Abstract
Summary
Using an integrated approach, we here report the identification of 67 novel histone marks, a discovery that increases the current number of known histone marks by about 70%. We verified one of the newly-identified marks, lysine crotonylation (Kcr), as a novel, evolutionarily-conserved histone post-translational modification. The unique structure and genomic localization of Kcr suggest that it is mechanistically and functionally different from lysine acetylation (Kac), a previously-described post-translational modification. Specifically, in both human somatic and mouse male germ cell genomes, histone Kcr marks either active promoters or potential enhancers. In male germinal cells immediately following meiosis, Kcr is enriched on sex chromosomes and specifically marks testis-specific genes, including a significant proportion of X-linked genes that escape sex chromosome inactivation in haploid cells. These results therefore dramatically extend the repertoire of histone PTM sites and designate Kcr as a specific mark of active sex chromosome-linked genes in post-meiotic male germ cells.
Keywords: Crotonyl-CoA, histones, lysine acetylation, lysine crotonylation, mass spectrometry, protein post-translational modifications
INTRODUCTION
Mounting evidence suggests that histone PTMs play a crucial role in diverse biological processes, such as cell differentiation and organismal development, and that aberrant modification of histones contributes to diseases such as cancer (Berdasco and Esteller, 2010; Fullgrabe et al., 2011). At least eleven types of PTMs have been reported at over 60 different amino acid residues on histones, including histone methylation, acetylation, propionylation, butyrylation, formylation, phosphorylation, ubiquitylation, sumoylation, citrullination, proline isomerization, and ADP ribosylation (Martin and Zhang, 2007; Ruthenburg et al., 2007).
Histone PTMs are thought to contribute to the regulation of chromatin-templated processes via two major mechanisms (Kouzarides, 2007; Ruthenburg et al., 2007). First, histone PTMs can directly modulate the packaging of chromatin either by altering the net charge of histone molecules or by altering inter-nucleosomal interactions, thereby regulating chromatin structure and the access of DNA-binding proteins such as transcription factors. Second, histone PTMs regulate chromatin structure and function by recruiting PTM-specific binding proteins, which recognize modified histones via specialized structural folds such as bromo-, chromo- and PHD domains (Wysocka et al., 2005; Wysocka et al., 2006; Zeng and Zhou, 2002). Alternatively, histone PTMs can also function by inhibiting the interaction of specific binders with chromatin. PTM-induced changes in protein interactions between chromatin and its binding partners are in turn translated into biological outcomes (Margueron et al., 2005).
While the majority of known histone PTMs are located within the N-terminal tail domain of core histones, PTMs of crucial importance for histone-DNA and histone-histone interactions have also been found in the globular domain of core histones (Cosgrove et al., 2004; Garcia et al., 2007c; Mersfelder and Parthun, 2006). Novel PTM sites occurring outside of the N-terminal tails continue to be discovered, generally with the aid of sequence and modification-specific antibodies or by unbiased mass spectrometry (MS) methods (Chu et al., 2006; Garcia et al., 2007b; Johnson et al., 2004; Wisniewski et al., 2007). The recent discovery of O-GlcNAc modification (Sakabe et al., 2010) suggests that additional histone PTMs may yet be discovered.
Here, we used an integrated, mass spectrometry-based proteomics approach, which takes advantage of in vitro propionylation, efficient peptide separation using isoelectric focusing (OFFGEL), and the high sensitivity of the LTQ Orbitrap Velos mass spectrometer to carry out a comprehensive analysis of histone PTMs. With this approach, we achieved high sequence coverage of peptide mapping in core and linker histones, ranging from 87% to 100%, which in turn resulted in the identification of 67 new PTM sites. These newly identified histone marks expand the total number of known histone PTMs by about 70%. Interestingly, our results show that histones are intensely modified at various residues not only in the N-terminal tail, but also within globular domains. Among the new modifications, we identified tyrosine hydroxylation (Yoh) and lysine crotonylation (Kcr) as two novel histone mark types. Finally, we demonstrated that histone Kcr is a robust indicator of active promoters and could be an important signal in the control of male germ cell differentiation.
RESULTS
Experimental design
Histone proteins are characterized by a high ratio of both lysine and arginine residues (Garcia et al., 2007a; Zee et al., 2010). As a result, tryptic digestions of histones tend to yield peptides that are relatively small and hydrophilic, which are difficult for subsequent detection by MS due to poor retention by the C18 RP-HPLC column. This problem can be addressed by chemical derivatization (e.g., lysine propionylation) of amine groups in the protein (N-terminal amines, and free and monomethylated lysine ε-amino groups) before or after tryptic digestion (Garcia et al., 2007a). Similarly, lysine propionylation of core histones, before or after tryptic digestion, can generate complementary peptide sequences that boost the sequence coverage of peptide mapping by MS. Additionally, IEF separation of the tryptic digest into 12 fractions will further reduce peptide complexity and improve dynamic range.
Using this rationale, we designed an integrated approach for the systematic analysis of histone PTMs which maximizes both sequence coverage and sensitivity, leading to the identification of many novel PTM sites. In this method, MS analysis was carried out in histone proteolytic peptides that were generated by four parallel methods (Figure 1A): (i) tryptic digestion of core histones without an in vitro chemical derivatization reaction, (ii) tryptic peptides that were in vitro propionylated after tryptic digestion, (iii) tryptic peptides that were generated by tryptic digestion of in vitro propionylated histone proteins, and (iv) tryptic peptides that were generated by tryptic in-gel digestion of the individual histone proteins.
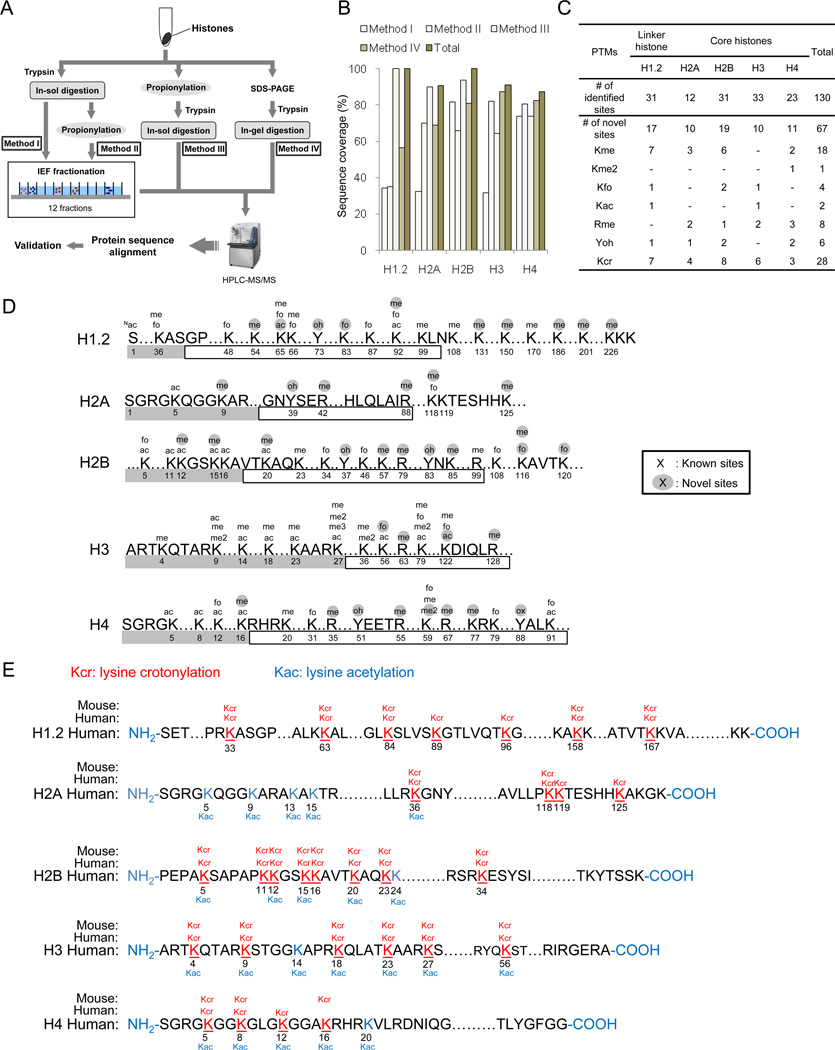
Figure 1.
Experimental strategy and results for identified histone PTM sites. (A) Schematic diagram of the experimental design for comprehensive mapping of PTM sites in linker and core histones from HeLa cells. Histone extracts were in-solution trypticly digested without chemical propionylation (Method I), chemically propionylated after in-solution tryptic digestion (Method II), chemically propionylated before in-solution tryptic digestion (Method III), and in-gel digested after SDS-PAGE gel separation. Samples from Methods I and II were further subjected to IEF fractionation to generate 12 fractions. (B) Peptide sequence coverage of linker and core histones in each of the four methods is shown. (C) A table summarizing all the PTM sites identified by this study. Abbreviations: me, monomethylation; me2, dimethylation; me3, trimethylation; fo, formylation; ac, acetylation; oh, hydroxylation; cr, crotonylation. (D) A diagram showing sites of histone PTMs other than Kcr identified in this study. Amino acid residue number is indicated below its sequence. Gray and blank boxes indicate N-terminal and globular core domains, respectively. (E) Illustrations of histone Kcr sites in human HeLa cells and mouse MEF cells. All Kcr sites are shown in red and underlined. Previously reported Kac sites are shown in blue.
We used PTMap, an algorithm capable of identifying all possible PTMs of a protein (Chen et al., 2009), to analyze all acquired MS/MS data and identify histone peptides with or without a PTM. As anticipated, sequence coverage by MS mapping was significantly improved after in vitro propionylation, either before or after tryptic digestion (Figure 1B). Among the four methods, Method III (in vitro propionylation before tryptic digestion of histones) achieved the highest sequence coverage of histones H1.2 (100%), H2A (90.7%) and H2B (94.4%). Method IV gave the best coverage for histones H3 (87.3%) and H4 (82.3%). In aggregate, we achieved sequence coverage of 100% of H1.2, 90.7% of H2A, 100% of H2B, 91% of H3, and 87.3% of H4. To our knowledge, this represents the highest reported sequence coverage for peptide mapping in histones.
Using this approach, we identified 130 unique PTM sites, which not only confirmed 63 previously known histone PTMs, but also revealed 67 novel ones, including 28 Kcr sites, 18 lysine monomethylation (Kme) sites, 1 lysine dimethylation (Kme2) site, 4 lysine formylation (Kfo) sites, 2 lysine acetylation (Kac) sites, 8 arginine monomethylation (Rme) sites and 6 tyrosine hydroxylation (Yoh) sites (Figure 1C).
A summary of the non-Kcr modification sites and Kcr sites identified in this study is shown in Figure 1D and 1E, respectively. All the MS/MS spectra for the identified histone PTM peptides were carefully verified as previously reported (Chen et al., 2005). We confirmed the identification of Kcr peptides and 10 novel non-Kcr PTM sites by MS/MS of their corresponding synthetic peptides or by high-resolution MS/MS (Figure S1). Identification and validation of these non-Kcr PTMs are included in Extended Experimental Procedures.
Characterization of the novel histone PTM sites
A core histone protein typically consists of an unstructured N-terminus, a globular core including a central histone-fold domain, and a conformationally mobile C-terminal tail (Garcia et al., 2007c; Mersfelder and Parthun, 2006). The central histone-fold domain consists of 3 α-helices and two loops that are known to be involved in histone pair-pair and histone-DNA interaction sites (McGhee and Felsenfeld, 1980). The majority of known histone PTM sites were previously identified in the N-terminal regions of histones.
In this study, 39 novel non-Kcr PTM sites were identified. Interestingly, among these sites, only 4 sites were mapped to the N-terminal domains, while 25 were mapped to central histone-fold domains and another 10 were mapped to the C-terminal domains (Figure 1D). Six PTM sites (including 3 monomethylated residues at H2BR79, H3R63 and H4K77, 1 acetylated residue at H3K122 and 2 hydroxylated sites at H2BY83 and H4Y88) are located at the histone-fold domains (Figure S1M). H3R63 and H4K77 participate in DNA interactions (Arents and Moudrianakis, 1993; Luger et al., 1997; Mersfelder and Parthun, 2006), while H2BR79, H2BY83 and H4Y88 participate in the H2B–H4 interaction. While the amino acid residues located on the outer nucleosome surface do not contact DNA, they are known to regulate chromatin structure (Mersfelder and Parthun, 2006). Five PTM sites were mapped at the outer nucleosome surface, among which we found 3 novel PTM sites, including monomethyllysine and formyllysine at H2BK116, and dimethyllysine at H4K59 (Figure S1M). Given the important roles performed by these residues in nucleosome structure and DNA binding, it is highly likely that these newly discovered PTM sites will have significant impacts on transcriptional and epigenetic regulation.
Identification and validation of Kcr residues in histones
A PTM will induce a structural change in the substrate residue and therefore a change of its molecular weight. Interestingly, on 28 lysine residues of core histone peptides, our analysis identified a mass shift of + 68 Da that does not match the shift associated with any known PTM (Figure 1E). This result suggested the possible presence of a previously unreported histone mark.
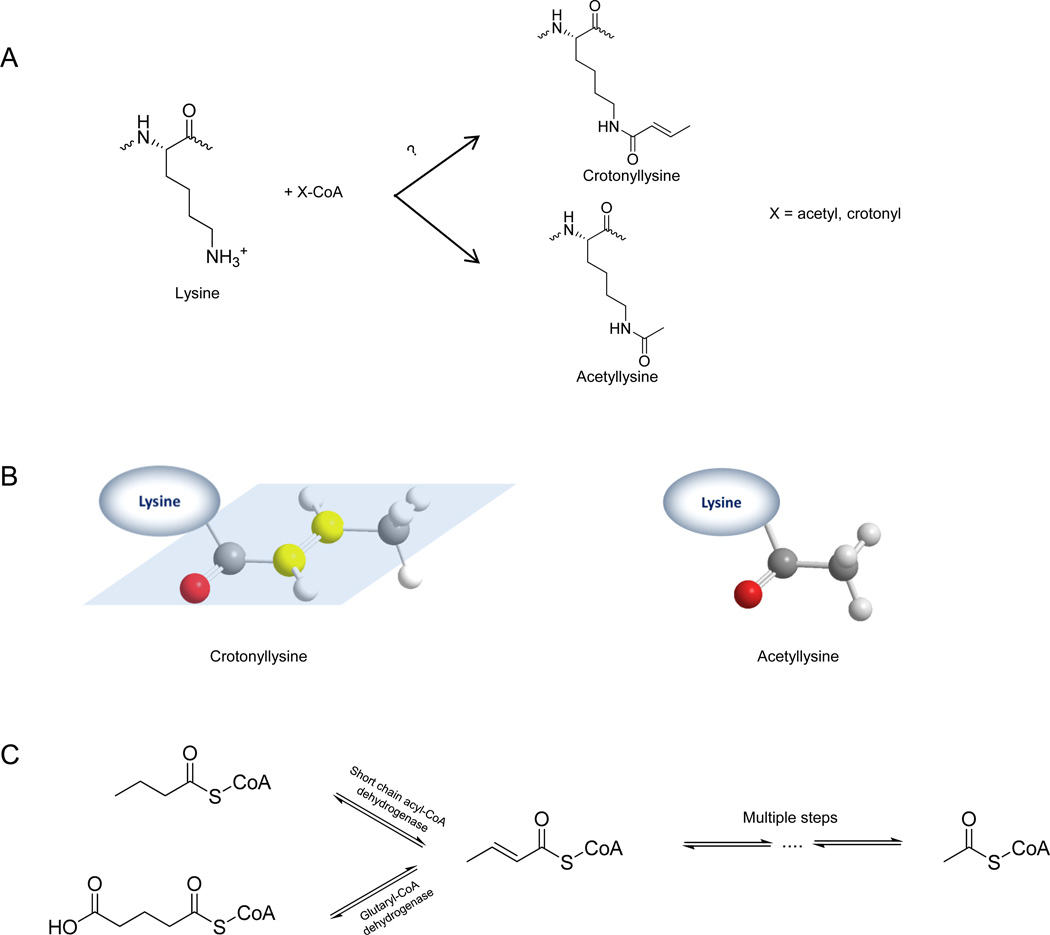
To determine the nature of this modification, we selected one of these peptides, PEPAK+68SAPAPK (modified at H2BK5), for further analysis. After manual inspection of the high-resolution MS data (precursor ion mass at m/z 580.8181), we determined the accurate mass shift of this modification was + 68.0230 Da. By setting the mass tolerance to ± 0.01 Da (∼ 9 ppm, which is within the mass accuracy of our mass spectrometer), and specifying a maximum of 2 nitrogen atoms, we were able to deduce the possible element compositions of the modification group as either C4H4O or H6NO3. The former, C4H5O (mass shift plus one proton), is the only reasonable molecular formula of this modification. There were 4 possible structures consistent with the element composition: Kcr (Figure 2A and 2B), vinylacetyllysine (3-butenoyllysine), methacryllysine, and cyclopropanecarboxyllysine (Figure S2A). As crotonyl-CoA is an important and abundant intermediate (Figure 2C) in metabolic pathways of butyryl-CoA and acetyl-CoA, we focused on Kcr as the PTM candidate most likely to cause the mass shift.
Figure 2.
Short-chain lysine acylations. (A) An illustration of the enzymatic reactions for lysine acetylation by lysine acetyltransferases (KATs) using acetyl-CoA as a cofactor, and a hypothesized mechanism for Kcr using crotonyl-CoA as a cofactor. (B) Ball-and-stick models of a crotonyl group and an acetyl group. The three-dimensional arrangement of four carbons and one oxygen of the crotonyl group are rigid and located in the same plane (left). The two olefinic carbons of the crotonyl group are shown in yellow. In contrast, the tetrahedral CH3 in the acetyl group (right) can be rotated such that it is structurally very different from the crotonyl group. (C) Crotonyl-CoA metabolism pathways. Crotonyl-CoA was generated from butyryl-CoA or glutaryl-CoA, and oxidized to acetyl-CoA through multiple steps.
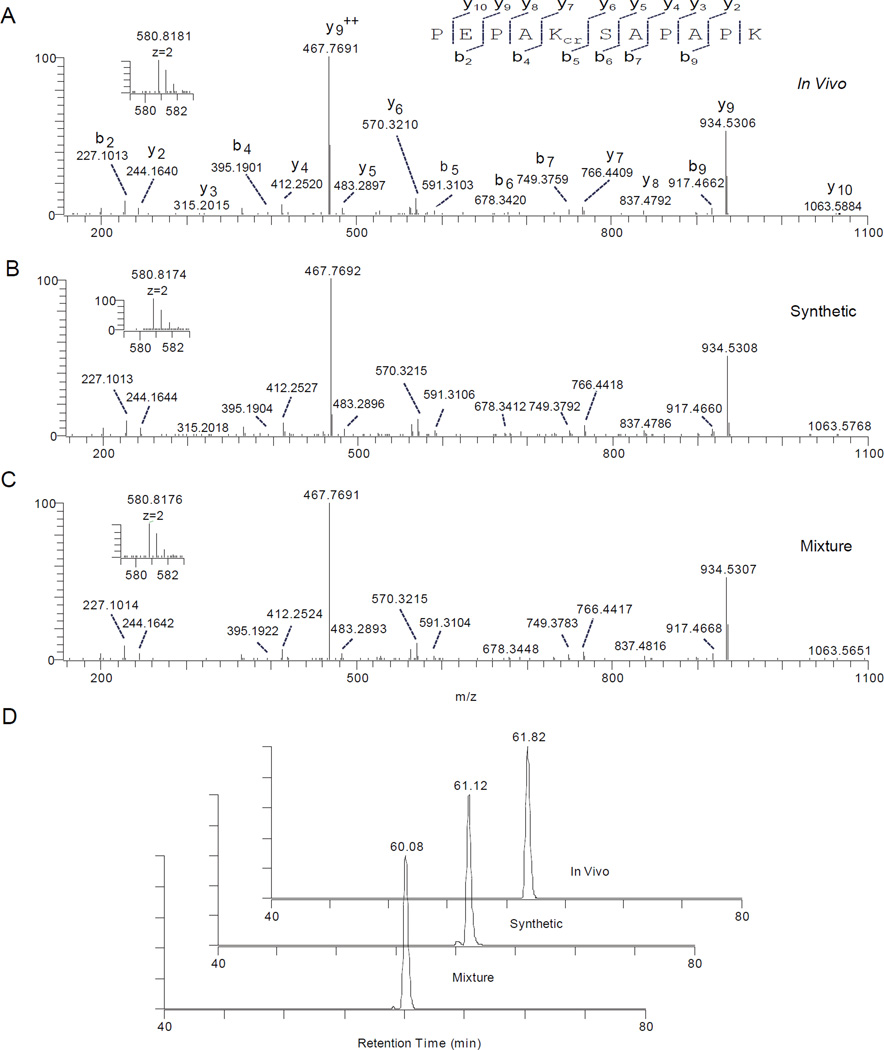
To test if the identified mass shift of + 68.0230 Da was caused by Kcr, we synthesized the Kcr peptide, PEPAKcrSAPAPK, and compared its MS/MS spectrum with that of the in vivo-derived peptide. The in vivo modified peptide bearing a lysine residue with a mass shift of +68.0230 Da, the synthetic Kcr peptide with the same peptide sequence (PEPAKcrSAPAPK), and the mixture of the two peptides exhibited almost identical parent masses and high-resolution MS/MS spectra (Figure 3A to C). In addition, the mixture of the in vivo and synthetic peptides co-eluted in HPLC/MS analysis (Figure 3D). These results indicated that the identified mass shift of + 68.0230 Da was very likely caused by Kcr.
Figure 3.
Identification and verification of a Kcr peptide, PEPAKcrSAPAPK (Kcr indicates a crotonyllysine residue). (A, B, C) High-resolution MS/MS spectrum of a tryptic peptide, PEPAKSAPAPK, with a mass of +68.0230 Da at its Lys5 residue identified from in vivo histone H2B (A), its synthetic Kcr counterpart (B), and a peptide mixture of the in vivo-derived tryptic peptide and its synthetic counterpart (C), each showing the same MS/MS fragmentation patterns and the same precursor ion mass. Inset shows their precursor ion masses. (D) Extracted ion chromatograms (XICs) of the in vivo-derived PEPAK+68.0230SAPAPK peptide, the synthetic Kcr counterpart, and their mixture by nano-HPLC/MS/MS analysis using a reversed-phase HPLC column, showing the co-elution of the two peptides.
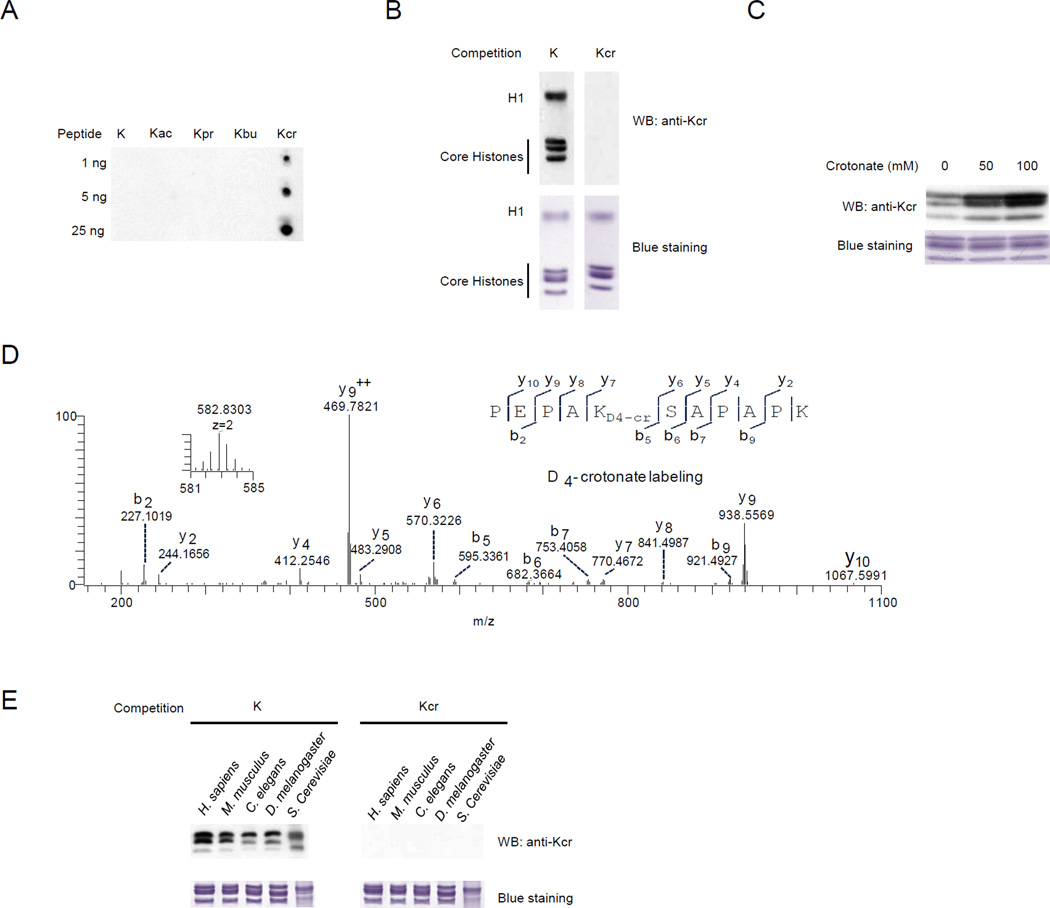
To further confirm Kcr in histones, we generated a pan antibody against Kcr. This pan anti-Kcr antibody specifically recognized a peptide library bearing Kcr, but not four other peptide libraries in which the fixed lysine residue was unmodified, acetylated, propionylated, or butyrylated (Figure 4A). The specificity of the pan anti-Kcr antibody was further demonstrated by Western blotting with three different bovine serum albumin (BSA) derivatives, where peptide lysines were chemically modified by a crotonyl, vinylacetyl or methacryl group, respectively. The result showed that pan anit-Kcr antibody recognized only the lysine crotonylated BSA, but not the unmodified, lysine vinylacetylated or lysine methacrylated BSA (Figure S2C). This pan anti-Kcr antibody was subsequently used for Western blotting and immunostaining of Kcr signal.
Figure 4.
Detection of Kcr in histones by Western blotting. (A) Specificity of pan anti-Kcr antibody demonstrated by dot-spot assay using five peptide libraries with indicated amount (ng). Each peptide library contains 13 residues CXXXXXKXXXXXX, where X is a mixture of 19 amino acids (excluding cysteine), C is cysteine, and the 7th residue is a fixed lysine residue: unmodified lysine (K), Kac, propionyllysine (Kpr), butyryllysine (Kbu), and Kcr. (B) Detection of Kcr in histones. Western blotting was carried out using the histones from HeLa cells with competition of a peptide library bearing a fixed unmodified lysine (K) or Kcr. (C) Dynamics of histone Kcr in response to crotonate. The histone proteins extracted from human prostate cancer cell line Du145 incubated with 0, 50 or 100 mM crotonate for 24 hours, were Western blotted with anti-Kcr pan antibody. (D) MS/MS spectrum of PEPA KD4-crSAPAPK identified from D4-crotonate-labeled sample. The mixture of D4-, D3- and D2-crotonyl groups was used for the identification of D4-crotonyl peptide. (E) Kcr signals in core histones of S. cerevisiae, C. elegans, D. melanogaster (S2), M. musculus (MEF), as well as H. sapiens (HeLa) cells by Western blotting analysis with competition.
The antibody detected a Kcr signal among all core histone proteins: H2A, H2B, H3, H4, and linker histone H1. In each protein, the signal could be efficiently competed away by a peptide library bearing a Kcr, but not peptide libraries bearing an unmodified lysine (Figure 4B), metharcryllysine (Figure S2D), acetyllysine, propionyllysine, or butyryllysine (Figure S2E). The strong Kcr signals detected in histones indicated that this novel modification is present in nuclei and is associated with chromosomes. Indeed, immunostaining using the pan antibody showed that Kcr mainly existed in nuclei (Figure S2F, see also the analysis below of spermatogenic cells).
Isotopic labeling is an established method to confirm in vivo protein modifications that was previously used for the study of histone Kac (Allis et al., 1985). We rationalized that D4-crotonate can be converted into crotonyl-CoA in vivo, which then functions as a lysine crotonylation donor. Consistent with this hypothesis, the histone Kcr signal from HeLa cells increased dramatically after cells were cultured with crotonate (Figure 4C). After tryptic digestion of histones from D4-crotonate labeled HeLa cells, we carried out peptide immunoprecipitation using the anti-Kcr antibody. The enriched Kcr peptides were then subjected to HPLC/MS/MS analysis and protein sequence alignment, which confirmed Kcr on histone H2BK5 by D4-crotonate labeling (Figure 4D). (Note: The D4-crotonic acid was mixed with D3- and D2-crotonic acid (Figure S2G). This characteristic isotopic distribution was used for the identification of D4-crotonyl peptide.)
Histone Kcr sites in human cells
To identify histone Kcr sites, we used a Mascot sequence alignment to analyze MS/MS data derived from HeLa histones, using Kcr (+ 68.02621 Da) as a variable modification. These analyses led to the identification of 28 Kcr sites in human histones (Figure 1E). In addition, 19 of these 28 identified sites were confirmed by in vivo D4-crotonate labeling experiments. The Kcr peptides and their corresponding MS/MS spectra are included in Figure S4 and S5.
In summary, we have used five independent methods – MS/MS and HPLC coelution of synthetic peptides, D4-crotonate labeling, Western blotting, and immunostaining – to show that histone Kcr exists in cells, and identified 28 lysines on various histones that are subject to this PTM.
Histone Kcr is an evolutionarily-conserved histone PTM
To test if lysine crotonylation is present in histones from other eukaryotic cells, we isolated histones from yeast S. cerevisiae, C. elegans, Drosophila S2 cells, mouse embryonic fibroblast (MEF) cells, as well as human HeLa cells. Kcr signals were detected among core histones from all five species by Western blotting (Figure 4E). Taking advantage of affinity enrichment using the pan anti-Kcr antibody and HPLC/MS/MS, we identified 24 Kcr sites on mouse MEF cells (Figure 1E). All of the annotated MS/MS spectra for Kcr mouse histone peptides are included in Figure S6. Taken together, our results revealed that Kcr is an evolutionarily conserved histone mark appearing in eukaryotic cells from a wide range of species.
Histone Kcr and Kac are mechanistically and functionally different
In order to examine whether histone Kcr is derived from promiscuous histone acetyltransferase (HAT) activity, we overexpressed two HATs, CBP and p300, in 293T cells and tested for histone crotonylation levels by Western blotting. We found that overexpression of CBP or p300 led to the enhancement of histone Kac, but did not significantly change the levels of histone Kcr (Figure S2H).
Using a fluorometric assay, we also demonstrated that histone lysine deacetylaseses (HDACs) 1, 2, 3, and 6 exhibit potent lysine deacetylation activities, but have very weak or no effect on lysine decrotonylation. For example, lysine decrotonylation activity was not detected with HDAC6 exposure, though the enzyme’s deacetylation activity was very strong in this assay (Figure S2I). These results suggest that enzymes required for the addition and removal of Kcr on histone proteins may be different from those for histone Kac regulation.
Genome-wide mapping of histone Kcr in human cells
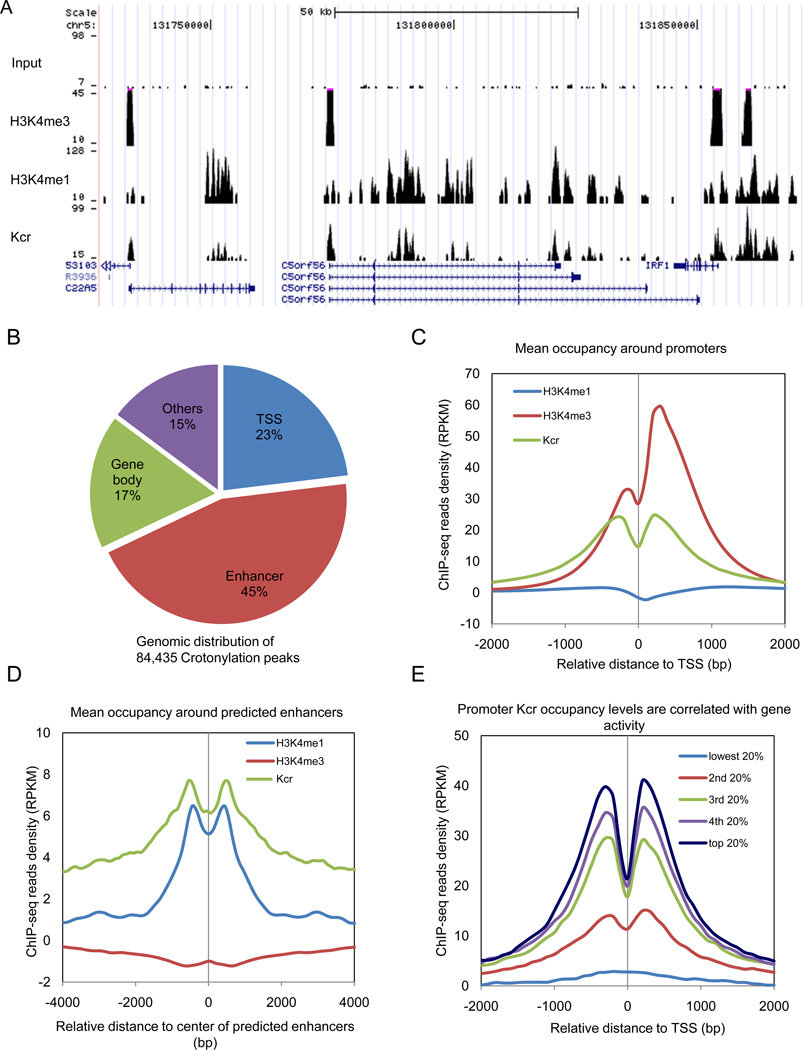
In order to explore the in vivo function of histone Kcr, we performed ChIP-seq analysis with the pan anti-Kcr antibody to determine the genomic distribution of histones with this modification in the human fetal lung fibroblast IMR90 cell line. As a control, we compared the Kcr distribution with previously obtained ChIP-seq results for H3K4me3 (marking gene promoters) and H3K4me1 (marking enhancers) distribution (Heintzman et al., 2007).
Strikingly, we found an abundance (totaling 84,435 peaks) of this novel histone modification in the human genome. Histone Kcr was largely associated with active chromatins, including both the transcription starting site (TSS) and regions previously predicted to be enhancers (Hawkins et al., 2010) (Figure 5A and B).
Figure 5.
Enrichment of histone Kcr on active chromatin. (A) ChIP-seq snapshots of input, H3K4me3, H3K4me1 and Kcr in IMR90 cells. (B) Pie chart showing the genomic distribution of all histone Kcr peaks with annotated genomic regions. TSS is defined as regions +/− 2.5kb around known transcription starting sites in RefSeq database. Enhancers are promoter distal regions associated with H3K4me1 as predictive mark. (C) Curves showing the mean reads density of indicated histone modification around all known TSS. Reads densities are calculated within a 100bp sliding window and normalized by subtracting reads density in the control input ChIP-seq data. RPKM is calculated as the number of reads which map per kilobase of genomic region per million mapped reads. (D) Average normalized read densities of H3K4me3, H3K4me1 and histone Kcr around predicted enhancers are plotted. (E) All RefSeq genes are divided into 5 groups based on their expression level calculated from mRNA-seq data, and the average normalized read densities of Kcr around each group of TSS are plotted.
The majority (68%) of histone Kcr peaks was associated with either promoter or predicted enhancer regions (Figure 5B). At promoters, histone Kcr showed the strongest enrichment flanking TSS, in contrast to the observation that H3K4me3 was more enriched downstream of TSS (Figure 5C). At predicted enhancers, we observed a strong enrichment of Kcr in agreement with the H3K4me1 pattern at these sites (Figure 5D). Furthermore, we also observed a strong correlation between gene expression and Kcr level at promoters (Figure 5E). Taken together, these data strongly support a role for histone Kcr in gene regulation, especially at promoters and enhancers.
In addition, we also compared Kcr sites with histone lysine acetylation sites revealed by ChIP-seq assay using a pan anti-Kac antibody. The result showed that histone crotonylation generally occupies similar locations as acetylation in IMR90 cells as there is a significant overlap between Kac and Kcr peaks (Figure S3A). These results suggested that in resting somatic cells, open chromatins are simultaneously marked by both histone Kac and Kcr. Given the finding that these two marks are catalyzed by different sets of enzymes, we assumed that the regulation of Kcr would likely have different spatial and temporal dynamics from Kac. To test this prediction, we explored the function of Kcr in the highly dynamic spermatogenesis process.
Histone Kcr marks testis-specific genes activated in post-meiotic cells
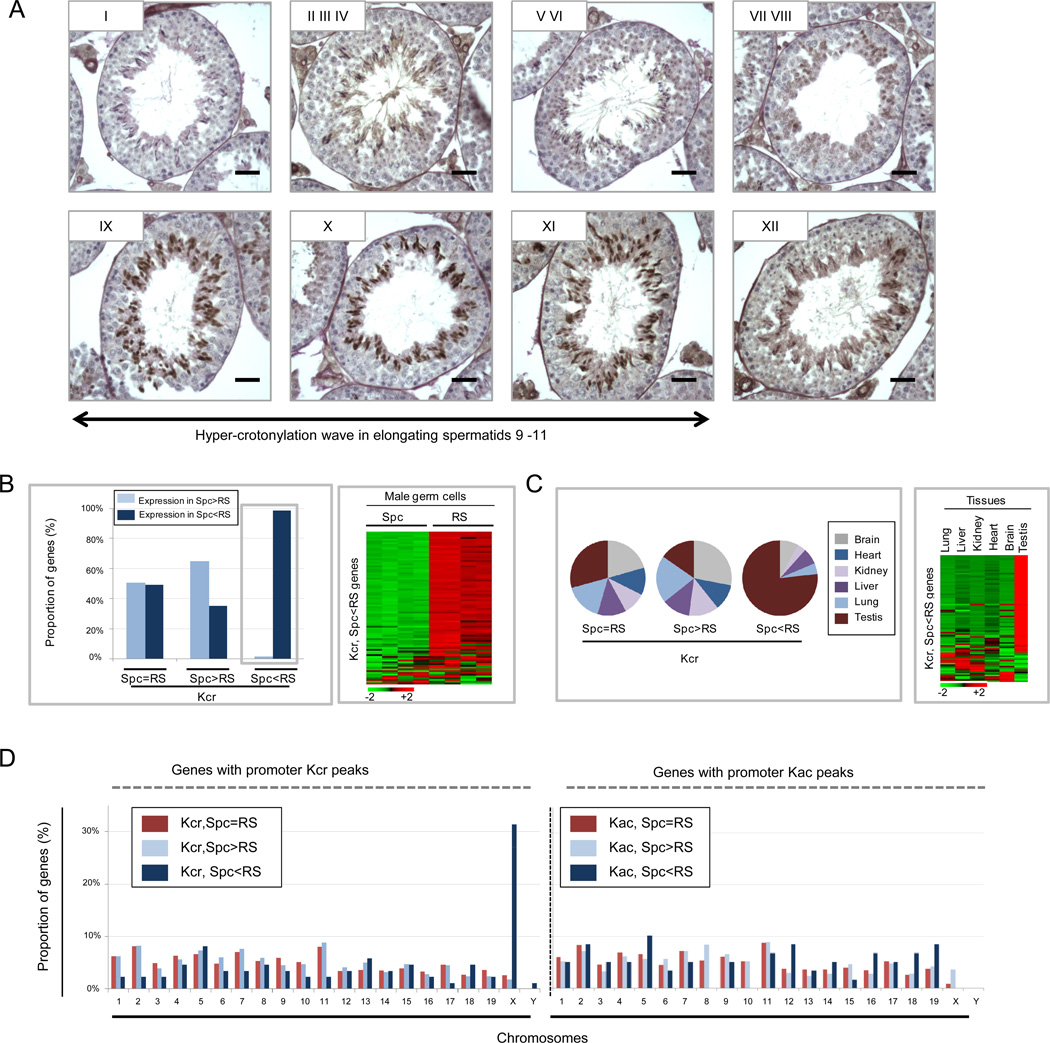
We next investigated the genomic distribution of histone Kcr in spermatogenic cells, where a very specific gene expression program directs key steps of differentiation (Boussouar et al., 2008; Gaucher et al., 2010). We first examined the global dynamics of Kcr during mouse spermatogenesis using immunohistochemistry. We observed an intense labeling of Kcr during elongating steps 9 to 11 in spermatids (Figure 6A), which coincides with a genome-wide histone hyperacetylation previously reported (Hazzouri et al., 2000). Since a general transcriptional shutdown occurs at these stages of germ-cell differentiation (Zhao et al., 2004), we investigated the relationship between Kcr and gene expression using a ChIP-seq experiment in earlier stages. Spermatogenic cells were fractionated to enrich spermatocytes (Spc) and post-meiotic round spermatids (RS), which were used for ChIP-Seq analysis. As shown in Figure S3B, the genomic regions associated with histone Kcr from mouse spermatogenic cells showed a pattern very similar to that of human somatic cells: histone Kcr is enriched at the TSS of genes (Figure S3C). Additionally, we identified many Kcr sites lacking histone Kac, suggesting that crotonylation can occur independently of Kac (Figure S3D).
Figure 6.
Correlation of histone Kcr with gene expression in meiotic, post-meiotic male germ cells, and in tissues. (A) Hyper-crotonylation wave in elongating spermatids. Kcr was detected on paraffin mouse testis tubule sections representing different stages of spermatogenesis by immunohistochemistry (IH) using an anti-Kcr antibody. Pre-meiotic spermatogonia (Spg) and meiotic spermatocytes (Spc) cells are present at the periphery and middle of the tubule sections, whereas post-meiotic round (RS), elongating (ES) and condensing (CS) spermatids are near the lumen. The nuclei of ES are positive for Kcr. (B, C) Genes associated with higher Kcr in RS than Spc are mostly post-meiotically activated and show a predominant expression in the testis. The genes associated with Kcr peaks were divided into three categories according to their Kcr levels in Spc and RS: i) Spc=RS, similar Kcr levels between Spc and RS; ii) Spc>RS, lower Kcr levels in RS than Spc (fold change >=2); iii) Spc<RS, higher Kcr levels in RS than Spc (fold change >=2). The expression of these genes in male germ cells (B) and tissues (C) was then compared among the three categories. (B) Expression in male germ cells. Left panel: respective proportions of genes (Y axis) with higher expression either in Spc or RS among the three gene categories (X axis). Right panel: heatmap showing the expression of the third category of genes (Kcr, Spc <RS) in Spc (4 samples) and in RS (4 samples). Color scale showing low expression in green to high expression in red. (C) Expression of genes in tissues. Left panel: pie charts showing the respective proportions of genes with the highest level of expression in the indicated. Genes with the highest tissue-specific expression are those whose levels of expression, in the indicated tissue, are elevated by at least two standard deviations above the mean expression in all tissues. Right panel: heatmap showing the expression of the third category of genes in the indicated tissues. (D) The X-linked genes are highly and specifically marked by histone Kcr in RS. The respective proportion (%) of genes associated with Kcr (left panel) or Kac peaks (right panel) among chromosomes in male germ cells is shown.
To gain insight into the biological significance of promoter histone Kcr, we classified the genes associated with histone Kcr into three categories, depending on the variation of Kcr levels before and after meiosis: i) genes associated with similar levels of promoter Kcr between spermatocytes (Spc) and round spermatids (RS), ii) genes associated with lower promoter Kcr levels in RS than Spc, and iii) genes associated with higher promoter Kcr levels in RS than Spc (Figure 6B). We then analyzed expression levels among the three categories of genes. To this end, transcriptomic data from staged mouse spermatogenic cells (available on GEO website: GSE217 and GSE4193) were downloaded. The respective proportions of genes showing higher expression in either Spc or RS were determined among the three categories of genes. This analysis revealed no significant connection between histone Kcr levels and higher gene expression in either meiotic (Spc) or post-meiotic (RS) cells for the first two categories of genes (Kcr: Spc=RS and Spc>RS). However, the vast majority of the genes with higher Kcr in RS (Kcr:Spc<RS) were found to be activated after meiosis (Figure 6B, left). This result indicates a tight correlation between post-meiotic histone Kcr and a specific haploid cell gene expression program.
To further characterize these genes, we analyzed their expression patterns in testis and several somatic tissues from adult mice. Here again, most of the genes associated with a post-meiotic increase of Kcr (Spc <RS), but not the other two gene categories, were highly expressed in testis (Figure 6C). A parallel ChIP-seq analysis of Kac showed that only 35% of these genes are associated with histone Kac at the promoters (data not shown). Therefore, the majority of the highly-expressed testis genes seemed to be predominantly labeled by histone Kcr but not by histone Kac.
Finally, we analyzed the genomic localizations of genes among the three categories of Kcr-marked genes. Our data showed that 31.4% and 1.2% of the third category of genes (Kcr, Spc<RS) were located on the X and Y chromosomes, respectively, while the other two categories of genes showed no particular bias in genomic distribution (Figure 6D, left). In contrast, three categories of Kac-marked genes showed no bias toward a particular chromosome (Figure 6D, right). This result therefore confirms distinct genomic distribution patterns for histone Kcr and histone Kac.
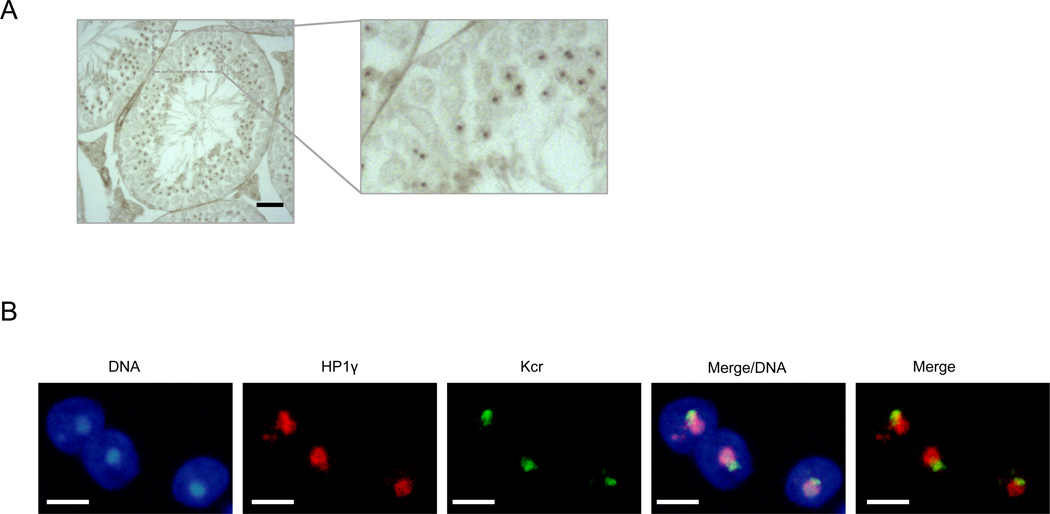
Given the post-meiotic enrichment of Kcr on sex chromosomes, we reasoned that an in situ approach may allow us to visualize a preferential association of Kcr with sex chromosomes. Remarkably, immunohistochemistry (IH) staining of testis sections (without counter-staining) revealed the presence of a single, dot-like structure exclusively found in round spermatids (Figure 7A and Figure S3E). To analyze the nature of this structure, we used sections of seminiferous tubules containing mostly round spermatids for the co-staining of either Kac and Kcr (Figure S3F), or Kcr and HP1γ (Figure 7B). HP1γ marks the heterochromatic chromocenter, as well as adjacent mostly-inactive sex chromosome, which is consistent with previously reported works (Namekawa et al., 2006; Turner, 2007). Kcr concentrates at the sex chromosomes and is visible as a DAPI-dense structure adjacent to the chromosome center, which itself is composed of centromeric and pericentric heterochromatin and devoid of Kac (Turner, 2007). The co-staining of Kcr and HP1γ confirmed these findings and supported the ChIP-seq results of an accumulation of Kcr on the sex chromosomes.
Figure 7.
Sex chromosomes are highly crotonylated in round spermatids. IH (A) on a testis tubule section and immunofluorescence (IF) (B) of male germ cells showing high crotonylation enrichment in the sex chromosome region (besides the HP1γ enriched chromocenter) in post-meiotic round spermatids. IH is shown without counterstaining for better visualization of the region detected by the antibody. IF: pan-Kcr was detected in green, and is shown in co-staining with HP1γ in bright red fluorescence. Scale bar for IF: 5 micrometers.
These data indicate that histone Kcr may play an important role in epigenetically marking sex chromosomes in the post-meiotic stages of spermatogenesis. On these chromosomes, this histone mark may be involved in specifically defining a subset of genes that escape sex chromosome inactivation after the completion of meiosis.
Discussion
In this study, we described an integrated approach for the systematic analysis of histone PTMs (Figure 1). With this unique approach, we identified 130 PTM sites on human histones, including 63 known and 67 novel histone marks (Figure 1C). There are about 93 previously described histone marks based on the UniProt database (http://www.uniprot.org; Accession number: P16403 for histone H1.2; P0C0S8 for histone H2A; P62807 for histone H2B; P84243 for histone H3.3; P62805 for histone H4) and 3 histone O-GlcNAc sites that were recently identified (Sakabe et al., 2010). Thus, our study expands the catalogue of known histone marks by around 70%, which affirms the unprecedented quality and sensitivity of our approach. In particular, we have identified Yoh and Kcr as two novel types of histone PTM. Therefore, this work dramatically extends the catalogue of histone PTM sites in mammalian cells and provides a platform for the discovery of novel mechanisms of histone regulation.
Five lines of evidence suggest that histone Kcr and histone Kac are different from each other and involve distinct pathways for addition and removal. First, the two PTMs are structurally very different. A crotonyl group has a more rigid structure, due to the presence of a flattened C-C π-bond (Figure 2B), while the methyl group of an acetyl group is tetrahedral and rotatable. In light of these obvious electronic and geometric differences, it is highly likely that Kcr uses distinct regulatory enzymes from Kac. Second, some lysine residues appear to be modified by crotonylation but not acetylation in core histones from specific cells of interest (e.g., H2A118, H2A119, H2A125, H2BK34, H1.2K89, and H1.2K167 in HeLa and MEF cells reported here). Third, our results show that KATs and HDACs exert very different effects upon Kac and Kcr. Fourth, the status of short-chain lysine acylation may be modulated in response to CoA concentration. In the case of crotonyl-CoA, it can be generated either from butyryl-CoA by short chain acyl-CoA dehydrogenase, or from glutaryl-CoA by glutaryl-CoA dehydrogenase. Once formed, crotonyl-CoA can be converted to acetyl-CoA for the TCA cycle. Finally, histone Kac and Kcr mark different sets of gene in some differentiated cells, as we report here in mouse sperm cells.
Strikingly, in both human somatic and mouse male germ cell genomes, histone Kcr specifically labels enhancers and, most precisely, the TSS of active genes. In post-meiotic male germ cells, a gain in histone Kcr is a consistent indicator of an X-linked haploid cell-specific gene expression program. Indeed, the identification of a subset of genes presenting increased Kcr in round spermatids allowed us to show that such genes are enriched on the X chromosome and are mostly predominantly expressed in the testis with a post-meiotic pattern of expression.
Meiosis is known to be associated with the inactivation of sex chromosomes, called meiotic sex chromosome inactivation (MSCI). MSCI initiates in pachytene spermatocytes, and sex chromosome gene silencing continues after meiosis in round spermatids (RS) until the general shutdown of transcription (Namekawa et al., 2006; Turner, 2007). However, in round spermatids, a significant number of X-linked genes have been shown to be specifically reactivated (Mueller et al., 2008; Namekawa et al., 2006). Interestingly, our results showed that histone Kcr specifically marks X-linked genes that are post-meiotically re-expressed. Consistent with this observation, our in situ experiment uncovered a remarkable post-meiotic labeling of Kcr on sex chromosomes. In addition, our result indicates that the addition of Kcr goes beyond the specific sets of genes identified above and also includes intergenic regions, a conclusion which has been confirmed by analysis of ChIP-seq data (data not shown).
Another interesting observation is the occurrence of genome-wide histone Kcr in elongating spermatids following the general transcriptional shut-down associated with histone replacement. The only genome-wide histone modification previously known to be associated with histone removal is histone hyperacetylation (Govin et al., 2004). The occurrence of global histone Kcr at the same time strongly suggests that, like histone Kac, Kcr is likely to affect chromatin structure and hence facilitate histone replacement. Our data indicate that there is a limited overlap between histone Kcr and Kac. Although both histone modifications are associated with gene expression and their occurrence at a larger scale could facilitate histone removal, they may differently orient the reorganization of the marked genomic regions after histone removal, and hence be important histone marks in the establishment of a region-specific male epigenome organization.
Given the fact that all the previously-studied histone lysine PTM pathways, such as lysine acetylation and methylation, have important biological functions, we anticipate that Kcr will also likely play a major role in the regulation of histone protein structure and function. Identification of histone Kcr raises many interesting questions. What enzymes regulate the addition or removal of Kcr? What role does histone Kcr play in the regulation of histone structure and function? Are there proteins that bind to histone Kcr? Is lysine crotonylation also present on non-nuclear proteins as well? Answers to these questions will require further investigation.
Experimental Procedures
In-solution proteolytic digestion and chemical derivatization of histone proteins
In-solution tryptic digestion of histone samples was carried out using a protocol previously described (Kim et al., 2006; Luo et al., 2008). In vitro lysine propionylation of histone extract and tryptic histone peptides was performed as previously described (Garcia et al., 2007a). Histone extracts were in-solution digested without chemical propionylation, chemically propionylated after in-solution digestion, or chemically propionylated before in-solution digestion.
Isoelectric focusing (IEF) fractionation
The histone proteolytic peptides were separated using an Agilent 3100 OFFGEL Fractionator (Agilent, Santa Clara, CA) according to the manufacturer’s instructions. Twelve fractions were obtained from each IEF fractionation experiment.
Nano-HPLC/mass spectrometric analysis
The tryptic digests were injected into a NanoLC-1D plus HPLC system (Eksigent Technologies, Dublin, CA), and analyzed by an LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific, Waltham, MA). Full scan MS spectra from m/z 350 – 1400 were acquired in the Orbitrap. Twenty of the most intense ions were isolated for MS/MS analysis.
Protein sequencing alignment
All MS/MS spectra were searched against the NCBInr human protein sequence database using Mascot and PTMap software (Chen et al., 2009). For histone samples that were generated by tryptic digestion of propionylated histones, the specific parameters included lysine propionylmethylation (+ 70.04187 Da) and lysine propionylation as variable modifications. For histone samples propionylated after trypsin digestion, N-terminal propionylation was included as a fixed modification. All the identified peptides were manually verified according to the rules described previously (Chen et al., 2005).
Generation of pan anti-Kcr antibody
The pan anti-Kcr antibody was generated and purified from rabbit with lysine-crotonylated bovine serum albumin (BSA) as an antigen. For more details, see Extended Experimental Procedures.
ChIP-seq
ChIP-seq for histone Kcr or Kac was carried out as previously described with 500 µg IMR90 chromatin (or 100 µg of fractionated germ cells chromatin) and 5 µg pan anti-Kcr or anti-Kac antibody (Hawkins et al., 2010). ChIP-seq libraries for sequencing were prepared following Illumina protocols (Illumina, San Diego, CA) with minor modifications. Libraries for input samples were generated using 20 ng corresponding input chromatin. Briefly, ChIPed DNA was first blunted with END-IT DNA repair kit (Epicenter Biotechnology, Madison, WI) and then incubated with Klenow (exo-) (New England Biolabs, MA) and dATP to generate single base 3'-dA overhang. Illumina sequencing adapter was then ligated to the resulting DNA, and followed by size selection (180–400bp) from a 8% acrylamide gel. This size-selection step was repeated after PCR amplification with DNA primers supplied by Illumina. Libraries were sequenced using Illumina GAII or HiSeq machine as per manufacturer's protocols. Following sequencing cluster imaging, base calling were conducted using the Illumina pipeline. Reads were mapped to human hg18 (for IMR90 data) or mouse mm9 (for sperm cell data) genome build with a bowtie software package. Total mapped tags were paired down to unique, monoclonal tags. These are tags that mapped to one location in the genome and each sequence is represented once.
For additional experimental materials and methods, see Extended Experimental Procedures, including methods for preparation of histones from HeLa cells, in-solution proteolytic digestion and chemical derivatization of histone proteins, HPLC/MS/MS analysis and protein sequence database searching, verification of lysine crotonylated peptides by HPLC/MS/MS analysis, synthesis of BSA derivatives, conjugation of Kcr-immobilized agarose beads, generation of pan anti-Kac and anti-Kcr antibodies, Western blotting with competition of a peptide library, affinity enrichment of Kcr peptides, synthesis of Boc-Lys(crotonyl)-AMC and Boc-Lys(ac)-AMC, in vitro lysine decrotonylation and lysine deacetylation reaction assays, transient transfection, in vivo D4-crotonate labeling of histones, immunofluorescence of HeLa cells and germ cells using anti-Kcr antibody, immunohistochemistry of testis tubule cross-sections using anti-Kcr antibody, mouse spermatogenic cell fractionation, chromatin immunoprecipitation in fractionated germ cells, expression analyses, multiple sequence alignment for linker histones (H1.1, H1.2, H1.3, H1.4 and H1.5) by CLUSTAL 2.0.12, identification of mono-, di-, and trimethylated lysine residues, identification of Nε-formylated and acetylated lysine residues, identification of monomethylated arginine residues, identification of hydroxylated tyrosine residues.
Annotated MS/MS spectra of all the modified peptides bearing a PTM other than Kcr were included in Figure S7.
Supplementary Material
Acknowledgement
This work was supported by NIH grants to B.R, J.W, and Y.Z., by INCa-DHOS, ANR blanche “EpiSperm” and ARC research programs to S.K..
Abbreviations
- MS
Mass spectrometry
- PTM
Protein post-translational modification
- HPLC
High-performance liquid chromatography
- ChIP
Chromatin Immunoprecipitation
- TSS
Transcription Starting Site
- MSCI
Meiotic Sex Chromosome Inactivation
- Spc
Spermatocytes
- RS
Round Spermatids
- IH
Immunohistology
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Allis CD, Chicoine LG, Richman R, Schulman IG. Deposition-related histone acetylation in micronuclei of conjugating Tetrahymena. Proc Natl Acad Sci U S A. 1985;82:8048–8052. doi: 10.1073/pnas.82.23.8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arents G, Moudrianakis EN. Topography of the histone octamer surface: repeating structural motifs utilized in the docking of nucleosomal DNA. Proc Natl Acad Sci U S A. 1993;90:10489–10493. doi: 10.1073/pnas.90.22.10489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdasco M, Esteller M. Aberrant epigenetic landscape in cancer: how cellular identity goes awry. Dev Cell. 2010;19:698–711. doi: 10.1016/j.devcel.2010.10.005. [DOI] [PubMed] [Google Scholar]
- Boussouar F, Rousseaux S, Khochbin S. A new insight into male genome reprogramming by histone variants and histone code. Cell Cycle. 2008;7:3499–3502. doi: 10.4161/cc.7.22.6975. [DOI] [PubMed] [Google Scholar]
- Chen Y, Chen W, Cobb MH, Zhao Y. PTMap--a sequence alignment software for unrestricted, accurate, and full-spectrum identification of post-translational modification sites. Proc Natl Acad Sci U S A. 2009;106:761–766. doi: 10.1073/pnas.0811739106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Kwon SW, Kim SC, Zhao Y. Integrated approach for manual evaluation of peptides identified by searching protein sequence databases with tandem mass spectra. J Proteome Res. 2005;4:998–1005. doi: 10.1021/pr049754t. [DOI] [PubMed] [Google Scholar]
- Chu F, Nusinow DA, Chalkley RJ, Plath K, Panning B, Burlingame AL. Mapping post-translational modifications of the histone variant MacroH2A1 using tandem mass spectrometry. Mol Cell Proteomics. 2006;5:194–203. doi: 10.1074/mcp.M500285-MCP200. [DOI] [PubMed] [Google Scholar]
- Cosgrove MS, Boeke JD, Wolberger C. Regulated nucleosome mobility and the histone code. Nat Struct Mol Biol. 2004;11:1037–1043. doi: 10.1038/nsmb851. [DOI] [PubMed] [Google Scholar]
- Fullgrabe J, Kavanagh E, Joseph B. Histone onco-modifications. Oncogene. 2011 doi: 10.1038/onc.2011.121. [DOI] [PubMed] [Google Scholar]
- Garcia BA, Mollah S, Ueberheide BM, Busby SA, Muratore TL, Shabanowitz J, Hunt DF. Chemical derivatization of histones for facilitated analysis by mass spectrometry. Nat Protoc. 2007a;2:933–938. doi: 10.1038/nprot.2007.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia BA, Pesavento JJ, Mizzen CA, Kelleher NL. Pervasive combinatorial modification of histone H3 in human cells. Nat Methods. 2007b;4:487–489. doi: 10.1038/nmeth1052. [DOI] [PubMed] [Google Scholar]
- Garcia BA, Shabanowitz J, Hunt DF. Characterization of histones and their post-translational modifications by mass spectrometry. Curr Opin Chem Biol. 2007c;11:66–73. doi: 10.1016/j.cbpa.2006.11.022. [DOI] [PubMed] [Google Scholar]
- Gaucher J, Reynoird N, Montellier E, Boussouar F, Rousseaux S, Khochbin S. From meiosis to postmeiotic events: the secrets of histone disappearance. FEBS J. 2010;277:599–604. doi: 10.1111/j.1742-4658.2009.07504.x. [DOI] [PubMed] [Google Scholar]
- Govin J, Caron C, Lestrat C, Rousseaux S, Khochbin S. The role of histones in chromatin remodelling during mammalian spermiogenesis. Eur J Biochem. 2004;271:3459–3469. doi: 10.1111/j.1432-1033.2004.04266.x. [DOI] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazzouri M, Pivot-Pajot C, Faure AK, Usson Y, Pelletier R, Sele B, Khochbin S, Rousseaux S. Regulated hyperacetylation of core histones during mouse spermatogenesis: involvement of histone deacetylases. Eur J Cell Biol. 2000;79:950–960. doi: 10.1078/0171-9335-00123. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Johnson L, Mollah S, Garcia BA, Muratore TL, Shabanowitz J, Hunt DF, Jacobsen SE. Mass spectrometry analysis of Arabidopsis histone H3 reveals distinct combinations of post-translational modifications. Nucleic Acids Res. 2004;32:6511–6518. doi: 10.1093/nar/gkh992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Luo H, Li Y, Mu JJ, Zhang J, Tonaka T, Hamamori Y, Jung SY, Wang Y, Qin J. Regulation of intra-S phase checkpoint by ionizing radiation (IR)-dependent and IR-independent phosphorylation of SMC3. J Biol Chem. 2008;283:19176–19183. doi: 10.1074/jbc.M802299200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Trojer P, Reinberg D. The key to development: interpreting the histone code? Curr Opin Genet Dev. 2005;15:163–176. doi: 10.1016/j.gde.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Martin C, Zhang Y. Mechanisms of epigenetic inheritance. Curr Opin Cell Biol. 2007;19:266–272. doi: 10.1016/j.ceb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- McGhee JD, Felsenfeld G. Nucleosome structure. Annu Rev Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- Mersfelder EL, Parthun MR. The tale beyond the tail: histone core domain modifications and the regulation of chromatin structure. Nucleic Acids Res. 2006;34:2653–2662. doi: 10.1093/nar/gkl338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller JL, Mahadevaiah SK, Park PJ, Warburton PE, Page DC, Turner JM. The mouse X chromosome is enriched for multicopy testis genes showing postmeiotic expression. Nat Genet. 2008;40:794–799. doi: 10.1038/ng.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namekawa SH, Park PJ, Zhang LF, Shima JE, McCarrey JR, Griswold MD, Lee JT. Postmeiotic sex chromatin in the male germline of mice. Curr Biol. 2006;16:660–667. doi: 10.1016/j.cub.2006.01.066. [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakabe K, Wang Z, Hart GW. Beta-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc Natl Acad Sci U S A. 2010;107:19915–19920. doi: 10.1073/pnas.1009023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JM. Meiotic sex chromosome inactivation. Development. 2007;134:1823–1831. doi: 10.1242/dev.000018. [DOI] [PubMed] [Google Scholar]
- Wisniewski JR, Zougman A, Kruger S, Mann M. Mass spectrometric mapping of linker histone H1 variants reveals multiple acetylations, methylations, and phosphorylation as well as differences between cell culture and tissue. Mol Cell Proteomics. 2007;6:72–87. doi: 10.1074/mcp.M600255-MCP200. [DOI] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- Zee BM, Levin RS, Xu B, LeRoy G, Wingreen NS, Garcia BA. In vivo residue-specific histone methylation dynamics. J Biol Chem. 2010;285:3341–3350. doi: 10.1074/jbc.M109.063784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Zhou MM. Bromodomain: an acetyl-lysine binding domain. FEBS Lett. 2002;513:124–128. doi: 10.1016/s0014-5793(01)03309-9. [DOI] [PubMed] [Google Scholar]
- Zhao M, Shirley CR, Hayashi S, Marcon L, Mohapatra B, Suganuma R, Behringer RR, Boissonneault G, Yanagimachi R, Meistrich ML. Transition nuclear proteins are required for normal chromatin condensation and functional sperm development. Genesis. 2004;38:200–213. doi: 10.1002/gene.20019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.