SUMMARY
In Escherichia coli, RNA degradation often begins with conversion of the 5′-terminal triphosphate to a monophosphate, creating a better substrate for internal cleavage by RNase E. Remarkably, no homologue of this key endonuclease is present in many bacterial species, such as Bacillus subtilis and various pathogens. Here we report that the degradation of primary transcripts in B. subtilis can nevertheless be triggered by an analogous process to generate a short-lived, monophosphorylated intermediate. Like its E. coli counterpart, the B. subtilis RNA pyrophosphohydrolase that catalyzes this event is a Nudix protein that prefers unpaired 5′ ends. However, in B. subtilis this modification exposes transcripts to rapid 5′-exonucleolytic degradation by RNase J, which is absent in E. coli but present in most bacteria lacking RNase E. This pathway, which closely resembles the mechanism by which deadenylated mRNA is degraded in eukaryotic cells, explains the stabilizing influence of 5′-terminal stem-loops in such bacteria.
Keywords: RppH, YtkD, 5′ exonuclease, RNA decay, RNA stability, yhxA, glpP, ermC
INTRODUCTION
Messenger RNA degradation plays a key role in the control of gene expression in all organisms by limiting the number of times that each mRNA molecule can be used as a template for protein synthesis. mRNA lifetimes are quite diverse, differing by as much as two orders of magnitude for distinct transcripts within the same cell. In bacteria, for example, mRNA half-lives can be as short as seconds or as long as an hour, while in vertebrates they range from minutes to days. However, despite the universal importance of mRNA turnover, the distinct mRNA termini and degradative activities in prokaryotes and eukaryotes, and even in different bacterial species, have raised questions about the mechanistic conservation and evolutionary origins of this process.
Most of what is known about bacterial mRNA degradation has been learned from studies in Escherichia coli. E. coli cells contain multiple endoribonucleases and 3′ exoribonucleases, but no 5′ exoribonucleases (Zuo and Deutscher, 2001; Even et al., 2005; Arraiano et al., 2010). Because these 3′ exonucleases are impeded by the stem-loop structures that typically are present at the 3′ ends of E. coli messages (Spickler and Mackie, 2000; Vincent and Deutscher, 2006), it appears that endonucleolytic cleavage generally precedes 3′ exonucleolytic attack (Belasco, 2010). The endonuclease most important for mRNA decay in E. coli is RNase E, an essential enzyme that cleaves RNA within single-stranded regions that are AU-rich (Ono and Kuwano, 1979; Mudd et al., 1990; Babitzke and Kushner, 1991; Melefors and von Gabain, 1991; Taraseviciene et al., 1991; McDowall et al., 1994). Although RNase E cuts RNA at internal sites, it greatly prefers substrates that bear only one phosphate rather than three at the 5′ end, a selectivity that results from a discrete RNase E pocket that can bind a monophosphorylated 5′ terminus while the active site binds and cleaves the RNA internally (Mackie, 1998; Callaghan et al., 2005). This property helps to explain why the monophosphorylated 3′-terminal fragments produced by RNase E cleavage usually are much more labile than the triphosphorylated primary transcripts from which they derive. By contrast, the 5′-terminal fragments that result from RNase E cleavage are susceptible to rapid 3′ exonucleolytic attack due to the lack of base pairing at their 3′ ends.
Interestingly, a stem-loop at the 5′-terminus can protect many primary transcripts from degradation in E. coli even though RNase E is not able to interact with their triphosphorylated 5′ ends (Emory et al., 1992; Bouvet and Belasco, 1992; Hansen et al., 1994; Arnold et al., 1998; Baker and Mackie, 2003). This observation suggested the existence of a pathway in which the 5′ end can govern access by RNase E to internal cleavage sites in primary transcripts. Recent studies have revealed the mechanism of that pathway by identifying a step that can precede RNase E cleavage in E. coli: the removal of pyrophosphate from the 5′ end so as to convert a triphosphate into a monophosphate (Celesnik et al., 2007). This rate-determining event, which triggers rapid RNase E cleavage of the resulting monophosphorylated intermediate, is catalyzed by the RNA pyrophosphohydrolase RppH, a member of the Nudix hydrolase enzyme family (Deana et al., 2008). Consistent with the stabilizing effect of a 5′-terminal stem-loop in E. coli, pyrophosphate removal by RppH is inhibited by base pairing at the 5′ end.
mRNA stabilization by a 5′-terminal stem-loop has also been reported in other bacterial species, such as Bacillus subtilis (Hambraeus et al., 2002; Sharp and Bechhofer, 2005). Remarkably, however, B. subtilis lacks a homologue of RNase E, as do about one-third of all bacterial species whose genomes have been sequenced (Shahbabian et al., 2009). Instead, it contains two other ribonucleases not present in E. coli, RNase Y and RNase J, both of which perform functions essential for cell growth and have been implicated in mRNA degradation (Even et al., 2005; Mäder et al., 2008; Shahbabian et al., 2009; Daou-Chabo et al., 2009; Yao and Bechhofer, 2010). RNase Y is a membrane-associated endonuclease (Hunt et al., 2006; Shahbabian et al., 2009; Commichau et al., 2009), while RNase J, a heterotetramer comprising J1 and J2 subunits, is a multifunction enzyme with both endonuclease and 5′ exonuclease activity (Even et al., 2005; Mathy et al., 2007; Li de la Sierra-Gallay et al., 2008; Mathy et al., 2010). The latter activity, which is foreign to E. coli, is a property of the J1 subunit, whereas both the J1 and J2 subunits have endonuclease activity (Mathy et al., 2010). Interestingly, in vitro experiments suggest that a monophosphorylated RNA 5′ end can potentiate both the endonuclease activity of RNase Y and the 5′ exonuclease activity of RNase J (Mathy et al., 2007; Li de la Sierra-Gallay et al., 2008; Shahbabian et al., 2009). These observations hint at possible ways in which 5′-terminal base pairing could protect primary transcripts from degradation in B. subtilis and other bacteria despite the absence of RNase E in those species. However, even if RNase Y or RNase J does participate in the critical early events of 5′-end-dependent RNA decay, it is not clear what would trigger its action, as none of the Nudix hydrolases encoded by the B. subtilis genome have a high degree of homology to E. coli RppH (<27% sequence identity).
We have now investigated the mechanism of 5′-end-dependent mRNA degradation in B. subtilis. Here we report the identification of an enzyme that can initiate mRNA decay in that species by converting triphosphorylated primary transcripts with unpaired 5′ termini into full-length intermediates bearing a single phosphate at the 5′ end. These intermediates are then rapidly degraded by a 5′-exonucleolytic mechanism dependent on RNase J1. Though distinct from 5′-end-dependent mRNA turnover in E. coli, this pathway for bacterial mRNA degradation closely resembles the principal mechanism of mRNA decay in eukaryotic cells.
RESULTS
RNA pyrophosphohydrolase activity of BsRppH in vitro
In E. coli, 5′-end-dependent RNA degradation is triggered by RppH (EcRppH), a Nudix hydrolase that deprotects the 5′ terminus of primary transcripts by removing pyrophosphate to generate a monophosphorylated decay intermediate that is rapidly destroyed (Deana et al., 2008). As a first step toward determining whether a similar deprotection event occurs in bacterial species that lack RNase E, we purified the six proteins in B. subtilis that have a canonical or near-canonical Nudix motif (GX5EX7REUXEEXGU, where U is a residue with a hydrophobic side chain (Mildvan et al., 2005)) and assayed each for RNA pyrophosphohydrolase activity in vitro. The triphosphorylated RNA substrate in these assays was the ΔermC transcript, which was previously shown to decay in B. subtilis by a 5′-end-dependent mechanism, as evidenced by the longer lifetime that results when a stem-loop is added to its 5′ terminus (Sharp and Bechhofer, 2005). This RNA was synthesized with a 5′-terminal γ-32P label and an internal fluorescein label by in vitro transcription. Each of the six Nudix hydrolases of B. subtilis was then tested for its ability to release the radiolabel from the 5′ end of the substrate while leaving the rest of the molecule intact (Figure 1A, Figure S1A). Only one, YtkD, here renamed BsRppH, had significant activity, releasing the 5′ radiolabel as a product with a high electrophoretic mobility. This activity was intrinsic to BsRppH, as it was abolished when a single glutamate residue of the Nudix motif was replaced with glutamine (BsRppH-E68Q).
Figure 1. Conversion of triphosphorylated to monophosphorylated RNA by purified BsRppH.
A, B) Release of orthophosphate from the 5′ end of triphosphorylated RNA by BsRppH. Triphosphorylated ΔermC RNA bearing a 5′-terminal γ-32P label and an internal fluorescein label was treated with purified BsRppH or BsRppH-E68Q (75 nM), and reaction samples isolated at time intervals were analyzed by (A) gel electrophoresis (with subsequent detection of radioactivity and fluorescence) or (B) thin layer chromatography beside the same RNA treated with EcRppH (with subsequent detection of radioactivity).
C) Conversion of triphosphorylated RNA to monophosphorylated RNA by BsRppH. Triphosphorylated, diphosphorylated, and monophosphorylated GA(CU)13 bearing a single 32P label between the first and second nucleotides was treated with purified BsRppH (75 nM), and the radiolabeled starting materials and reaction products were subjected to alkaline hydrolysis and analyzed by thin layer chromatography (bottom) or examined by gel electrophoresis without hydrolysis to confirm RNA integrity (top).
D) Inhibition of BsRppH by a 5′-terminal stem-loop. Triphosphorylated rpsT P1 and rpsT P1+hp RNA bearing a 5′-terminal γ-32P label and an internal fluorescein label were treated with purified BsRppH or BsRppH-E68Q (7.5 nM), reaction samples were isolated periodically and analyzed by gel electrophoresis, and the radioactivity remaining in the RNA was plotted as a function of time.
See also Figure S1.
To identify the form in which the γ-32P label was released, the radiolabeled BsRppH reaction product was examined at higher resolution by thin layer chromatography. Unlike EcRppH, which released the radiolabel primarily as pyrophosphate, the principal 32P-labeled reaction product of BsRppH was orthophosphate (Figure 1B). This observation called into question how many phosphates remain on the 5′ end of RNA after reaction with BsRppH, which was addressed by examining the RNA reaction product of another substrate, GA(CU)13. GA(CU)13 bearing a 5′-terminal triphosphate, diphosphate, or monophosphate and a single 32P label between the first and second nucleotide was synthesized by in vitro transcription and treated with BsRppH. The RNA reaction product was then subjected to alkaline hydrolysis, and the 5′-terminal nucleotide was examined by thin layer chromatography and autoradiography. BsRppH-catalyzed hydrolysis of the triphosphorylated substrate generated a monophosphorylated RNA product without the detectable accumulation of a diphosphorylated intermediate (Figure 1C). The diphosphorylated substrate likewise yielded a monophosphorylated product. These findings indicate that BsRppH converts triphosphorylated transcripts to monophosphorylated RNA by releasing the γ and β phosphates as orthophosphate ions.
Prior studies have shown that the presence of a 5′-terminal stem-loop can prolong RNA lifetimes in E. coli, in part by hindering pyrophosphate removal by EcRppH (Deana et al., 2008). We therefore investigated whether BsRppH requires access to an unpaired 5′-terminal nucleotide by comparing the reactivity of two RNAs previously shown to differ in their reactivity toward EcRppH: rpsT P1 and a variant thereof (P1+hp) to which a 5′-terminal stem-loop beginning with the same three nucleotides had been added. Phosphate removal by purified BsRppH was much faster for the P1 transcript than for P1+hp (Figure 1D, Figure S1B). We conclude that the RNA pyrophosphohydrolase activity of BsRppH is greatly impeded when the 5′ end of RNA is sequestered by base pairing.
Effect of BsRppH on the decay rate of mRNA in B. subtilis
To determine whether the RNA pyrophosphohydrolase activity of BsRppH influences mRNA degradation in B. subtilis, we first examined the decay of ΔermC mRNA, a synthetic plasmid-encoded transcript known to decay by a 5′-end-dependent mechanism (Sharp and Bechhofer, 2005). Consistent with a role for BsRppH in ΔermC mRNA turnover, deleting the BsRppH gene (ΔrppH) slowed this process, doubling the average measured half-life of the ΔermC transcript in B. subtilis from 3.2 ± 0.2 min to 6.4 ± 0.5 min (Figure 2A and C, Table S1). To identify a natural B. subtilis transcript whose degradation is BsRppH-dependent, we screened several chromosomally encoded mRNAs for an increase in longevity in ΔrppH versus wild-type cells. In this manner, the dicistronic yhxA-glpP transcript was identified as a likely BsRppH target. This 2.2-kb mRNA encodes a protein of unknown function (YhxA) and a transcription antiterminator (GlpP) important for regulating the synthesis of glycerol-3-phosphate dehydrogenase (Glatz et al., 1996). In the absence of BsRppH, the average measured half-life of the yhxA-glpP transcript increased from 3.7 ± 0.6 min to 8.3 ± 0.6 min (Figure 2B and C, Table S1).
Figure 2. Effect of BsRppH on the decay rate and phosphorylation state of mRNA in B. subtilis.
The decay of ΔermC and yhxA-glpP mRNA in wild-type and ΔrppH B. subtilis cells was monitored by Northern blot analysis of total RNA extracted at time intervals after inhibiting transcription with rifampicin. A probe specific for tRNACys was used to confirm that an equal amount of total RNA was loaded in each lane. In addition, the 5′ phosphorylation state of ΔermC or yhxA-glpP mRNA in the same strains was examined by PABLO analysis with oligonucleotides X90 and Yerm1 or, after cleavage with DNAzyme YhxA1, oligonucleotides X32 and Y0, respectively. The PABLO ligation product of yhxA-glpP comigrated with an electrophoretic marker 214 nt long, equal to the sum of the lengths of the 5′ yhxA-glpP cleavage product and X32 (data not shown).
A) Decay of plasmid-encoded ΔermC mRNA.
B) Decay of chromosomally encoded yhxA-glpP mRNA.
C) Semi-log plots of mRNA concentration as a function of time after rifampicin addition.
D) PABLO analysis of plasmid-encoded ΔermC mRNA (left) and chromosomally encoded yhxA-glpP mRNA (right) in wild-type and ΔrppH cells.
Representative experiments are shown; mean values and standard deviations from multiple half-life and PABLO measurements are provided in Table S1. See also Figure S2.
Effect of BsRppH on the phosphorylation state of mRNA in B. subtilis
To ascertain whether the BsRppH-dependent degradation of yhxA-glpP mRNA occurs through the generation of a monophosphorylated intermediate, we first had to identify the 5′-terminal nucleotide of the transcript. The 5′ end was mapped at medium resolution to a site 85–105 nucleotides upstream of the probable yhxA initiation codon (AUG) and 70–90 nucleotides upstream of the likely Shine-Dalgarno element (AAUGGAG) by examining the length of the 5′-terminal RNA fragments produced upon cleavage of yhxA-glpP mRNA with a set of three target-specific 10–23 DNAzymes (Figure S2A) (Santoro and Joyce, 1997). A short distance upstream of that region is a sequence element (TTGACT – 17 bp – TATATT) that resembles a typical B. subtilis σA promoter (Haldenwang, 1995). Mutating either the − 10 or − 35 region of that putative promoter abolished its activity in mRNA synthesis (Figure S2B), indicating that it directs transcription of the yhxA-glpP operon.
Precise identification of the 5′-terminal nucleotide of yhxA-glpP mRNA was accomplished by PABLO analysis, a splinted ligation assay for detecting monophosphorylated RNA 5′ ends (Celesnik et al., 2007). This assay takes advantage of the ability of T4 DNA ligase to covalently join a DNA oligonucleotide (oligo X) to the 5′ end of a monophosphorylated RNA when the two are juxtaposed by simultaneous base pairing to a bridging DNA oligonucleotide (oligo Y). Previous studies have shown that PABLO ligation can occur only when the 3′ end of oligo X and the 5′ end of the RNA are positioned directly adjacent to one another upon base pairing with oligo Y or, with lower efficiency, when they are separated by one nucleotide (Celesnik et al., 2007). Total RNA extracted from wild-type B. subtilis cells was first treated with tobacco acid pyrophosphatase (TAP) to convert 5′-terminal triphosphates to ligatable monophosphates and cleaved within yhxA codon 30 with a DNAzyme to improve the electrophoretic resolution of the assay. The RNA was then examined by PABLO with a nested set of four bridging oligonucleotides differing in length by one nucleotide each at the expected site of yhxA-glpP transcription initiation. Only two of these oligonucleotides mediated ligation of DNAzyme-cleaved yhxA-glpP RNA to oligo X (Figure S2C). One (Y0) perfectly juxtaposed the 3′ end of oligo X and the expected 5′-terminal nucleotide of the transcript, resulting in a high ligation yield, while the other (Y+1) left a one-nucleotide gap between those two ends, resulting in a lower ligation yield. In this manner, we determined that transcription of the yhxA-glpP operon begins at a unique site seven nucleotides downstream of the promoter (TTGACT – 17 bp – TATATT – 6 bp – A) and 93 nucleotides upstream of the probable yhxA translation initiation codon.
Having precisely mapped the 5′ end of the yhxA-glpP transcript, we were next able to examine its natural 5′ phosphorylation state by PABLO analysis without prior TAP treatment. In wild-type B. subtilis cells, a ligation yield of 5.6 ± 1.3% was observed for yhxA-glpP mRNA in the presence of oligo Y0 (Figure 2D, Table S1), indicating that the γ and β phosphates are removed from the 5′ end of the transcript to generate a monophosphorylated decay intermediate. The PABLO ligation yield fell to only 1.3 ± 0.3% in ΔrppH cells, suggesting the importance of BsRppH for 5′-terminal phosphate removal and implying a causal relationship between this modification and mRNA decay. In view of the yhxA-glpP ligation yield of 5.6% and the 80% ligation yield obtained for fully monophosphorylated yhxA-glpP mRNA after TAP treatment (Figure S2D), we estimate that in wild-type cells about 7% of yhxA-glpP mRNA is monophosphorylated at steady state. Similarly, a substantial fraction of ΔermC mRNA from wild-type cells underwent PABLO ligation (17 ± 4%), whereas the relative abundance of the monophosphorylated form of ΔermC mRNA was halved to 8.0 ± 0.5% in ΔrppH cells (Figure 2D, Table S1), suggesting that BsRppH participates in 5′ phosphate removal from this transcript as well.
Complementation of an rppH null mutant
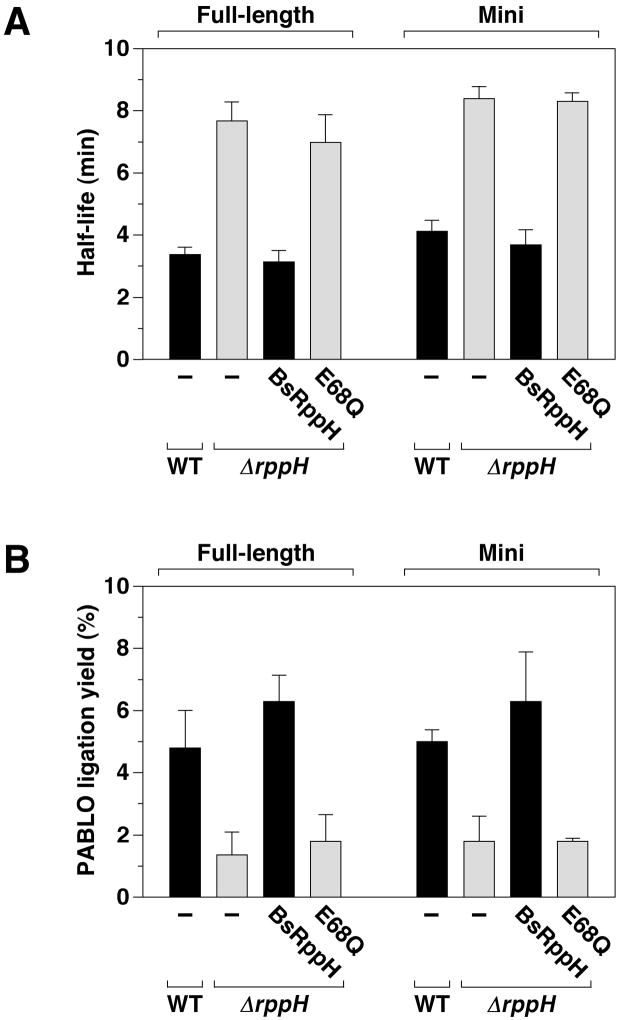
To make it easier to investigate the role of BsRppH in mRNA decay, we enhanced detection of the yhxA-glpP transcript by cloning the operon on a multicopy shuttle plasmid. In addition, we created an in-frame deletion within the cloned yhxA-glpP operon so as to join the first 34 codons of yhxA to the last 8 codons of glpP, resulting in a mini transcript 0.35 kb in length. The plasmid-encoded full-length and mini transcripts each decayed with approximately the same BsRppH-dependent half-life and PABLO ligation yield as the chromosomally encoded transcript (Figure 3, Tables S2 and S3), indicating that the degradation of yhxA-glpP mRNA is not affected by its cellular concentration or by internal elements within the 1.8-kb region absent from the mini transcript. The diminished ligation yield and prolonged half-life of each of the plasmid-encoded transcripts in ΔrppH cells were restored to normal by complementation with a wild-type copy of the B. subtilis rppH gene inserted elsewhere on the B. subtilis chromosome but not by a catalytically inactive rppH-E68Q allele, demonstrating that those abnormal phenotypes in ΔrppH cells resulted from a lack of BsRppH pyrophosphohydrolase activity.
Figure 3. Comparison of the decay rate and phosphorylation state of full-length and mini yhxA-glpP mRNA and complementation of a ΔrppH mutation.
Half-lives and PABLO ligation yields were measured for plasmid-encoded full-length and mini yhxA-glpP mRNA in wild-type and ΔrppH B. subtilis cells and in ΔrppH cells in which BsRppH or BsRppH-E68Q was ectopically produced. Bar heights and error bars correspond to means and standard deviations from multiple experiments, the numerical values of which are provided in Tables S2 and S3. Black bars, cells containing active BsRppH; gray bars, cells lacking active BsRppH.
A) mRNA half-lives.
B) PABLO yields.
Effect of 5′-terminal base pairing on mRNA destabilization by BsRppH in B. subtilis
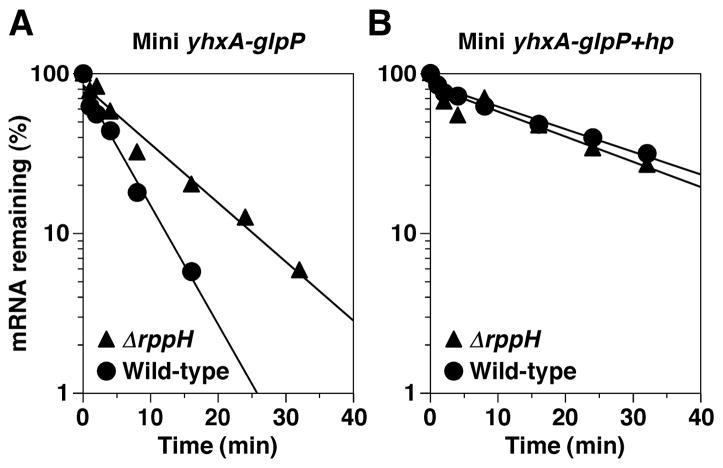
The inhibitory effect of 5′-terminal base pairing on the activity of purified BsRppH suggested that RNA degradation triggered by γ and β phosphate removal could be blocked in vivo by structurally sequestering the RNA 5′ end. This hypothesis was tested in B. subtilis by adding a 5′-terminal stem-loop to the mini yhxA-glpP transcript, generating mini yhxA-glpP+hp mRNA. The first four nucleotides of the resulting transcript were identical in sequence to those present at the 5′ end of its progenitor but were base-paired. In wild-type cells, this modification increased the half-life of the mini transcript to 21 ± 2 min, markedly longer than the 4.1 ± 0.4 min half-life of its unmodified counterpart (Figure 4, Figure S3). Moreover, in contrast to the unmodified mini transcript, the mini transcript bearing a 5′ stem-loop was not further stabilized in ΔrppH cells. This epistatic effect of a 5′-terminal stem-loop indicates that the destabilizing influence of BsRppH in B. subtilis requires access to an unsequestered 5′ end. We conclude that the RNA pyrophosphohydrolase activity of BsRppH acts directly to trigger the decay of yhxA-glpP mRNA in B. subtilis by converting the triphosphorylated 5′ end to a monophosphate, thereby making the transcript more susceptible to degradation.
Figure 4. Effect of a 5′-terminal stem-loop on the decay rate and BsRppH sensitivity of mini yhxA-glpP mRNA.
The decay of (A) mini yhxA-glpP mRNA or (B) mini yhxA-glpP+hp mRNA in wild-type and ΔrppH B. subtilis cells was monitored by Northern blot analysis of total RNA extracted at time intervals after inhibiting transcription, and band intensities were plotted on a semi-log graph. These two transcripts were identical except that mini yhxA-glpP+hp mRNA contained a 5′-terminal stem-loop (AGGGCCGAAGCTTCGGCCCT) that did not change the sequence of the first four transcribed nucleotides.
See also Figure S3.
Identification of the ribonuclease that degrades monophosphorylated yhxA-glpP mRNA in B. subtilis
To identify the ribonuclease that degrades yhxA-glpP mRNA after 5′ phosphate removal by BsRppH, we examined the consequence of depleting two B. subtilis ribonucleases that reportedly prefer monophosphorylated RNA as a substrate: the 5′ exonuclease/endonuclease RNase J1 and the endonuclease RNase Y. Because both of these ribonucleases are essential for cell growth, their genes (rnjA and rny, respectively) were each placed under the control of an IPTG-inducible Pspac promoter, and the effect of IPTG withdrawal was observed in each of the two resulting strains.
When the Pspac-rnjA strain was grown in the presence of IPTG, the half-life of the chromosomally encoded yhxA-glpP transcript (4.2 ± 0.4 min) was similar to that seen in wild-type cells (Figure S4A). Depletion of RNase J1 by IPTG removal caused the half-life to increase more than four-fold to 18 ± 1 minutes (Figure 5A). In a parallel experiment, the mini yhxA-glpP transcript was likewise stabilized upon depletion of RNase J1 (data not shown). By contrast, the decay rate of yhxA-glpP mRNA was not affected by depleting RNase Y from the Pspac-rny strain (Figure 5A), unlike rpsO mRNA, a known RNase Y target (Yao and Bechhofer, 2010) that was stabilized more than six-fold (Figure S4B). These findings indicate that RNase J1 is the 5′-monophosphate-dependent ribonuclease that degrades the yhxA-glpP transcript in B. subtilis and that RNase Y does not contribute significantly to this process. Consistent with the conclusion that RNase J1 is principally responsible for degrading monophosphorylated yhxA-glpP mRNA, the concentration of that decay intermediate increased threefold relative to its triphosphorylated precursor when RNase J1 was depleted (Figure 5B). Prior evidence that RNase J1 depletion prolongs the lifetime of ΔermC mRNA (Yao et al., 2009) suggests that the monophosphorylated form of that message is degraded by a similar mechanism.
Figure 5. Degradation of monophosphorylated yhxA-glpP mRNA by RNase J.
A) Effect of RNase J1 or RNase Y depletion on the longevity of yhxA-glpP mRNA. The half-life of the chromosomal yhxA-glpP transcript was measured in wild-type B. subtilis cells and cells in which RNase J1 or RNase Y produced under the control of an IPTG-inducible promoter had been depleted by removing IPTG from the growth medium. Bar heights and error bars correspond to means and standard deviations, respectively.
B) Accumulation of monophosphorylated yhxA-glpP mRNA upon depletion of RNase J1. PABLO analysis was performed on the chromosomal yhxA-glpP transcript extracted from wild-type B. subtilis cells and cells in which RNase J1 had been depleted. In each case, the transcript was cleaved site-specifically with DNAzyme YhxA1 to improve the electrophoretic resolution of the assay.
See also Figure S4.
Exonucleolytic degradation of monophosphorylated mini yhxA-glpP RNA
RNase J1 functions as both a 5′ exonuclease and an endonuclease. To determine which of these activities degrades yhxA-glpP mRNA after removal of the γ and β phosphates, we compared the degradation of triphosphorylated and monophosphorylated mini yhxA-glpP RNA by purified RNase J1, either alone or in a complex with RNase J2. In each case, the RNA substrate was 5′-end-labeled, making it possible to determine whether the 5′-terminal nucleotide was released as a mononucleotide, indicating exonucleolytic degradation, or as part of an oligonucleotide, indicating endonucleolytic cleavage at an internal site. Both RNase J1 alone and the RNase J1-J2 complex degraded the monophosphorylated transcript much more rapidly than its triphosphorylated counterpart (Figure 6, Figure S5). Even at early reaction times, more than 90% of the 5′ radiolabel released by either enzyme from the monophosphorylated transcript was in the form of a mononucleotide, whereas degradation of the triphosphorylated transcript generated mainly oligonucleotides as the initial 5′-terminal products.
Figure 6. Degradation of mini yhxA-glpP RNA by purified RNase J.
A) Degradation of 5′-end-labeled monophosphorylated and triphosphorylated mini yhxA-glpP RNA by RNase J (an equimolar mixture of the J1 and J2 subunits). Reaction samples were quenched at time intervals and analyzed by electrophoresis beside radiolabeled ATP, radiolabeled AMP, and a 60-min mini yhxA-glpP RNA sample to which no RNase J had been added (Mock).
B) Rates of exonucleolytic and endonucleolytic degradation by RNase J. For each lane of panel A, the percentage of the total radioactivity that corresponded to exonucleolytic products (mononucleotides) or endonucleolytic products (oligonucleotides) was plotted as a function of time after correcting for any such products already present at time 0.
See also Figure S5.
These findings indicate that RNase J1 and the RNase J1-J2 complex both prefer monophosphorylated RNA as a substrate and degrade it principally by a 5′-exonucleolytic mechanism. Whereas monophosphorylated RNA is a much better substrate for exonucleolytic degradation than its triphosphorylated counterpart, endonucleolytic cleavage by these ribonucleases appears to be slow irrespective of the 5′ phosphorylation state.
DISCUSSION
The ability of RNase E to preferentially cleave monophosphorylated RNA decay intermediates is of critical importance to the 5′-end-dependent pathway for degrading primary transcripts in E. coli. However, the absence of that key ribonuclease and a close sequence homologue of the E. coli RNA pyrophosphohydrolase EcRppH in a substantial fraction of bacterial species left unexplained how 5′-terminal base pairing governs mRNA turnover in such organisms. We have now determined the mechanism of 5′-end-dependent mRNA degradation in one such species, B. subtilis, where our findings reveal that BsRppH can trigger decay by converting the triphosphorylated 5′ termini of primary transcripts to monophosphorylated ends, thereby exposing them to attack by the monophosphate-dependent 5′ exonuclease activity of RNase J (Figure 7). This pathway differs significantly from its counterpart in E. coli, which lacks RNase J and relies instead on endonucleolytic cleavage by RNase E to destroy RNA after pyrophosphate is removed from the 5′ end (Deana et al., 2008).
Figure 7. Mechanism of a 5′-end-dependent pathway for RNA degradation in B. subtilis.
BsRppH (hatchet) removes the γ and β phosphates from the 5′ end of a triphosphorylated primary transcript, either simultaneously or sequentially. Once deprotected in this manner, the monophosphorylated decay intermediate is rapidly degraded by the 5′ exonuclease activity of theJ1 subunit of RNase J (Pac-Man).
The biological function of BsRppH in B. subtilis has not previously been identified, although the purified enzyme was shown to convert (deoxy)nucleoside triphosphates into (deoxy)nucleoside monophosphates (Xu et al., 2004). Except for the Nudix motif, it has limited sequence similarity to EcRppH; indeed, their overall sequence identity (23%) is less than that of another B. subtilis Nudix hydrolase that appears to lack RNA pyrophosphohydrolase activity (MutT). As previously reported for its hydrolytic activity on mononucleotide substrates (Xu et al., 2004), BsRppH removes the γ and β phosphates from the 5′ end of RNA almost exclusively as orthophosphate, whereas EcRppH releases them primarily (~85%) as pyrophosphate but also (~15%) as orthophosphate (Deana et al., 2008). Whether diphosphorylated RNA or pyrophosphate is transiently produced by BsRppH as a short-lived reaction intermediate is not clear. The purified enzyme is much more effective at removing phosphates from an unpaired 5′ end than from a 5′ end sequestered by base pairing, suggesting that BsRppH binds not only to the triphosphate on which it acts but also to at least one unpaired 5′-terminal nucleotide. This substrate preference helps to explain how a 5′-terminal stem-loop can protect mRNA from rapid degradation in B. subtilis by rendering it resistant to phosphate removal by BsRppH. By contrast, the susceptibility of mRNA to BsRppH-mediated degradation appears not to be influenced by features of the protein-coding region, as an in-frame deletion that almost completely removed the two translational units of the yhxA-glpP transcript had little or no effect on its phosphorylation state, half-life, or mechanism of decay. Prior evidence that BsRppH is produced during vegetative growth and sporulation (Ramírez et al., 2004) implies that it has the potential to initiate mRNA degradation during both of those growth stages.
Two riboucleases in B. subtilis have been reported to preferentially degrade RNAs that bear a single phosphate at the 5′ end: RNase J and RNase Y (Mathy et al., 2007; Li de la Sierra-Gallay et al., 2008; Shahbabian et al., 2009). Our findings indicate that, after phosphate removal by BsRppH, monophosphorylated yhxA-glpP mRNA is degraded by the 5′ exonuclease activity of the J1 subunit of RNase J and not by RNase Y. Furthermore, our studies have confirmed that the 5′ exonuclease activity of RNase J degrades monophosphorylated RNA much faster than triphosphorylated RNA and have shown that it can do so at a rate well in excess of that for internal cleavage by its endonuclease activity, which appears to be indifferent to the 5′ phosphorylation state of RNA. The low percentage of yhxA-glpP mRNA that is monophosphorylated at steady state in B. subtilis indicates that this decay intermediate is very rapidly degraded once it forms. Consistent with this interpretation, the relative abundance of monophosphorylated yhxA-glpP mRNA increases markedly when the J1 subunit is depleted from cells. The 5′ exonuclease activity of RNase J is also thought to degrade 3′-terminal mRNA fragments that are generated by endonucleolytic cleavage (Mathy et al., 2007; Collins et al., 2007; Deikus et al., 2008; Daou-Chabo et al., 2009).
Various properties of the rppH deletion mutant suggest that BsRppH may not be the only RNA pyrophosphohydrolase in B. subtilis. For example, deleting the B. subtilis rppH gene reduces the percentage of ΔermC mRNA that is monophosphorylated but does not eliminate that decay intermediate from cells. Moreover, the same deletion impedes the degradation of mini yhxA-glpP mRNA to a lesser degree than does the depletion of RNase J1 or the addition of a 5′-terminal stem-loop, which may hinder 5′ phosphate removal not only by BsRppH but also by any redundant activity that may be present. Finally, the level of RNA pyrophosphohydrolase activity in crude extracts of B. subtilis is reduced but not abolished when the rppH gene is deleted (data not shown). Although the source of the redundant activity remains to be determined, it is not likely to be one of the other Nudix hydrolases, none of which contribute detectably to the RNA pyrophosphohydrolase activity in crude cell extracts or exhibit significant levels of such activity when purified and assayed in the presence of magnesium ions.
The discovery that Nudix hydrolases trigger mRNA decay in B. subtilis and E. coli via distinct pathways involving unrelated ribonucleases that operate by entirely different mechanisms implies that the evolutionary imperative for conserving the 5′-terminal triggering event was greater than that for the subsequent steps in degradation. For this reason, and because Nudix hydrolases are present in almost all living organisms (McLennan, 2006), a 5′-end-dependent degradation pathway similar to the one described here for B. subtilis is likely to be important for governing mRNA lifetimes in many other Gram-positive and Gram-negative bacterial species that contain RNase J, such as Staphylococcus aureus, Streptococcus pyogenes, and Helicobacter pylori. This pathway bears a striking resemblance to the principal mechanism of mRNA decay in eukaryotic cells, where poly(A) tail removal facilitates cleavage of the m7Gppp cap by Dcp2 or Nudt16 to generate a monophosphorylated decay intermediate that is then rapidly degraded by the 5′ exonuclease Xrn1 (Muhlrad et al., 1994; Dunckley and Parker, 1999; Song et al., 2010). Interestingly, although RNase J and Xrn1 are not sequence homologues, the bacterial RNA pyrophosphohydrolases BsRppH and EcRppH and the eukaryotic decapping enzymes Dcp2 and Nudt16 are all Nudix hydrolases, suggesting divergent evolution from a common progenitor. Thus, it appears that the deprotection of RNA 5′ ends by a Nudix hydrolase is an ancient mechanism for triggering RNA degradation.
EXPERIMENTAL PROCEDURES
Strains
Measurements of the lifetime and phosphorylation state of mRNA were performed in B. subtilis strain 1A2 or 168 (Zeigler et al., 2008) and derivatives thereof in which the B. subtilis rppH gene (previously denoted ytkD) had been deleted or the RNase J1 gene (rnjA) had been placed under the control of an IPTG-inducible promoter (Pspac-rnjA) (Britton et al., 2007). The rppH gene was deleted by transforming B. subtilis 1A2 with a DNA fragment comprising a spectinomycin resistance gene flanked on one side by a 1-kb B. subtilis chromosomal segment ending 112 bp upstream of the rppH start codon and on the other side by a 1-kb chromosomal segment beginning 36 bp upstream of the rppH stop codon. Recombinants were selected on plates containing 100 μg/ml spectinomycin, and the deletion was confirmed by PCR amplification and DNA sequencing. Complementation of the B. subtilis rppH deletion mutant was achieved by integrating a wild-type or inactive copy of the rppH gene (rppH or rppH-E68Q) into the amyE locus. In each case, the complementing rppH translational unit was first inserted into integration vector pDR66 (Ireton et al., 1993) so as to place it under the control of an IPTG-inducible Pspac promoter. The resulting plasmids were then introduced into B. subtilis, and chromosomal integrants were selected on plates containing 5 μg/ml chloramphenicol and confirmed by PCR.
Plasmids and oligonucleotides
Plasmids pPlacBsRppH6, pPlacNudFH6, pPlacYcvIH6, pPlacMutTH6, pPlacYtbEH6 and pPlacYjhBH6 are derivatives of pRNGL3 (Lee et al., 2002) that were used to produce each of the six B. subtilis Nudix hydrolases as an amino-terminally hexahistidine-tagged protein in E. coli under the control of an IPTG-inducible lacUV5 promoter. Plasmid pPlacBsRppH6-E68Q was constructed from pPlacBsRppH6 by a substitution at codon 68 (GAA → CAA). Plasmid pSD285, encoding ΔermC mRNA, was described previously (Yao et al., 2008). Plasmid pBYGfl was constructed by inserting the entire B. subtilis yhxA-glpP operon and a fragment of pDG148-Stu (Joseph et al., 2001) bearing the B. subtilis pUB110 replication origin and a kanamycin resistance gene into the E. coli vector pPM30 (Meacock and Cohen, 1980). Plasmid pBYGmini is a deletion variant of pBYGfl in which the first 34 codons of the yhxA were fused in-frame to the last 8 codons of glpP. Plasmid pBYGmini+hp was constructed from pBYGmini by inserting the self-complementary sequence AGGGCCGAAGCTTCGGCCCT precisely at the beginning of the transcribed region of the mini yhxA-glpP gene. Plasmids pBYGmini35 and pBYGmini10 are identical to pBYGmini except for a modification altering the −35 (TTGACT → CCCCCC) or −10 (TATATT → CCCCCC) region, respectively, of the mini yhxA-glpP promoter. The sequences of the oligodeoxynucleotides used for Northern blotting, PABLO analysis, PCR amplification, in vitro transcription, and RNA cleavage are provided in Table S4.
In vitro assays of RNA pyrophosphohydrolase activity
B. subtilis Nudix hydrolases bearing an amino-terminal hexahistidine tag were produced in E. coli, purified by affinity chromatography, and assayed for RNA pyrophosphohydrolase activity in the presence of magnesium ions, as described previously (Deana et al., 2008). Triphosphorylated ΔermC, rpsT P1, and rpsT P1+hp RNA bearing a 5′-terminal γ-32P label and an internal fluorescein label and triphosphorylated, diphosphorylated, and monophosphorylated GA(CU)13 bearing a single 32P label between the first and second nucleotide were synthesized by in vitro transcription (Deana et al., 2008) and used as substrates in these assays.
RNA extraction from B. subtilis
Total RNA was isolated from B. subtilis by one of two methods: either treatment with lysozyme followed by the hot-phenol extraction procedure as described previously (Celesnik et al., 2008) or by the glass beads method as described previously (Bechhofer et al., 2008).
Measurement of RNA lifetimes
All B. subtilis strains were grown at 37°C in defined BLM medium (Stülke et al., 1993) containing 0.75% fructose (w/v) as the carbon source and 0.4 mM KH2PO4 but no additional amino acids, except in the RNase J1 and RNase Y depletion experiments, which were performed as described previously (Britton et al., 2007). Total cellular RNA was extracted from B. subtilis at time intervals after inhibiting transcription with rifampicin (0.2 mg/ml), and equal amounts were subjected to gel electrophoresis on 6% polyacrylamide containing 8 M urea or 1.2% agarose containing 2.4% formaldehyde. RNA was transferred to a Hybond-XL membrane (GE Healthcare) by electroblotting for polyacrylamide gels or overnight capillary transfer in 20xSSC for agarose gels. The chromosomally encoded yhxA-glpP transcript was detected by probing with an internally radiolabeled DNA or RNA probe complementary to glpP, whereas the plasmid-encoded yhxA-glpP and mini yhxA-glpP transcripts, ΔermC mRNA, and tRNACys were detected by probing with a complementary 5′-end-labeled oligodeoxynucleotide (Table S4). Radioactive bands were visualized with a Storm 820 PhosphorImager (Molecular Dynamics) or a Typhoon Trio Imager (GE Healthcare), and the band intensities were quantified by using ImageQuant software (Molecular Dynamics). RNA half-lives were calculated by linear regression analysis of data.
Cleavage of yhxA-glpP mRNA by DNAzymes
Total RNA from B. subtilis (12 μg) was combined in a final volume of 36 μl with 400 pmoles of one of three oligodeoxynucleotides (Table S4) designed to act as 10–23 DNAzymes (Santoro and Joyce, 1997) that cleave yhxA-glpP mRNA within yhxA codon 30 (YhxA1), 73 (YhxA2), or 92 (YhxA3), where yhxA codon 1 is assumed to be the AUG triplet located nine nucleotides downstream of a likely Shine-Dalgarno element (AAUGGAG). The mixtures were heated to 75°C for 5 min, slowly cooled to 30°C, and chilled on ice for at least 1 min. T4 DNA ligase buffer (New England Biolabs) was added to a final concentration of 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 10 mM dithiothreitol, and 1 mM ATP, and the cleavage reactions were allowed to proceed at 37°C for 4 hr. The reaction products were concentrated by ethanol precipitation and analyzed by Northern blot analysis alongside a 5′-end-labeled 500-bp DNA ladder (New England Biolabs).
PABLO analysis
To map the 5′-terminal nucleotide of yhxA-glpP mRNA, total cellular RNA (120 μg) was treated for 2 hr at 37°C with tobacco acid pyrophophatase (TAP, 12 units, Epicentre Biotechnologies) in 50 mM sodium acetate (pH 6.0), 1 mM EDTA, 0.1% β-mercaptoethanol, and 0.01% Triton X-100. The reaction was stopped by EDTA addition and phenol-chloroform extraction, and the reaction products were recovered by ethanol precipitation. Samples (20 μg) were then analyzed by PABLO as described previously (Celesnik et al., 2008), except that each was combined with DNAzyme YhxA1, oligodeoxynucleotide X32, and one of four bridging oligodeoxynucleotides (Y−1, Y0, Y+1,Y+2) that differed in length from one another by one nucleotide (Table S4). Subsequent PABLO analyses of yhxA-glpP mRNA without TAP treatment were performed with oligonucleotides YhxA1, X32, and Y0. The PABLO ligation yield for fully monophosphorylated yhxA-glpP mRNA was determined by first treating total cellular RNA (20 μg) with TAP (0.3, 1.0, or 3.0 units) to remove γ and β phosphates. PABLO analysis of mini yhxA-glpP mRNA made use of oligonucleotides X90 and Y0 but omitted YhxA1. PABLO analysis of the ΔermC transcript was performed with oligonucleotides X90 and Yerm1 (Table S4). In each case, the PABLO ligation products were detected by Northern blot analysis with transcript-specific probes (Table S4).
RNA degradation by purified RNase J
The DNA template for in vitro synthesis of mini yhxA-glpP RNA was prepared from plasmid pBYGmini by PCR amplification, and RNA synthesis was performed by in vitro transcription with T7 RNA polymerase (Promega) according to the manufacturer’s instructions. Triphosphorylated mini yhxA-glpP RNA bearing a single radiolabeled phosphate at the 5′ end was prepared by in vitro transcription in the presence of [γ–32P]ATP. Monophosphorylated mini yhxA-glpP RNA bearing a single radiolabeled phosphate at the 5′ end was prepared by first synthesizing unlabeled monophosphorylated mini yhxA-glpP RNA by in vitro transcription in the presence of an 8-fold molar excess of AMP over ATP; the resulting transcript was then treated with Antarctic phosphatase (New England Biolabs) and 5′-end-labeled with T4 polynucleotide kinase (New England Biolabs) and [γ–32P]ATP. In each case, the RNA was purified by electrophoresis on 5% polyacrylamide and quantified spectrophotometrically. To equalize the specific radioactivity of the 5′-triphosphorylated and 5′-monophosphorylated RNAs (3000 cpm/pmol), the radiolabeled monophosphorylated transcript was mixed with unlabeled monophosphorylated transcript. Enzyme-catalyzed reactions were performed at 37°C by combining the RNA substrate (10 pmol) with purified B. subtilis RNase J1 or RNase J1+J2 tetramers (4 pmol) in 50 μl of buffer containing 20 mM Tris-HCl (pH 8.0), 8 mM MgCl2, 100 mM NH4Cl, and 0.1 mM dithiothreitol, as described previously (Mathy et al., 2010). At time intervals, 5 μl reaction samples were quenched with an equal volume of stop buffer (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, and 0.05% xylene cyanol) and analyzed by electrophoresis on a 20% polyacrylamide/7 M urea gel beside radiolabeled ATP and AMP.
Supplementary Material
Acknowledgments
We are grateful to Patrick Eichenberger for helpful advice. This research was supported by grants from the National Institutes of Health to J.G.B. (GM35769) and D.H.B. (GM48804) and from the CNRS (UPR 9073), Université Paris VII-Denis Diderot, and the Agence Nationale de la Recherche (subtilRNA2) to C.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arnold TE, Yu J, Belasco JG. mRNA stabilization by the ompA 5′ untranslated region: two protective elements hinder distinct pathways for mRNA degradation. RNA. 1998;4:319–330. [PMC free article] [PubMed] [Google Scholar]
- Arraiano CM, Andrade JM, Domingues S, Guinote IB, Malecki M, Matos RG, Moreira RN, Pobre V, Reis FP, Saramago M, et al. The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol Rev. 2010;34:883–923. doi: 10.1111/j.1574-6976.2010.00242.x. [DOI] [PubMed] [Google Scholar]
- Babitzke P, Kushner SR. The Ams (altered mRNA stability) protein and ribonuclease E are encoded by the same structural gene of Escherichia coli. Proc Natl Acad Sci USA. 1991;88:1–5. doi: 10.1073/pnas.88.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker KE, Mackie GA. Ectopic RNase E sites promote bypass of 5′-end-dependent mRNA decay in Escherichia coli. Mol Microbiol. 2003;47:75–88. doi: 10.1046/j.1365-2958.2003.03292.x. [DOI] [PubMed] [Google Scholar]
- Bechhofer DH, Oussenko IA, Deikus G, Yao S, Mathy N, Condon C. Analysis of mRNA decay in Bacillus subtilis. Methods Enzymol. 2008;447:259–276. doi: 10.1016/S0076-6879(08)02214-3. [DOI] [PubMed] [Google Scholar]
- Belasco JG. All things must pass: contrasts and commonalities in eukaryotic and bacterial mRNA decay. Nat Rev Mol Cell Biol. 2010;11:467–478. doi: 10.1038/nrm2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet P, Belasco JG. Control of RNase E-mediated RNA degradation by 5′-terminal base pairing in E. coli. Nature. 1992;360:488–491. doi: 10.1038/360488a0. [DOI] [PubMed] [Google Scholar]
- Britton RA, Wen T, Schaefer L, Pellegrini O, Uicker WC, Mathy N, Tobin C, Daou R, Szyk J, Condon C. Maturation of the 5′ end of Bacillus subtilis 16S rRNA by the essential ribonuclease YkqC/RNase J1. Mol Microbiol. 2007;63:127–138. doi: 10.1111/j.1365-2958.2006.05499.x. [DOI] [PubMed] [Google Scholar]
- Callaghan AJ, Marcaida MJ, Stead JA, McDowall KJ, Scott WG, Luisi BF. Structure of Escherichia coli RNase E catalytic domain and implications for RNA turnover. Nature. 2005;437:1187–1191. doi: 10.1038/nature04084. [DOI] [PubMed] [Google Scholar]
- Celesnik H, Deana A, Belasco JG. Initiation of RNA decay in Escherichia coli by 5′ pyrophosphate removal. Mol Cell. 2007;27:79–90. doi: 10.1016/j.molcel.2007.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celesnik H, Deana A, Belasco JG. PABLO analysis of RNA 5′-phosphorylation state and 5′-end mapping. Methods Enzymol. 2008;447:83–98. doi: 10.1016/S0076-6879(08)02205-2. [DOI] [PubMed] [Google Scholar]
- Collins JA, Irnov I, Baker S, Winkler WC. Mechanism of mRNA destabilization by the glmS ribozyme. Genes Dev. 2007;21:3356–3368. doi: 10.1101/gad.1605307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commichau FM, Rothe FM, Herzberg C, Wagner E, Hellwig D, Lehnik-Habrink M, Hammer E, Volker U, Stulke J. Novel activities of glycolytic enzymes in Bacillus subtilis: interactions with essential proteins involved in mRNA processing. Mol Cell Proteomics. 2009;8:1350–1360. doi: 10.1074/mcp.M800546-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daou-Chabo R, Mathy N, Benard L, Condon C. Ribosomes initiating translation of the hbs mRNA protect it from 5′-to-3′ exoribonucleolytic degradation by RNase J1. Mol Microbiol. 2009;71:1538–1550. doi: 10.1111/j.1365-2958.2009.06620.x. [DOI] [PubMed] [Google Scholar]
- Deana A, Celesnik H, Belasco JG. The bacterial enzyme RppH triggers messenger RNA degradation by 5′ pyrophosphate removal. Nature. 2008;451:355–358. doi: 10.1038/nature06475. [DOI] [PubMed] [Google Scholar]
- Deikus G, Condon C, Bechhofer DH. Role of Bacillus subtilis RNase J1 endonuclease and 5′-exonuclease activities in trp leader RNA turnover. J Biol Chem. 2008;283:17158–17167. doi: 10.1074/jbc.M801461200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunckley T, Parker R. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 1999;18:5411–5422. doi: 10.1093/emboj/18.19.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emory SA, Bouvet P, Belasco JG. A 5′-terminal stem-loop structure can stabilize mRNA in Escherichia coli. Genes Dev. 1992;6:135–148. doi: 10.1101/gad.6.1.135. [DOI] [PubMed] [Google Scholar]
- Even S, Pellegrini O, Zig L, Labas V, Vinh J, Brechemmier-Baey D, Putzer H. Ribonucleases J1 and J2: two novel endoribonucleases in B. subtilis with functional homology to E. coli RNase E. Nucleic Acids Res. 2005;33:2141–2152. doi: 10.1093/nar/gki505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatz E, Nilsson RP, Rutberg L, Rutberg B. A dual role for the Bacillus subtilis glpD leader and the GlpP protein in the regulated expression of glpD: antitermination and control of mRNA stability. Mol Microbiol. 1996;19:319–328. doi: 10.1046/j.1365-2958.1996.376903.x. [DOI] [PubMed] [Google Scholar]
- Haldenwang WG. The sigma factors of Bacillus subtilis. Microbiol Rev. 1995;59:1–30. doi: 10.1128/mr.59.1.1-30.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambraeus G, Karhumaa K, Rutberg B. A 5′ stem-loop and ribosome binding but not translation are important for the stability of Bacillus subtilis aprE leader mRNA. Microbiology. 2002;148:1795–1803. doi: 10.1099/00221287-148-6-1795. [DOI] [PubMed] [Google Scholar]
- Hansen MJ, Chen LH, Fejzo MLS, Belasco JG. The ompA 5′ untranslated region impedes a major pathway for mRNA degradation in Escherichia coli. Mol Microbiol. 1994;12:707–716. doi: 10.1111/j.1365-2958.1994.tb01058.x. [DOI] [PubMed] [Google Scholar]
- Hunt A, Rawlins JP, Thomaides HB, Errington J. Functional analysis of 11 putative essential genes in Bacillus subtilis. Microbiology. 2006;152:2895–2907. doi: 10.1099/mic.0.29152-0. [DOI] [PubMed] [Google Scholar]
- Ireton K, Rudner DZ, Siranosian KJ, Grossman AD. Integration of multiple developmental signals in Bacillus subtilis through the Spo0A transcription factor. Genes Dev. 1993;7:283–294. doi: 10.1101/gad.7.2.283. [DOI] [PubMed] [Google Scholar]
- Joseph P, Fantino JR, Herbaud ML, Denizot F. Rapid orientated cloning in a shuttle vector allowing modulated gene expression in Bacillus subtilis. FEMS Microbiol Lett. 2001;205:91–97. doi: 10.1111/j.1574-6968.2001.tb10930.x. [DOI] [PubMed] [Google Scholar]
- Lee K, Bernstein JA, Cohen SN. RNase G complementation of rne null mutation identifies functional interrelationships with RNase E in Escherichia coli. Mol Microbiol. 2002;43:1445–1456. doi: 10.1046/j.1365-2958.2002.02848.x. [DOI] [PubMed] [Google Scholar]
- Li de la Sierra-Gallay I, Zig L, Jamalli A, Putzer H. Structural insights into the dual activity of RNase. J Nat Struct Mol Biol. 2008;15:206–212. doi: 10.1038/nsmb.1376. [DOI] [PubMed] [Google Scholar]
- Mackie GA. Ribonuclease E is a 5′-end-dependent endonuclease. Nature. 1998;395:720–723. doi: 10.1038/27246. [DOI] [PubMed] [Google Scholar]
- Mäder U, Zig L, Kretschmer J, Homuth G, Putzer H. mRNA processing by RNases J1 and J2 affects Bacillus subtilis gene expression on a global scale. Mol Microbiol. 2008;70:183–196. doi: 10.1111/j.1365-2958.2008.06400.x. [DOI] [PubMed] [Google Scholar]
- Mathy N, Bénard L, Pellegrini O, Daou R, Wen T, Condon C. 5′-to-3′ exoribonuclease activity in bacteria: role of RNase J1 in rRNA maturation and 5′ stability of mRNA. Cell. 2007;129:681–692. doi: 10.1016/j.cell.2007.02.051. [DOI] [PubMed] [Google Scholar]
- Mathy N, Hebert A, Mervelet P, Benard L, Dorleans A, Li de la Sierra-Gallay I, Noirot P, Putzer H, Condon C. Bacillus subtilis ribonucleases J1 and J2 form a complex with altered enzyme behaviour. Mol Microbiol. 2010;75:489–498. doi: 10.1111/j.1365-2958.2009.07004.x. [DOI] [PubMed] [Google Scholar]
- McDowall KJ, Lin-Chao S, Cohen SN. A+U content rather than a particular nucleotide order determines the specificity of RNase E cleavage. J Biol Chem. 1994;269:10790–10796. [PubMed] [Google Scholar]
- McLennan AG. The Nudix hydrolase superfamily. Cell Mol Life Sci. 2006;63:123–143. doi: 10.1007/s00018-005-5386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacock PA, Cohen SN. Partitioning of bacterial plasmids during cell division: a cis-acting locus that accomplishes stable plasmid inheritance. Cell. 1980;20:529–542. doi: 10.1016/0092-8674(80)90639-x. [DOI] [PubMed] [Google Scholar]
- Melefors Ö, von Gabain A. Genetic studies of cleavage-initiated mRNA decay and processing of ribosomal 9S RNA show that the Escherichia coli ams and rne loci are the same. Mol Microbiol. 1991;5:857–864. doi: 10.1111/j.1365-2958.1991.tb00759.x. [DOI] [PubMed] [Google Scholar]
- Mildvan AS, Xia Z, Azurmendi HF, Saraswat V, Legler PM, Massiah MA, Gabelli SB, Bianchet MA, Kang LW, Amzel LM. Structures and mechanisms of Nudix hydrolases. Arch Biochem Biophys. 2005;433:129–143. doi: 10.1016/j.abb.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Mudd EA, Krisch HM, Higgins CF. RNase E, an endoribonuclease, has a general role in the chemical decay of E. coli mRNA: evidence that rne and ams are the same genetic locus. Mol Microbiol. 1990;4:2127–2135. doi: 10.1111/j.1365-2958.1990.tb00574.x. [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Decker CJ, Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′-3′ digestion of the transcript. Genes Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- Ono M, Kuwano M. A conditional lethal mutation in an Escherichia coli strain with a longer chemical lifetime of mRNA. J Mol Biol. 1979;129:343–357. doi: 10.1016/0022-2836(79)90500-x. [DOI] [PubMed] [Google Scholar]
- Ramírez MI, Castellanos-Juárez FX, Yasbin RE, Pedraza-Reyes M. The ytkD (mutTA) gene of Bacillus subtilis encodes a functional antimutator 8-oxo-(dGTP/GTP)ase and is under dual control of sigma A and sigma F RNA polymerases. J Bacteriol. 2004;186:1050–1059. doi: 10.1128/JB.186.4.1050-1059.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro SW, Joyce GF. A general purpose RNA-cleaving DNA enzyme. Proc Natl Acad Sci USA. 1997;94:4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbabian K, Jamalli A, Zig L, Putzer H. RNase Y, a novel endoribonuclease, initiates riboswitch turnover in Bacillus subtilis. EMBO J. 2009;28:3523–3533. doi: 10.1038/emboj.2009.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp JS, Bechhofer DH. Effect of 5′-proximal elements on decay of a model mRNA in Bacillus subtilis. Mol Microbiol. 2005;57:484–495. doi: 10.1111/j.1365-2958.2005.04683.x. [DOI] [PubMed] [Google Scholar]
- Song MG, Li Y, Kiledjian M. Multiple mRNA decapping enzymes in mammalian cells. Mol Cell. 2010;40:423–432. doi: 10.1016/j.molcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spickler C, Mackie GA. Action of RNase II and polynucleotide phosphorylase against RNAs containing stem-loops of defined structure. J Bacteriol. 2000;182:2422–2427. doi: 10.1128/jb.182.9.2422-2427.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stülke J, Hanschke R, Hecker M. Temporal activation of β-glucanase synthesis in Bacillus subtilis is mediated by the GTP pool. J Gen Microbiol. 1993;139:2041–2045. doi: 10.1099/00221287-139-9-2041. [DOI] [PubMed] [Google Scholar]
- Taraseviciene L, Miczak A, Apirion D. The gene specifying RNase E (rne) and a gene affecting mRNA stability (ams) are the same gene. Mol Microbiol. 1991;5:851–855. doi: 10.1111/j.1365-2958.1991.tb00758.x. [DOI] [PubMed] [Google Scholar]
- Vincent HA, Deutscher MP. Substrate recognition and catalysis by the exoribonuclease RNase R. J Biol Chem. 2006;281:29769–29775. doi: 10.1074/jbc.M606744200. [DOI] [PubMed] [Google Scholar]
- Xu W, Jones CR, Dunn CA, Bessman MJ. Gene ytkD of Bacillus subtilis encodes an atypical nucleoside triphosphatase member of the Nudix hydrolase superfamily. J Bacteriol. 2004;186:8380–8384. doi: 10.1128/JB.186.24.8380-8384.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S, Bechhofer DH. Initiation of decay of Bacillus subtilis rpsO mRNA by endoribonuclease RNase Y. J Bacteriol. 2010;192:3279–3286. doi: 10.1128/JB.00230-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S, Blaustein JB, Bechhofer DH. Erythromycin-induced ribosome stalling and RNase J1-mediated mRNA processing in Bacillus subtilis. Mol Microbiol. 2008;69:1439–1449. doi: 10.1111/j.1365-2958.2008.06370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S, Sharp JS, Bechhofer DH. Bacillus subtilis RNase J1 endonuclease and 5′ exonuclease activities in the turnover of ΔermC mRNA. RNA. 2009;15:2331–2339. doi: 10.1261/rna.1749109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeigler DR, Prágai Z, Rodriguez S, Chevreux B, Muffler A, Albert T, Bai R, Wyss M, Perkins JB. The origins of 168, W23, and other Bacillus subtilis legacy strains. J Bacteriol. 2008;190:6983–6995. doi: 10.1128/JB.00722-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y, Deutscher MP. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001;29:1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.