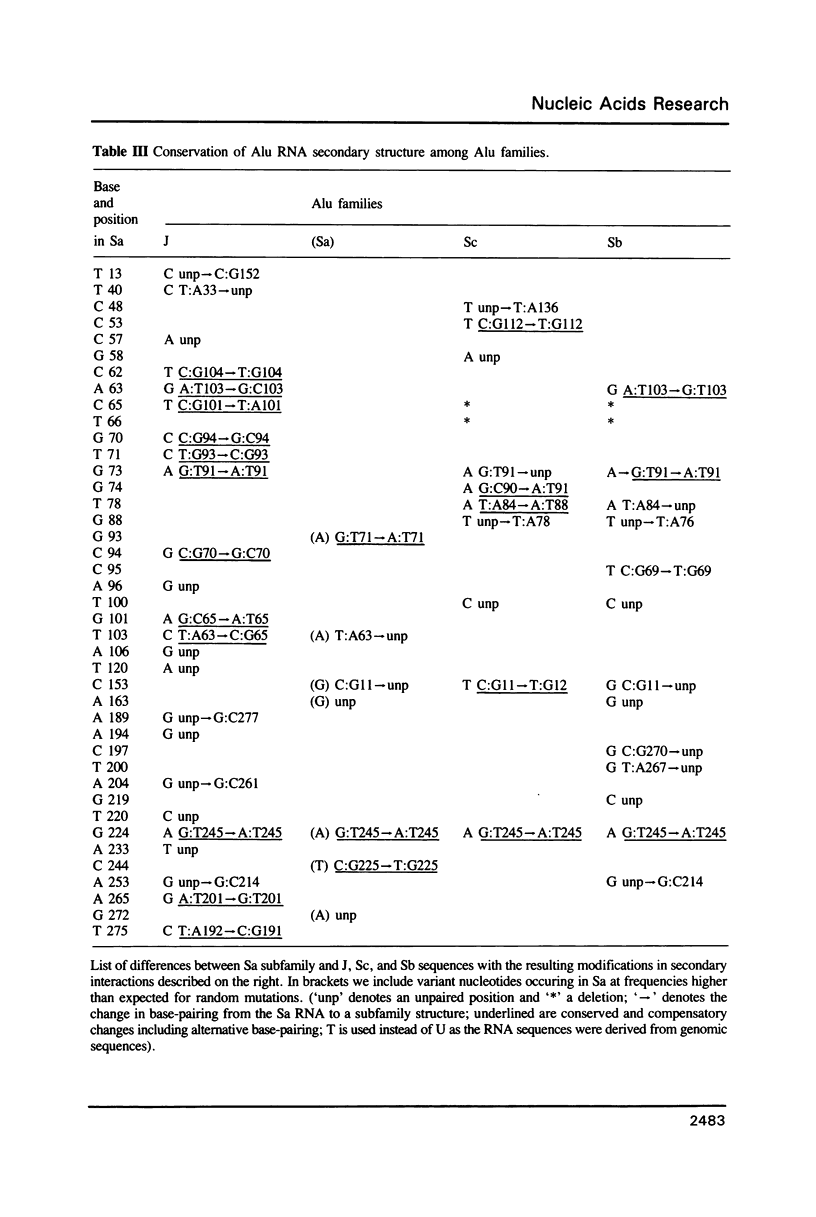
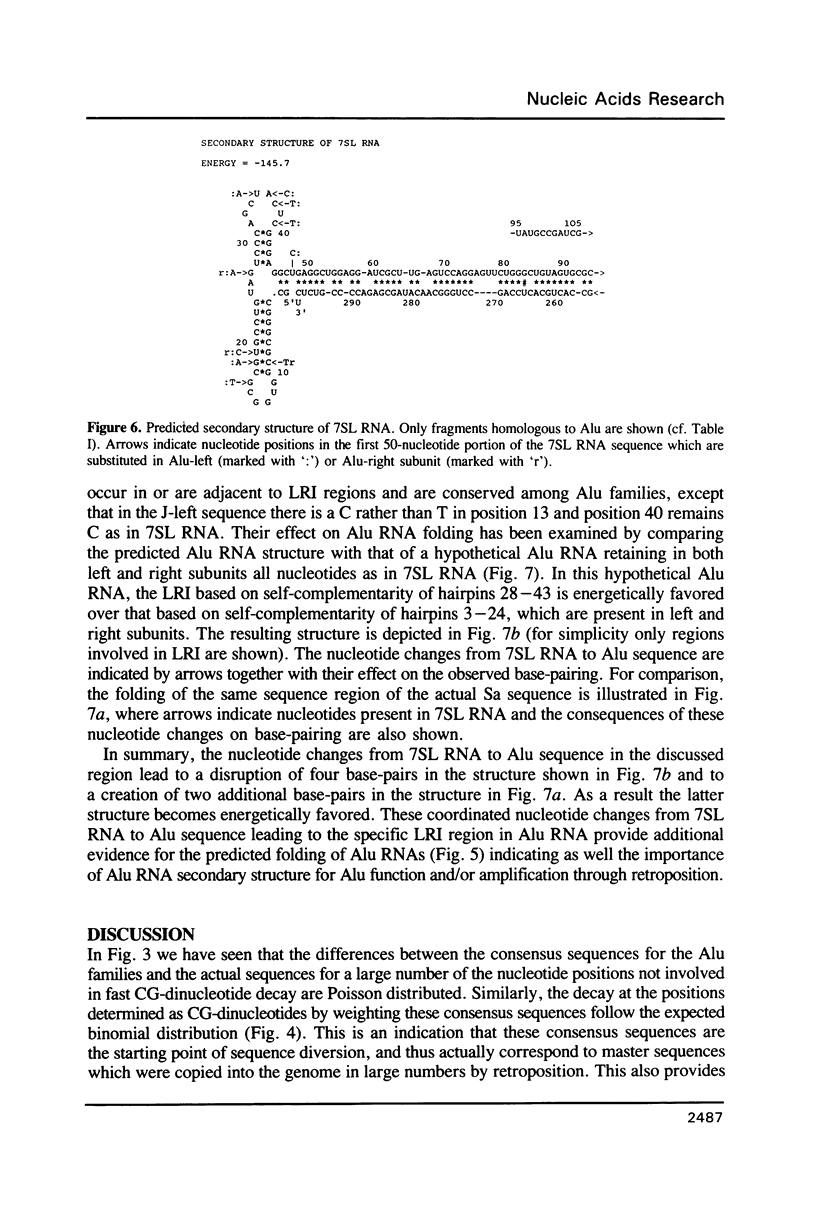
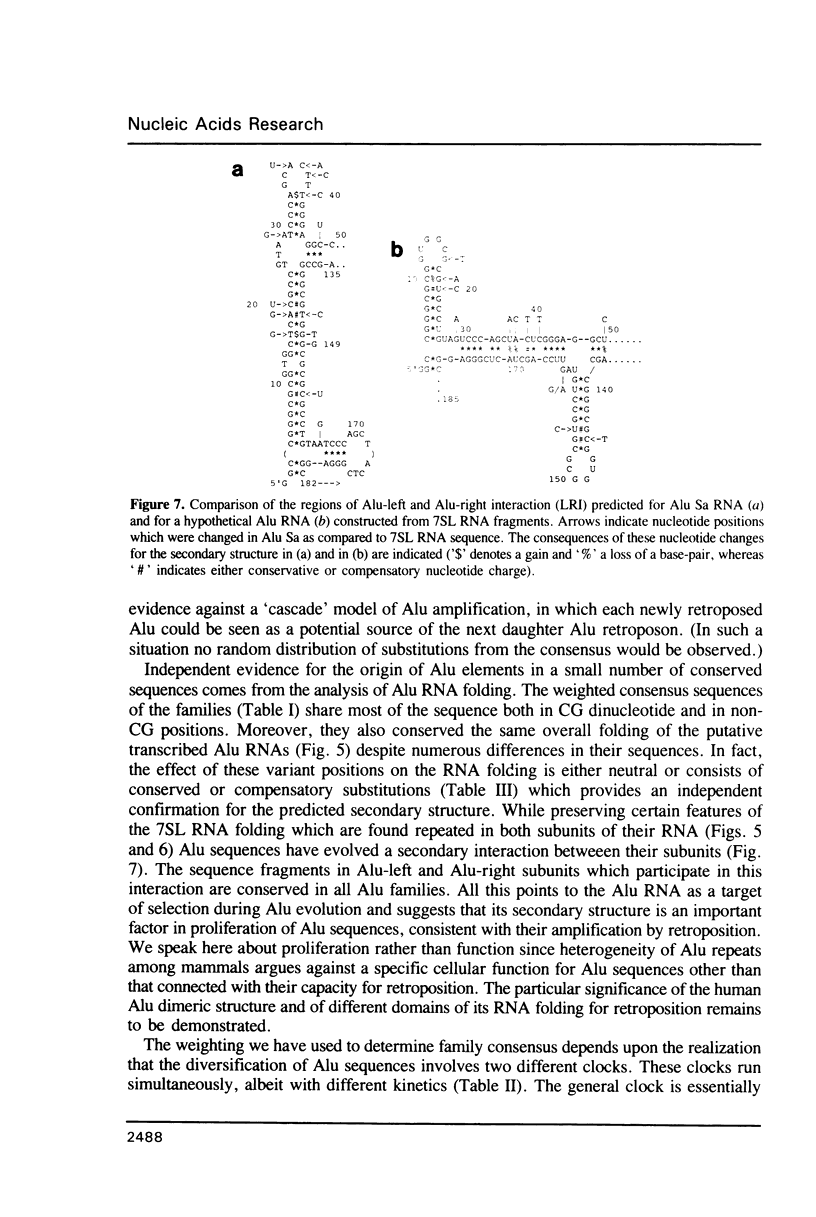
Abstract
A statistical analysis of a set of genomic human Alu elements is based on a published alignment and a recent classification of these sequences. After separation of the Alu sequences into families, the consensus sequences of these families are determined, using the correct weighting of the unidirectional decay of CG-dinucleotides. For, the tenfold greater mutation rate at CG's requires separate consideration of an independent clock at every stage of analysis. The distributions of the substitutions with respect to the new consensus sequences, taking the CG and the non-CG-nucleotide positions separately, lie far closer to the expected distributions than the total diversity. Computer analysis of the folding of RNAs derived from these sequences indicates that RNA secondary structure is conserved among Alu families, suggesting its importance for Alu proliferation and/or function. The folding pattern, further substantiated by a number of compensatory mutations, includes secondary structure domains which are homologous to those observed in 7SL RNA and a defined region of interaction between the two Alu subunits. These results are consistent with a model in which a small number of conserved Alu master genes give rise via retroposition to the numerous copies of Alu pseudogenes, that then diversify by random substitution. The master genes appeared at different periods during evolution giving rise to different families of Alu sequences.
Full text
PDF











Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Britten R. J., Baron W. F., Stout D. B., Davidson E. H. Sources and evolution of human Alu repeated sequences. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4770–4774. doi: 10.1073/pnas.85.13.4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten R. J. Rates of DNA sequence evolution differ between taxonomic groups. Science. 1986 Mar 21;231(4744):1393–1398. doi: 10.1126/science.3082006. [DOI] [PubMed] [Google Scholar]
- Brown T. C., Jiricny J. Different base/base mispairs are corrected with different efficiencies and specificities in monkey kidney cells. Cell. 1988 Aug 26;54(5):705–711. doi: 10.1016/s0092-8674(88)80015-1. [DOI] [PubMed] [Google Scholar]
- Bulmer M. Neighboring base effects on substitution rates in pseudogenes. Mol Biol Evol. 1986 Jul;3(4):322–329. doi: 10.1093/oxfordjournals.molbev.a040401. [DOI] [PubMed] [Google Scholar]
- Gingerich P. D. Temporal scaling of molecular evolution in primates and other mammals. Mol Biol Evol. 1986 May;3(3):205–221. doi: 10.1093/oxfordjournals.molbev.a040391. [DOI] [PubMed] [Google Scholar]
- Gundelfinger E. D., Di Carlo M., Zopf D., Melli M. Structure and evolution of the 7SL RNA component of the signal recognition particle. EMBO J. 1984 Oct;3(10):2325–2332. doi: 10.1002/j.1460-2075.1984.tb02134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwu H. R., Roberts J. W., Davidson E. H., Britten R. J. Insertion and/or deletion of many repeated DNA sequences in human and higher ape evolution. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3875–3879. doi: 10.1073/pnas.83.11.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J., Smith T. A fundamental division in the Alu family of repeated sequences. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4775–4778. doi: 10.1073/pnas.85.13.4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariya Y., Kato K., Hayashizaki Y., Himeno S., Tarui S., Matsubara K. Revision of consensus sequence of human Alu repeats--a review. Gene. 1987;53(1):1–10. doi: 10.1016/0378-1119(87)90087-4. [DOI] [PubMed] [Google Scholar]
- Lehrman M. A., Russell D. W., Goldstein J. L., Brown M. S. Alu-Alu recombination deletes splice acceptor sites and produces secreted low density lipoprotein receptor in a subject with familial hypercholesterolemia. J Biol Chem. 1987 Mar 5;262(7):3354–3361. [PubMed] [Google Scholar]
- Markert M. L., Hutton J. J., Wiginton D. A., States J. C., Kaufman R. E. Adenosine deaminase (ADA) deficiency due to deletion of the ADA gene promoter and first exon by homologous recombination between two Alu elements. J Clin Invest. 1988 May;81(5):1323–1327. doi: 10.1172/JCI113458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myerowitz R., Hogikyan N. D. A deletion involving Alu sequences in the beta-hexosaminidase alpha-chain gene of French Canadians with Tay-Sachs disease. J Biol Chem. 1987 Nov 15;262(32):15396–15399. [PubMed] [Google Scholar]
- Perez-Stable C., Shen C. K. Competitive and cooperative functioning of the anterior and posterior promoter elements of an Alu family repeat. Mol Cell Biol. 1986 Jun;6(6):2041–2052. doi: 10.1128/mcb.6.6.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quentin Y. The Alu family developed through successive waves of fixation closely connected with primate lineage history. J Mol Evol. 1988;27(3):194–202. doi: 10.1007/BF02100074. [DOI] [PubMed] [Google Scholar]
- Rinehart F. P., Ritch T. G., Deininger P. L., Schmid C. W. Renaturation rate studies of a single family of interspersed repeated sequences in human deoxyribonucleic acid. Biochemistry. 1981 May 26;20(11):3003–3010. doi: 10.1021/bi00514a003. [DOI] [PubMed] [Google Scholar]
- Rogers J. H. The origin and evolution of retroposons. Int Rev Cytol. 1985;93:187–279. doi: 10.1016/s0074-7696(08)61375-3. [DOI] [PubMed] [Google Scholar]
- Rouyer F., Simmler M. C., Page D. C., Weissenbach J. A sex chromosome rearrangement in a human XX male caused by Alu-Alu recombination. Cell. 1987 Nov 6;51(3):417–425. doi: 10.1016/0092-8674(87)90637-4. [DOI] [PubMed] [Google Scholar]
- Sawada I., Willard C., Shen C. K., Chapman B., Wilson A. C., Schmid C. W. Evolution of Alu family repeats since the divergence of human and chimpanzee. J Mol Evol. 1985;22(4):316–322. doi: 10.1007/BF02115687. [DOI] [PubMed] [Google Scholar]
- Slagel V., Flemington E., Traina-Dorge V., Bradshaw H., Deininger P. Clustering and subfamily relationships of the Alu family in the human genome. Mol Biol Evol. 1987 Jan;4(1):19–29. doi: 10.1093/oxfordjournals.molbev.a040422. [DOI] [PubMed] [Google Scholar]
- Trabuchet G., Chebloune Y., Savatier P., Lachuer J., Faure C., Verdier G., Nigon V. M. Recent insertion of an Alu sequence in the beta-globin gene cluster of the gorilla. J Mol Evol. 1987;25(4):288–291. doi: 10.1007/BF02603112. [DOI] [PubMed] [Google Scholar]
- Ullu E., Murphy S., Melli M. Human 7SL RNA consists of a 140 nucleotide middle-repetitive sequence inserted in an alu sequence. Cell. 1982 May;29(1):195–202. doi: 10.1016/0092-8674(82)90103-9. [DOI] [PubMed] [Google Scholar]
- Ullu E., Tschudi C. Alu sequences are processed 7SL RNA genes. Nature. 1984 Nov 8;312(5990):171–172. doi: 10.1038/312171a0. [DOI] [PubMed] [Google Scholar]
- Ullu E., Weiner A. M. Upstream sequences modulate the internal promoter of the human 7SL RNA gene. 1985 Nov 28-Dec 4Nature. 318(6044):371–374. doi: 10.1038/318371a0. [DOI] [PubMed] [Google Scholar]
- Watson J. B., Sutcliffe J. G. Primate brain-specific cytoplasmic transcript of the Alu repeat family. Mol Cell Biol. 1987 Sep;7(9):3324–3327. doi: 10.1128/mcb.7.9.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willard C., Nguyen H. T., Schmid C. W. Existence of at least three distinct Alu subfamilies. J Mol Evol. 1987;26(3):180–186. doi: 10.1007/BF02099850. [DOI] [PubMed] [Google Scholar]
- Youssoufian H., Antonarakis S. E., Bell W., Griffin A. M., Kazazian H. H., Jr Nonsense and missense mutations in hemophilia A: estimate of the relative mutation rate at CG dinucleotides. Am J Hum Genet. 1988 May;42(5):718–725. [PMC free article] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwieb C. The secondary structure of the 7SL RNA in the signal recognition particle: functional implications. Nucleic Acids Res. 1985 Sep 11;13(17):6105–6124. doi: 10.1093/nar/13.17.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]



