Summary
In response to stress, eukaryotic cells accumulate mRNAs and proteins at discrete sites, or foci, in the cytoplasm. However, the mechanisms regulating foci formation, and the biological function of the larger ribonucleoprotein (RNP) assemblies, remain poorly understood. Here, we show that the cAMP-dependent protein kinase (PKA) in Saccharomyces cerevisiae is a key regulator of the assembly of Processing-bodies (P-bodies), an RNP complex implicated in mRNA processing and translation. The data suggest that PKA specifically inhibits the formation of the larger P-body aggregates by directly phosphorylating Pat1, a conserved constituent of these foci that functions as a scaffold during the assembly process. Finally, we present evidence indicating that P-body foci are required for the long-term survival of stationary phase cells. This work therefore highlights the general relevance of RNP foci in quiescent cells, and provides a framework for the study of the many RNP assemblies that form in eukaryotic cells.
Introduction
When an essential nutrient or growth factor is lacking, eukaryotic cells arrest growth and may enter into a distinct resting state, known as G0 or stationary phase (Gray et al., 2004; Pardee, 1989). A number of characteristic changes are associated with this transition, including a decreased rate of metabolism and a heightened resistance to stress (Gray et al., 2004; Werner-Washburne et al., 1993). These growth-arrested cells can also accumulate particular proteins and mRNAs at discrete sites, or foci, in the cytoplasm (Narayanaswamy et al., 2009; Teixeira et al., 2005). Two of the best-characterized of these RNP foci are the P-bodies and stress granules that have been suggested to play a role in mRNA decay and/or storage (Anderson and Kedersha, 2008, 2009; Buchan et al., 2008; Eulalio et al., 2007a; Lui et al., 2010). These RNP particles are found in all eukaryotes, from the single-celled yeasts to humans, suggesting that their biological functions have been conserved through evolution.
P-bodies were initially identified as cytoplasmic foci that contain non-translating mRNAs and proteins important for the decapping and degradation of these transcripts (Balagopal and Parker, 2009; Eulalio et al., 2007a). For example, the Dcp1-Dcp2 decapping enzyme and the Xrn1 exonuclease are both associated with P-bodies (Bashkirov et al., 1997; Cougot et al., 2004; Eystathioy et al., 2003; Ingelfinger et al., 2002; Sheth and Parker, 2003; van Dijk et al., 2002). The maintenance of these foci requires the continued presence of the associated mRNA and there appears to be a dynamic equilibrium between P-body formation and active translation (Anderson and Kedersha, 2009; Balagopal and Parker, 2009; Brengues et al., 2005; Kedersha et al., 2005; Teixeira et al., 2005). Based on these data, the assembly of the large P-body foci has been proposed to occur in two distinct stages (Figure S1) (Decker et al., 2007; Franks and Lykke-Andersen, 2008). In the first step, mRNAs associate with the protein constituents of P-bodies to form RNP monomers. Several of these P-body proteins likely contribute to the translationally-repressed state of these mRNAs (Coller and Parker, 2005; Franks and Lykke-Andersen, 2008). The P-body monomers then aggregate in the second stage to form the larger cytoplasmic foci detected in stressed cells. This aggregation may be mediated by the prion-like domains present in a number of P-body proteins (Decker et al., 2007; Gilks et al., 2004; Reijns et al., 2008). However, the physiological role of the larger foci remains unclear as mRNA decapping and decay, and the inhibition of translation, have all been found to proceed normally in cells lacking these larger assemblies (Decker et al., 2007; Eulalio et al., 2007b; Stoecklin et al., 2006). Moreover, a recent study has suggested that decapping and decay occur while the mRNA is still associated with actively translating ribosomes (Hu et al., 2009).
A variety of stress conditions induce P-body formation but the signaling pathways mediating these responses have not been identified. In this study, we examined the signaling requirements for P-body formation in response to glucose deprivation in S. cerevisiae. These RNP structures form rapidly upon glucose starvation and dissipate upon re-addition of this carbon source (Teixeira et al., 2005). The work here demonstrates that the cAMP-dependent protein kinase (PKA) is a key regulator of this P-body assembly process. In contrast, the inactivation of other pathways important for nutrient sensing, including the Target of Rapamycin Complex 1 (TORC1) and AMP-activated protein kinase (AMPK) pathways, did not influence the formation of these RNP structures (Hedbacker and Carlson, 2008; Wullschleger et al., 2006). The results indicate that these effects of PKA are due, at least in part, to the direct phosphorylation of Pat1. This protein is a conserved core constituent of eukaryotic P-bodies that has been suggested to act as a scaffolding molecule during the assembly process (Braun et al., 2010; Marnef and Standart, 2010; Pilkington and Parker, 2008). Finally, we present evidence that suggests that the larger P-body foci are required for the long-term survival of stationary phase cells. In all, this work identifies a key regulator of P-body assembly in S. cerevisiae, a primary target of this signaling pathway in the P-body machinery and a potential biological function for the larger assemblies in quiescent cells.
Results
The inactivation of PKA signaling was both necessary and sufficient for P-body formation
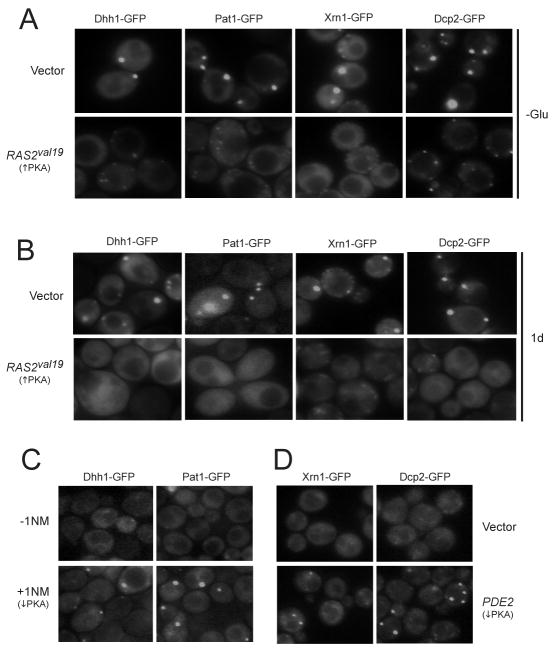
Since PKA is a key effector of glucose signaling in S. cerevisiae, we tested whether this pathway had a role in the regulation of P-body assembly (Santangelo, 2006; Slattery et al., 2008; Zaman et al., 2009; Zaman et al., 2008). Four different GFP-tagged reporters for P-body foci were used for these studies. In addition to Dcp2, Xrn1 and Pat1, we examined the localization of Dhh1, an RNA helicase that is also associated with P-bodies (Teixeira et al., 2005). Using these reporters, we found large P-body foci in the cytoplasm of wild-type cells following either an acute or gradual deprivation of glucose (Figure 1A, B). In general, one, or perhaps two, such foci were observed in a fraction of the glucose-starved cells. Interestingly, the number of these large foci was diminished significantly in cells with constitutively-elevated levels of PKA activity (Figure 1A, B; 2SA-D). PKA signaling was up-regulated either moderately by the introduction of the dominant positive RAS2val19 allele, or more dramatically by deletion of the BCY1 locus that encodes the regulatory subunit of PKA (Toda et al., 1987; Toda et al., 1985). In this yeast, PKA activity is positively regulated by the Ras proteins, Ras1 and Ras2 (Toda et al., 1985). We found that the severity of the P-body defect was proportional to the PKA activity present. For example, large P-body foci were rarely detected in bcy1Δ cells (Fig. S2A, B). Instead a disperse cytoplasmic GFP pattern was generally observed and the number of cells with foci was reduced by more than ten-fold for each reporter tested. In the presence of the RAS2val19 allele, the number of cells containing large P-body foci was reduced by three- to seven-fold depending upon the reporter being examined (Figure 1A, B; S2C, D). In addition, RAS2val19 cells often contained numerous, smaller foci in their cytoplasm (see Figure 1A). Finally, we found that diminished signaling through the PKA pathway was sufficient to induce P-body formation in glucose-replete conditions (Figure 1C, D). PKA activity was lowered either by inactivating an analog-sensitive version of PKA or by the over-expression of the cAMP phosphodiesterase, Pde2 (Bishop et al., 2001; Sass et al., 1986; Stephan et al., 2009). Therefore, the inactivation of PKA activity was both a necessary and sufficient condition for P-body formation in S. cerevisiae.
Figure 1.
P-body formation was regulated by Ras/PKA signaling activity.
(A) Elevated signaling through the Ras/PKA pathway inhibited P-body formation in response to an acute glucose starvation. Strains expressing GFP-tagged versions of Dcp2, Dhh1, Pat1 and Xrn1 were transformed with either a vector control or a MET3-RAS2val19 plasmid. Cells were grown to mid-log phase in SC-glucose medium and expression from the RAS2val19 locus was induced by transferring cells to a medium lacking methionine for 2 hr. The cells were then starved for glucose for 15 min and visualized by fluorescence microscopy.
(B) The formation of P-bodies in early stationary phase was inhibited by elevated Ras/PKA signaling. Wild-type yeast with either a control vector or a MET3-RAS2val19 plasmid were grown for 24 hr in an SC-glucose medium lacking methionine.
(C, D) The down-regulation of PKA activity was sufficient to induce P-body formation in glucose-replete conditions. In C, cells containing an analog-sensitive allele of TPK1, tpk1-as, were treated with 5 μM 1NM-PP1 for 15 min. In D, the cells were transformed with either a control vector or a high-copy PDE2 plasmid.
P-body formation was not influenced by either TORC1 or AMPK signaling activity
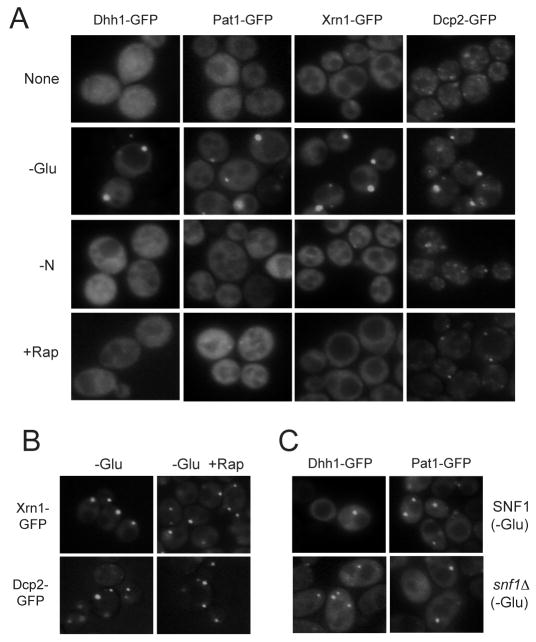
The TORC1 complex is sensitive to the drug, rapamycin, and has been implicated in the growth response to nutrients in a number of eukaryotes, including S. cerevisiae (De Virgilio and Loewith, 2006; Sengupta et al., 2010; Wullschleger et al., 2006). We therefore investigated whether the inactivation of TORC1 would also result in P-body formation. In contrast to the above results with PKA, we did not observe P-bodies in cells that were treated with rapamycin (Figure 2A; S3A). Moreover, P-body foci were not detected in cells that were deprived of a nitrogen source, a condition that has been shown to result in diminished TORC1 signaling (Figure 2A) (Sengupta et al., 2010; Wullschleger et al., 2006). Neither rapamycin treatment nor the lack of a nitrogen source interfered with the formation of P-bodies induced by glucose starvation (Figure 2B; S3B). However, as noted previously, P-body formation was disrupted by the presence of the translation elongation inhibitor, cycloheximide (Figure S3C). Finally, we found no significant decrease in the number or size of P-body foci in cells lacking the AMPK ortholog, Snf1 (Figure 2C). Altogether, these data indicated that P-body assembly in S. cerevisiae was regulated by PKA signaling, but not TORC1 or Snf1 activity.
Figure 2.
P-body formation was not influenced by either TORC1 or Snf1 signaling activity.
(A) Neither nitrogen deprivation or rapamycin treatment resulted in P-body formation. Strains expressing GFP-tagged versions of the indicated P-body markers were grown to mid-log phase in SC-glucose medium. The cells were then transferred to a medium lacking either a nitrogen source (-N) or glucose (-Glu), or treated with 200 ng/ml rapamycin (+Rap) for 15 min.
(B) The presence of rapamycin did not interfere with the P-body formation induced by glucose starvation. Cells were grown to mid-log phase in an SC-glucose medium and then transferred for 15 min to a medium lacking glucose that contained either 0 or 200 ng/ml rapamycin.
(C) Snf1 activity was not required for P-body formation in response to glucose starvation. Wild-type and snf1Δ cells were grown to mid-log phase in an SC-glucose medium and transferred to a medium lacking glucose (SC-D) for 15 min before fluorescence microscopy.
Pat1 is a substrate for PKA
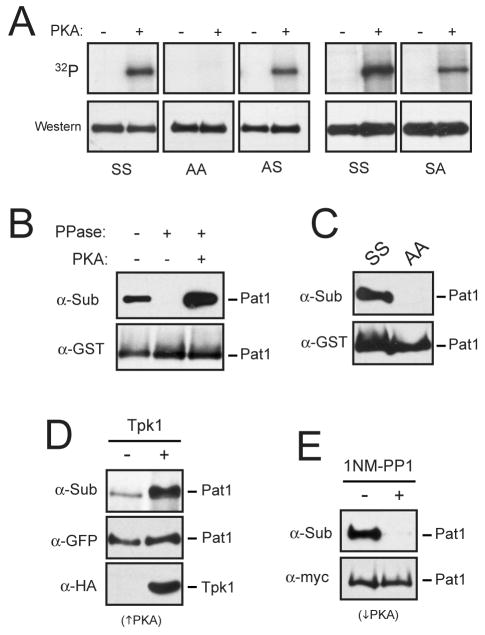
A bioinformatics study from our laboratory identified Pat1 as a candidate substrate for PKA based on the evolutionary conservation of a PKA consensus site in this protein (Budovskaya et al., 2005). This sequence, R-R-R-S456-S457-Y, was found to be conserved in all of the Saccharomyces species examined. Moreover, recent phosphoproteome studies have indicated that these two serine residues, Ser-456 and Ser-457, are phosphorylated in vivo (Albuquerque et al., 2008; Chi et al., 2007). We tested whether this phosphorylation was catalyzed by PKA and found that Pat1 was indeed phosphorylated by PKA in vitro at both Ser-456 and Ser-457 (Figure 3A). A Pat1 variant where both of these serines were replaced by an alanine, Pat1-AA, was not recognized by PKA (Figure 3A). To determine whether this site was also phosphorylated by PKA in vivo, we used an antibody that specifically recognizes phosphorylated PKA sites (see Experimental Procedures). We have used this reagent previously to analyze a number of different PKA substrates (Budovskaya et al., 2005; Chang et al., 2004; Deminoff et al., 2006; Deminoff et al., 2009; Stephan et al., 2009). Here, we found that this antibody recognized a Pat1 fusion protein that was precipitated from log phase cultures of wild-type cells (Figure 3B). This recognition was lost upon phosphatase treatment and was restored upon a subsequent incubation with PKA and ATP (Figure 3B). In contrast, the Pat1-AA protein was not recognized by this antibody (Figure 3C). Finally, the signal with this anti-substrate antibody was elevated in cells with increased PKA activity and dramatically reduced following the inactivation of PKA (Figure 3D, E). Therefore, both the PKA site in Pat1 and the presence of PKA activity in cells were required for recognition by this antibody. Taken together, these data demonstrated that Pat1 was both an in vivo and in vitro substrate for PKA.
Figure 3.
The Pat1 protein was a substrate for PKA.
(A) PKA phosphorylated Pat1 at both Ser-456 and Ser-457 in vitro. The indicated Pat1 variants were precipitated from cell extracts and either mock treated (-) or incubated with PKA and [γ-32P] ATP. The reaction products were then separated on SDS-polyacrylamide gels and the level of PKA phosphorylation was assessed by autoradiography (32P).
(B) Pat1 was phosphorylated by PKA in vivo. A GST-tagged Pat1 protein was precipitated and treated with λ phosphatase, as indicated. An aliquot of the beads from this latter reaction was washed and incubated with PKA and 2.5 mM ATP. The relative level of PKA phosphorylation was assessed by Western blotting with an anti-PKA substrate antibody (α-Sub), as described in the Experimental Procedures. PPase, λ phosphatase.
(C) Pat1 recognition by the anti-PKA substrate antibody required the presence of the two serine residues, Ser-456 and Ser-457. The indicated Pat1 variants were precipitated from cell extracts and the relative level of PKA phosphorylation was assessed by Western blotting with the anti-PKA substrate antibody.
(D) The in vivo level of the PKA phosphorylation on Pat1 was elevated in a strain over-expressing the PKA catalytic subunit, Tpk1.
(E) Recognition by the anti-PKA substrate antibody was lost following the inactivation of PKA. The tpk1-as strain was incubated with 5 μM 1NM-PP1 for 15 min to inactivate PKA.
The phosphorylation of Pat1 by PKA regulates P-body formation
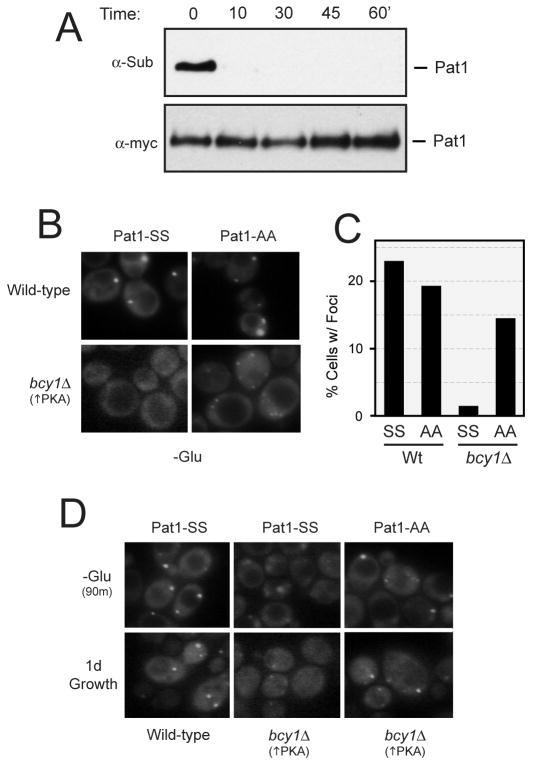
If Pat1 was the target responsible for the PKA effects on P-body assembly, we would expect that this phosphorylation would be rapidly lost upon glucose deprivation. We used the anti-substrate antibody described above to test this possibility, and found that this phosphorylation was significantly reduced within 10 min of glucose starvation (Figure 4A). Thus, the kinetics of dephosphorylation were consistent with Pat1 being a physiologically-relevant target for PKA. We also tested whether the presence of the nonphosphorylatable variant, Pat1-AA, would suppress the P-body defects associated with constitutive PKA activity. For these experiments, we introduced a GFP-tagged Pat1-AA into bcy1Δ cells and assessed P-body assembly after glucose deprivation. With both the acute and gradual deprivation regimens, we found that the Pat1-AA variant was more efficiently incorporated into P-bodies in bcy1Δ cells than the wild-type Pat1 protein (Figure 4B-D). These data therefore suggest that the inhibition of P-body foci formation by PKA is due, at least in part, to the direct phosphorylation of Pat1.
Figure 4.
The PKA-mediated inhibition of P-body formation was suppressed by the presence of a non-phosphorylatable form of Pat1.
(A) The PKA-dependent phosphorylation of Pat1 was rapidly lost upon glucose starvation. Cells expressing a myc-tagged Pat1 were grown to mid-log phase in an SC-glucose medium and then transferred to a medium lacking glucose (SC-D) for the indicated time. The level of PKA phosphorylation was assessed by Western blotting with the anti-PKA substrate antibody.
(B, C) The presence of the non-phosphorylatable variant, Pat1-AA, resulted in an elevated number of P-body foci in bcy1Δ cells. Wild-type and bcy1Δ cells expressing the indicated GFP-tagged Pat1 proteins were grown to mid-log phase and then transferred to a medium lacking glucose for 15 min. Representative fluorescence microscopy images are shown in B and the quantification of these data in C.
(D) The presence of the non-phosphorylatable Pat1 also suppressed the inhibitory effects of elevated PKA signaling on P-body formation during an extended glucose starvation (-Glu, 90 min) or growth into early stationary phase (1d).
Multiple activities have been attributed to Pat1 including a role in the repression of translation, in the stimulation of mRNA decapping (and hence decay) and as a scaffolding element during the assembly of P-body foci (Bonnerot et al., 2000; Braun et al., 2010; Coller and Parker, 2005; Holmes et al., 2004; Marnef and Standart, 2010; Pilkington and Parker, 2008). PKA phosphorylation could be influencing any one of these activities. However, previous work has clearly demonstrated that elevated PKA activity does not prevent the decrease in translation initiation that accompanies an acute starvation for glucose (Ashe et al., 2000). Consistent with this result, we found that neither elevated PKA activity or alterations of the Pat1 PKA site influenced the growth defect caused by the over-expression of Pat1 (Figure S4A; data not shown). We therefore tested whether PKA activity might be affecting mRNA decay. A block to mRNA decay, like that associated with an xrn1Δ strain, has been shown to result in an increased number of P-body foci, even in glucose-rich conditions (Cougot et al., 2004; Eulalio et al., 2007a; Sheth and Parker, 2003). Therefore, the PKA inhibition of foci formation could be due to an increased rate of mRNA decay that would result in fewer mRNA templates upon which to build the P-body structures. However, in contrast to this prediction, we found that there were similar levels of total polyA+ RNA in wild-type and RAS2val19 cells following a 15 minute period of glucose starvation (Figure S4B). In addition, the half-life of the PGK1pG transcript was not significantly affected by the presence of elevated PKA activity (Figure S4C) (Hatfield et al., 1996). Finally, we found that the introduction of the RAS2val19 allele into an xrn1Δ strain led to a significant decrease in the number of P-body foci present (Figure S4D). This latter result indicated that the inhibitory effects of PKA on P-body assembly did not require the activity of the primary 5′-to-3′ exonuclease in eukaryotic cells. In all, these data suggest that PKA is not influencing earlier events in the P-body assembly pathway, like the inhibition of translation or mRNA decay. Instead, the data are more consistent with PKA affecting a later step in P-body foci formation.
Dhh1 recruitment into P-bodies is disrupted by the PKA phosphorylation of Pat1
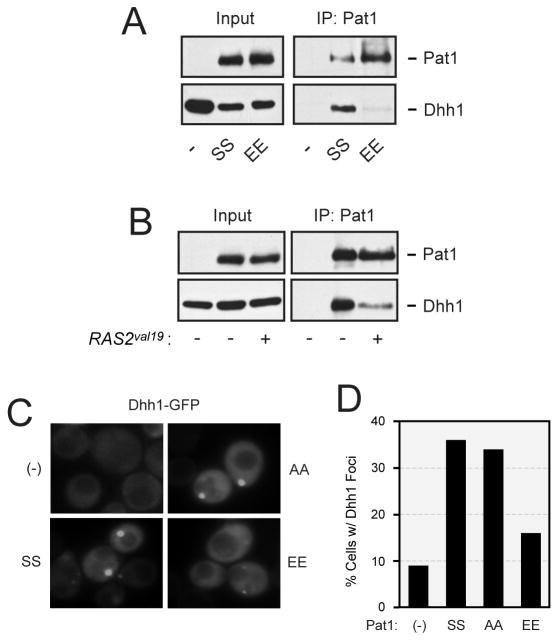
Pat1 binds to a number of P-body proteins, including Dhh1, and has been proposed to act as a scaffold during P-body assembly (Braun et al., 2010; Nissan et al., 2010; Pilkington and Parker, 2008). Here, we tested whether this interaction with Dhh1 was influenced by the presence of either elevated PKA activity or the Pat1-EE variant. The presence of a glutamic acid can functionally substitute for a phosphorylated serine, and thus the Pat1-EE variant may mimic the PKA phosphorylated form of Pat1. We found that the amount of Dhh1 co-precipitating with Pat1 was dramatically lower in both of these conditions (Figure 5B, C; S5A). Moreover, the frequency of Dhh1 foci was significantly decreased in cells containing only the Pat1-EE variant (Figure 5C, D). This latter effect was not specific to Dhh1 as the frequency of Edc3 and Dcp2 recruitment to cytoplasmic foci was also diminished in cells containing the Pat1-EE variant (Figure S5B). The localization of Pat1-EE to foci was itself impaired by a factor of two-fold; the fraction of cells with large foci was 20% for Pat1-EE as compared to 39% for the wild-type protein, Pat1-SS. In all, these data suggested that PKA influences P-body assembly by directly phosphorylating Pat1 and thereby disrupting particular protein-protein interactions important for foci formation.
Figure 5.
The presence of the Pat1-EE variant disrupted Dhh1 recruitment to P-bodies.
(A) The Pat1-EE variant exhibited a weaker interaction with Dhh1 than the wild-type Pat1. The indicated myc-tagged Pat1 proteins were precipitated from yeast cell extracts and the relative level of the associated Dhh1-GFP was assessed by Western blotting.
(B) The presence of elevated Ras/PKA signaling activity resulted in less Pat1 binding to Dhh1. Wild-type cells expressing a myc-tagged Pat1 and Dhh1-GFP fusion protein were transformed with either a vector control (-) or RAS2val19 plasmid (+). The myc-tagged Pat1 was immunoprecipitated from cell extracts and the relative level of the associated Dhh1-GFP was assessed by Western blotting.
(C) The formation of Dhh1 foci upon glucose starvation was impaired in strains containing the Pat1-EE variant. A pat1Δ strain expressing the GFP-tagged Dhh1 reporter and the indicated versions of Pat1 was grown to mid-log phase and transferred to a medium lacking glucose (SC-D) for 15 min.
(D) A histogram showing the fraction of cells in panel C that contained at least one large P-body focus following glucose starvation.
PKA phosphorylation of Pat1 is important for cell survival in stationary phase
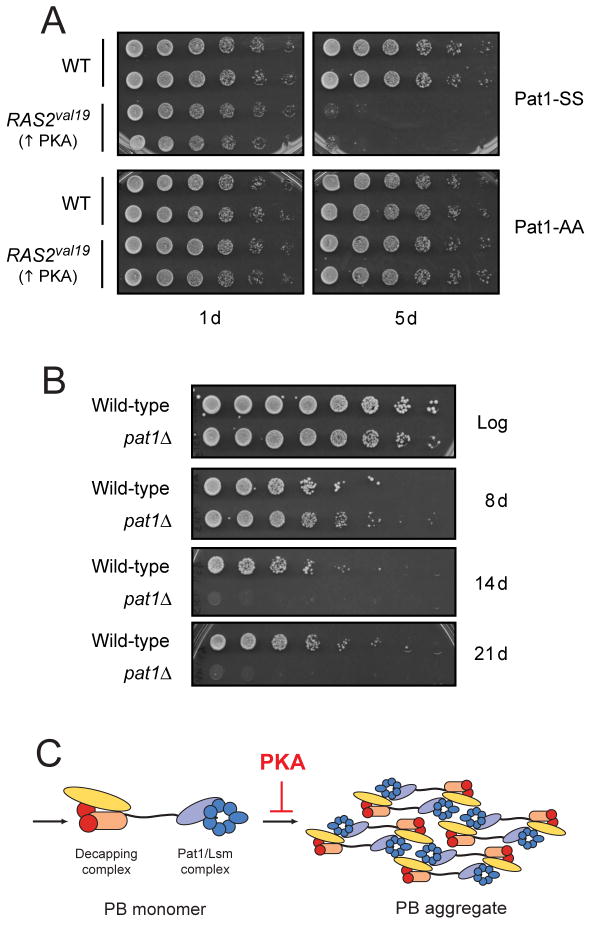
The Ras/PKA signaling pathway in S. cerevisiae is thought to regulate the entry into stationary phase and thus the survival of cells within this resting state (Dechant and Peter, 2008; Herman, 2002). For example, mutants with constitutive PKA activity, like RAS2val19, fail to enter into a normal stationary phase and rapidly lose viability upon nutrient deprivation (Figure 6A) (Toda et al., 1985). However, the key PKA substrates responsible for these viability defects have not yet been identified. Remarkably, we found that the presence of the nonphosphorylatable Pat1-AA was able to strongly suppress the loss of viability associated with stationary phase cultures of the RAS2val19 mutant (Figure 6A). This result suggested that Pat1, and perhaps the formation of P-body foci, might be important for the long-term survival of quiescent cells. Consistent with this possibility, pat1Δ and dhh1Δ mutants were found to lose viability more rapidly in stationary phase than a wild-type strain (Figure 6B; S6). Therefore, Pat1 and its phosphorylation by PKA appear to be important determinants of cell survival in stationary phase.
Figure 6.
The PKA phosphorylation of Pat1 may influence stationary phase cell survival.
(A) The presence of the nonphosphorylatable Pat1-AA variant suppressed the loss of viability observed in stationary phase cultures of the RAS2val19 mutant. pat1Δ cells containing either a control vector or a RAS2val19 plasmid were grown for the indicated number of days in a YM-glucose minimal medium. The number of viable cells present was determined by plating out increasing dilutions of the cultures. The cells also contained a plasmid expressing either Pat1-SS or Pat1-AA, as indicated.
(B) Cells lacking Pat1 exhibited a decreased rate of survival in stationary phase. Wild-type or pat1Δ strains were grown for the indicated number of days and the number of viable cells in each culture was assessed.
(C) A model proposing that PKA activity specifically inhibits the aggregation step of P-body assembly. See text for further details.
Discussion
The formation of RNP foci in eukaryotic cells appears to be part of a general response to stress. However, it is typically not known how the assembly of these RNP structures is regulated and what function the larger assemblies serve in the stressed cells (Eulalio et al., 2007a; Parker and Sheth, 2007). In this study, we show that the Ras/PKA signaling pathway is a key regulator of P-body formation in S. cerevisiae. In particular, our data demonstrate that the inactivation of this pathway is both a necessary and sufficient condition for the formation of large P-body foci. Moreover, our findings identify Pat1, an evolutionarily-conserved component of these RNP structures, as a critical target of this control. PKA directly phosphorylates Pat1 and this phosphorylation was found to disrupt the interactions occurring between Pat1 and other components of the P-body, including the Dhh1 helicase. These observations are intriguing in light of recent work indicating that Pat1 might function as a central scaffold upon which P-bodies are assembled (Braun et al., 2010; Marnef and Standart, 2010; Pilkington and Parker, 2008). PKA phosphorylation may therefore influence key protein interactions required for the formation of the larger foci.
The assembly of P-body foci has been proposed to occur in two discrete stages (Figure 6C). We feel that the data here are most consistent with PKA affecting the second step in this model and specifically inhibiting the formation of the larger aggregate structures. This premise is consistent with elevated PKA activity preventing the formation of P-body foci without affecting the block to translation initiation that accompanies an acute glucose starvation (Ashe et al., 2000). In addition, moderately elevated PKA activity resulted in the presence of smaller but more numerous P-body foci suggesting that PKA is regulating the final aggregation of these smaller structures. Finally, our data also indicate that the mere presence of a translationally-repressed pool of mRNA is not sufficient for the formation of the larger P-body foci. For example, neither rapamycin treatment nor nitrogen deprivation was able to induce P-body formation. Both of these conditions result in diminished TORC1 signaling and a dramatically reduced rate of translation initiation (Barbet et al., 1996; Hay and Sonenberg, 2004; Zaman et al., 2008). A recent study with secretory pathway mutants also found that the extent of P-body formation was not correlated with the strength of the protein translation defect (Kilchert et al., 2010). Therefore, the data cumulatively indicate that it is diminished PKA signaling that is the key signal for P-body foci formation in S. cerevisiae.
This study also suggests that Pat1, and its phosphorylation by PKA, are important for the normal control of cell survival during periods of quiescence. In particular, we found that mutants lacking Pat1 exhibited a diminished capacity to survive in stationary phase and that the presence of Pat1-AA was able to effectively suppress the loss of viability associated with RAS2val19 stationary phase cultures. Although Pat1 has multiple functions, several observations suggest that its role in P-body foci formation is responsible for these survival phenotypes. First, our data here suggest that PKA activity specifically regulates the formation of the larger P-body assemblies. This control appears to be mediated by Pat1 as the introduction of the Pat1-AA variant restores full-sized foci to cells with elevated PKA activity. Second, recent studies indicate that the other activities associated with the Pat1 protein, including the stimulation of decapping and translational repression, occur either co-translationally or within the context of a P-body monomer (or smaller RNP complex) (Decker et al., 2007; Eulalio et al., 2007b; Hu et al., 2009; Reijns et al., 2008). In addition, the work here and elsewhere indicates that PKA does not influence either global mRNA decay or the block to translation associated with glucose starvation (Ashe et al., 2000). Finally, the PKA site in Pat1 is located within a C-terminal domain that has been shown to be both necessary and sufficient for P-body foci formation (Pilkington and Parker, 2008). In contrast, the Pat1 domain required for efficient mRNA decapping is located at the N-terminus of this protein. In all, we feel that these observations are most consistent with the presence of P-body foci being responsible for the efficient long-term survival of G0 cells. P-body assemblies therefore may function in a manner similar to the RNP granules in oocytes that store maternal mRNA transcripts for later use (Anderson and Kedersha, 2006). This possibility will need to be examined further, but it is interesting to note that recent work has identified a large number of different RNP and protein foci in stationary phase cells (Narayanaswamy et al., 2009; Noree et al., 2010). The sequestration of mRNA and protein into discrete sites in the cytoplasm may thus be a general feature of quiescent cells.
The data here also suggest that there is a second PKA substrate, apart from Pat1, that is important for the regulation of P-body assembly. This assertion follows from the observation that elevated PKA activity results in a more severe defect in P-body formation than does either the loss of Pat1 or the presence of the Pat1-EE variant. Identifying this second target could provide additional insights into the mechanisms governing P-body foci formation. Finally, it is important to point out that stationary phase survival in S. cerevisiae has been used a model for the study of the mechanisms governing eukaryotic aging (Kennedy, 2008). It is therefore interesting to speculate that the ability to form RNP foci might be generally important for the normal aging process. Determining how and why these RNP structures form could therefore increase our understanding of the cellular basis of aging and diseases, like cancer, that are associated with an aberrant control of cell growth. A continued analysis of the role of PKA signaling in the regulation of P-bodies, and other stationary phase foci in S. cerevisiae, would represent an important step towards this broader goal.
Experimental Procedures
Additional methods, including a description of growth conditions and plasmid construction, are included in the Supporting Information available online.
Fluorescence Microscopy
Cells expressing the fusion constructs were grown to early log phase (0.3-0.6 OD600 units/ml) before exposure to the appropriate starvation regimen or drug treatment. For carbon and nitrogen starvation, cells were collected by centrifugation and washed with SC-D or SD-N media before being re-suspended in the same medium. Rapamycin was added to a final concentration of 200 ng/ml to inactivate TORC1. For the tpk1-as strain, the inhibitor, 1NM-PP1, was added to a final concentration 2-5 μM. Cells were incubated at 30°C for the indicated period of time, collected by centrifugation, and re-suspended and spotted onto microscope slides. The samples were then imaged as described (Budovskaya et al., 2005). For the quantification shown, the data represent the average of two or three experiments that examined at least 100 cells. In each case, the variation between replicates was less than 10%.
Western immunoblotting and immunoprecipitations
Protein samples for Western blotting were prepared with a glass bead lysis method, separated on 7.5-10% SDS-polyacrylamide gels and transferred to nitrocellulose membranes (Hybond ECL, GE Healthcare) as described (Budovskaya et al., 2002; Budovskaya et al., 2004). The membranes were then probed with the appropriate primary and secondary antibodies. The Supersignal chemiluminescent substrate (Pierce) was subsequently used to detect the reactive bands. Cell extracts for immunoprecipitation were prepared by re-suspending log-phase cells in Lysis buffer (25 mM Tris-HCl pH 7.4, 140 mM NaCl, 0.1% Tween-20, 1 mM PMSF) and lysing by agitation with glass beads. Myc- and GST-tagged proteins were immunoprecipitated with the appropriate monoclonal antibodies (Cell Signaling), and the immunoprecipitates were collected on Protein A-Sepharose (GE Healthcare). To monitor PKA phosphorylation in vivo, substrate proteins were precipitated under denaturing conditions as described with protease and phosphatase inhibitors present at all steps (Budovskaya et al., 2005; Deminoff et al., 2006). The level of PKA phosphorylation was then assessed by Western blotting with the anti-PKA substrate antibody (Cell Signaling) as described (Chang et al., 2004; Deminoff et al., 2006).
In vitro kinases assays
The immunoprecipitated substrate proteins were incubated with λ phosphatase (NEB) in λ phosphatase reaction buffer supplemented with 6 mM MnCl2 for 1 hr and washed three times with wash buffer (25 mM Tris-HCl pH 7.4, 140 mM NaCl, 0.1% (v/v) Tween-20, 1 mM PMSF). The in vitro kinase assay (IVKA) was then performed by incubating the immunoprecipitated material with 10 U of bPKA (Sigma) and 10 μCi [γ-32P] ATP or 2.5 mM unlabeled ATP in a 40 μl reaction (50 mM potassium phosphate, 5 mM NaF, 10 mM MgCl2, 4.5 mM DTT and both protease and phosphatase inhibitors). The proteins were separated by SDS-polyacrylamide gel electrophoresis and the level of phosphorylation was assessed by either autoradiography or Western blotting with the anti-PKA substrate antibody, respectively.
Supplementary Material
Acknowledgments
We thank Anita Hopper, James Broach, Tsien-Hien Chang, Michael Hampsey and Roy Parker for reagents used in this study, Rebecca Hurto for helpful discussions and Venkat Gopalan and members of the Herman lab for comments on the manuscript. We are especially grateful to Helen Chamberlin and Jian-Qiu Wu for access to their fluorescent microscopes. This work was supported by a grant from the National Institutes of Health (GM65227) to P.K.H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albuquerque CP, Smolka MB, Payne SH, Bafna V, Eng J, Zhou H. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol Cell Proteomics. 2008;7:1389–1396. doi: 10.1074/mcp.M700468-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. RNA granules. J Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10:430–436. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- Ashe MP, De Long SK, Sachs AB. Glucose depletion rapidly inhibits translation initiation in yeast. Mol Biol Cell. 2000;11:833–848. doi: 10.1091/mbc.11.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagopal V, Parker R. Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr Opin Cell Biol. 2009;21:403–408. doi: 10.1016/j.ceb.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbet NC, Schneider U, Helliwell SB, Stansfield I, Tuite MF, Hall MN. TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashkirov VI, Scherthan H, Solinger JA, Buerstedde JM, Heyer WD. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J Cell Biol. 1997;136:761–773. doi: 10.1083/jcb.136.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop AC, Buzko O, Shokat KM. Magic bullets for protein kinases. Trends Cell Biol. 2001;11:167–172. doi: 10.1016/s0962-8924(01)01928-6. [DOI] [PubMed] [Google Scholar]
- Bonnerot C, Boeck R, Lapeyre B. The two proteins Pat1p (Mrt1p) and Spb8p interact in vivo, are required for mRNA decay, and are functionally linked to Pab1p. Mol Cell Biol. 2000;20:5939–5946. doi: 10.1128/mcb.20.16.5939-5946.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun JE, Tritschler F, Haas G, Igreja C, Truffault V, Weichenrieder O, Izaurralde E. The C-terminal alpha-alpha superhelix of Pat is required for mRNA decapping in metazoa. EMBO J. 2010;29:2368–2380. doi: 10.1038/emboj.2010.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues M, Teixeira D, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Muhlrad D, Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J Cell Biol. 2008;183:441–455. doi: 10.1083/jcb.200807043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budovskaya YV, Hama H, DeWald DB, Herman PK. The C terminus of the Vps34p phosphoinositide 3-kinase is necessary and sufficient for the interaction with the Vps15p protein kinase. J Biol Chem. 2002;277:287–294. doi: 10.1074/jbc.M109263200. [DOI] [PubMed] [Google Scholar]
- Budovskaya YV, Stephan JS, Deminoff SJ, Herman PK. An evolutionary proteomics approach identifies substrates of the cAMP-dependent protein kinase. Proc Natl Acad Sci U S A. 2005;102:13933–13938. doi: 10.1073/pnas.0501046102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budovskaya YV, Stephan JS, Reggiori F, Klionsky DJ, Herman PK. The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J Biol Chem. 2004;279:20663–20671. doi: 10.1074/jbc.M400272200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YW, Howard SC, Herman PK. The Ras/PKA signaling pathway directly targets the Srb9 protein, a component of the general RNA polymerase II transcription apparatus. Mol Cell. 2004;15:107–116. doi: 10.1016/j.molcel.2004.05.021. [DOI] [PubMed] [Google Scholar]
- Chi A, Huttenhower C, Geer LY, Coon JJ, Syka JE, Bai DL, Shabanowitz J, Burke DJ, Troyanskaya OG, Hunt DF. Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc Natl Acad Sci U S A. 2007;104:2193–2198. doi: 10.1073/pnas.0607084104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N, Babajko S, Seraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Virgilio C, Loewith R. The TOR signalling network from yeast to man. Int J Biochem Cell Biol. 2006;38:1476–1481. doi: 10.1016/j.biocel.2006.02.013. [DOI] [PubMed] [Google Scholar]
- Dechant R, Peter M. Nutrient signals driving cell growth. Curr Opin Cell Biol. 2008;20:678–687. doi: 10.1016/j.ceb.2008.09.009. [DOI] [PubMed] [Google Scholar]
- Decker CJ, Teixeira D, Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol. 2007;179:437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deminoff SJ, Howard SC, Hester A, Warner S, Herman PK. Using substrate-binding variants of the cAMP-dependent protein kinase to identify novel targets and a kinase domain important for substrate interactions in Saccharomyces cerevisiae. Genetics. 2006;173:1909–1917. doi: 10.1534/genetics.106.059238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deminoff SJ, Ramachandran V, Herman PK. Distal recognition sites in substrates are required for efficient phosphorylation by the cAMP-dependent protein kinase. Genetics. 2009;182:529–539. doi: 10.1534/genetics.109.102178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol. 2007a;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol. 2007b;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eystathioy T, Jakymiw A, Chan EK, Seraphin B, Cougot N, Fritzler MJ. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. Rna. 2003;9:1171–1173. doi: 10.1261/rna.5810203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks TM, Lykke-Andersen J. The control of mRNA decapping and P-body formation. Mol Cell. 2008;32:605–615. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JV, Petsko GA, Johnston GC, Ringe D, Singer RA, Werner-Washburne M. “Sleeping beauty”: quiescence in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2004;68:187–206. doi: 10.1128/MMBR.68.2.187-206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield L, Beelman CA, Stevens A, Parker R. Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5830–5838. doi: 10.1128/mcb.16.10.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- Hedbacker K, Carlson M. SNF1/AMPK pathways in yeast. Front Biosci. 2008;13:2408–2420. doi: 10.2741/2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman PK. Stationary phase in yeast. Curr Opin Microbiol. 2002;5:602–607. doi: 10.1016/s1369-5274(02)00377-6. [DOI] [PubMed] [Google Scholar]
- Holmes LE, Campbell SG, De Long SK, Sachs AB, Ashe MP. Loss of translational control in yeast compromised for the major mRNA decay pathway. Mol Cell Biol. 2004;24:2998–3010. doi: 10.1128/MCB.24.7.2998-3010.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Sweet TJ, Chamnongpol S, Baker KE, Coller J. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature. 2009;461:225–229. doi: 10.1038/nature08265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingelfinger D, Arndt-Jovin DJ, Luhrmann R, Achsel T. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. Rna. 2002;8:1489–1501. [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy BK. The genetics of ageing: insight from genome-wide approaches in invertebrate model organisms. J Intern Med. 2008;263:142–152. doi: 10.1111/j.1365-2796.2007.01903.x. [DOI] [PubMed] [Google Scholar]
- Kilchert C, Weidner J, Prescianotto-Baschong C, Spang A. Defects in the secretory pathway and high Ca2+ induce multiple P-bodies. Mol Biol Cell. 2010;21:2624–2638. doi: 10.1091/mbc.E10-02-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui J, Campbell SG, Ashe MP. Inhibition of translation initiation following glucose depletion in yeast facilitates a rationalization of mRNA content. Biochem Soc Trans. 2010;38:1131–1136. doi: 10.1042/BST0381131. [DOI] [PubMed] [Google Scholar]
- Marnef A, Standart N. Pat1 proteins: a life in translation, translation repression and mRNA decay. Biochem Soc Trans. 2010;38:1602–1607. doi: 10.1042/BST0381602. [DOI] [PubMed] [Google Scholar]
- Narayanaswamy R, Levy M, Tsechansky M, Stovall GM, O'Connell JD, Mirrielees J, Ellington AD, Marcotte EM. Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc Natl Acad Sci U S A. 2009;106:10147–10152. doi: 10.1073/pnas.0812771106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan T, Rajyaguru P, She M, Song H, Parker R. Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Mol Cell. 2010;39:773–783. doi: 10.1016/j.molcel.2010.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noree C, Sato BK, Broyer RM, Wilhelm JE. Identification of novel filament-forming proteins in Saccharomyces cerevisiae and Drosophila melanogaster. J Cell Biol. 2010;190:541–551. doi: 10.1083/jcb.201003001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee AB. G1 events and regulation of cell proliferation. Science. 1989;246:603–608. doi: 10.1126/science.2683075. [DOI] [PubMed] [Google Scholar]
- Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Pilkington GR, Parker R. Pat1 contains distinct functional domains that promote P-body assembly and activation of decapping. Mol Cell Biol. 2008;28:1298–1312. doi: 10.1128/MCB.00936-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijns MA, Alexander RD, Spiller MP, Beggs JD. A role for Q/N-rich aggregation-prone regions in P-body localization. J Cell Sci. 2008;121:2463–2472. doi: 10.1242/jcs.024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santangelo GM. Glucose signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2006;70:253–282. doi: 10.1128/MMBR.70.1.253-282.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sass P, Field J, Nikawa J, Toda T, Wigler M. Cloning and characterization of the high-affinity cAMP phosphodiesterase of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1986;83:9303–9307. doi: 10.1073/pnas.83.24.9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, Sabatini DM. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol Cell. 2010;40:310–322. doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery MG, Liko D, Heideman W. Protein kinase A, TOR, and glucose transport control the response to nutrient repletion in Saccharomyces cerevisiae. Eukaryot Cell. 2008;7:358–367. doi: 10.1128/EC.00334-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan JS, Yeh YY, Ramachandran V, Deminoff SJ, Herman PK. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc Natl Acad Sci U S A. 2009;106:17049–17054. doi: 10.1073/pnas.0903316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G, Mayo T, Anderson P. ARE-mRNA degradation requires the 5′-3′ decay pathway. EMBO Rep. 2006;7:72–77. doi: 10.1038/sj.embor.7400572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T, Cameron S, Sass P, Zoller M, Scott JD, McMullen B, Hurwitz M, Krebs EG, Wigler M. Cloning and characterization of BCY1, a locus encoding a regulatory subunit of the cyclic AMP-dependent protein kinase in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:1371–1377. doi: 10.1128/mcb.7.4.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T, Uno I, Ishikawa T, Powers S, Kataoka T, Broek D, Cameron S, Broach J, Matsumoto K, Wigler M. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell. 1985;40:27–36. doi: 10.1016/0092-8674(85)90305-8. [DOI] [PubMed] [Google Scholar]
- van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Seraphin B. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. Embo J. 2002;21:6915–6924. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner-Washburne M, Braun E, Johnston GC, Singer RA. Stationary phase in the yeast Saccharomyces cerevisiae. Microbiol Rev. 1993;57:383–401. doi: 10.1128/mr.57.2.383-401.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Zaman S, Lippman SI, Schneper L, Slonim N, Broach JR. Glucose regulates transcription in yeast through a network of signaling pathways. Mol Syst Biol. 2009;5:245. doi: 10.1038/msb.2009.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman S, Lippman SI, Zhao X, Broach JR. How Saccharomyces responds to nutrients. Annu Rev Genet. 2008;42:27–81. doi: 10.1146/annurev.genet.41.110306.130206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.