 10.1093/nar/14.23.9339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goad W. B., Kanehisa M. I. Pattern recognition in nucleic acid sequences. I. A general method for finding local homologies and symmetries. Nucleic Acids Res. 1982 Jan 11;10(1):247–263. doi: 10.1093/nar/10.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler U., Hoffman B. J. A simple and very efficient method for generating cDNA libraries. Gene. 1983 Nov;25(2-3):263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- Hatch C. L., Bonner W. M. Sequence of cDNAs for mammalian H2A.Z, an evolutionarily diverged but highly conserved basal histone H2A isoprotein species. Nucleic Acids Res. 1988 Feb 11;16(3):1113–1124. doi: 10.1093/nar/16.3.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Humphrey T., Proudfoot N. J. A beginning to the biochemistry of polyadenylation. Trends Genet. 1988 Sep;4(9):243–245. doi: 10.1016/0168-9525(88)90028-5. [DOI] [PubMed] [Google Scholar]
- Khandjian E. W., Gauchat J. F. One single burst of SV40 gene expression induces cell proliferation in transforming infection of mouse cells. Oncogene. 1988 Aug;3(2):195–200. [PubMed] [Google Scholar]
- Khandjian E. W., Matter J. M., Léonard N., Weil R. Simian virus 40 and polyoma virus stimulate overall cellular RNA and protein synthesis. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1476–1480. doi: 10.1073/pnas.77.3.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandjian E. W. UV crosslinking of RNA to nylon membrane enhances hybridization signals. Mol Biol Rep. 1986;11(2):107–115. doi: 10.1007/BF00364822. [DOI] [PubMed] [Google Scholar]
- Lau L. F., Nathans D. Expression of a set of growth-related immediate early genes in BALB/c 3T3 cells: coordinate regulation with c-fos or c-myc. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1182–1186. doi: 10.1073/pnas.84.5.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau L. F., Nathans D. Identification of a set of genes expressed during the G0/G1 transition of cultured mouse cells. EMBO J. 1985 Dec 1;4(12):3145–3151. doi: 10.1002/j.1460-2075.1985.tb04057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linzer D. I., Nathans D. Growth-related changes in specific mRNAs of cultured mouse cells. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4271–4275. doi: 10.1073/pnas.80.14.4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff W. F., Pandey N. B. Multiple regulatory steps control histone mRNA concentrations. Trends Biochem Sci. 1988 Feb;13(2):49–52. doi: 10.1016/0968-0004(88)90027-8. [DOI] [PubMed] [Google Scholar]
- Matrisian L. M., Glaichenhaus N., Gesnel M. C., Breathnach R. Epidermal growth factor and oncogenes induce transcription of the same cellular mRNA in rat fibroblasts. EMBO J. 1985 Jun;4(6):1435–1440. doi: 10.1002/j.1460-2075.1985.tb03799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matter J. M., Tiercy J. M., Weil R. Sequential stimulation of cellular RNA synthesis in polyoma-infected mouse kidney cell cultures. Nucleic Acids Res. 1983 Oct 11;11(19):6611–6629. doi: 10.1093/nar/11.19.6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May E., May P., Weil R. "Early" virus-specific RNA may contain information necessary for chromosome replication and mitosis induced by Simian Virus 40. Proc Natl Acad Sci U S A. 1973 Jun;70(6):1654–1658. doi: 10.1073/pnas.70.6.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melli M., Spinelli G., Arnold E. Synthesis of histone messenger RNA of HeLa cells during the cell cycle. Cell. 1977 Sep;12(1):167–174. doi: 10.1016/0092-8674(77)90194-5. [DOI] [PubMed] [Google Scholar]
- Milbrandt J. A nerve growth factor-induced gene encodes a possible transcriptional regulatory factor. Science. 1987 Nov 6;238(4828):797–799. doi: 10.1126/science.3672127. [DOI] [PubMed] [Google Scholar]
- Murphy D., Brickell P. M., Latchman D. S., Willison K., Rigby P. W. Transcripts regulated during normal embryonic development and oncogenic transformation share a repetitive element. Cell. 1983 Dec;35(3 Pt 2):865–871. doi: 10.1016/0092-8674(83)90119-8. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutzbank T., Robinson R., Oren M., Levine A. J. SV40 large tumor antigen can regulate some cellular transcripts in a positive fashion. Cell. 1982 Sep;30(2):481–490. doi: 10.1016/0092-8674(82)90245-8. [DOI] [PubMed] [Google Scholar]
- Schümperli D. Cell-cycle regulation of histone gene expression. Cell. 1986 May 23;45(4):471–472. doi: 10.1016/0092-8674(86)90277-1. [DOI] [PubMed] [Google Scholar]
- Schümperli D. Multilevel regulation of replication-dependent histone genes. Trends Genet. 1988 Jul;4(7):187–191. doi: 10.1016/0168-9525(88)90074-1. [DOI] [PubMed] [Google Scholar]
- Scott M. R., Westphal K. H., Rigby P. W. Activation of mouse genes in transformed cells. Cell. 1983 Sep;34(2):557–567. doi: 10.1016/0092-8674(83)90388-4. [DOI] [PubMed] [Google Scholar]
- Singh K., Carey M., Saragosti S., Botchan M. Expression of enhanced levels of small RNA polymerase III transcripts encoded by the B2 repeats in simian virus 40-transformed mouse cells. Nature. 1985 Apr 11;314(6011):553–556. doi: 10.1038/314553a0. [DOI] [PubMed] [Google Scholar]
- Singh K., Saragosti S., Botchan M. Isolation of cellular genes differentially expressed in mouse NIH 3T3 cells and a simian virus 40-transformed derivative: growth-specific expression of VL30 genes. Mol Cell Biol. 1985 Oct;5(10):2590–2598. doi: 10.1128/mcb.5.10.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittman D. B., Chiu I. M., Pan C. J., Cohn R. H., Kedes L. H., Marzluff W. F. Isolation of two clusters of mouse histone genes. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4078–4082. doi: 10.1073/pnas.78.7.4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittman D. B., Graves R. A., Marzluff W. F. Structure of a cluster of mouse histone genes. Nucleic Acids Res. 1983 Oct 11;11(19):6679–6697. doi: 10.1093/nar/11.19.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein G. S., Stein J. L., Park W. D., Detke S., Lichtler A. C., Shephard E. A., Jansing R. L., Phillips I. R. Regulation of histone gene expression in HeLa S3 cells. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):1107–1120. doi: 10.1101/sqb.1978.042.01.112. [DOI] [PubMed] [Google Scholar]
- Türler H., Salomon C. Small and middle T antigens contribute to lytic and abortive polyomavirus infection. J Virol. 1985 Feb;53(2):579–586. doi: 10.1128/jvi.53.2.579-586.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. W., Robins A. J., d'Andrea R., Wells J. R. Inverted duplication of histone genes in chicken and disposition of regulatory sequences. Nucleic Acids Res. 1985 Feb 25;13(4):1369–1387. doi: 10.1093/nar/13.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil R. Viral 'tumor antigens': A novel type of mammalian regulator protein. Biochim Biophys Acta. 1978 Nov 17;516(3):301–388. doi: 10.1016/0304-419x(78)90012-4. [DOI] [PubMed] [Google Scholar]
- Wells D., Bains W., Kedes L. Codon usage in histone gene families of higher eukaryotes reflects functional rather than phylogenetic relationships. J Mol Evol. 1986;23(3):224–241. doi: 10.1007/BF02115579. [DOI] [PubMed] [Google Scholar]
- Wells D., Hoffman D., Kedes L. Unusual structure, evolutionary conservation of non-coding sequences and numerous pseudogenes characterize the human H3.3 histone multigene family. Nucleic Acids Res. 1987 Apr 10;15(7):2871–2889. doi: 10.1093/nar/15.7.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells D., Kedes L. Structure of a human histone cDNA: evidence that basally expressed histone genes have intervening sequences and encode polyadenylylated mRNAs. Proc Natl Acad Sci U S A. 1985 May;82(9):2834–2838. doi: 10.1073/pnas.82.9.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E. M., Shapiro D. L., Allis C. D., Gorovsky M. A. Sequence and properties of the message encoding Tetrahymena hv1, a highly evolutionarily conserved histone H2A variant that is associated with active genes. Nucleic Acids Res. 1988 Jan 11;16(1):179–198. doi: 10.1093/nar/16.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R. S., Bonner W. M. Separation of basal histone synthesis from S-phase histone synthesis in dividing cells. Cell. 1981 Dec;27(2 Pt 1):321–330. doi: 10.1016/0092-8674(81)90415-3. [DOI] [PubMed] [Google Scholar]
- Wu R. S., Tsai S., Bonner W. M. Patterns of histone variant synthesis can distinguish G0 from G1 cells. Cell. 1982 Dec;31(2 Pt 1):367–374. doi: 10.1016/0092-8674(82)90130-1. [DOI] [PubMed] [Google Scholar]
- Yamamoto M., Maehara Y., Takahashi K., Endo H. Cloning of sequences expressed specifically in tumors of rat. Proc Natl Acad Sci U S A. 1983 Dec;80(24):7524–7527. doi: 10.1073/pnas.80.24.7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Roeder R. G., Heintz N. The primary structure and expression of four cloned human histone genes. Nucleic Acids Res. 1983 Nov 11;11(21):7409–7425. doi: 10.1093/nar/11.21.7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Daal A., White E. M., Gorovsky M. A., Elgin S. C. Drosophila has a single copy of the gene encoding a highly conserved histone H2A variant of the H2A.F/Z type. Nucleic Acids Res. 1988 Aug 11;16(15):7487–7497. doi: 10.1093/nar/16.15.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]