Abstract
Since the establishment of a canonical animal microRNA biogenesis pathway driven by the RNase III enzymes Drosha and Dicer, an unexpected variety of alternative mechanisms that generate functional microRNAs have emerged. We review here the many Drosha-independent and Dicer-independent microRNA biogenesis strategies characterized over the past few years. Beyond reflecting the flexibility of small RNA machineries, the existence of non-canonical pathways has consequences for interpreting mutants in the core microRNA machinery. Such mutants are commonly used to assess the consequences of “total” microRNA loss, and indeed, they exhibit many overall phenotypic similarities. Nevertheless, ongoing studies reveal a growing number of settings in which alternative microRNA pathways contribute to distinct phenotypes amongst core microRNA biogenesis mutants.
Introduction
microRNAs (miRNAs) are abundant ~22 nucleotide (nt) regulatory RNAs, derived from endogenous short hairpin transcripts, that collectively play key roles in diverse developmental and physiological processes in most eukaryotes (Flynt and Lai, 2008). The general defining features of miRNA genes are cleavage of their precursor transcripts by one or more RNase III enzymes, and sorting of mature species into Argonaute proteins of the Ago-subfamily (Axtell et al., 2011). As with other classes of Argonaute-bound small RNAs, miRNAs serve as antisense guides to identify regulatory targets (Czech and Hannon, 2010).
Plant miRNAs frequently pair extensively with one or a few targets, and these interactions have reliably proven to mediate their key functions (Axtell et al., 2011). In contrast, animal miRNAs have propensity to recognize targets via ~7 nt complements to their 5′ ends (preferentially nucleotides 2-8, the miRNA “seed”) (Bartel, 2009). Computational and experimental strategies provide evidence for 100s-1000 direct conserved targets for individual human miRNAs, such that a majority of human transcripts carry conserved binding sites for multiple miRNAs (Bartel, 2009). The broad nature of animal miRNA target networks has made it difficult to infer phenotypically relevant aspects of miRNA biology. Moreover, many individual miRNA mutants have subtle phenotypes, and relatively few miRNA knockouts have yet been reported in many species, including most vertebrates (Smibert and Lai, 2008).
Instead, knockouts of core miRNA factors are commonly used as a proxy to assess the phenotypic effects of removing miRNA-mediated regulation. Over a hundred studies have studied straight or conditional knockouts of mouse dicer (Bernstein et al., 2003; Harfe et al., 2005; Kanellopoulou et al., 2005; Yi et al., 2006), and they collectively show this enzyme to be required for normal development, differentiation, and/or physiology of most tissues. In some cases dicer phenotypes can be causally linked to removal of specific miRNAs. For example, conditional knockout of dicer in the B cell lineage blocks the transition from pro-B to pre-B cells, accompanied by global upregulation of many targets of the mir-17-92 cluster, including the propapoptotic miR-17-92 target Bim (Koralov et al., 2008). These phenotypes were shared by knockout of the mir-17-92 cluster, which is highly expressed in the B cell lineage (Ventura et al., 2008), suggesting that it is responsible for a substantial aspect of this aspect of the dicer mutant phenotype. In zebrafish, a compelling illustration was the rescue of early embryogenesis in dicer mutants by injection of a single small RNA duplex for miR-430, the major early-expressed miRNA in this species (Giraldez et al., 2005).
While it is generally reasonable to infer that phenotypes of mutants such as dicer are due to miRNA loss, functional connections to individual miRNAs is often correlative, especially in the intact animal. Phenotypes may stem from concomitant loss of multiple miRNAs, and there are technical challenges to re-expressing functional miRNAs in biogenesis mutants. Furthermore, the existence of alternative miRNA pathways raises the possibility that core biogenesis mutants maintain subclasses of active miRNAs, or conversely that their phenotypes do not simply reflect the removal of miRNAs.
The goals of this review are thus twofold. First, we describe the diversity of alternative miRNA biogenesis mechanisms, which reflect evolutionary flexibility in the acceptance and routing of different sources of double-stranded RNA by RNase III enzymes and Ago proteins. Second, we discuss biological settings where loss of different core miRNA machinery has distinct consequences for depletion of miRNAs and related regulatory RNAs, and how this may impact the interpretation of organismal phenotype. The extensive literature on plant miRNAs notwithstanding, we focus this review on animal systems, for which alternate miRNA pathways are more abundant and the biological roles of miRNAs less well-understood.
Part I: Canonical and alternative miRNA biogenesis pathways
The canonical miRNA biogenesis pathway
A canonical pathway driven by RNase III enzymes generates the majority of animal miRNAs (Ghildiyal and Zamore, 2009) (Figure 1A). Primary miRNA (pri-miRNA) transcripts are typically products of RNA Polymerase II, and the hairpins are usually contained within non-coding RNAs or the introns of messenger RNAs (mRNAs); frequently, multiple pri-miRNA hairpins are encoded by an individual transcript. Their biogenesis begins with cleavage near the base of each pri-miRNA hairpin by the nuclear Drosha/DGCR8 heterodimer. DGCR8 (known as Pasha in invertebrates) is a dsRNA binding protein that recognizes the proximal ~10 bp of stem of the pri-miRNA hairpin, positioning the catalytic sites of the RNase III enzyme Drosha (Han et al., 2006). Cleavage releases a pre-miRNA hairpin that is typically ~55-70 nt in length.
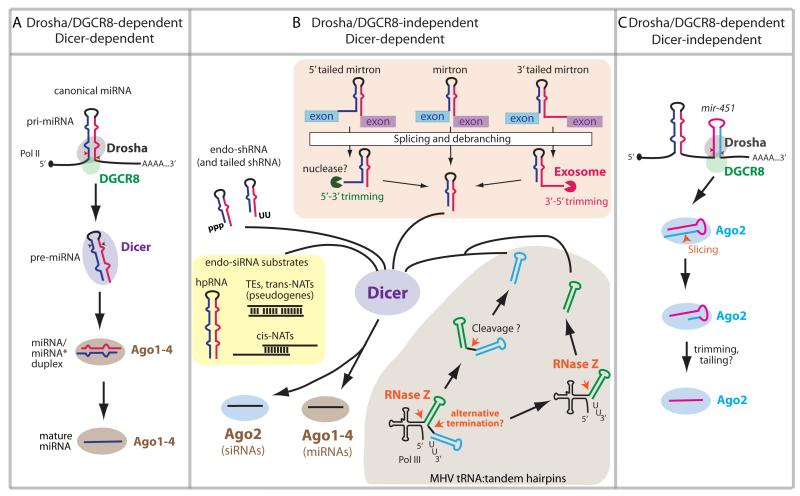
Figure 1. Canonical and major alternative miRNA biogenesis pathways in animals.
(A) Canonical animal miRNAs are generated through consecutive cleavages of hairpin precursors by two RNase III enzymes. In the nucleus, the single strand-double strand junction of the pri-miRNA hairpin is recognized by DGCR8, which positions the catalytic site of the RNase III enzyme Drosha. This cleavage generates a ~55-70 pre-miRNA hairpin that is exported to the cytoplasm, where it is cleaved towards the terminal loop end by the RNase III enzyme Dicer. The miRNA/miRNA* duplexes are loaded into miRNA-class Argonaute effectors (in mammals, Ago1-4). One of the duplex strands is preferentially retained in Ago to form the functional RNA-induced silencing complex.
(B) Many Drosha/DGCR8-independent pathways can generate pre-miRNA-like hairpins that serve as Dicer substrates. Mirtrons are short intronic hairpins that are excised by splicing and linearized by lariat debranching; tailed mirtrons require further resection by nucleases, e.g. 3′-5′ resection of 3′ tailed mirtrons by the RNA exosome. RNA pol III-transcribed MHV68 tRNA-shRNA fusions are processed into pre-miRNA-like hairpins with defined 5′ and 3′ ends as a result of RNaseZ cleavage and pol III termination, respectively. Endo-shRNAs without lower stems for Drosha/DGCR8 processing may derive from pol III transcription or cleavage by as yet unknown endo- or exonucleases. These non-canonical miRNAs, like canonical miRNAs, are incorporated to Ago1-4. Endogenous substrates with extended dsRNA character, including hpRNAs, transposable elements (TEs), antisense pseudogenes and natural antisense transcripts (NATs), are directly cleaved by Dicer to generate siRNAs. These may potentially sort to all of the mammalian Agos, but presumably only those that load Ago2 can fulfill target slicing.
(C) Pri-mir-451 is processed by Drosha/DGCR8, and the resulting ~18-bp pre-mir-451 is directly incorporated to Ago2. The Slicer activity of Ago2 cleaves the 3′ arm of pre-mir-451, giving rise to ac-pre-mir-451, which is further resected by an as yet unknown mechanism to generate mature miR-451.
The ~2 nt 3′ overhangs of pre-miRNA hairpins are recognized by Exportin-5 (Exp-5) and its partner Ran-GTP, enabling their nuclear export. In the cytoplasm, the Dicer RNase III enzyme cleaves pre-miRNAs ~2 helical turns into the hairpin, yielding ~22 nt small RNA duplexes. One of the strands is usually preferentially incorporated into an effector Ago protein and guides it to targets (Czech and Hannon, 2010). Select miRNA:target pairs in animals exhibit extensive complementarity permitting their cleavage by Slicer-class Ago proteins (Karginov et al., 2010; Shin et al., 2010; Yekta et al., 2004). However, the bulk of miRNA targets lack sufficient pairing for slicing, and are instead repressed by deadenylation, mRNA degradation, and/or translational suppression (Fabian et al., 2010).
While Dicer and Ago proteins are central to miRNA biogenesis in all species, certain homologs have distinct properties (Ghildiyal and Zamore, 2009). For example, C. elegans and vertebrates encode a single Dicer that generates both miRNAs and siRNAs, but Drosophila encodes two Dicers, of which Dcr-1 is specialized for pre-miRNA cleavage and Dcr-2 is selective for siRNA biogenesis. Argonaute proteins also exhibit specialization. Drosophila has two Ago-class effectors, of which AGO1 is dominant for miRNAs and AGO2 for siRNAs; these are further specialized in that AGO2 is a more effective Slicer than AGO1, and AGO2-resident species are modified by 2′O-methylation. All four vertebrate Ago-class effectors participate in miRNA-mediated regulation and carry similar miRNA contents; thus, they lack comparable sorting mechanisms that distinguish Drosophila AGO1 and AGO2 cargoes. However, only Ago2 amongst vertebrate Ago proteins has Slicer activity, implying that it has unique activities for small RNA biogenesis and/or function.
The mirtron pathway
Deep sequencing of D. melanogaster revealed short RNA duplexes mapped to short hairpin introns, termed “mirtrons”, where the mature small RNA termini coincided with splice acceptor and donor sites (Okamura et al., 2007; Ruby et al., 2007). This suggested that splicing might substitute for Drosha cleavage, and this indeed proved to be the case (Figure 1B). As with other introns, the splicing reaction generates a non-linear intermediate that must be resolved by the lariat debranching enzyme before the hairpin structure can be adopted. At this step, mirtron products appear as pre-miRNA mimics and enter the canonical biogenesis pathway as Exp-5 and Dcr-1 substrates, yielding mature products that populate AGO1 and can regulate typical seed-matching targets.
Mirtrons are prevalent in both D. melanogaster and C. elegans (Chung et al., 2011), perhaps exploiting the fact that their genomes contain an abundant class of short introns that overlaps the length of pre-miRNAs (Lim and Burge, 2001). In fact, one of the earliest annotated worm miRNA genes (mir-62) was later recognized as a mirtron (Ruby et al., 2007). Subsequently, mirtrons were recognized in diverse vertebrates (Babiarz et al., 2008; Berezikov et al., 2007; Glazov et al., 2008). Cloning of murine small RNAs from dgcr8 or drosha knockout cells verified near-complete loss of canonical miRNAs, but maintained expression of mirtron-derived miRNAs (Babiarz et al., 2008; Chong et al., 2010; Yi et al., 2009). Therefore, vertebrate mirtrons likely follow a similar maturation pathway as in invertebrates.
Tailed mirtrons: 5′ vs. 3′ tails
With conventional mirtrons, both ends of the pre-miRNA are defined by splicing. However, in the atypical locus Drosophila mir-1017, only the 5′ hairpin end matches the splice donor site, followed by a ~100 nt unstructured tail before the splice acceptor site (Ruby et al., 2007). Conversely, there exist vertebrate introns where 3′ hairpin ends coincide with splice acceptor sites, but are preceded by unstructured tails following their splice donor sites (Babiarz et al., 2008; Glazov et al., 2008). Presumably such “tailed mirtrons” are processed by splicing, but require additional biogenesis steps (Figure 1B).
The biogenesis of Drosophila 3′ tailed mirtrons was recently reported to utilize the RNA exosome (Flynt et al., 2010), the major 3′-5′ exoribonuclease in eukaryotes. In this pathway, the 3′ tail of the spliced and debranched full-length intron is removed by the exosome to yield the pre-miRNA. In vitro assays showed that the exosome was inhibited by the hairpin structure, allowing for pre-mir-1017 release. As with conventional mirtrons, pre-mir-1017 is then cleaved by Dcr-1 and loaded into AGO1 to function as a typical miRNA. The biogenesis of mammalian 5′ tailed mirtrons has not been elucidated, but their configuration suggests potential involvement of 5′-3′ exoribonucleases, such as the XRN family. Thus far, 3′ tailed mirtrons have only been described in Drosophila, while 5′ tailed mirtrons have only been annotated in vertebrates, suggesting adoption of distinct hybrid pathways for splicing-mediated miRNA biogenesis in different animal clades.
box H/ACA- and box C/D snoRNA-derived miRNAs
Small nucleolar RNAs (snoRNAs) have analogies to miRNAs in that they are also abundant, deeply conserved short RNAs that serve as antisense guides. In their best-known roles, snoRNAs guide post-transcriptional modifications of rRNA and snRNA targets. The presence of sub-motifs permits snoRNAs to be categorized as C/D box or H/ACA box classes, which typically mediate 2′-O-ribose methylation and pseudouridylation, respectively. As well, many “orphan snoRNAs” lack apparent rRNA or snRNA targets, perhaps suggesting other regulatory targets.
Small RNA libraries usually contain a population of reads, sometimes quite substantial in number, from rRNAs, tRNAs, and snoRNAs. As routine turnover of these abundant ncRNAs generates shorter species, most miRNA annotators set aside reads matching known ncRNAs. On the other hand, the presence of reads from known ncRNAs in Ago immunoprecipitates (IP) can provide a rationale to consider them further. For example, analysis of human Ago1-IP and Ago2-IP revealed enrichment of duplex reads derived from a hairpin in the ACA45 snoRNA/mir-1839, which were established as Drosha/DGCR8-independent and Dicer-dependent (Babiarz et al., 2008; Ender et al., 2008). Similarly, the snoRNA GlsR17 in Giardia lamblia generates a Dicer-dependent functional miRNA (Saraiya and Wang, 2008). These studies prompted re-evaluation of other snoRNA-derived (sdRNA) reads, and it is now documented that sdRNAs are frequently recovered from both C/D and H/ACA box classes (Babiarz et al., 2011; Brameier et al., 2011; Ono et al., 2011; Scott et al., 2009; Taft et al., 2009). In some cases, these have been further shown to be dependent on Dicer, to associate with Ago complexes, and to direct detectable repression of complementary targets, generalizing the notion of dual-function snoRNAs that have miRNA activity (Figure 2).
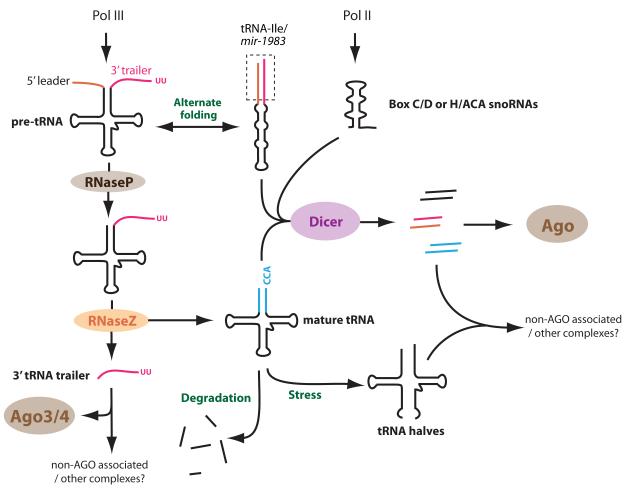
Figure 2. Summary of tRNA and snoRNA-derived small RNAs.
During typical tRNA maturation, the 5′ leader and 3′ trailer of the pre-tRNA are removed by RNase P and RNase Z cleavages, respectively, followed by 3′ CCA addition. Subfragments of tRNAs are frequently observed, many of which reflect routine tRNA turnover and are not regulatory in nature. However, a number of specific pathways have been observed that extend the regulatory range of tRNA loci. In the case of tRNA-Ile/mir-1983, an alternative fold of the pre-tRNA adopts an extensive hairpin that permits cleavage by Dicer. Some mature tRNA cloverleafs may also serve as Dicer substrates and/or generate subfragments that load Ago proteins; this is most prominent in Tetrahymena where the Piwi protein Twi12 carries 3′ tRNA fragments exclusively. Some RNaseZ-generated tRNA 3′ trailers associate preferentially with Ago3/4 and regulatory activity of these trailers was reported, although this is not necessarily mediated by Ago complexes. Under stress conditions, mature tRNA are cut into halves, which may associate with unknown complexes to exert regulatory roles. A number of box C/D and box H/ACA snoRNAs can also give rise to Ago-associated, miRNA-like species in a Drosha/DGCR8-independent, Dicer-dependent manner.
Still, there remains good reason to be wary in the functional interpretation of sdRNAs. Although many sdRNAs map with regional preference across snoRNA precursors, this alone is not definitive evidence for a specific biogenesis pathway, as opposed to reflecting more stable degradation fragments. In addition, the simple presence of sdRNAs in Ago-IP libraries may not necessarily reflect genuine residence in Ago, as some abundant cellular species may simply fail to be sufficiently depleted in IP reactions. Nevertheless, there are now clearly many compelling sdRNA substrates that provide a basis for future detailed biochemical analyses of their biogenesis or function.
miRNAs from tRNAs
Analogous to cases of snoRNA-derived miRNAs, some tRNA-derived RNAs (tdRNAs) also contribute to the miRNA pool. One of the first examples came from deep sequencing of mouse embryonic stem (mES) cells deleted for dgcr8 or dicer (Babiarz et al., 2008). With the tRNA-Ile/mir-1983 locus, a population of fairly heterogeneous reads was recovered, including reads that spanned its intron or included the untemplated 3′ CCA seen in mature tRNAs. However, a species from the 3′ end of the pre-tRNA (miR-1983) was Dicer-dependent but DGCR8-independent, suggesting its identity as a non-canonical miRNA (Babiarz et al., 2008). Interestingly, the tRNA-Ile precursor was predicted to adopt different folds (Figure 2). One formed a typical tRNA cloverleaf, which is cleaved near its 5′ end by RNase P and near its 3′ end by tRNase Z, prior to CCA addition. However, the terminal sequences normally removed by RNase P/Z can also basepair, thereby extending the duplex base of the tRNA hairpin to present a plausible Dicer substrate. Therefore, alternative conformations can determine entry into different biogenesis pathways.
Further study provided additional evidence for other Dicer-dependent tdRNAs, tdRNA accumulation in Ago complexes, and/or modulation of tdRNA levels by Ago availability (Cole et al., 2009; Haussecker et al., 2010). As with snoRNAs, caution is warranted in the general interpretation of tRNA fragments that appear in small RNA libraries, and the population of any specific read may actually be contributed through a combination of generic degradation and specific Dicer processing. Moreover, as the Dicer-cleaved product of a tRNA-Gln is 3′ modified and inefficiently loaded in Ago complexes (Cole et al., 2009), Dicer processing does not guarantee Ago loading. Still, the collected studies provide ample precedent that some abundant tdRNAs comprise miRNAs.
Other tRNA fragments have been cloned, including tRNA halves that accumulate during starvation or oxidative stress (Pederson, 2010). Relatively little is known about their function, but they seem unlikely to be via Argonaute proteins owing to their large size (>35 nt) and their existence in S. cerevisiae, which lacks RNAi/miRNA pathways altogether. However, the Tetrahymena Piwi protein Twi12 carries smaller, 18-22 nt species that derive nearly exclusively from the 3′ ends of mature tRNAs (Couvillion et al., 2010). Their function is not known, but Twi12 itself is an essential gene. Thus, intersections between tdRNAs and Argonaute pathways deserve further study.
tRNaseZ-derived miRNAs
Maturation of canonical miRNAs generates several byproduct species, including flanking miRNA offset reads (moRs) produced by Drosha cleavage and free terminal loops produced by Dicer cleavage (Berezikov et al., 2011; Shi et al., 2009). In analogous fashion, other small RNA species are released during tRNA maturation (Pederson, 2010). For example, tRNase Z cleavage of pre-tRNA transcripts releases ~18-25 nt 3′ tRNA trailers (Lee et al., 2009). While these may be byproducts, their non-stoichiometric accumulation relative to cognate mature tRNAs may suggest potential functional activity. While this is not necessarily mediated by Ago proteins, a number of 3′ tRNA trailers are responsive to Ago levels (Haussecker et al., 2010) (Figure 2).
These observations set a precedent that tRNase Z may define the ends of some miRNA species, independently of Drosha. This proved to be the case for miRNAs encoded by murine γ-herpesvirus 68 (MHV68). Each of the ~20 MHV68 miRNAs maps to tandem hairpins located immediately downstream of a tRNA moiety, suggesting their expression as tRNA-fusions from Pol III promoters (Pfeffer et al., 2005; Reese et al., 2010). In some cases miRNA were cloned from both of the tandem hairpins, but in other cases one of the hairpins is preferentially processed into stable small RNAs. These hairpins lack additional “lower stem” pairing indicative of Drosha/DGCR8 binding. Indeed, the production of MHV68 miRNAs is Drosha-independent and instead dependent on tRNase Z, which cleaves the 5′ ends of the MHV68 pre-miRNA hairpins (Bogerd et al., 2010) (Figure 1B). The mechanism that defines the 3′ end of the internal hairpin has not been definitively established, and could involve an endonuclease that separates the tandem hairpins. However, the existence of U-rich stretches at the end of both foldbacks in these tandem arrangements suggests that alternative pol III termination may define the 3′ ends. The resulting pre-miRNAs are further processed into mature miRNAs by Dicer (Bogerd et al., 2010).
The tRNA-miRNA system is flexible, since the tRNA, miRNA hairpin and the pol III promoter can be readily exchanged (Bogerd et al., 2010). Remarkably, artificial tRNA-shRNA chimeric expression cassettes were optimized to generate functional siRNAs before elucidation of the MHV68 pathway (Scherer et al., 2007). In these constructs, only a single shRNA hairpin is introduced after the tRNA, and the shRNA ends are thus defined by tRNase Z on the 5′ end and by the pol III terminator on the 3′ end. The reason for the tRNA-tandem hairpin layout in the MHV68 genome is unclear (Pfeffer et al., 2005; Reese et al., 2010), but its pervasive nature suggests that it has been selected for some functional reason.
Endo-shRNAs
The first strategies for transgenic RNAi in mammalian cells used pol III-driven short hairpin RNAs (shRNAs), for which direct definition of hairpins by transcription permits their processing by Dicer (Medina and Joshi, 1999). Years later, the concept that transcription might determine the ends of some pre-miRNAs was extended to endogenous shRNAs (endo-shRNAs). This is currently a catch-all category for DGCR8-independent, Dicer-dependent loci where at least one pre-miRNA hairpin end is generated by transcriptional initiation or termination (Figure 1B). Multiple mechanisms may be involved in their biogenesis, just as with the different flavors of mirtrons.
The earlier mentioned tRNA-Ile/mir-1983 is analogous to synthetic shRNAs, in which both pre-miRNA ends are determined by transcription. In the case of mir-320, its 5p species are strongly under-representated relative to 3p reads. While this might be influenced by highly asymmetric loading of miRNA/star duplexes, 5′ RACE detected a processed end corresponding to the 5′ end of the hairpin (Babiarz et al., 2008). In principle, if this reflected a genuine transcription start, the resulting 5′ triphosphates of 5p reads would be inefficiently ligated by the standard miRNA cloning protocol and thus depleted from libraries. However, a mechanism to determine the 3′ end of pre-mir-320 has not yet been elucidated. As well, 5′ tailed endo-shRNAs exist (Babiarz et al., 2008), for which the removal of 5′ flanks may potentially be analogous to biogenesis of 5′ tailed mirtrons.
siRNAs (and miRNAs) from endo-siRNA pathways
In many invertebrates and plants, the RNA interference (RNAi) pathway mediates antiviral defense by generating small interfering RNAs (siRNAs) from dsRNA aspects of viral life cycles (Ding and Voinnet, 2007). However, nematode and fly mutants that specifically impair RNAi are otherwise viable, fertile, and exhibit fairly normal morphology. In mammals, the execution of antiviral defense by the interferon pathway suggested for some time that endogenous RNAi might not even be permissible. However, a broader appreciation of endo-siRNA pathways emerged from deep sequencing studies (Okamura and Lai, 2008).
In Drosophila, both somatic and germline tissues are broadly competent to utilize Dcr-2 to cleave endo-siRNAs from transposable elements (TEs), cis-natural antisense transcripts (cis-NATs) typically comprising convergently transcribed 3′ UTRs, and from long hairpin RNAs (hpRNAs) comprising extensive duplex structure (Chung et al., 2008; Czech et al., 2008; Ghildiyal et al., 2008; Kawamura et al., 2008; Okamura et al., 2008a; Okamura et al., 2008b) (Figure 1B). Although endo-siRNAs from all of these substrates are preferentially loaded into AGO2, a subset load into AGO1 and effectively comprise a subpopulation of miRNA.
The capacity for endo-siRNA biogenesis in vertebrate cells is more limited than in Drosophila, due to the propensity for dsRNA to activate the interferon response. However, certain celltypes such as murine ES cells, oocytes, and preimplantation embryos are tolerant of dsRNA and can use these triggers to mount specific RNAi responses (Paddison et al., 2002; Svoboda et al., 2000; Yang et al., 2001). In yet another example of experimental manipulation preceding the elucidation of underlying endogenous pathways, ES cells and oocytes were later found to express diverse endo-siRNAs from long duplexed precursors (Babiarz et al., 2008; Tam et al., 2008; Watanabe et al., 2008). In addition to the endo-siRNA classes reported in fly, mouse oocytes express abundant siRNAs from dsRNA formed by antisense transcribed pseudogenes hybridized to their sense counterparts (Tam et al., 2008; Watanabe et al., 2008). Interestingly, endo-siRNAs from many pseudogene:sense pairs were inferred to be functional, based on broad upregulation of cognate target mRNAs in microarray analysis of dicer−/− oocytes (Tam et al., 2008). Although it is not known whether mammalian endo-siRNAs are sorted to a specific Ago protein, as in Drosophila, it is presumed that endo-siRNAs resident in mammalian Ago2 mediate the bulk of target regulation via slicing.
miR-451 and Dicer-independent miRNA biogenesis
Initial computational studies of Drosophila canonical miRNAs revealed a characteristic pattern of evolutionary divergence for conserved miRNAs, in that the terminal loop evolves much more quickly than does either miRNA or miRNA* species on the hairpin arms (Lai et al., 2003). Although this was defined on the basis of pairwise alignment of two Drosophila species, it was later found to apply across canonical miRNAs and mirtrons amongst Drosophilid and vertebrate genomes (Berezikov et al., 2007; Berezikov et al., 2005; Flynt et al., 2010; Okamura et al., 2007).
A prominent exception to the rule of preferred loop divergence occurs with vertebrate mir-451. Its terminal loop, like its mature products, is completely conserved across all vertebrates from human to fish. In contrast, its presumed miRNA* species, that is, the hairpin sequence complementary to mature miR-451, contains multiple divergent positions in mir-451 orthologs (Yang et al., 2010). Moreover, its mature cloned species extend over the terminal loop instead of being confined to a hairpin arm, and longer cloned products sharing a 5′ end but extending to 30 nt could be recovered in small RNA libraries. All of these properties suggested that miR-451 is not generated by the canonical miRNA pathway.
Detailed study of mir-451 homologs from human, mouse and zebrafish revealed its maturation by an unexpected pathway, the first known to be independent of Dicer (Cheloufi et al., 2010; Cifuentes et al., 2010; Yang et al., 2010). pri-mir-451 is initially cleaved by Drosha/DGCR8 to generate a short pre-miRNA with only ~18 bp of duplex stem, too short to serve as a Dicer substrate. Instead, pre-mir-451 is loaded directly into Ago proteins (Figure 1C). Those hairpins that enter non-slicing Ago proteins (e.g. Ago1) cannot be matured further, while those that load Ago2 are sliced on their 3′ hairpin arm, as guided by the 5′ end of the hairpin, yielding a 30 nt Ago-cleaved species. This is subject to a 3′ resection activity that trims ~7 nt to leave the dominantly cloned 23 nt miR-451; the relevant nuclease(s) remains to be identified.
Altogether, the collected studies reveal diverse Drosha-independent and Dicer-independent strategies for miRNA biogenesis…and there is more. For example, the vault non-coding RNA generates a Drosha-independent miRNA (Persson et al., 2009). Curiously, insertion of canonical pri-mir-124 into the Sindbis RNA virus yields functional miR-124 (Shapiro et al., 2010). Sindbis-mir-124 matures cytoplasmically, since miR-124 accumulated in dgcr8 null cells, and cells depleted for Exportin-5 (Shapiro et al., 2010). The strategy by which a cellular miRNA matures when inserted into an RNA virus is currently a mystery.
Part II: Biological implications of alternative miRNA pathways
Use of miRNA pathway mutants to infer miRNA-dependent phenotypes
A hallmark of genetic analysis is that mutants with similar phenotypes can often be ordered within a common pathway. Although the molecular consequences of lacking the panel of core miRNA pathway components have been extensively characterized, only been a few biological settings have been subjected to detailed phenotypic comparison. In some cases, mutants of different core miRNA components do present similar phenotypes. For example, conditional knockout of dicer and dgcr8 during skin development were indistinguishable, causing rough flaky skin, defects in hair follicle downgrowth, ectopic apoptosis, and lethality by 5-6 days after birth (Yi et al., 2009). Similarly, conditional knockout of drosha and dicer within the T cell compartment induced highly overlapping phenotypes, including loss of Foxp3+ cells and lethality due to spontaneous inflammatory diseases by ~3 weeks (Chong et al., 2008).
Such studies logically support the notion that the major phenotypes of core miRNA biogenesis mutants are attributable to miRNA loss, and certainly the canonical pathway generates the strong majority of miRNA species. Nevertheless, as more studies are conducted, phenotypic differences amongst core miRNA pathway members have begun to emerge (Table 1). Interpretation of their differences is limited by the fact that vertebrate studies have focused heavily on dicer mutants, and have rarely been performed in parallel with drosha, dgcr8, and/or ago knockouts. However, given the variety of Drosha/DGCR8-independent pathways, e.g. mirtrons and endo-siRNAs, it is expected that loss of Dicer should exhibit some differences with drosha or dgcr8 mutants. Reciprocally, substrates that are uniquely cleaved by Drosha or by Dicer may cause additional differences. Finally, Ago2 processing of Dicer-independent species may underlie yet other phenotypic distinctions. We discuss here several examples, which collectively suggest that differences amongst mutants in canonical miRNA machinery may continue to grow as they are scrutinized further.
Table 1.
The impact of non-canonical miRNA pathways and core factor activities on knockout phenotypes.
| Phenotypic comparison | Biological setting | Relevant non-canonical pathways |
|---|---|---|
| Drosha/Dgcr8-KO = Dicer-KO | many places | bulk phenotypes due to loss of canonical miRNAs |
| Drosha/Dgcr8-KO < Dicer-KO | ES cells | roles for Drosha/DGCR8-independent miRNAs (endo-shRNAs, hp-siRNAs?) |
| brain | roles for Drosha/DGCR8-independent miRNAs (snoRNAs, mirtrons?) | |
| oocytes | miRNA activity suppressed; endo-siRNAs functional | |
| Dicer-KO < Drosha-KO | thymocytes | possible roles for direct mRNA cleavages by Drosha/DGCR8 |
| Drosha/Dgcr8/Ago2-KO < Dicer-KO | retinal epithelium | roles for direct Dicer cleavage of Alu dsRNA |
| Dicer-KO < Ago2-KO (or Slicer-dead) | erythropoiesis | Dicer-independent miR-451 and/or other miRNAs? |
| dAGO1+dAGO2-KO < Dgcr8-KO = Dicer1-KO | fly olfactory neurons | potential Ago-independent functions of Dgcr8/Dicer-1? |
dicer vs. dgcr8 mutants in ES cells and brain: Microprocessor-independent Dicer substrates?
Comparison between mouse ES (mES) cells conditionally deleted for dgcr8 and dicer revealed general similarities, including strong defects in cell proliferation and differentiation (Kanellopoulou et al., 2005; Murchison et al., 2005; Wang et al., 2007). Rescue experiments using miRNA mimics introduced into mutant cells attributed some phenotypes to specific canonical miRNAs, such as control of G1-S transition by members of the mir-290 cluster that target cell cycle factors (Wang et al., 2008). On the other hand, dicer knockout cells exhibited noticeably stronger defects than dgcr8 knockouts. For example, compared to dgcr8 knockout cells, dicer knockout cells are much more difficult to grow out following Cre-mediated excision, and loss of dicer causes a more complete block in directed differentiation assays (Kanellopoulou et al., 2005; Murchison et al., 2005; Wang et al., 2007). While the mechanistic basis for these differences remain to be understood, there exists a population of DGCR8-independent mirtrons, endo-shRNAs, and hp-siRNAs in ES cells (Babiarz et al., 2008). The rescue approach, searching for small RNA mimics that preferentially improve the ability to differentiate dicer−/− cells, relative to dgcr8−/− cells, may prove informative in uncovering biological activities of non-canonical ES miRNAs. In particular, the relatively high abundance of the endo-shRNAs mir-320 and mir-484 and the hp-siRNA SINE locus suggests them as candidates for functional study.
Comparison of dicer and dgcr8 conditional knockout in post-mitotic neurons revealed further phenotypic differences. Both genotypes caused microcephaly and lethality, but deletion of dicer resulted in earlier lethality and more severe morphological abnormalities (Babiarz et al., 2011). Small RNA analysis revealed diverse DGCR8-independent, Dicer-dependent small RNAs in brain, of which a majority (by total abundance) derived from snoRNAs, followed by mirtrons. By comparison, these classes comprised only a small minority of non-canonical miRNAs in ES cells (Babiarz et al., 2008). Therefore, cell-specific deployment of different classes of non-canonical miRNAs may underlie distinct underpinnings to phenotypic differences between dicer and dgcr8 conditional knockouts in different settings.
dicer vs. dgcr8 mutants in oocytes: suppression of miRNA-mediated regulation?
The differences between loss of DGCR8 and Dicer are unexpectedly more profound in oocytes. dicer knockout oocytes exhibit strong phenotypes, including defects in meiotic spindle assembly and chromosome condensation, inability to complete even the first cell division, and broad alterations across the transcriptome (Murchison et al., 2007; Tang et al., 2007). While this might plausibly reflect early and broad roles for maternally inherited miRNAs, attempts to identify a signature of miRNA target site enrichment in transcripts upregulated in dicer mutant oocytes notably did not succeed. Instead, evidence was obtained that TEs, especially mouse transposon (MTs) and short interspersed repetitive elements (SINEs), were deregulated in this condition (Murchison et al., 2007). These observations helped motivate the search for endo-siRNAs in oocytes, which as mentioned are generated by select TEs and complementary gene:pseudogene pairs (Tam et al., 2008; Watanabe et al., 2008). Curiously, the mRNA targets that are functionally repressed by endo-siRNAs are enriched for genes involved in microtubule dynamics, suggesting a possible connection to the spindle defects of dicer mutant oocytes.
A surprise came with the analysis of dgcr8 mutant oocytes. While these exhibited the same strong and complete loss of canonical miRNAs as dicer mutant oocytes, the loss of maternal DGCR8 was compatible with normal dynamics and morphology of oocyte maturation, and paternally rescued embryos were subsequently viable and fertile (Suh et al., 2010). Although zygotic DGCR8 is obviously required for embryonic development, due to general roles for miRNAs, maternal-zygotic dgcr8 mutants developed normally until the blastocyst stage. Absent morphological defects, a molecular signature of dgcr8 loss might still have been detectable. However, the transcriptomes of wildtype and dgcr8−/− oocytes were essentially identical, with a scant three transcripts differing significantly (one being the floxed dgcr8 transcript itself). This suggested that miRNAs are dispensable for oocyte maturation and early mammalian development. Microarray analysis showed that known mirtron and endo-shRNA seeds were not enriched amongst deregulated transcripts in dicer−/− oocytes, pointing to endo-siRNAs as likely the key small RNA regulators in this setting.
Explicit tests of regulatory activity of miRNAs during oocyte maturation provided another surprise. The accumulation of mature miRNAs is dynamic during this process (Tang et al., 2007), and their function was reflected by the repression of reporters bearing perfectly-matched sites to abundant miRNAs, presumably by slicing. However, their capacity to repress bulged sensors was progressively lost during oocyte maturation, as was the localization of miRNA-targeted transcripts to P-bodies (Ma et al., 2010). Understanding how general miRNA effector activity might be antagonized is a challenge for the future, but the retention of slicing in oocytes implies a selective defect in Argonaute-associated effectors. One wonders whether aberrant activity of such a miRNA-suppressing pathway in later development may underlie disease or cancer.
mRNA cleavage by Drosha: direct regulation of dgcr8, and of other mRNAs?
A current mystery in miRNA biogenesis regards how Drosha/DGCR8 complex specifically recognizes pri-miRNA substrates. The main determinant reported is that DGCR8 recognizes the junction between the single-stranded and double-stranded region of the pri-miRNA hairpin base (Han et al., 2006). However, as presumably millions of transcript structures juxtapose single-stranded and double-stranded regions, it is unclear how pri-miRNAs are selectively identified.
Perhaps Drosha is not entirely selective for miRNAs. Bacterial RNase III enzymes mature ribosomal RNA, and yeast RNase III not only shares this activity but also matures certain snRNAs and snoRNAs (Drider and Condon, 2004). Consistent with this, mammalian Drosha was reported to be involved in ribosomal RNA biogenesis (Fukuda et al., 2007; Wu et al., 2000). Note, though, that defective rRNA processing was not observed following drosha knockout in the lymphoid system (Chong et al., 2008).
As the depth of sequencing catalogs increases, small RNA reads mapped to mRNA hairpins have begun to emerge (Berezikov et al., 2011). The best-characterized mRNA target of Drosha happens to encode its cofactor DGCR8. Here, hairpins within its 5′ UTR and coding region are cleaved by Drosha, yielding a low level of small RNAs. However, bulk dgcr8 “pre-miRNA” hairpins are not destined for miRNA production, since cell fractionation showed them to be nuclearly restricted (Han et al., 2009). Instead, the main function of dgcr8 cleavage is to repress accumulation of DGCR8 protein (Figure 3A). Reciprocally, Drosha protein is unstable in the absence of DGCR8. Together, cross-regulation tunes their respective levels for appropriate heterodimer function (Han et al., 2009; Triboulet et al., 2009). Cleavage of dgcr8 5′ UTR hairpins by Drosha is conserved in D. melanogaster (Han et al., 2009; Kadener et al., 2009), indicating that it is an ancient regulatory strategy.
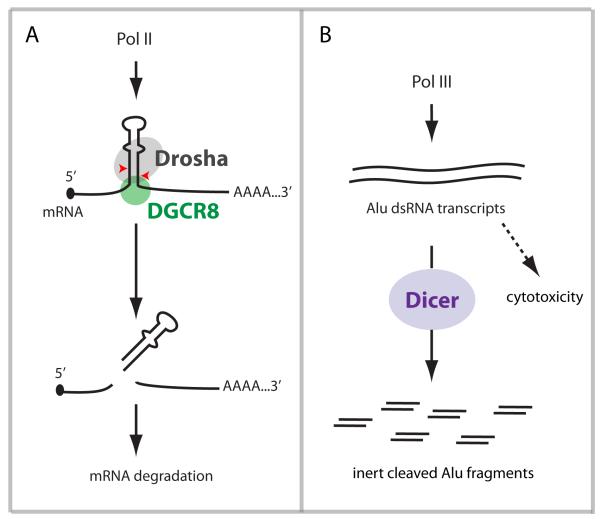
Figure 3. Direct cleavage of RNA substrates by RNase III enzymes.
RNase III enzymes may have regulatory roles that are independent of miRNA production. (A) mRNAs bearing short hairpin structures in untranslated or coding regions can be recognized and cleaved by Drosha/DGCR8, resulting in mRNA destabilization. The best-characterized example of this is cleavage of the dgcr8 mRNA by Drosha. (B) The pol III-transcribed long double-stranded Alu transcripts are toxic to cells unless they are cleaved by Dicer to short, inert fragments. In addition, direct dicing of viral replication intermediates may contribute to antiviral defense in Drosophila.
With this precedent, one may wonder whether Drosha has a more general role in mRNA cleavage. One study concluded that dgcr8 is a fairly specific Drosha mRNA target (Shenoy and Blelloch, 2009). However, another study reported certain stronger phenotypes upon conditional knockout of drosha during T-cell development, compared to dicer (Chong et al., 2010). Amongts transcripts uniquely upregulated in DN3 cells upon drosha knockout, a number bore potentially structured regions that generated ~20-25 nt small RNA reads and could be cleaved by Drosha in vitro (Chong et al., 2010). At present, such mRNA cleavage has not been causally linked to phenotypes. Moreover, observed changes in gene expression may have arisen from differential representation of celltypes, since substantial canonical miRNAs were still sequenced in the DN3 knockouts of drosha and dicer (Chong et al., 2010). Still, “degradome” sequencing of mRNA fragments bearing 5′ phosphates revealed Drosha-dependent, Ago2-independent cleaved mRNAs that may include direct Drosha targets (Karginov et al., 2010). Finally with Kaposi’s Sarcoma-associated Herpesvirus, Drosha cleavage of miRNAs located in the 3′ UTR of the viral transcript KapB downregulates this mRNA in cis, independent of activity of the miRNAs as trans-regulators (Lin and Sullivan, 2011). It will be a challenge for the future to clarify the extent to which mRNA cleavage by Drosha influences gene expression, and if so, how these targets are selected appropriately.
dicer-specific function in macular degeneration: direct dicing of Alu RNAs?
If Drosha can regulate messages by direct cleavage, one may wonder whether Dicer might do the same. A recent study of human patients with geographic atrophy (GA), an age-related macular degeneration disease of the retinal pigmented epithelium (RPE), showed reduction of dicer mRNA and protein but little change in other core miRNA pathway components (Kaneko et al., 2011). This led them to systematically analyze conditional knockouts for most of the major miRNA pathway members (drosha, dgcr8, dicer, ago2, ago1, ago3, ago4 and tarbp2). Impressively, only ablation of dicer recapitulated the GA phenotype, suggesting that this disease is not due to general alteration in miRNA activity.
Using an antibody that recognizes long dsRNA, the authors found that dsRNA accumulates in human GA eyes, as well as in human RPE cells and mouse eyes depleted of Dicer. The dsRNA population was cloned and found to include Alu repeat RNAs of ~300 nt. Their accumulation was a specific property of Dicer-depleted cells and not other genotypes, and loss of Dicer deregulated Alu but not other retrotransposon transcripts. In fact, functional tests demonstrated that accumulation of Alu RNAs is cytotoxic and that injection of in vitro transcribed Alu induced GA. On the other hand, injection of Dicer-cleaved Alu small RNAs, or other non-coding RNAs, had no effect. Together, these tests supported a model in which Dicer exerts a miRNA-independent function in cleaving Alu dsRNAs to render them inert (Kaneko et al., 2011) (Figure 3B).
Although no single Ago gene (including Ago2-Slicer) is needed to prevent GA, it remains to be seen whether cleaved Alu siRNAs also function as Ago-loaded species. However, the notion of direct dicing as a biological function bears similarity to studies of persistent viral infection of Drosophila cultured cells. It is well-established that a Dicer-2/AGO2-mediated siRNA pathway executes antiviral defense in flies (Wang et al., 2006). However, bulk viral siRNAs generated by Dicer-2 in latently infected cells appear to associate poorly with effector complexes. Those that are successfully loaded enter AGO2 (Czech et al., 2008), but bulk viral siRNAs did not associate with either AGO2 or AGO1 (Flynt et al., 2009). One interpretation of these findings is that direct dicing of the viral replication intermediate plays a substantial role in controlling persistent viral infection of Drosophila cells.
Can the theme of non-canonical substrates of core miRNA factors be extended beyond Drosha and Dicer? Mammalian Exp-5 was recently reported to directly transport dicer mRNA to the cytoplasm (Bennasser et al., 2011). Given the topology of preferred Exp-5 binding to hairpins with 3′ overhangs, it is not clear how Exp-5 binds dicer transcripts. Nevertheless, this example suggests it may be worth considering whether Exp-5 transports other non-miRNA substrates.
ago2 vs. dicer mutants in hematopoietic system: Slicer-mediated functions?
The elucidation of miR-451 biogenesis raises a new wrinkle, in that Dicer knockout cells do not universally remove all miRNAs. Phenotypes of knock-in mice bearing the Ago2 Slicer-deficient allele are provocative, including full perinatal lethality and prominent anemia (Cheloufi et al., 2010). While miR-451 is clearly deficient in this genetic condition, the loss of miR-451 per se does not explain the Ago2-Slicer mutant phenotype. Instead, mir-451 deletion mutants are fully viable and exhibit only mild anemia, although this presents a more substantial problem upon oxidative challenge (Patrick et al., 2010; Rasmussen et al., 2010; Yu et al., 2010). This may be taken to support the existence of essential miRNA-directed cleavage events. Certainly there are a number of documented endogenous mRNA cleavages programmed by miRNAs, although none are known to be required for hematopoiesis or viability (Karginov et al., 2010; Shin et al., 2010; Yekta et al., 2004).
Additional scenarios for the requirement of Ago2 Slicer activity include that it may process other Dicer-independent miRNAs yet to be identified, is required more generally for miRNA biogenesis (Diederichs and Haber, 2007), or potentially regulates targets independently of mature miRNA guides. Consistent with the latter possibility, Ago2 is capable of using guides larger than mature siRNAs/miRNAs to direct target cleavage (Tan et al., 2009). Very recently, the direct association of Ago2 with murine transcripts was studied genomewide, and compared between normal and dicer−/− ES cells (Leung et al., 2011). As expected, a dominant Dicer-dependent signature of target sites complementary to the seeds of highly expressed ES miRNAs was seen. Less expected, though, was the observation of Dicer-independent Ago2 targeting signatures, which included G-rich motifs. Although the mechanistic significance of this remains to be explored further, evidence was presented that these motifs correlate with preferential conservation and target de-repression in ago2−/− ES cells.
These data may support the notion that Ago2 is targeted to certain transcripts independently of mature miRNAs, perhaps reflecting intrinsic RNA affinity, or perhaps guided by RNAs independent of Dicer. Most profilings of Argonaute-associated RNAs have focused exclusively on <30 nt species and mRNAs. While miRNA-sized species are the predominant contents of Ago proteins in the short RNA fraction, this could be biased by the fact that their size is very stereotyped. In principle, larger heterogenously-sized cargoes might not be appear distinct from background in total RNA labelings. Therefore, it may be informative to broaden sequencing surveys to search for intermediate-sized RNAs that might associate with Ago proteins.
Drosophila ago1/ago2 mutants vs. pasha/dcr-1 mutants in the olfactory system
To date, most efforts to infer miRNA functions from miRNA pathway components have focused on the biogenesis factors, but certainly analysis of Ago mutants should be informative. For example, Drosophila ago1/ago2 double mutant embryos exhibited phenotypes more severe than either single mutant (Meyer et al., 2006). There are four mammalian Argonautes, but their genetic analysis is aided by the close linkage of ago1/3/4, which can be deleted in a single event. Such triple knockout ES cells maintain normal levels of miRNA-mediated silencing, indicating that Ago2 is capable of supporting siRNA and miRNA activity (Su et al., 2009). Deletion of ago2 in this background yielded quadruple ago1-4 knockout cells that exhibit strong growth defects (Su et al., 2009), although such mutant cells remain to be examined in intact mice.
In principle, as removal of the effector proteins should effectively abolish all mi/siRNAs, regardless of their biogenesis history, one might intuit that this situation should be “worse” than any individual biogenesis factor. However, a few years ago, a genetic screen in Drosophila olfactory projection neurons revealed two mutants exhibiting a distinct set of dendritic and axonal mistargeting phenotypes (Berdnik et al., 2008). These mutants disrupted dcr-1 and pasha (dgcr8), implying a common function of the miRNA pathway in controlling olfactory wiring. Surprisingly, neither ago1[k08121] nor ago2[414], as single or double mutants, recapitulated the olfactory system defect. It is possible that this ago1 mutant is not null; however, the insertion alleles isolated for dcr-1 and pasha were not necessarily null either. Moreover, ago1[k08121] is known to have strongly decreased mRNA and protein levels (Kataoka et al., 2001; Okamura et al., 2004). Whether this implies that a very small amount of Ago effector complex suffices for the morphogenesis of projection neurons, or whether there are Ago-independent activities of the miRNA pathway, remains to be clarified.
Does loss of core miRNA machinery reflect the cumulative phenotypes of individual miRNA mutants?
These many examples provide substantial evidence that no single core miRNA pathway component is essential to generate all animal miRNAs. However, a final consideration regards the very expectation that core biogenesis mutants should reflect the cumulative phenotypes of all individual miRNA mutants. In Drosophila, induction of dcr-1 and pasha null clones during wing development results in blistering of the adult wing, but otherwise the integrity of the wing margin remains fully intact (Bejarano et al., 2010). This might be taken as evidence that miRNAs are not required to specify the wing margin, were it not for the fact that deletion of a single miRNA--mir-9a--alone confers fully penetrant wing notching (Li et al., 2006).
A trivial explanation might be that residual proteins or miRNAs in mutant clones suffice for normal wing development. However, in vivo sensor assays showed that miR-9a function was lost to a similar extent in dcr-1, pasha and mir-9a clones (Bejarano et al., 2010). Moreover, similar mutant clones of mir-9a still yielded wing notching. Perhaps most compelling were observations using a 3′ UTR sensor for dLMO, a key miR-9a target during wing development (Bejarano et al., 2010; Biryukova et al., 2009): the dLMO sensor was de-repressed in dcr-1, pasha and mir-9a clones, but endogenous dLMO protein was de-repressed only in mir-9a clones (Bejarano et al., 2010).
The simplest interpretation is that there exist other miRNAs whose activity is antagonistic to miR-9a during wing development. More generally, as a majority of animal transcripts may be targeted by miRNAs, and many processes are typically under positive and negative control, it may not be so unexpected for the loss of miRNA biogenesis factors not to phenocopy the loss of specific miRNAs. For example, both positive and negative regulators of peripheral neurogenesis contain target sites for the same miRNAs (Lai et al., 1998; Lai and Posakony, 1997), perhaps explaining why phenotypes associated with loss of miRNA binding sites from individual neural regulators are not recapitulated by dcr-1 clones. In fact during early zebrafish development, both positive and negative regulators of Nodal signaling are repressed by miR-430, such that the inhibition of target sites from individual transcripts is more severe than loss of miR-430 or Dicer (Choi et al., 2007). Therefore, the absence of phenotypes in core miRNA pathway mutants cannot reliably be taken to mean the absence of compelling miRNA functions in a given setting.
Conclusion and Outlook
The evolutionary flexibility of small RNA pathways is clearly evident from the diversity of animal miRNA and siRNA pathways, and further illustrated by recent studies in fungal systems. For example, certain budding yeasts encode a clear Argonaute ortholog but lack a recognizable Dicer. Detailed investigation of S. castellii revealed that an orphan RNase III enzyme executes dicing, even though this protein has only one RNase III domain instead of the two seen in canonical Dicers, and entirely lacks the usual helicase and PAZ domains (Drinnenberg et al., 2009). IP cloning from the Neurospora crassa Argonaute QDE-2 revealed a diversity of miRNA-like species (Lee et al., 2010), at least one of which is Dicer-independent but instead requires the RNase III enzyme MRPL3. More surprisingly, some siRNA loci in Neurospora require neither Dicer nor MRPL3, implying yet another nuclease in their biogenesis. Finally, the study of Ago1 complexes from S. pombe dcr1 mutants revealed a system of Dicer-independent “primal RNAs” that prime the RNAi machinery (Halic and Moazed, 2010).
Going even further “out of the box”, cell death in C. elegans was found to involve a caspase-dependent cleavage of Dicer, converting it from an RNase into a DNase that fragments chromosomes (Nakagawa et al., 2010). This may cause one to wonder whether other miRNA/RNAi factors may have DNA-directed functions. For example, some archaeabacterial Argonaute proteins preferentially utilize a DNA guide strand (Yuan et al., 2005). Altogether, the collected studies suggest that small RNA researchers have not yet fully appreciated the inventiveness of Nature in defining non-canonical functions of small RNA-associated proteins, which can be incorporated into unexpected pathways and mediate unexpected biology.
Acknowledgements
We apologize to authors whose work was not cited due to length restrictions. We thank Katsutomo Okamura and Robert Blelloch for helpful discussion, and members of the Lai laboratory for work that inspired this perspective. Work in E.C.L.’s group was supported by the Burroughs Wellcome Fund, the Alfred Bressler Scholars Fund, the Starr Cancer Consortium (I3-A139) and the NIH (R01-GM083300).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Axtell MJ, Westholm JO, Lai EC. Vive la différence: biogenesis and evolution of microRNAs in plants and animals. Genome Biol. 2011;12:221–221. 213. doi: 10.1186/gb-2011-12-4-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz JE, Hsu R, Melton C, Thomas M, Ullian EM, Blelloch R. A role for noncanonical microRNAs in the mammalian brain revealed by phenotypic differences in Dgcr8 versus Dicer1 knockouts and small RNA sequencing. Rna. 2011;17 doi: 10.1261/rna.2442211. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejarano F, Smibert P, Lai EC. miR-9a prevents apoptosis during wing development by repressing Drosophila LIM-only. Dev Biol. 2010;338:63–73. doi: 10.1016/j.ydbio.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennasser Y, Chable-Bessia C, Triboulet R, Gibbings D, Gwizdek C, Dargemont C, Kremer EJ, Voinnet O, Benkirane M. Competition for XPO5 binding between Dicer mRNA, pre-miRNA and viral RNA regulates human Dicer levels. Nat Struct Mol Biol. 2011;18:323–327. doi: 10.1038/nsmb.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdnik D, Fan AP, Potter CJ, Luo L. MicroRNA processing pathway regulates olfactory neuron morphogenesis. Curr Biol. 2008;18:1754–1759. doi: 10.1016/j.cub.2008.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Chung W-J, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Berezikov E, Robine N, Samsonova A, Westholm JO, Naqvi A, Hung JH, Okamura K, Dai Q, Bortolamiol-Becet D, Martin R, et al. Deep annotation of Drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence. Genome Res. 2011;21:203–215. doi: 10.1101/gr.116657.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Biryukova I, Asmar J, Abdesselem H, Heitzler P. Drosophila mir-9a regulates wing development via fine-tuning expression of the LIM only factor, dLMO. Dev Biol. 2009;327:487–496. doi: 10.1016/j.ydbio.2008.12.036. [DOI] [PubMed] [Google Scholar]
- Bogerd HP, Karnowski HW, Cai X, Shin J, Pohlers M, Cullen BR. A mammalian herpesvirus uses noncanonical expression and processing mechanisms to generate viral MicroRNAs. Mol Cell. 2010;37:135–142. doi: 10.1016/j.molcel.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brameier M, Herwig A, Reinhardt R, Walter L, Gruber J. Human box C/D snoRNAs with miRNA like functions: expanding the range of regulatory RNAs. Nucleic Acids Res. 2011;39:675–686. doi: 10.1093/nar/gkq776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science. 2007;318:271–274. doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]
- Chong MM, Rasmussen JP, Rudensky AY, Littman DR. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. J Exp Med. 2008;205:2005–2017. doi: 10.1084/jem.20081219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong MM, Zhang G, Cheloufi S, Neubert TA, Hannon GJ, Littman DR. Canonical and alternate functions of the microRNA biogenesis machinery. Genes Dev. 2010;24:1951–1960. doi: 10.1101/gad.1953310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WJ, Agius P, Westholm JO, Chen M, Okamura K, Robine N, Leslie CS, Lai EC. Computational and experimental identification of mirtrons in Drosophila melanogaster and Caenorhabditis elegans. Genome Res. 2011;21:286–300. doi: 10.1101/gr.113050.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WJ, Okamura K, Martin R, Lai EC. Endogenous RNA interference provides a somatic defense against Drosophila transposons. Current Biology. 2008;18:795–802. doi: 10.1016/j.cub.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson N, et al. A Novel miRNA Processing Pathway Independent of Dicer Requires Argonaute2 Catalytic Activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW, Green PJ, Barton GJ, Hutvagner G. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15:2147–2160. doi: 10.1261/rna.1738409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvillion MT, Sachidanandam R, Collins K. A growth-essential Tetrahymena Piwi protein carries tRNA fragment cargo. Genes Dev. 2010;24:2742–2747. doi: 10.1101/gad.1996210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet. 2010;12:19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel J, Sachidanandam R, et al. An endogenous siRNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichs S, Haber DA. Dual Role for Argonautes in MicroRNA Processing and Posttranscriptional Regulation of MicroRNA Expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drider D, Condon C. The continuing story of endoribonuclease III. J Mol Microbiol Biotechnol. 2004;8:195–200. doi: 10.1159/000086700. [DOI] [PubMed] [Google Scholar]
- Drinnenberg IA, Weinberg DE, Xie KT, Mower JP, Wolfe KH, Fink GR, Bartel DP. RNAi in budding yeast. Science. 2009;326:544–550. doi: 10.1126/science.1176945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- Flynt AS, Chung WJ, Greimann JC, Lima CD, Lai EC. microRNA biogenesis via splicing and exosome-mediated trimming in Drosophila. Mol Cell. 2010;38:900–907. doi: 10.1016/j.molcel.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet. 2008;9:831–842. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynt AS, Liu N, Lai EC. Dicing of viral replication intermediates during silencing of latent Drosophila viruses. Proc Natl Acad Sci U S A. 2009;106:5270–5275. doi: 10.1073/pnas.0813412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T, Yamagata K, Fujiyama S, Matsumoto T, Koshida I, Yoshimura K, Mihara M, Naitou M, Endoh H, Nakamura T, et al. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat Cell Biol. 2007;9:604–611. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, Zamore PD. Endogenous siRNAs Derived from Transposons and mRNAs in Drosophila Somatic Cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Glazov EA, Cottee PA, Barris WC, Moore RJ, Dalrymple BP, Tizard ML. A microRNA catalog of the developing chicken embryo identified by a deep sequencing approach. Genome Res. 2008;18:957–964. doi: 10.1101/gr.074740.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halic M, Moazed D. Dicer-independent primal RNAs trigger RNAi and heterochromatin formation. Cell. 2010;140:504–516. doi: 10.1016/j.cell.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Han J, Pedersen JS, Kwon SC, Belair CD, Kim YK, Yeom KH, Yang WY, Haussler D, Blelloch R, Kim VN. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci U S A. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S, Rodriguez J, Abruzzi KC, Khodor YL, Sugino K, Marr MT, 2nd, Nelson S, Rosbash M. Genome-wide identification of targets of the drosha-pasha/DGCR8 complex. RNA. 2009;15:537–545. doi: 10.1261/rna.1319309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko H, Dridi S, Tarallo V, Gelfand BD, Fowler BJ, Cho WG, Kleinman ME, Ponicsan SL, Hauswirth WW, Chiodo VA, et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011 doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes & Development. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karginov FV, Cheloufi S, Chong MM, Stark A, Smith AD, Hannon GJ. Diverse endonucleolytic cleavage sites in the mammalian transcriptome depend upon microRNAs, Drosha, and additional nucleases. Mol Cell. 2010;38:781–788. doi: 10.1016/j.molcel.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka Y, Takeichi M, Uemura T. Developmental roles and molecular characterization of a Drosophila homologue of Arabidopsis Argonaute1, the founder of a novel gene superfamily. Genes Cells. 2001;6:313–325. doi: 10.1046/j.1365-2443.2001.00427.x. [DOI] [PubMed] [Google Scholar]
- Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada T, Siomi MC, Siomi H. Drosophila endogenous small RNAs bind to Argonaute2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, Jensen K, Cobb BS, Merkenschlager M, Rajewsky N, Rajewsky K. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132:860–874. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- Lai EC, Burks C, Posakony JW. The K box, a conserved 3′ UTR sequence motif, negatively regulates accumulation of Enhancer of split Complex transcripts. Development. 1998;125:4077–4088. doi: 10.1242/dev.125.20.4077. [DOI] [PubMed] [Google Scholar]
- Lai EC, Posakony JW. The Bearded box, a novel 3′ UTR sequence motif, mediates negative post-transcriptional regulation of Bearded and Enhancer of split Complex gene expression. Development. 1997;124:4847–4856. doi: 10.1242/dev.124.23.4847. [DOI] [PubMed] [Google Scholar]
- Lai EC, Tomancak P, Williams RW, Rubin GM. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4:R42.41–R42.20. doi: 10.1186/gb-2003-4-7-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC, Li L, Gu W, Xue Z, Crosthwaite SK, Pertsemlidis A, Lewis ZA, Freitag M, Selker EU, Mello CC, Liu Y. Diverse pathways generate microRNA-like RNAs and Dicer-independent small interfering RNAs in fungi. Mol Cell. 2010;38:803–814. doi: 10.1016/j.molcel.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Young AG, Bhutkar A, Zheng GX, Bosson AD, Nielsen CB, Sharp PA. Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nat Struct Mol Biol. 2011 doi: 10.1038/nsmb.1991. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang F, Lee JA, Gao FB. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes & Development. 2006;20:2793–2805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Burge CB. A computational analysis of sequence features involved in recognition of short introns. Proc Natl Acad Sci U S A. 2001;98:11193–11198. doi: 10.1073/pnas.201407298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YT, Sullivan CS. Expanding the role of Drosha to the regulation of viral gene expression. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1105799108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Flemr M, Stein P, Berninger P, Malik R, Zavolan M, Svoboda P, Schultz RM. MicroRNA activity is suppressed in mouse oocytes. Curr Biol. 2010;20:265–270. doi: 10.1016/j.cub.2009.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina MF, Joshi S. RNA-polymerase III-driven expression cassettes in human gene therapy. Curr Opin Mol Ther. 1999;1:580–594. [PubMed] [Google Scholar]
- Meyer WJ, Schreiber S, Guo Y, Volkmann T, Welte MA, Muller HA. Overlapping functions of argonaute proteins in patterning and morphogenesis of Drosophila embryos. PLoS Genet. 2006;2:e134. doi: 10.1371/journal.pgen.0020134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci U S A. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Stein P, Xuan Z, Pan H, Zhang MQ, Schultz RM, Hannon GJ. Critical roles for Dicer in the female germline. Genes Dev. 2007;21:682–693. doi: 10.1101/gad.1521307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa A, Shi Y, Kage-Nakadai E, Mitani S, Xue D. Caspase-dependent conversion of dicer ribonuclease into a death-promoting deoxyribonuclease. Science. 2010;328:327–334. doi: 10.1126/science.1182374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Balla S, Martin R, Liu N, Lai EC. Two distinct mechanisms generate endogenous siRNAs from bidirectional transcription in Drosophila. Nat Struct Mol Biol. 2008a;15:581–590. doi: 10.1038/nsmb.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Chung W-J, Ruby JG, Guo H, Bartel DP, Lai EC. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008b;453:803–806. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Lai EC. Endogenous small interfering RNAs in animals. Nat Rev Mol Cell Biol. 2008;9:673–678. doi: 10.1038/nrm2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono M, Scott MS, Yamada K, Avolio F, Barton GJ, Lamond AI. Identification of human miRNA precursors that resemble box C/D snoRNAs. Nucleic Acids Res. 2011;39:3879–3891. doi: 10.1093/nar/gkq1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddison PJ, Caudy AA, Hannon GJ. Stable suppression of gene expression by RNAi in mammalian cells. Proc Natl Acad Sci U S A. 2002;99:1443–1448. doi: 10.1073/pnas.032652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick DM, Zhang CC, Tao Y, Yao H, Qi X, Schwartz RJ, Jun-Shen Huang L, Olson EN. Defective erythroid differentiation in miR-451 mutant mice mediated by 14-3-3zeta. Genes Dev. 2010;24:1614–1619. doi: 10.1101/gad.1942810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T. Regulatory RNAs derived from transfer RNA? RNA. 2010;16:1865–1869. doi: 10.1261/rna.2266510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson H, Kvist A, Vallon-Christersson J, Medstrand P, Borg A, Rovira C. The non-coding RNA of the multidrug resistance-linked vault particle encodes multiple regulatory small RNAs. Nat Cell Biol. 2009;11:1268–1271. doi: 10.1038/ncb1972. [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, et al. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- Rasmussen KD, Simmini S, Abreu-Goodger C, Bartonicek N, Di Giacomo M, Bilbao-Cortes D, Horos R, Von Lindern M, Enright AJ, O’Carroll D. The miR-144/451 locus is required for erythroid homeostasis. J Exp Med. 2010;207:1351–1358. doi: 10.1084/jem.20100458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese TA, Xia J, Johnson LS, Zhou X, Zhang W, Virgin HW. Identification of novel microRNA-like molecules generated from herpesvirus and host tRNA transcripts. J Virol. 2010;84:10344–10353. doi: 10.1128/JVI.00707-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraiya AA, Wang CC. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 2008;4:e1000224. doi: 10.1371/journal.ppat.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer LJ, Frank R, Rossi JJ. Optimization and characterization of tRNA-shRNA expression constructs. Nucleic Acids Res. 2007;35:2620–2628. doi: 10.1093/nar/gkm103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MS, Avolio F, Ono M, Lamond AI, Barton GJ. Human miRNA precursors with box H/ACA snoRNA features. PLoS Comput Biol. 2009;5:e1000507. doi: 10.1371/journal.pcbi.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro JS, Varble A, Pham AM, Tenoever BR. Noncanonical cytoplasmic processing of viral microRNAs. RNA. 2010;16:2068–2074. doi: 10.1261/rna.2303610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy A, Blelloch R. Genomic analysis suggests that mRNA destabilization by the microprocessor is specialized for the auto-regulation of Dgcr8. PLoS ONE. 2009;4:e6971. doi: 10.1371/journal.pone.0006971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Hendrix D, Levine M, Haley B. A distinct class of small RNAs arises from pre-miRNA-proximal regions in a simple chordate. Nat Struct Mol Biol. 2009;16:183–189. doi: 10.1038/nsmb.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin C, Nam JW, Farh KK, Chiang HR, Shkumatava A, Bartel DP. Expanding the MicroRNA Targeting Code: Functional Sites with Centered Pairing. Mol Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert P, Lai EC. Lessons from microRNA mutants in worms, flies and mice. Cell Cycle. 2008:7. doi: 10.4161/cc.7.16.6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H, Trombly MI, Chen J, Wang X. Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Genes Dev. 2009;23:304–317. doi: 10.1101/gad.1749809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh N, Baehner L, Moltzahn F, Melton C, Shenoy A, Chen J, Blelloch R. MicroRNA Function Is Globally Suppressed in Mouse Oocytes and Early Embryos. Current Biology. 2010;20:271–277. doi: 10.1016/j.cub.2009.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda P, Stein P, Hayashi H, Schultz RM. Selective reduction of dormant maternal mRNAs in mouse oocytes by RNA interference. Development. 2000;127:4147–4156. doi: 10.1242/dev.127.19.4147. [DOI] [PubMed] [Google Scholar]
- Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS. Small RNAs derived from snoRNAs. RNA. 2009;15:1233–1240. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, Hannon GJ. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan GS, Garchow BG, Liu X, Yeung J, Morris J.P.t., Cuellar TL, McManus MT, Kiriakidou M. Expanded RNA-binding activities of mammalian Argonaute 2. Nucleic Acids Res. 2009;37:7533–7545. doi: 10.1093/nar/gkp812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Kaneda M, O’Carroll D, Hajkova P, Barton SC, Sun YA, Lee C, Tarakhovsky A, Lao K, Surani MA. Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 2007;21:644–648. doi: 10.1101/gad.418707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triboulet R, Chang HM, Lapierre RJ, Gregory RI. Post-transcriptional control of DGCR8 expression by the Microprocessor. RNA. 2009;15:1005–1011. doi: 10.1261/rna.1591709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XH, Aliyari R, Li WX, Li HW, Kim K, Carthew R, Atkinson P, Ding SW. RNA interference directs innate immunity against viruses in adult Drosophila. Science. 2006;312:452–454. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet. 2008;40:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Wu H, Xu H, Miraglia LJ, Crooke ST. Human RNase III is a 160-kDa protein involved in preribosomal RNA processing. J Biol Chem. 2000;275:36957–36965. doi: 10.1074/jbc.M005494200. [DOI] [PubMed] [Google Scholar]
- Yang JS, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, Papapetrou EP, Sadelain M, O’Carroll D, Lai EC. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc Natl Acad Sci U S A. 2010;107:15163–15168. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Tutton S, Pierce E, Yoon K. Specific double-stranded RNA interference in undifferentiated mouse embryonic stem cells. Mol Cell Biol. 2001;21:7807–7816. doi: 10.1128/MCB.21.22.7807-7816.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- Yi R, O’Carroll D, Pasolli HA, Zhang Z, Dietrich FS, Tarakhovsky A, Fuchs E. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat Genet. 2006;38:356–362. doi: 10.1038/ng1744. [DOI] [PubMed] [Google Scholar]
- Yi R, Pasolli HA, Landthaler M, Hafner M, Ojo T, Sheridan R, Sander C, O’Carroll D, Stoffel M, Tuschl T, Fuchs E. DGCR8-dependent microRNA biogenesis is essential for skin development. Proc Natl Acad Sci U S A. 2009;106:498–502. doi: 10.1073/pnas.0810766105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, dos Santos CO, Zhao G, Jiang J, Amigo JD, Khandros E, Dore LC, Yao Y, D’Souza J, Zhang Z, et al. miR-451 protects against erythroid oxidant stress by repressing 14-3-3zeta. Genes Dev. 2010;24:1620–1633. doi: 10.1101/gad.1942110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan YR, Pei Y, Ma JB, Kuryavyi V, Zhadina M, Meister G, Chen HY, Dauter Z, Tuschl T, Patel DJ. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol Cell. 2005;19:405–419. doi: 10.1016/j.molcel.2005.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]