Abstract
RNA molecules play diverse functional roles in natural biological systems. There has been growing interest in designing synthetic RNA counterparts for programming biological function. The design of synthetic RNA molecules that exhibit diverse activities, including sensing, regulatory, information processing, and scaffolding activities, has highlighted the advantages of RNA as a programmable design substrate. Recent advances in implementing these engineered RNA molecules as key control elements in synthetic genetic networks are highlighting the functional relevance of this class of synthetic elements in programming cellular behaviors.
Introduction
Engineered biological systems have exciting potential in developing solutions to many global challenges, including environmental remediation, sustainability, scalable manufacturing, and health and medicine. Our ability to design and build synthetic biological systems is a key technology to improving the human condition. In addition, the redesign of biological systems can be used as an effective strategy to test, and thereby strengthen, our understanding of natural systems. Synthetic biology is an emerging research field with a primary goal of making the engineering of biology faster, less expensive, and more reliable. As such, core activities in synthetic biology have been focused on the development of foundational tools and technologies that assist in the design, construction, and characterization of biological systems (Endy, 2005; Smolke and Silver, 2011). Recent advances in construction and fabrication technologies are supporting synthesis of large pieces of DNA including entire pathways and genomes (Carr and Church, 2009). While progress has been made in the design of complex genetic circuits (Purnick and Weiss, 2009), current capabilities for constructing large genetic systems surpass our ability to design such systems. This growing `design gap' has highlighted the need to develop methods that support the generation of new functional biological components and scalable design strategies for complex genetic circuits that will lay the foundation for integrated biological devices and systems.
The vast majority of genetic systems engineered to-date have utilized protein-based transcriptional control strategies (Purnick and Weiss, 2009). However, as the examples of functional RNA molecules playing key roles in the behavior of natural biological systems have grown over the past decade, there has been growing interest in the design and implementation of synthetic counterparts. Researchers have taken advantage of the relative ease with which RNA molecules can be modeled and designed to engineer functional RNA molecules that act as diverse components including sensors, regulators, controllers (ligand-responsive RNA regulators), and scaffolds. More recently, researchers have begun to move beyond molecular design and integrate these synthetic RNA molecules as key elements in genetic circuits to program cellular behavior, highlighting the relevance and advantages of RNA-based control strategies. We will review the rapidly growing field of RNA synthetic biology as it transitions from the molecular design of RNA-based genetic parts and devices to the implementation of these elements in genetic systems for programming complex biological behaviors.
RNA as a natural regulatory molecule
The growing interest in using RNA to build synthetic controllers is due in large part to the steadily increasing examples of natural RNA regulators that control gene expression through diverse mechanisms in different organisms. One of the earliest examples is the regulation of gene expression through RNA secondary structure. The study of differential expression of genes in phage genomes led to the discovery that secondary structure of a mRNA transcript can restrict access to the ribosome binding site (RBS), thereby inhibiting translation (Kozak, 2005). Similarly, bacteria utilize the formation of tight hairpins in mRNA transcripts to stall and attenuate translation in the regulation of amino acid biosynthesis (Yanofsky, 1981). In addition, RNA structure is highly temperature sensitive, such that in certain cases hairpin structures that inhibit translation and can be modulated by temperature have been found to have functional roles in the heat and cold shock responses of several bacteria (Kozak, 2005). Finally, it has been shown that strong secondary structures on the 5' and 3' ends of a mRNA strand can protect the transcript from degradation by exoribonucleases and endoribonucleases (Alifano et al., 1994). The resulting extended half-lives of the transcripts can significantly increase protein production and have functional roles in processes such as photosynthesis and bacterial cell adhesion (Alifano et al., 1994; Ehretsmann et al., 1992).
In addition to structural mechanisms, the discovery that RNA can exhibit catalytic activity opened the door to a wider array of regulatory functions (Kruger et al., 1982). These catalytic RNAs, or ribozymes, typically catalyze cleavage or ligation of the RNA backbone through a reversible phosphodiester cleavage reaction (Serganov and Patel, 2007). Ribozymes have functional roles in alternative splicing, RNA replication, translation, and transcript stability and function in both prokaryotes and eukaryotes (Serganov and Patel, 2007). Furthermore, the discovery that ribozyme cleavage of the glmS transcript in bacteria is inhibited by binding of the metabolite GlcN6P has led to several discoveries of ribozymes acting as key components in riboswitches, a class of RNA regulators that respond to cellular metabolites and cofactors to modulate enzyme levels in related biosynthesis (Mandal and Breaker, 2004). Finally, RNase P is a catalytic RNA that functions in trans and can carry out multiple turnover cleavage events in the processing of 5' leader sequences from tRNA (Serganov and Patel, 2007). The discovery of ribozymes with natural gene regulatory activity in trans presents an intriguing proof of principle that a single catalytic RNA can be used to regulate several different genes in a biological system.
The last major mechanism that RNA uses to regulate protein synthesis is through antisense-mediated regulation of translation. Trans-acting small RNAs in bacteria are generally transcribed from their own independent transcripts and can range from 50–514 nucleotides (nts) in length (Gottesman, 2004). These RNAs often act in concert with the RNA binding protein Hfq and can promote or inhibit translation of their target mRNAs by relieving secondary structural elements or inhibiting ribosome initiation or processivity (Schluter et al., 2010). Similarly, in higher eukaryotes, RNA interference (RNAi) pathways use small RNAs, siRNA and microRNAs, to guide protein complexes to complementary mRNAs, leading to silencing of those targets (Carthew and Sontheimer, 2009). MicroRNAs have diverse roles in almost every cellular process and they are currently thought to regulate up to a third of human genes (Bartel, 2009). RNAi, particularly the siRNA pathway, is widely used as a tool in biological research for genetic loss of function studies and is currently being explored for therapeutic and biotechnological uses (Nicoloso et al., 2009).
Finally, RNA is involved in gene regulation in several other less widespread or well-studied mechanisms. For example, noncoding RNAs have been characterized that have roles in quality control of translation (Gottesman, 2004), binding to and inhibiting proteins involved in protein synthesis, and epigenetic DNA modification (Mattick et al., 2009). Furthermore, the recent discovery of the widespread regulatory activity and conservation of long noncoding RNAs represents an exciting area for further research in RNA-based gene regulation (Mercer et al., 2009).
RNA as a programmable and efficient substrate for engineering biological controllers
In addition to the diversity of mechanisms by which RNA can act as a regulatory molecule in nature, RNA exhibits several properties that make it an attractive design substrate in synthetic systems. RNA is composed of four building blocks that interact through well-characterized hydrogen-bond, base-stacking, and electrostatic interactions. The folding of RNA is primarily dictated by its secondary structure, in contrast to the folding of proteins, which involves a large degree of tertiary interactions. Models that predict RNA secondary structure have been developed based on the optimization of energies contributed by the Watson-Crick AU and GC base pairs as well as the GU wobble pair (Hofacker, 2003; Mathews et al., 2004; Zadeh et al., 2011). Progress in RNA 3-dimensional structural studies have revealed a set of non-canonical base pairing interactions, or Hoogsteen base pairs, that are key in the formation of the RNA tertiary structure. Early modeling frameworks that predict RNA tertiary structure based on a primary sequence have been developed and demonstrated to generate native-like structure predictions (Das and Baker, 2007; Parisien and Major, 2008). In addition to structure prediction based on thermodynamics, RNA kinetic folding program has also been developed to capture the stochastic nature of the RNA folding process (Flamm et al., 2000). These computational tools can be utilized to access structural information encoded in the primary RNA sequence, thereby aiding the rational design of genetic controllers based on hybridization schemes or structural elements.
RNA-based controllers exhibit additional advantages as functional elements in synthetic biological systems as the field moves toward more complex genetic systems design. RNA controllers generally exhibit more compact genetic footprints than their protein counterparts. In addition, RNA controllers generally place less of an energetic and resource load on the host cell as functional RNA molecules do not require the translation process to synthesize the functional elements. Another important consideration in genetic system design is timing of coupled control processes, where RNA-based posttranscriptional control strategies will generally act at faster time scales than transcription-based control strategies. The energetic, resource, and space efficiencies of RNA-based control strategies present important features supporting scaling to large-scale genetic system designs.
Approaches to generate functional RNA components
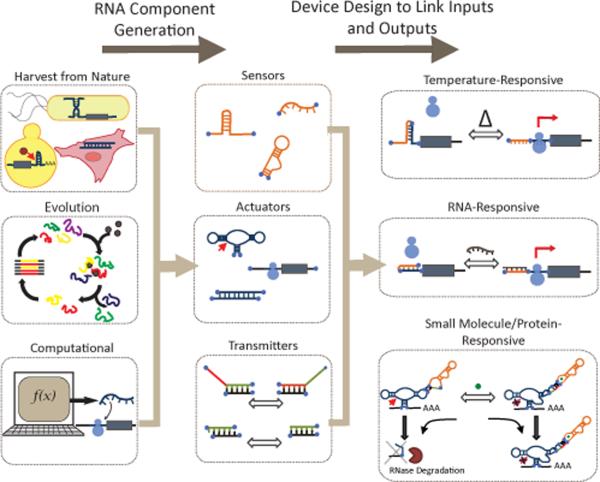
Functional RNA components are the basic building blocks for constructing genetic devices that encode human-defined functions. A fundamental challenge in the field then is the efficient generation of new component functions, such as sensing, information transmission, and actuation functions, that can be assembled into more complex devices. Three general approaches are taken to engineer new functional RNA components: harvesting from nature, computational design, and molecular evolution (Figure 1).
Figure 1.
RNA components used to engineer synthetic control functions can be harvested from natural systems or generated using molecular evolution and computational approaches. These components, encoding sensing, actuation, and information transmission activities, can then be assembled into RNA devices using various molecular engineering strategies to link one or more inputs of interest, such as temperature, RNA, small molecules, or proteins, to desired regulatory activities.
Harvesting and refining RNA components from nature
Many functional RNA components are derived from naturally-occurring elements, as advances in RNA biology have led to discoveries of natural RNA molecules that exhibit diverse functional activities. In certain instances the RNA component can be `harvested' from its native context and used in a synthetic genetic context where it will exhibit the desired activity. In one example, researchers isolated a ligand-binding RNA sequence to thiamine pyrophosphate (TPP) from a natural riboswitch and linked this natural RNA ligand-binding component to a hammerhead ribozyme to construct a synthetic RNA-based sensing-actuation element that responded to TPP in E. coli (Wieland et al., 2009). The native function of a naturally-occurring RNA element can also be altered to generate new functions or `refined' to make the element compatible with modular integration into broader genetic device or system platforms. Combined rational and evolutionary strategies have been applied to alter the native function of natural RNA elements. In one example, the native activity of an RNase III hairpin substrate was altered in yeast by modifying key sequences that are essential to protein-binding and cleavage activities within the regulatory element (Babiskin and Smolke, 2011a, b). The resulting hairpin libraries were inserted in the 3' UTR of the target transcript, and an in vivo screen was performed to identify a set of new hairpin sequences exhibiting a wide-range of regulatory activities. Similar approaches have been applied to alter the native activities of diverse RNA regulatory elements, including ribozymes (Chen et al., 2009), RNase cleavage sites (Pfleger et al., 2006), stabilizing elements (Smolke et al., 2000), RBS sequences (Anderson et al., 2006), and riboswitches (Dixon et al., 2010).
Computational tools for the design of RNA components
Computational tools have been developed to aid researchers in designing functional RNA components. For example, a computational method for designing synthetic RBS sequences was recently described based on predicting the energies of secondary structures around and including the RBS and the energies associated with the RBS:rRNA interaction (Salis et al., 2009). A two state thermodynamic model was devised, the initial state being the free 30S ribosomal subunit and folded mRNA transcript and the final state being the 30S complex bound to the transcript. The difference in free energy between these two states was used to predict a translation initiation rate for a given mRNA sequence. The computational method was applied to generate RBS sequences in E. coli that resulted in fluorescent reporter levels that spanned several orders of magnitude. This tool was then applied to design and optimize the expression of an input to a genetic AND gate. As another example, design tools for microRNA and shRNA elements have been developed by companies such as Life Technologies and Thermo Scientific. Given the sequence of a target gene of interest, these RNAi design tools can predict targeting sequences that will allow for efficient RNAi-mediated knockdown of that gene. These programs will design a shRNA or miRNA that incorporate the predicted targeting sequences, including the structural elements necessary for correct biogenesis and silencing efficiency, and primer sequences for construction and cloning of the regulatory element.
Forward engineering with current computational tools is not precise, and strategies for most effectively using these tools require some level of generating multiple functional RNA elements and then screening for those sequences that exhibit desired activities. For example, the RBS design tool has a probability of 0.47 of achieving a target protein expression level within two-fold. Further development of design tools that more accurately predict the precise DNA sequence needed to achieve a quantitative functional activity for different RNA components will allow for the efficient design, construction, and implementation of functional components tailored for various networks and systems.
Evolving new RNA component functions
While many functional RNA components can be harvested from natural biological systems and then refined and enhanced through rational design and evolutionary methods, such strategies can be limited to generating functions that are closely-related to the native activities. To generate new RNA component functions de novo, researchers have turned to in vitro selection strategies. The ability to readily interconvert between genetic information (DNA) and functional molecule (RNA) through transcription and reverse transcription (RT) processes and the ability to readily amplify DNA through polymerase chain reaction (PCR) processes allows efficient sampling of large RNA sequence space in vitro. Thus, large RNA libraries can be searched for rare functional sequences, which can subsequently be recovered, amplified, and searched again.
RNA presents a unique advantage over protein as a design substrate for control devices, as new sensing functions can be generated de novo through an in vitro selection strategy, systematic evolution of ligands by exponential enrichment (SELEX) (Ellington and Szostak, 1990; Tuerk and Gold, 1990). SELEX typically starts with an initial RNA library of ~1014–1015 molecules, each composed of a randomized region spanning ~30–70 nts flanked by constant sequences. Ligand-binding sequences are isolated by partitioning the RNA library through any of a number of different strategies, although affinity chromatography-based methods are most commonly used. The recovered sequences are subsequently amplified through RT-PCR and used as the starting library for the next round of selection. Iterative rounds of selection are performed, and the selection stringency and counterselections can be tailored to enrich for RNA sequences with high affinities and specificities to the target ligand. As one example, by incorporating counterselections against caffeine in the later selection rounds, an aptamer was selected to theophylline that exhibits a 10,000-fold lower affinity for caffeine, which differs from theophylline by a single methyl group (Jenison et al., 1994). In vitro selection strategies have also been applied to the generation of novel ribozymes capable of RNA ligation (Bartel and Szostak, 1993).
When utilizing RNA components generated through in vitro selection strategies for cellular applications, the activities of the in vitro optimized components may not translate directly to the complex cellular environment. Cell-based selection and screening strategies have been used to perform a secondary screen on in vitro enriched RNA component libraries by the component function to a measurable gene expression output. In one example, a yeast three-hybrid system was developed to allow screening of aptamer libraries to transcription factor NF-κB for in vivo activities (Cassiday and Maher, 2003). This system takes advantage of the modular nature of a yeast transcription activator, GAL4, by splitting it into a DNA binding and a transcription activation domain. The DNA binding domain is fused a known RNA-binding protein, MS2, and the transcription activation domain is fused to the target protein ligand, NF-κB. The in vitro enriched aptamer library is fused to the known MS2-binding RNA sequence, such that library sequences that can bind to the protein ligand will recruit the transcriptional activation domain to the DNA binding domain, resulting in expression of a reporter gene and enabling screening of RNA aptamer sequences in yeast. In another example, an in vitro enriched aptamer pool to a small molecule ligand, atrazine, was screened in E. coli through a cell-based motility assay (Sinha et al., 2010). The RNA aptamer library was coupled to an RBS through a randomized linker region, where the resulting device library was linked to a gene that controls cell motility, such that functional aptamer sequences could be recovered from cells exhibiting greatest mobility in the presence of atrazine on solid medium. A similar approach has also been taken by directly inserting an in vitro enriched aptamer pool to neomycin in the 5' UTR of a GFP reporter gene in E. coli and screening for in vivo functional neomycin aptamers (Weigand et al., 2008). The combined in vitro and in vivo approaches provide a powerful strategy for generating and tailoring new component functions for the cellular environment.
Design strategies for synthetic RNA devices
General description of RNA devices and design approaches
Construction of RNA control devices generally starts by functionally linking sensor and actuator components to support transmission of information detected by the sensor into regulated activity of the actuator (Figure 1). Two strategies are generally taken in the design of RNA control devices: (i) direct linkage of sensor and actuator components; (ii) linkage of sensor and actuator components through a distinct information transmission component. In the first strategy, the regulatory effect is imparted by the resulting conformational change in the sensor component in the presence of the input signal, which directly affects the activity of the actuator component. In the second strategy, the transmitter component guides secondary structure changes in the sensor and actuator components, which direct these components between active and inactive conformations. Both modular and non-modular design strategies have been adopted in the construction of RNA controllers. Modular device design strategies introduce standardized communication interfaces between the sensor and actuator components to insulate the specific sequences of these components from one another, resulting in design platforms that support the interchange of functional components (i.e., sensor, actuator) without significant device redesign. Utilizing these design approaches researchers have developed RNA control devices that respond to diverse input signals, including temperature, nucleic acids, small molecules, and proteins.
Temperature-responsive control devices
Temperature can serve as an input to RNA control devices by taking advantage of the temperature-sensitive nature of RNA secondary structure. RNA sensors can detect changes in temperature through conformational rearrangement of RNA secondary structures, such as RNA hairpin structures. For example, a natural RNA temperature sensor is located in the 5' UTR of a mRNA transcript in many Gram-negative bacteria and modulates the accessibility of an RBS and thus expression of the encoded gene through temperature-dependent melting of secondary structure (Chowdhury et al., 2006). Synthetic RNA temperature sensors that mimic this natural system have been generated though both rational in silico design and a combined rational/screening approach, and coupled to an RBS actuator to generate temperature-responsive RNA control devices in E. coli (Neupert et al., 2008; Waldminghaus et al., 2008).
RNA-responsive control devices
RNA itself can serve as an input to RNA control devices by taking advantage of the ability of RNA molecules to interact through simple Watson-Crick base pairing rules (Benenson, 2009). Such device designs can potentially use RNA molecules as both the input and output signals, which allows for consistent signal carriers and simple extension to multi-layered signal processing and computational schemes (Choi et al., 2010; Lucks et al., 2011; Venkataraman et al., 2010). One of the earliest examples of an RNA-responsive control device is the riboregulator, an elegant genetic device that relies on interacting cis-repressor and trans-activator RNA (taRNA) sequences (Isaacs et al., 2004). The cis-repressor is located on the 5' end of the mRNA of interest and binds through Watson-Crick base pairing to the RBS, thereby occluding ribosome binding and inhibiting protein translation. A taRNA is designed to preferentially bind to the cis-repressor, such that in its presence, the RBS is accessible and protein translation occurs. This RNA control device has been shown to have several features important to system implementation, including a simple design, tunability of the quantitative regulatory response through modulation of the thermodynamic binding properties of the cis-repressor and taRNA, and the ability to simultaneously regulate multiple genes with the same taRNA. These properties have supported the implementation of this RNA-responsive control device in more complex genetic systems, including a genetically-encoded counter in E. coli (Friedland et al., 2009) and a bacterial kill switch responsive to multiple taRNA inputs (Callura et al., 2010).
Other examples of RNA-responsive control devices have used RNAi substrates as the input RNA signals. RNA devices that detect RNAi substrate inputs are generally designed by placing RNAi target sites in the 3' UTR of an output gene in mammalian cells (Brown et al., 2007; Kumar et al., 2011; Leisner et al., 2010; Rinaudo et al., 2007). This architecture is particularly useful in designing Boolean logic evaluators that integrate multiple inputs by simply placing multiple RNAi target sites in the same 3' UTR. Early examples of such RNAi-based logic evaluators utilized synthetic siRNAs (Rinaudo et al., 2007) and endogenous miRNAs (Brown et al., 2007; Brown et al., 2006) as inputs, where the latter allow for logical assessment of cellular state. As these devices support detecting RNAi substrates as input signals, their architectures can be readily extended to detecting other molecular inputs that themselves regulate the production of the direct RNAi inputs, such as transcription factors (Leisner et al., 2010). By placing synthetic microRNAs under the transcriptional control of promoters responsive to synthetic transcription factors, researchers applied these RNAi-responsive devices to the detection of combinations of the transcription factor inputs.
A more recent example of an RNA-responsive control device utilized an antisense RNA-based input and examined circuit architectures that employed RNA signals as both inputs and outputs in E. coli (Lucks et al., 2011). Researchers developed RNA devices that utilize antisense RNA inputs to modulate transcriptional attenuation of a target gene based on a natural system from the Staphylococcus aureus plasmid pT181. The authors utilized a rational mutagenesis strategy to engineer two orthogonal variants of this RNA device by altering the specificity of the antisense/attenuator base pairing interactions. These device variants were implemented simultaneously to independently control expression of two fluorescent reporters in the same cell and were placed in series on the same transcript to control the transcription of the same gene in response to two inputs. Finally, these devices were implemented in a genetic circuit to propagate an input RNA signal through a three-layer transcriptional cascade. The transcription of a fluorescent reporter gene was controlled by the interaction between an attenuator and antisense RNA interaction. The transcription of this first antisense RNA was placed under the control of a second, orthogonal antisense-attenuator pair so that the input of the second antisense regulatory signal was propagated through a double inversion, resulting in transcription of the reporter gene.
Small molecule and protein-responsive control devices
RNA control devices can also be designed to respond to other biological molecules, including small molecules and proteins. While sensors that detect temperature and RNA act through base pairing interactions, RNA sensors interact with small molecules and proteins through more complex chemical interactions that cannot be predicted through RNA secondary structure, thereby typically requiring generation of RNA aptamers to target small molecule or protein ligands through in vitro selection strategies. Small molecule and protein-responsive control devices have been constructed by coupling RNA aptamers (sensors) to various gene-regulatory (actuator) components, whose mechanism of action is generally specific to the gene expression machinery associated with the host cell.
i. Prokaryotes and Chloroplasts
Small molecule-responsive control devices have been engineered in bacteria by modifying or adding component functions in natural riboswitch or regulatory elements. In one example, researchers modified the information transmission function encoded in a natural TPP-responsive riboswitch, which exhibits TPP-dependent gene repression, by randomizing a sequence within the riboswitch and screening the resulting library for new synthetic devices that exhibit TPP-dependent gene activation (Nomura and Yokobayashi, 2007). In another example, a theophylline aptamer was coupled to a natural riboswitch through a randomized linker sequence and the resulting library was screened for devices that exhibit logic operations (AND, NAND) by modulating ribosome access to the RBS (Sharma et al., 2008). RNA control devices have also been built by direct integration of RNA sensors into natural regulatory elements. A theophylline aptamer was integrated into a stem in the group I self-splicing intron from the bacteriophage T4 thymidylate synthase gene to construct a theophylline-dependent RNA splicing device that regulated E. coli growth (Thompson et al., 2002).
The majority of small molecule-responsive RNA control devices in bacteria have been designed to control translation initiation, due to the relative ease with which ribosome loading can be modulated through structural rearrangement of the RBS. For example, RBS-based devices were built in E. coli by coupling a theophylline aptamer to the RBS through a linker sequence capable of structural rearrangement through strand-displacement (Desai and Gallivan, 2004) or helix-slipping (Suess et al., 2004) mechanisms. The accessibility of the RBS (related to its single-stranded state) is altered by ligand-dependent changes in base pairing interactions between the sensor and linker (strand-displacement) or local nucleotide shifts within the linker (helix-slipping). In the device design strategy based on strand displacement, part of the aptamer sequence directly interacts with the RBS sequence, such that independent modification of either component may disrupt these interactions and thus require redesign of the linker sequence to maintain device function. Therefore, cell-based screening strategies have been developed to support the identification of new linker sequences through colorimetric (Lynch et al., 2007), flow cytometry (Lynch and Gallivan, 2009), and motility assays (Topp and Gallivan, 2008). Other examples of RBS-based devices were designed by coupling a theophylline aptamer to an indirect actuator, a hammerhead ribozyme, which in turn was coupled to a direct actuator, a RBS (Ogawa and Maeda, 2008; Wieland and Hartig, 2008). In this device design, the RBS is sequestered within the ribozyme stem, such that ligand-induced ribozyme cleavage results in unwinding of the ribozyme stem, thereby increasing ribosomal access to the RBS and thus gene expression levels.
Bacterial RNA control devices have been applied in plants, by implementing these devices in chloroplasts, intracellular organelles that evolved from endosymbiosis of cyanobacteria (Verhounig et al., 2010). As such, these organelles have separate gene expression machinery that is largely compatible with the machinery found in prokaryotes. A theophylline-responsive RBS-based control device previously engineered in E. coli (Suess et al., 2004) was modified through an in silico design strategy to optimize the translation initiation signals in plastids. The device was integrated into the tobacco chloroplast genome and demonstrated modulation of gene expression through translational regulation.
ii. Yeast
Different design approaches have been taken to engineer small molecule-responsive RNA control devices in eukaryotes, as fewer examples of natural riboswitch elements are available to build from and more sophisticated gene-regulatory mechanisms are available than in prokaryotes. RNA control devices have been built by integrating the RNA sensor directly into part of the actuator component, such that the conformational change associated with ligand binding to the aptamer affects the activity of the actuator. In one example, a tetramethylrosamine (TMR) aptamer and a synthetic RNA transcriptional activator were joined through the stem of the transcription activator, where part of the stem was randomized to allow screening for TMR-responsive gene-regulatory activity in Saccharomyces cerevisiae (Buskirk et al., 2004). In another example, a theophylline aptamer was integrated into the core cleavage region of a synthetic RNase III hairpin, which when placed in the 3' UTR of a target transcript, directed cleavage and subsequent inactivation of that transcript in yeast (Babiskin and Smolke, 2011a). However, the binding of theophylline to the integrated aptamer restricts cleavage activity of the RNase III enzyme, thereby activating gene expression. Other small molecule-responsive RNA control devices have been built by integrating an aptamer in proximity to a 5' splice site (Weigand and Suess, 2007) or a branch point (Kim et al., 2008). As an example, a tetracycline aptamer in close proximity to a 5' splice site, where the consensus sequence of the splice site is integrated into the aptamer stem (Weigand and Suess, 2007). The binding of tetracycline to the aptamer results in a conformation that potentially prevents access to the splice site, thereby achieving ligand-dependent regulation of splicing.
As a second design approach, composition frameworks have been developed to support the modular assembly of RNA control devices from underlying functional components in yeast (Win and Smolke, 2007). In the proposed framework, ribozyme-based devices were constructed by linking an aptamer to a hammerhead ribozyme through a distinct transmitter sequence designed to both encode the information processing function of the device and insulate the sensor and actuator components. This design strategy was demonstrated to support independent swapping of the sensor and transmitter components and thus tailoring of the encoded sensing and information processing functions without significant device redesign. The composition framework also supported extension to the modular assembly of multiple sensor, transmitter, and actuator domains to construct devices that exhibit higher-order information processing functions, including logic operations, signal and bandpass filters, and programmed cooperativity (Win and Smolke, 2008). The utilization of ribozymes as actuators in this RNA device platform also allows for transportability of the device across different organisms, because the ribozyme activity is independent of cell-specific gene-regulatory machinery (Chen et al., 2010).
iii. Mammalian cells
RNA control devices in mammalian cells have been designed through numerous strategies ranging from directly adapting regulatory mechanisms that were developed in simpler organisms to taking advantage of the rich complexity of RNA processing pathways unique to higher organisms. Several RNA control devices have been developed through design strategies that directly integrate an RNA aptamer to a gene-regulatory element. These approaches have been applied to the design of small molecule and protein-responsive RNA control devices that conditionally silence target genes through RNAi-mediated silencing mechanisms (An et al., 2006; Beisel et al., 2011; Saito et al., 2011). These devices have been designed by integrating an RNA aptamer directly within the basal segments of a miRNA or in the loop region of a shRNA, such that binding of the input molecule prevents correct biogenesis of the RNAi substrate and thus results in increased target gene expression levels. Conditional RNAi silencing in response to a wide range of ligands, including theophylline, tetracycline, xanthine, and L7Ae, have been demonstrated with these RNAi-based devices. While direct integration strategies generally limit the tuning of device regulatory activity that one can achieve through molecular alteration strategies, researchers demonstrated a tuning strategy for the miRNA-based device based on rational design of device clusters (Beisel et al., 2011).
A similar design approach was recently used to develop a protein-responsive control device based on modulating alternative RNA splicing in mammalian cells (Culler et al., 2010). In this RNA device, a three-exon, two-intron mini-gene was fused to the 5' end of a gene of interest, where the alternatively-spliced middle exon encoded a stop codon such that inclusion of this exon resulted in low target protein levels. Aptamers against endogenous (β-catenin, NF-κB) and heterologous (MS2 coat protein) proteins were placed in several intronic locations within the transcript, such that binding of the protein input altered the inclusion of the middle exon resulting in either increased or decreased expression levels. The regulatory effect of the input protein on the alternative splicing pattern was dependent on the location of the integrated sensor component and the input protein itself. By integrating multiple aptamers into different intronic integration sites, the researchers demonstrated combinatorial processing of multiple protein inputs.
Other mammalian RNA control devices have been developed through design strategies that couple the sensor and actuator components through a distinct transmitter component. In one example, a transmitter and RNA aptamer were integrated into the loop region of a shRNA (Beisel et al., 2008). In the active ligand-unbound conformation, the aptamer was incorrectly formed and the RNAi machinery correctly processed the device, resulting in silencing of the target gene. The inactive conformation of the device coincided with correct formation of the aptamer, such that binding of the small molecule input to the aptamer stabilized this conformation. The RNAi machinery did not correctly process this inactive conformation, thus resulting in increased target protein levels in the presence of input. As another example, the previously-described ribozyme-based devices, originally demonstrated in yeast, were shown to retain small molecule-responsive gene-regulatory activity in mammalian cell culture and in vivo (Chen et al., 2010). RNA devices that incorporate a distinct transmitter component can generally be tuned by altering the sequence of the transmitter component to optimize the thermodynamics of the competing hybridization reactions underlying the strand-displacement mechanism. For both the shRNA- and ribozyme-based devices, sequence alterations within the transmitter components were used to optimize the regulatory performance of the resulting devices in mammalian cells (Beisel et al., 2008; Chen et al., 2010).
Programming quantitative regulatory performance of RNA devices
The ability to quantitatively tailor regulatory performance of RNA control devices is essential for their broad integration in biological systems. The regulatory function of an RNA control device depends on the component linkage strategy and the activities of individual components within the host cell. Both evolutionary and rational strategies have been employed to quantitatively tune device regulatory properties. Cell-based screening and selection strategies have been applied to optimize the information transmission function encoded within randomized linker sequences placed between the sensor and actuator components by linking the regulatory activities of RNA device libraries to various measurable outputs, including enzyme reporter activities (Lynch et al., 2007), motility (Topp and Gallivan, 2008), and fluorescence reporter levels (Fowler et al., 2008; Lynch and Gallivan, 2009; Wieland and Hartig, 2008). High-throughput methods, such as plate-based reporter assays, growth assays, and fluorescence-activated cell sorting (FACS), can be employed to efficiently isolate devices with desired regulatory activities. Rational design strategies have been developed that utilize RNA structure prediction programs to guide sequence changes in the transmitter component to alter the energetic values associated with alternate device conformations, thereby changing the energetic partitioning between functional states of the device and the resulting device regulatory activities (Win and Smolke, 2007; Win and Smolke, 2008). The rational design strategies can also be complemented by the development of computational modeling frameworks to examine the effects of various kinetic rates associated with the device regulatory mechanism and gene expression processes on the quantitative performance of the device (Beisel et al., 2008; Beisel and Smolke, 2009). The reliable construction and quantitative programming of RNA control devices are key to their effective implementation in biological systems.
Applications of synthetic regulatory RNAs in biological systems
Implementation in genetic networks directed to biomanufacturing / biosynthesis applications
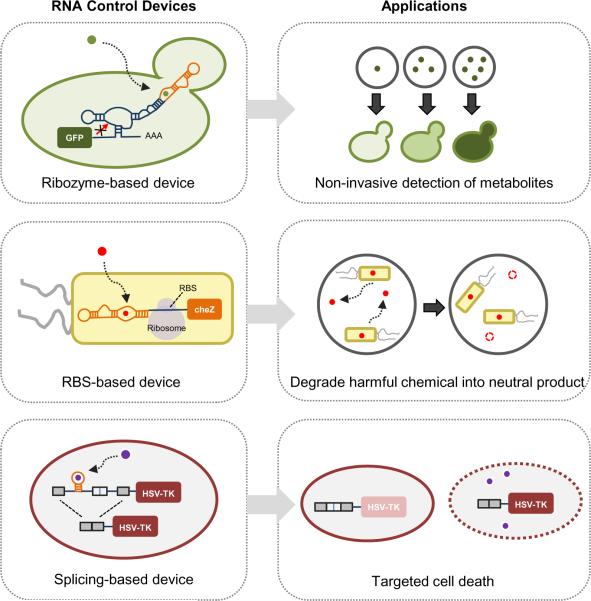
RNA devices have important applications in biosynthesis processes, where they can be implemented as noninvasive sensors of metabolite accumulation and controllers for optimizing flux and product yield. In one example, an RNA control device was utilized for noninvasive detection of metabolite accumulation in yeast (Win and Smolke, 2007) (Figure 2). Cells were engineered to express a construct harboring a xanthine-responsive ribozyme-based device regulating a GFP reporter gene. Xanthosine was fed to the yeast cells, which converted this fed substrate to xanthine through an endogenous enzyme activity. The conversion of xanthosine to xanthine was monitored indirectly as an increase in GFP levels, where increases in fluorescence correlated with increased product accumulation as measured by LC-MS. In a recent example, RNA devices that self-assemble into nanostructures inside bacterial cells were used to design intricate scaffolding systems for spatially organizing biosynthetic enzymes (Delebecque et al., 2011). RNA aptamers were used to engineer docking sites on the scaffolds, where the corresponding aptamer target proteins were fused to biosynthetic pathway enzymes to recruit the enzymes to the scaffolding docking sites. Researchers applied this RNA scaffolding system to a hydrogen synthesis pathway in E. coli and demonstrated a 48-fold increase in hydrogen production from the scaffolded system relative to the unscaffolded enzyme system. This latter system highlights an exciting role for RNA devices in engineering spatial organization of cellular molecules.
Figure 2.
RNA-based controllers have been integrated into engineered biological systems for applications spanning biosynthesis, bioremediation, to health and medicine. A metabolite-responsive ribozyme-based device linked to a fluorescent reporter output was demonstrated in yeast as a noninvasive sensor of metabolite concentration (top panel). A pollutant-responsive RBS-based device linked to a motility gene output was demonstrated in bacteria to program the cells to move along a gradient of the pollutant (middle panel). A disease marker-responsive alternative splicing-based device linked to a suicide gene output was demonstrated in human cells to target cell death to cells exhibiting increased signaling through disease pathways (bottom panel). These examples highlight the power of RNA controllers that enable researchers to access, transmit, and act on information within biological systems.
Implementation in genetic networks directed to environmental / agricultural applications
RNA devices also have important applications in agricultural biotechnology and environmental remediation. In the latter case, engineered organisms that may be released into the environment will need precise control over designed functions and safety mechanisms in place to prevent uncontrolled release. In one example, an RBS-based RNA control device was implemented to detect a toxic environmental pollutant, atrazine (Sinha et al., 2010) (Figure 2). The atrazine-responsive device was coupled to the cheZ gene to control the motility of an E. coli strain engineered to express an atrazine-catabolizing enzyme activity. The presence of atrazine activates the expression the cheZ gene, which allows cells to move along the source of the pollutant and convert atrazine into less harmful product, hydroxyatrazine.
Implementation in genetic networks directed to health and medicine applications
One of the most intriguing applications for RNA synthetic biology is in the area of health and medicine (Khalil and Collins, 2010). RNA displays several properties that provide distinct advantages for the design of genetically-encoded therapeutic activities (Chen et al., 2010). First, challenges associated with stable, long-term expression of multiple heterologous proteins in primary cells and the non-specific immunogenicity of these protein components represent a significant hurdle to implementing these systems in clinical applications. In contrast, RNA-only systems are compatible with size limitations of viral vectors and do not need additional transgenic proteins or transcriptional regulators required for inducible promoter systems. Moreover, RNA control devices can be built on modular, tunable design platforms, which allow the researcher to program a therapeutic activity as an output in response to specific levels of an input relevant to the disease or application of interest.
Importantly, in genetic devices designed for therapeutic applications, changes in gene expression resulting from the device must lead to eradication of the disease phenotype or elimination of the diseased cells. For example, an RNA device was designed by placing an RNA aptamer to the L7Ae protein in the 5' UTR of a transcript encoding a recombinant pro-apoptotic protein Bim (Saito et al., 2011). In this device design, binding of L7Ae to its RNA binding motif repressed translation of the downstream gene. The researchers also built a shRNA-based device targeted to the anti-apoptotic protein Bcl-xl, which incorporated the L7Ae aptamer in the loop region of the shRNA, such that binding of L7Ae reduced shRNA silencing and increased Bcl-xl expression. The simultaneous implementation of these two RNA control devices induced apoptosis in cultured HeLa cells by over two-fold when recombinant L7Ae was transiently expressed from a plasmid or stably expressed from the chromosome.
Synthetic RNA control devices have also been designed to interface with native cellular pathways, including pathways associated with human disease, and output specific therapeutic activities in response to the detection of molecular signals of disease. In one example, small RNA hairpins were injected into human cell lines (Venkataraman et al., 2010). Upon detection of an mRNA cancer marker, the small RNAs were designed to hybridize into long pieces of double-stranded RNA, which in turn activated the innate immune response causing the cells to undergo apoptosis. These chain-hybridizing RNA molecules were shown to selectively kill over 95% of cells in three model cancer cell lines using this mechanism. In another example, protein-responsive alternative splicing-based devices were designed to detect disease-associated markers NF-κB and β-catenin (Culler et al., 2010) (Figure 2). These devices were used to control the expression of the suicide gene herpes simplex virus-thymidine kinase (HSV-TK), which confers sensitivity to the prodrug Ganciclovir to induce apoptosis. Almost 80 percent of human cells harboring these RNA devices underwent apoptosis in response to increased signaling through the NF-κB or β-catenin pathways and the presence of the prodrug. The construction of tailored RNA control devices that can sense changes in cellular state and program specific behaviors in response to disease markers represents a significant advance towards translatable, “smart” molecular therapies.
Finally, RNA control devices can be used to improve the safety or efficacy of cell-based therapeutics. Recent progress has been made in the application of RNA devices to adoptive T-cell therapy, an approach currently in clinical trials, in which a patient's own T cells are harvested and trained against tumor-specific antigens or engineered to express chimeric antigen receptors before being injected back into the body. A major challenge associated with this strategy is ensuring that the T cells can survive and proliferate sufficiently when engrafted into the host to eradicate the diseased cells. To overcome this problem, small molecule-responsive RNA devices were applied to control T-cell proliferation in response to the application of drug molecules (Chen et al., 2010). Researchers placed ribozyme-based devices in the 3' UTR of transcripts encoding the proliferative cytokines, IL-15 and IL-2, such that in the absence of the small molecule input the transcript was destabilized through ribozyme cleavage, resulting in reduced T-cell survival. In the presence of the drug input, IL-2 or IL-15 was produced, resulting in increased T-cell proliferation and survival. The RNA-based control system was demonstrated in murine and primary human cell lines and translated in vivo by performing a 14-day mouse study, in which an average ~15-fold increase in T-cell proliferation was observed in immunocompromised mice with the administration of theophylline. While still in proof of concept stage, this platform was shown to be modular in that aptamers to different small molecules (theophylline, tetracycline) were used to control the expression of different proliferative cytokines and hence proliferation of the T cells. Therefore, as RNA sensors to clinically relevant molecules are generated, they can be integrated into the ribozyme device platform and translated to the clinic without major redesign of the control system.
Future perspectives and challenges
Synthetic RNA molecules hold much promise for building programmable genetic regulators, leading to transformative advances in how we interact with and program biology, providing access to otherwise inaccessible information on cellular state, and allowing sophisticated exogenous and embedded control over cellular functions. Realizing the next-generation of RNA synthetic biology will require advances that address several immediate challenges faced in the field.
The first challenge is associated with the limited diversity of existing functional RNA parts. Methods have been developed that allow for the de novo generation of RNA components exhibiting new regulatory and sensing activities (Bartel and Szostak, 1993; Ellington and Szostak, 1990; Tuerk and Gold, 1990). However, scalable strategies are needed to transform the existing landscape of available parts and support the rapid generation of diverse functional RNA activities. In particular, high-throughput, high-efficiency in vitro selection methods coupled to in vivo screening/selection methods are needed to search large sequence spaces effectively for novel activities. Once established, these strategies can be used to develop parts libraries that will support the tailored design of genetic systems.
The second challenge is associated with the lack of computational tools supporting rational RNA device and system design. Computational models and tools have been developed that move toward the much-needed CAD tools that can support RNA device design (Beisel and Smolke, 2009). However, improved RNA structure prediction tools are necessary that can address complex structures, folding kinetics, ligand binding, and in vivo folding mechanisms. These improved structure prediction tools will form the foundation of CAD tools supporting the rational design and tuning of RNA devices.
The third challenge is associated with the lack of design strategies and approaches supporting integrated genetic systems. As researchers move beyond the design of first-generation circuits that solely focus on transcriptional regulation, the unique roles of posttranscriptional, posttranslational, and spatial control strategies in genetic circuits have been highlighted (Khalil and Collins, 2010). Key questions remain in the field in terms of how to best combine different layers of regulation, gene control, and spatial organization encoded within RNA- and protein-based elements that act at different time scales, exhibit different regulatory activities, and place different energetic loads on the cell to develop large-scale genetic systems that operate reliably and predictably. In particular, strategies that effectively integrate RNA-based controllers with protein-based components (Deans et al., 2007) to leverage the relative advantages of RNA-based components, such as programmability, tunability, and energetic efficiency, with the potential disadvantages, such as weaker or more limited activities, in genetic circuits are critical to advance complex system design. Thus, the combined advances of RNA biology and engineering will support the incorporation of synthetic RNA controllers into more sophisticated genetic circuits that utilize diverse regulatory strategies, balance energetic load, and dynamically modulate systems behavior.
ACKNOWLEDGEMENTS
C.D.S. is funded by the NIH, NSF, and DARPA. R.J.B. receives funding through a NSF graduate fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alifano P, Bruni CB, Carlomagno MS. Control of mRNA processing and decay in prokaryotes. Genetica. 1994;94:157–172. doi: 10.1007/BF01443430. [DOI] [PubMed] [Google Scholar]
- An CI, Trinh VB, Yokobayashi Y. Artificial control of gene expression in mammalian cells by modulating RNA interference through aptamer-small molecule interaction. RNA. 2006;12:710–716. doi: 10.1261/rna.2299306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JC, Clarke EJ, Arkin AP, Voigt CA. Environmentally controlled invasion of cancer cells by engineered bacteria. J Mol Biol. 2006;355:619–627. doi: 10.1016/j.jmb.2005.10.076. [DOI] [PubMed] [Google Scholar]
- Babiskin AH, Smolke CD. Engineering ligand-responsive RNA controllers in yeast through the assembly of RNase III tuning modules. Nucleic Acids Res. 2011a;39:5299–5311. doi: 10.1093/nar/gkr090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiskin AH, Smolke CD. A synthetic library of RNA control modules for predictable tuning of gene expression in yeast. Mol Syst Biol. 2011b;7:471. doi: 10.1038/msb.2011.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP, Szostak JW. Isolation of new ribozymes from a large pool of random sequences [see comment] Science. 1993;261:1411–1418. doi: 10.1126/science.7690155. [DOI] [PubMed] [Google Scholar]
- Beisel CL, Bayer TS, Hoff KG, Smolke CD. Model-guided design of ligand-regulated RNAi for programmable control of gene expression. Mol Syst Biol. 2008;4:224. doi: 10.1038/msb.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel CL, Chen YY, Culler SJ, Hoff KG, Smolke CD. Design of small molecule-responsive microRNAs based on structural requirements for Drosha processing. Nucleic Acids Res. 2011;39:2981–2994. doi: 10.1093/nar/gkq954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel CL, Smolke CD. Design principles for riboswitch function. PLoS Comput Biol. 2009;5:e1000363. doi: 10.1371/journal.pcbi.1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benenson Y. RNA-based computation in live cells. Curr Opin Biotechnol. 2009;20:471–478. doi: 10.1016/j.copbio.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BD, Gentner B, Cantore A, Colleoni S, Amendola M, Zingale A, Baccarini A, Lazzari G, Galli C, Naldini L. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat Biotechnol. 2007;25:1457–1467. doi: 10.1038/nbt1372. [DOI] [PubMed] [Google Scholar]
- Brown BD, Venneri MA, Zingale A, Sergi Sergi L, Naldini L. Endogenous microRNA regulation suppresses transgene expression in hematopoietic lineages and enables stable gene transfer. Nat Med. 2006;12:585–591. doi: 10.1038/nm1398. [DOI] [PubMed] [Google Scholar]
- Buskirk AR, Landrigan A, Liu DR. Engineering a ligand-dependent RNA transcriptional activator. Chem Biol. 2004;11:1157–1163. doi: 10.1016/j.chembiol.2004.05.017. [DOI] [PubMed] [Google Scholar]
- Callura JM, Dwyer DJ, Isaacs FJ, Cantor CR, Collins JJ. Tracking, tuning, and terminating microbial physiology using synthetic riboregulators. Proc Natl Acad Sci U S A. 2010;107:15898–15903. doi: 10.1073/pnas.1009747107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr PA, Church GM. Genome engineering. Nat Biotechnol. 2009;27:1151–1162. doi: 10.1038/nbt.1590. [DOI] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassiday LA, Maher LJ., 3rd Yeast genetic selections to optimize RNA decoys for transcription factor NF-kappa B. Proc Natl Acad Sci U S A. 2003;100:3930–3935. doi: 10.1073/pnas.0736013100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Denison L, Levy M, Ellington AD. Direct selection for ribozyme cleavage activity in cells. RNA. 2009;15:2035–2045. doi: 10.1261/rna.1635209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YY, Jensen MC, Smolke CD. Genetic control of mammalian T-cell proliferation with synthetic RNA regulatory systems. Proc Natl Acad Sci U S A. 2010;107:8531–8536. doi: 10.1073/pnas.1001721107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HM, Chang JY, Trinh le A, Padilla JE, Fraser SE, Pierce NA. Programmable in situ amplification for multiplexed imaging of mRNA expression. Nat Biotechnol. 2010;28:1208–1212. doi: 10.1038/nbt.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S, Maris C, Allain FH, Narberhaus F. Molecular basis for temperature sensing by an RNA thermometer. EMBO J. 2006;25:2487–2497. doi: 10.1038/sj.emboj.7601128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culler SJ, Hoff KG, Smolke CD. Reprogramming cellular behavior with RNA controllers responsive to endogenous proteins. Science. 2010;330:1251–1255. doi: 10.1126/science.1192128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Baker D. Automated de novo prediction of native-like RNA tertiary structures. Proc Natl Acad Sci U S A. 2007;104:14664–14669. doi: 10.1073/pnas.0703836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deans TL, Cantor CR, Collins JJ. A tunable genetic switch based on RNAi and repressor proteins for regulating gene expression in mammalian cells. Cell. 2007;130:363–372. doi: 10.1016/j.cell.2007.05.045. [DOI] [PubMed] [Google Scholar]
- Delebecque CJ, Lindner AB, Silver PA, Aldaye FA. Organization of Intracellular Reactions with Rationally Designed RNA Assemblies. Science. 2011 doi: 10.1126/science.1206938. [DOI] [PubMed] [Google Scholar]
- Desai SK, Gallivan JP. Genetic screens and selections for small molecules based on a synthetic riboswitch that activates protein translation. J Am Chem Soc. 2004;126:13247–13254. doi: 10.1021/ja048634j. [DOI] [PubMed] [Google Scholar]
- Dixon N, Duncan JN, Geerlings T, Dunstan MS, McCarthy JE, Leys D, Micklefield J. Reengineering orthogonally selective riboswitches. Proc Natl Acad Sci U S A. 2010;107:2830–2835. doi: 10.1073/pnas.0911209107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehretsmann CP, Carpousis AJ, Krisch HM. mRNA degradation in procaryotes. FASEB J. 1992;6:3186–3192. doi: 10.1096/fasebj.6.13.1397840. [DOI] [PubMed] [Google Scholar]
- Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Endy D. Foundations for engineering biology. Nature. 2005;438:449–453. doi: 10.1038/nature04342. [DOI] [PubMed] [Google Scholar]
- Flamm C, Fontana W, Hofacker IL, Schuster P. RNA folding at elementary step resolution. RNA. 2000;6:325–338. doi: 10.1017/s1355838200992161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler CC, Brown ED, Li Y. A FACS-based approach to engineering artificial riboswitches. Chembiochem. 2008;9:1906–1911. doi: 10.1002/cbic.200700713. [DOI] [PubMed] [Google Scholar]
- Friedland AE, Lu TK, Wang X, Shi D, Church G, Collins JJ. Synthetic gene networks that count. Science. 2009;324:1199–1202. doi: 10.1126/science.1172005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S. The small RNA regulators of Escherichia coli: roles and mechanisms*. Annu Rev Microbiol. 2004;58:303–328. doi: 10.1146/annurev.micro.58.030603.123841. [DOI] [PubMed] [Google Scholar]
- Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs FJ, Dwyer DJ, Ding C, Pervouchine DD, Cantor CR, Collins JJ. Engineered riboregulators enable post-transcriptional control of gene expression. Nat Biotechnol. 2004;22:841–847. doi: 10.1038/nbt986. [DOI] [PubMed] [Google Scholar]
- Jenison RD, Gill SC, Pardi A, Polisky B. High-resolution molecular discrimination by RNA. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- Khalil AS, Collins JJ. Synthetic biology: applications come of age. Nat Rev Genet. 2010;11:367–379. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DS, Gusti V, Dery KJ, Gaur RK. Ligand-induced sequestering of branchpoint sequence allows conditional control of splicing. BMC Mol Biol. 2008;9:23. doi: 10.1186/1471-2199-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene. 2005;361:13–37. doi: 10.1016/j.gene.2005.06.037. [DOI] [PubMed] [Google Scholar]
- Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- Kumar D, Kim SH, Yokobayashi Y. Combinatorially inducible RNA interference triggered by chemically modified oligonucleotides. J Am Chem Soc. 2011;133:2783–2788. doi: 10.1021/ja1107436. [DOI] [PubMed] [Google Scholar]
- Leisner M, Bleris L, Lohmueller J, Xie Z, Benenson Y. Rationally designed logic integration of regulatory signals in mammalian cells. Nat Nanotechnol. 2010;5:666–670. doi: 10.1038/nnano.2010.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucks JB, Qi L, Mutalik VK, Wang D, Arkin AP. Versatile RNA-sensing transcriptional regulators for engineering genetic networks. Proc Natl Acad Sci U S A. 2011;108:8617–8622. doi: 10.1073/pnas.1015741108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch SA, Desai SK, Sajja HK, Gallivan JP. A high-throughput screen for synthetic riboswitches reveals mechanistic insights into their function. Chem Biol. 2007;14:173–184. doi: 10.1016/j.chembiol.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch SA, Gallivan JP. A flow cytometry-based screen for synthetic riboswitches. Nucleic Acids Res. 2009;37:184–192. doi: 10.1093/nar/gkn924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M, Breaker RR. Gene regulation by riboswitches. Nat Rev Mol Cell Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci U S A. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS, Amaral PP, Dinger ME, Mercer TR, Mehler MF. RNA regulation of epigenetic processes. Bioessays. 2009;31:51–59. doi: 10.1002/bies.080099. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- Neupert J, Karcher D, Bock R. Design of simple synthetic RNA thermometers for temperature-controlled gene expression in Escherichia coli. Nucleic Acids Res. 2008;36:e124. doi: 10.1093/nar/gkn545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura Y, Yokobayashi Y. Reengineering a natural riboswitch by dual genetic selection. J Am Chem Soc. 2007;129:13814–13815. doi: 10.1021/ja076298b. [DOI] [PubMed] [Google Scholar]
- Ogawa A, Maeda M. An artificial aptazyme-based riboswitch and its cascading system in E. coli. Chembiochem. 2008;9:206–209. doi: 10.1002/cbic.200700478. [DOI] [PubMed] [Google Scholar]
- Parisien M, Major F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature. 2008;452:51–55. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- Pfleger BF, Pitera DJ, Smolke CD, Keasling JD. Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nat Biotechnol. 2006;24:1027–1032. doi: 10.1038/nbt1226. [DOI] [PubMed] [Google Scholar]
- Purnick PE, Weiss R. The second wave of synthetic biology: from modules to systems. Nat Rev Mol Cell Biol. 2009;10:410–422. doi: 10.1038/nrm2698. [DOI] [PubMed] [Google Scholar]
- Rinaudo K, Bleris L, Maddamsetti R, Subramanian S, Weiss R, Benenson Y. A universal RNAi-based logic evaluator that operates in mammalian cells. Nat Biotechnol. 2007;25:795–801. doi: 10.1038/nbt1307. [DOI] [PubMed] [Google Scholar]
- Saito H, Fujita Y, Kashida S, Hayashi K, Inoue T. Synthetic human cell fate regulation by protein-driven RNA switches. Nat Commun. 2011;2:160. doi: 10.1038/ncomms1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotechnol. 2009;27:946–950. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Patel DJ. Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nat Rev Genet. 2007;8:776–790. doi: 10.1038/nrg2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V, Nomura Y, Yokobayashi Y. Engineering complex riboswitch regulation by dual genetic selection. J Am Chem Soc. 2008;130:16310–16315. doi: 10.1021/ja805203w. [DOI] [PubMed] [Google Scholar]
- Sinha J, Reyes SJ, Gallivan JP. Reprogramming bacteria to seek and destroy an herbicide. Nat Chem Biol. 2010;6:464–470. doi: 10.1038/nchembio.369. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Smolke CD, Carrier TA, Keasling JD. Coordinated, differential expression of two genes through directed mRNA cleavage and stabilization by secondary structures. Appl Environ Microbiol. 2000;66:5399–5405. doi: 10.1128/aem.66.12.5399-5405.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolke CD, Silver PA. Informing biological design by integration of systems and synthetic biology. Cell. 2011;144:855–859. doi: 10.1016/j.cell.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suess B, Fink B, Berens C, Stentz R, Hillen W. A theophylline responsive riboswitch based on helix slipping controls gene expression in vivo. Nucleic Acids Res. 2004;32:1610–1614. doi: 10.1093/nar/gkh321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson KM, Syrett HA, Knudsen SM, Ellington AD. Group I aptazymes as genetic regulatory switches. BMC Biotechnol. 2002;2:21. doi: 10.1186/1472-6750-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topp S, Gallivan JP. Random walks to synthetic riboswitches--a high-throughput selection based on cell motility. Chembiochem. 2008;9:210–213. doi: 10.1002/cbic.200700546. [DOI] [PubMed] [Google Scholar]
- Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Venkataraman S, Dirks RM, Ueda CT, Pierce NA. Selective cell death mediated by small conditional RNAs. Proc Natl Acad Sci U S A. 2010;107:16777–16782. doi: 10.1073/pnas.1006377107. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Verhounig A, Karcher D, Bock R. Inducible gene expression from the plastid genome by a synthetic riboswitch. Proc Natl Acad Sci U S A. 2010;107:6204–6209. doi: 10.1073/pnas.0914423107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldminghaus T, Kortmann J, Gesing S, Narberhaus F. Generation of synthetic RNA-based thermosensors. Biol Chem. 2008;389:1319–1326. doi: 10.1515/BC.2008.150. [DOI] [PubMed] [Google Scholar]
- Weigand JE, Sanchez M, Gunnesch EB, Zeiher S, Schroeder R, Suess B. Screening for engineered neomycin riboswitches that control translation initiation. RNA. 2008;14:89–97. doi: 10.1261/rna.772408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigand JE, Suess B. Tetracycline aptamer-controlled regulation of pre-mRNA splicing in yeast. Nucleic Acids Res. 2007;35:4179–4185. doi: 10.1093/nar/gkm425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland M, Benz A, Klauser B, Hartig JS. Artificial ribozyme switches containing natural riboswitch aptamer domains. Angew Chem Int Ed Engl. 2009;48:2715–2718. doi: 10.1002/anie.200805311. [DOI] [PubMed] [Google Scholar]
- Wieland M, Hartig JS. Improved aptazyme design and in vivo screening enable riboswitching in bacteria. Angew Chem Int Ed Engl. 2008;47:2604–2607. doi: 10.1002/anie.200703700. [DOI] [PubMed] [Google Scholar]
- Win MN, Smolke CD. A modular and extensible RNA-based gene-regulatory platform for engineering cellular function. Proceedings of the National Academy of Sciences. 2007;104:14283–14288. doi: 10.1073/pnas.0703961104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win MN, Smolke CD. Higher-order cellular information processing with synthetic RNA devices. Science. 2008;322:456–460. doi: 10.1126/science.1160311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky C. Attenuation in the control of expression of bacterial operons. Nature. 1981;289:751–758. doi: 10.1038/289751a0. [DOI] [PubMed] [Google Scholar]
- Zadeh JN, Steenberg CD, Bois JS, Wolfe BR, Pierce MB, Khan AR, Dirks RM, Pierce NA. NUPACK: Analysis and design of nucleic acid systems. J Comput Chem. 2011;32:170–173. doi: 10.1002/jcc.21596. [DOI] [PubMed] [Google Scholar]