Abstract
Guanine nucleotide regulatory proteins (G-proteins) are central to normal hepatocyte function and are implicated in hepatic disease initiation and progression. Regulators of G-protein signaling (RGS) are critical to defining G-protein-dependent signal fidelity, yet the role of RGS proteins in the liver is poorly defined. The aims of this study were to determine RGS17 expression in normal and transformed hepatic tissue and cells, and address the function of RGS17 in hepatic tumorgenicity. RGS17 expression was determined in human and rat HCC tissue and cell lines. Molecular approaches were used to alter RGS17 expression in HCC cells, effects on cell function measured, and RGS17 association with specific Gα-subunits determined. Using these approaches RGS17 mRNA, but not protein, was detectable in human and rat HCC tissue and cells. Conversely, RGS17 mRNA was not detected in normal tissue, isolated hepatocytes, or non-tumorigenic hepatic cells. Subsequent studies using transfected cells demonstrated RGS17 proteins were not post-translationally modified in HCC cells, and RGS17 expression is governed by protein degradation and not via miRNAs. Notwithstanding inherently low RGS17 protein levels, altering RGS17 expression profoundly affected HCC cell mitogenesis and migration. Analysis of RGS17-G-protein interaction demonstrated RGS17 associates with both Giα- and Gqα-subunits in HCC cells of human and rat origin. In conclusion, these data demonstrate that, despite difficulties in measuring endogenous RGS protein expression, RGS17 is differentially expressed in HCC and plays a central role in regulating transformed hepatocyte tumorgenicity.
Keywords: Hepatocellular carcinoma, G-protein, Regulator of G-protein signaling, mitogenesiss, migration
1.1 INTRODUCTION
Heterotrimeric guanine nucleotide regulatory proteins (G-proteins) are localized at the inner plasma membrane surface and act as intermediates between cell-surface receptors (G-protein coupled receptors; GPCRs) and intracellular effectors [1, 2]. G-proteins exist as functional dimers consisting of α-subunits (Gα), and βγ-dimer complexes (Gβγ). Upon GPCR activation, Gα exchanges guanosine diphosphate (GDP) for guanosine triphosphate (GTP) and the α-βγ complexes dissociate [1–3]. Both Gα and Gβγ regulate activity of downstream effectors including adenylyl cyclase (AC), phospholipase C, and ras-raf-mitogen activated protein kinase (MAPK) signaling [3–6]. Within the liver a central role for G-protein signaling has been reported during normal hepatic growth and regeneration [7–9]. Conversely, G-protein expression and function has been reported to be unbalanced in human and animal models of HCC [10, 11].
G-protein activation is initiated by GPCRs and deactivation accelerated by GTPase activating proteins (GAPs) [2, 12, 13]. However, intrinsic GTPase activity of Gα subunits is relatively low, and often insufficient to match the rate at which G protein-regulated responses are demanded [12, 14–16]. Regulators of G protein signaling (RGS) have been identified as critical intermediates in G protein dependent signaling [15, 17, 18]. RGS proteins are GAP proteins for Giα and Gqα classes of protein that accelerate GTP hydrolysis up to 2000 times that of inherent GTPase activity [15, 17, 19]. In doing so RGS proteins effectively regulate frequency and duration of G protein signaling. Consequently, distribution and regulation of RGS protein activity within the cell is central to determining rate of G-protein signaling, as well as balancing effects of different G protein-signaling pathways [15, 17, 18].
Despite the fact that more than 20 mammalian RGS homologs have been identified, relatively little is known regarding tissue distribution, cell localization, and involvement in tumorigenesis [20]. Most of the published data concerning RGS is derived from studies confined to the nervous system [21, 22]. To date the literature addressing RGS distribution/ function in other major organs, including liver, is limited to a few, mostly descriptive, papers [20, 23–25]. In light of the importance of G-protein expression and function in normal and transformed hepatocytes [7–11], we hypothesized that RGS proteins may play an equally important role in regulating hepatic tumorigenesis.
METHODS
Institutional assurances
Institutional review board approval was granted to analyze surgical specimens of patients undergoing HCC resection (n=7 patients; 4 male (mean age = 66.8 ± 6.5yrs) 3 female (mean age = 61.3 ± 7.3yrs). The Institutional Animal Care and Use Committee approved all animal experiments. Adenoviral particle production was performed in accordance with NIH Guidelines, and approved by the Institutional Biosafety Committee.
Materials
H4IIE, HepG2 and Clone 9 cells were purchased from ATCC (Bethesda, MD). HuH-7 cells were purchased from the Japan Health Sciences Foundation HSSRB (Osaka, Japan). WBF344 cells were a kind gift from Dr Coleman (UNC at Chapel Hill, NC). The MTT proliferation assay and pcDNA3.1 were purchased from Invitrogen (Carlsbad, CA). RNA/ miRNA extraction was performed using a ZR RNA MiniPrep Kit (Zymo Research, Orange, CA) and an miRNeasy Mini Kit (QIAGEN, Valencia, CA). IMPROM II™ transcription system, psiLentGene vector, GoTaq green master mix, DualGlo Luciferase Assay, and the cAMP-Glo kit were purchased from Promega (Madison, WI). The pGLuc plasmid (possessing Gaussian luciferase) and a BioLux Gaussia Luciferase Assay Kit were purchased from New England Biolabs (Ipswich, MA). Antibodies against SUMO-1, SUMO-1/ 3, β-actin and Gqα were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). An anti-Giα1–2 antibody was purchased from EMD Chemicals (Gibbstown, N J).
Cell culture
Human (HepG2 and HuH-7) and rat (H4IIE) hepatoma cells, and non-tumorigenic hepatic cells (WBF344 and Clone 9) were cultured in high glucose, complete DMEM supplemented with fetal bovine serum (FBS; 10% (v/v)) and antibiotics as previously reported [26–29]. Freshly isolated rat hepatocytes were obtained from male ACI rats (225–250g) using a two-step collagenase perfusion technique as previously reported [10]. Following isolation, hepatocyte cell purity was determined, and cells snap -frozen and stored (−80°C) prior to analysis.
In vivo rat model of hepatocellular carcinoma
The rat model of HCC was generated in male ACI rats (175–225g) as previously reported [30]. Briefly, H4IIE cells (passage 1–3) were cultured to 70% confluence, detached with trypsin-EDTA, and inoculated (1×107 cells in 0.1ml sterile PBS) directly into the right hepatic parenchyma. This resulted in reproducible focal tumor mass formation 14–16 days post-inoculation. The absence of intrahepatic metastasis was confirmed histologically, a single tumor mass (≈8–10mm in diameter) forming in the setting of normal underlying hepatic architecture [30].
Expression constructs
The complete coding sequence of RGS17 was amplified by RT-PCR from HepG2 RNA and cloned into pBluescript. A pcDNA3 plasmid harboring Giα2 dominant negative mutant (Giα2DN) sequence was purchased from Missouri S&T cDNA Resource Center (Rolla, MO). Full size coding sequences were sub-cloned into a pAdTrack-CMV shuttle vector that also possesses the GFP gene under an independent promoter. After recombination with adenoviral backbone core genome in BJ5183 E.coli strain, recombinants were expanded. Plasmids possessing adenoviral genome and gene of interest were isolated and used to transfect AD293 producer cells. Two weeks after cell incubation, viral particles were harvested and used in an amplification round with the producer cell line. Resulting viral stock was titered and stored (−80°C). To transduce HCC cells, viral stock was thawed and added to medium at different MOIs (5–8 hours).
mRNA analysis
RNA extraction from cells and tissue was performed using a ZR RNA MiniPrep Kit. To perform RT-PCR, 1 µg of total RNA was reverse transcribed using an IMPROM II™ transcription system with random hexamers to generate cDNA. Primers against rat RGS17 were 5’ GGAAACCAAAGGCCCAACAATAC 3’ (Forward), 5’ATCATCCTGGCCTTTTCTTCAACA 3’ (Reverse). Primers against human RGS17 were 5’ TGGGTCCTGAAGTAGCTGAAATGCGA AAA 3’ (Forward), 5’CCCATCTCAGCCCTCCAAAATGATTGTT 3’ (Reverse). PCR reactions consisted of 95°C for 2 minutes followed by 35 cycles of 95°C, 55°C and 72°C for 45 s each with a final elongation at 72°C for 2 minutes. Amplified products were resolved using a 1.5% (w/v) agarose gel and stained with ethidium bromide before conversion to digital images.
miRNA target predict ion and luciferase reporter assay
TargetScan (www.targetscan.com) predicts 4 potential highly conservative miRNA targets with in the 3’untranslated region (UTR) of the human RGS17 mRNA transcript: miR-27a/ 27b, miR-182, miR-25/ 32 and miR-34a/ 34b. The 3’-UTR of RGS17 mRNA was amplified by RT-PCR from HuH-7 RNA. A reporting construct w as design ed by inserting this 3’UTR after the firefly luciferase gene in pcDNA3 vector to produce a pcDNA3-Lu c/ 3’UTR construct. The four conserved potential miRNA binding sites were mutated by PCR and constructs bearing native or mutated 3’UTR were transfected in to HuH-7 cells. The pGLuc plasmid possessing Gaussian luciferase was co-transfected for normalization. 48 hours post-transfection firefly luciferase expression and Gaussian luciferase levels were measured.
RGS17 knockdown
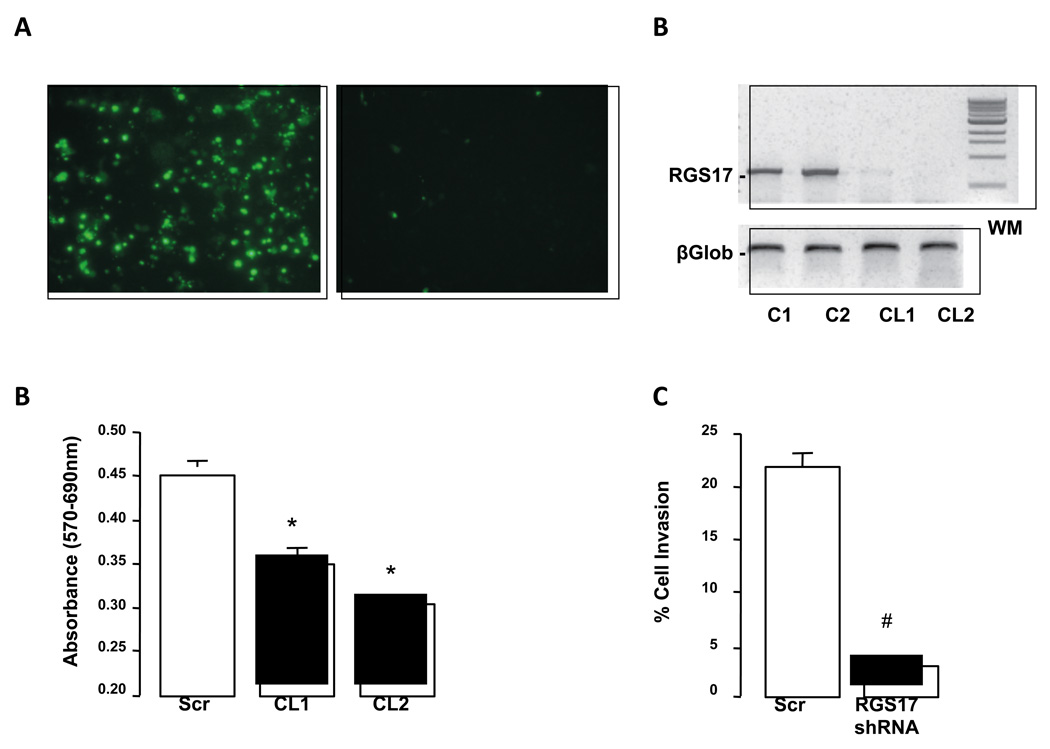
An shRNA for RGS17 was designed using shRNA Explorer software (GeneLink, Hawthorne, NY). Primers corresponding to target sequences were used to amplify an expressing cassette consisting of a U6 promoter, corresponding shRNA sequence, and termination signal. shRNA cassettes were co-transfected in AD293 cells with pcDNA3-RGS17-GFP fusion construct and silencing efficiency determined by fluorescence (48 hours post-transfection). Using this approach, 2 of 5 cassettes significantly reduced RGS17-GFP fusion expression, and were cloned into a psiLentGene vector and used to transfect HuH-7 cells. In a parallel series of experiments, HuH-7 cells were transfected with a control construct (scrambled sequence). Transfectants were selected for puromycin resistance, tested, and expanded.
Cell proliferation
Cells transduced with either RGS17+GFP or GFP only ad enoviruses were plated (5×104 cells/ well; 12-well plates), grown for 10 days, detached using trypsin, and proliferation measured using the MTT assay as per the manufacturer’s instructions.
Cell invasion
HuH-7 cells stably transfected with anti-RGS17 shRNA or control shRNA were plated in matrigel invasion chambers, or control chambers (8 µm pore size). DMEM culture medium containing 0.1% (w/v) bovine serum albumin (BSA) was added to the inserts and wells of the plates. 10µg/ ml of human fibronectin was then added to the wells and 24 hours after incubation inserts were retrieved, wiped inside, and cells on the outer surface were fixed and stained (hematoxylin and eosin), counted, and expressed as ratio of cells invading through matrigel inserts versus cells migrating through control inserts.
cAMP measurement
RGS17 deficient (or control) HuH-7 cells were made quiescent for 24 hours and pretreated with lysophosphatidic acid (LPA-non-specific GPCR-activator; 10µg/ ml) or mastoparan M7 (M7-Gi-specific activator; 10µM) for 30 minutes. cAMP levels were measured using a cAMP-Glo kit as per the manufacturer’s instructions.
miRNA microarray data
Total mRNA, with enriched miRNA fraction, was isolated using an miRNeasy Mini Kit. miRNA array hybridization was performed in the Molecular Core Facility (CMC).
Image analysis and statistics
Images were converted to digital format and integrated optical density performed using ImageJ Software (NIH, Bethesda, MD). Data are presented as means ± SEM. Statistical analysis was performed using JMP 8.0.2 software (Cary, NC). Group wide analysis was performed by one-way ANOVA. Pair-wise combinations within a group were analyzed by Paired Student’s t-test.p<0.05 was considered significant.
RESULTS
RGS17 expression is altered in HCC tissue and cells
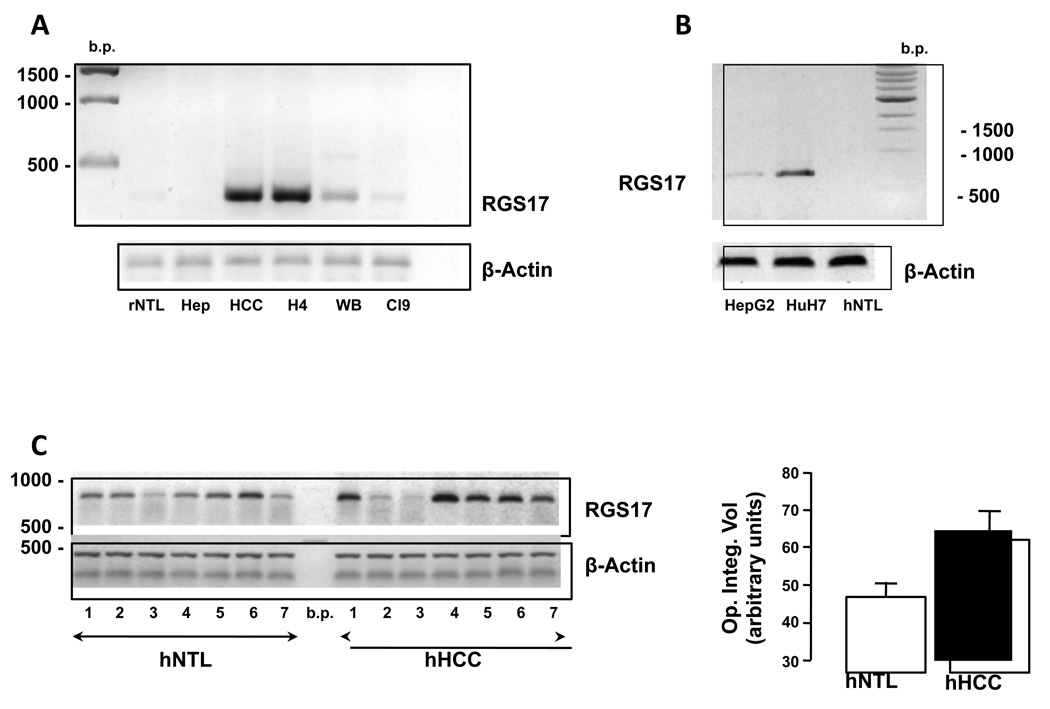
Using RT-PCR analysis we failed to detect RGS17 mRNA in whole rat liver or freshly isolated rat hepatocytes (Figure 1A). In contrast, RGS17 mRNA was readily detectable in cultured rat H4IIE cells and rat HCC tissue (Figure 1A). Conversely RGS17 mRNA expression was barely detectable in immortalized, non-tumorigenic rat hepatic cell lines (WBF344 and Clone 9) as compared to H4IIE cells under identical amplification conditions (Figure 1A).
Figure 1. RGS17 mRNA expression is altered in HCC.
A. Representative RT-PCR analysis of rat non-tumor liver (rNTL), isolated hepatocytes (Hep), rat HCC tissue (HCC), H4IIE HCC cells (H4) and non-transformed hepatic cells (WBF344; WB, and Clone 9; Cl9). Equal loading was confirmed using primers for β-actin. B. Representative RT-PCR analysis of human HCC cells (HepG2 and HuH-7) and human non-tumor liver (hNTL). C. RT-PCR analysis of hNTL and pair-matched, HCC (hHCC) (left panel). Cumulative densitometric analysis of signal intensity from RT-PCR analysis of human hNTL and hHCC (n=7).
Analysis of mRNA from human HCC cell lines (HepG2 and HuH-7) demonstrated readily detectable RGS17 mRNA expression (Figure 1B). In contrast, RGS17 mRNA was undetectable in a non-tumorigenic, non-transplanted human liver specimen (Figure 1B). Analysis of mRNA from 7 human HCC and pair-matched, non-tumor liver (NTL) samples failed to identify a significant difference in overall RGS17 mRNA expression (49.7 ± 3.7 (NTL) vs 64.8 ± 7.6 (HCC) (values are optical integrated density (arbitrary units)), p=0.061, n=7, Figure 1C). However, in 5 of 7 samples analyzed RGS17 expression was significantly increased compared to pair-matched NTL (49.3 ± 3.7 (NTL) vs 75.7 ± 3.6 (HCC), optical integrated density (arbitrary units), p=0.011; HCC vs. NTL). Of further note, all of the samples in which increased RGS17 mRNA was detected in, HCC arose in the setting of underlying hepatic fibrosis/ cirrhosis or hepatic steatosis (Table 1). Conversely, while HCC arose in the setting of cirrhosis in sample 2, cirrhotic damage was classified as micronodular by an independent pathologist. In sample 3, HCC arose in the setting of ostensibly normal underlying hepatic architecture.
Table 1.
Hepatocellular carcinoma (HCC) patient demographics and pathology
| Sample # | M/F | Tumor size (cm) |
HBV +/− |
HCV +/− |
Alcohol | Fibrosis or Cirrhosis |
Steatosis +/− |
|---|---|---|---|---|---|---|---|
| 1 | M | 1.0×0.7 | − | + | + | + | − |
| 2 | M | 12.5×4.0 | − | − | − | + | − |
| 3 | F | 8.0×11.0 | − | − | − | − | − |
| 4 | F | 7.0×5.0 | − | − | − | − | + |
| 5 | M | 5.0×3.0 | − | + | − | + | + |
| 6 | F | 4.6×3.2 | − | − | − | − | + |
| 7 | M | 3.5×3.5 | − | + | + | + | − |
Despite using 5 different, commercially available antibodies against RGS17 (ProSci Inc. (Poway, CA); US Biological (Swampscott, MA); ProteinTech Group (Chicago, IL); and 2 different clones from Santa Cruz Biotechnology (Santa Cruz, CA)), we failed to detect endogenous RGS17 protein by immunoblot in any of the cell lines or tissue previously analyzed for RGS17 mRNA (data not shown). Similarly, immunohistochemistry of rat HCC/ NTL failed to detect specific staining (above background) using any of these antibodies (data not shown). In an attempt to determine discrepancies between protein expression, as compared to antibody specificity, transient transfections were performed using plasmid vectors containing the cDNA for RGS17. Using this approach we again failed to detect RGS17 protein expression in transfected cells by immunoblot (H4IIE, WBF344, Clone 9, HepG2, or HuH-7) (data not shown).
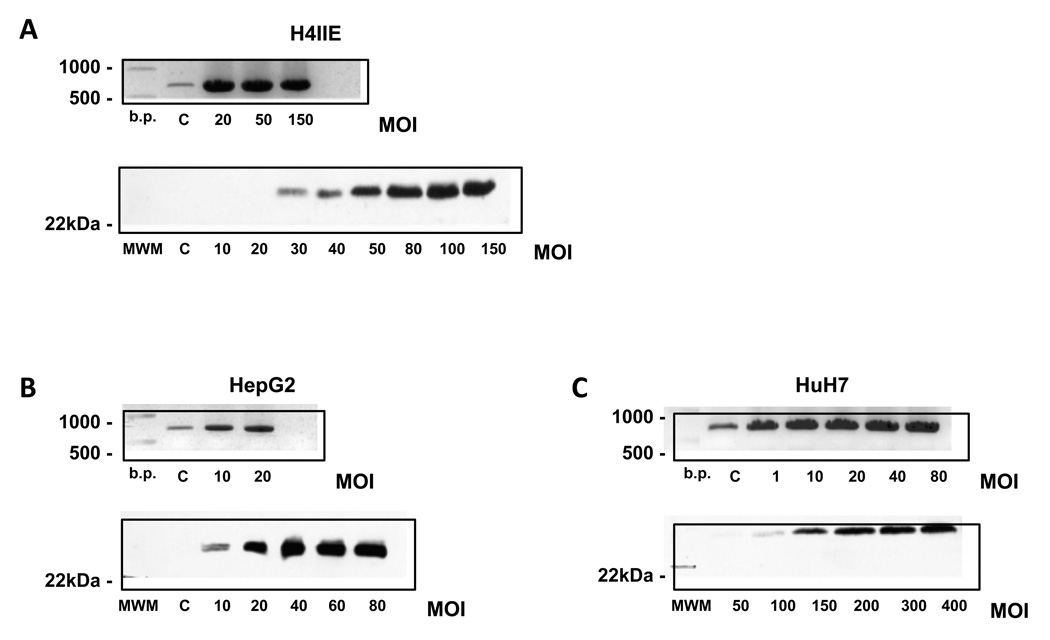
To conclusively determine whether RGS17 protein is detectable in cells of hepatic origin, RGS17 was over-expressed in rat (H4IIE) and human (HepG2 and HuH-7) HCC cells using an adenoviral vector system. The system design allows adenovirus development and amplification to be tracked, as well as infection efficiency. Using this approach, low dose infection of H4IIE cells (MOI <20) resulted in significant increases in RGS17 mRNA expression (Figure 2A). However to detect RGS17 protein, a considerably higher MOI was required (MOI >30, Figure 2A). This was similarly the case for both human HCC cell lines (HepG2 and HuH-7), the MOI required to detect protein being dependent on basal (non - transfected) RGS17 mRNA levels. That is, in HepG2 cells, a line expressing relatively low RGS17 m RN A, RGS protein was detected at an MOI ≤10 (Figure 2B). In contrast, while changes in RGS17 mRNA were detected in HuH-7 cells at an MOI of 1, RGS17 protein detection was not evident at MOIs <100 (Figure 2C).
Figure 2. Over-expression of RGS17 differentially affects RGS17 mRNA and protein expression.
Representative RT-PCR analysis of RGS17 mRNA expression (upper panels) and protein (lower panels) in A. rat H4IIE, B. Human HepG2, and C. Human HuH-7 HCC cell lines following adenoviral infection to over-express RGS17 with increasing multiplicity of infections (MOIs).
miRNAs do not significantly affect RGS17 protein expression in HuH-7 cells
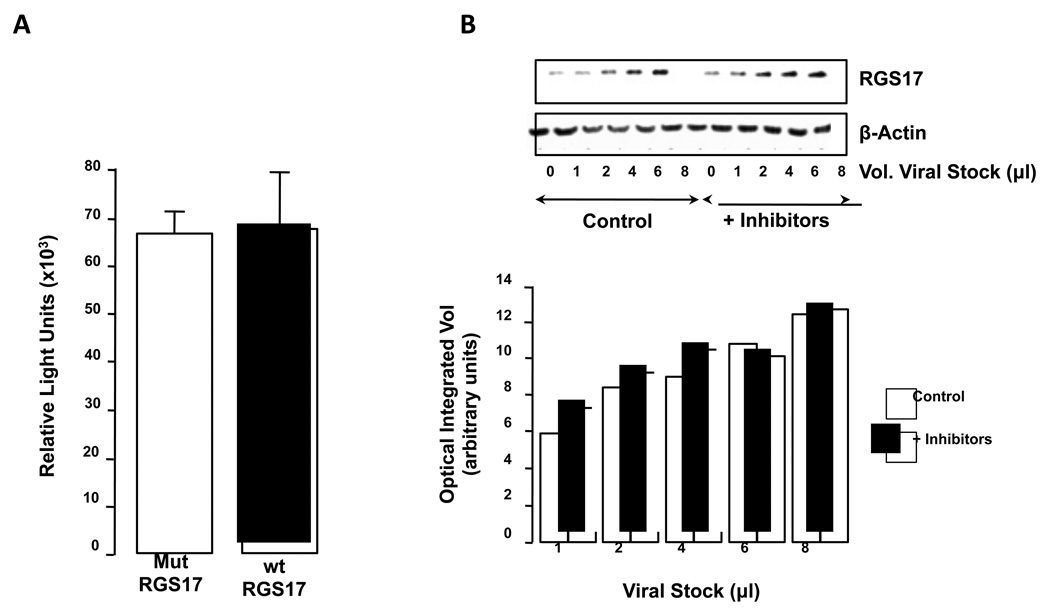
The identification of miRNAs as modulators of mRNA translation led us to hypothesize that high resistance to RGS17 protein expression following adenoviral transfection in HuH-7 cells may be due to miRNA(s). Using a reporter construct (based on the luciferase gene and the RGS17 3’UTR) the 4 potential most conservative miRNA target sites were mutated and assembled in a pcDNA3 expression vector and transfected into HuH-7 cells. Using this approach no significant difference in luciferase reporter activity was detected between mutated and control (wt) 3’UTR transfected cells (Figure 3A).
Figure 3. RGS17 mRNA expression is regulated by proteosome degradation not miRNAs.
A. HuH-7 cells were transfected to over-express mutations of the 4 potential most conserved miRNA sites (Mut RGS17) or control 3’ UTR transfected (wt RGS17) and luciferase activity measured as a marker of RGS17 mRNA expression. n = 12 independent experiments. B. HuH-7 cells were infected with increasing amounts of RGS17 bearing adenovirus in the absence or presence of proteosome inhibitors (Epoxomicin and MG132). Protein expression was determined by immunoblot (upper panel) and quantified by optical integrated volume (lower panel).
RGS17 is predominantly regulated by protein degradation in HCC cells
In HuH-7 cells infected with (different MOI) RGS17 bearing adenovirus concomitant with proteasome inhibitors (Epoxomicin and MG132), amount of RGS17 protein increased after proteasome treatment. This effect was particularly evident at low MOIs where heterologous protein concentration is closer to non-transfected level (Figure 3B).
Previous studies using neural tissue report RGS17 is conjugated to SUMO protein [31]]. Using the HuH-7 line infected to over-express RGS17, RGS17 was detected as a single band by immunoblot at ≈25kDa, corresponding to the predicted molecular weight of unmodified RGS17 (Figure 2C). Nevertheless, to confirm the absence of possible SUMO modifications in this system, RGS17 protein was next immunoprecipitated from RGS17 transfected H4IIE cells and immunoblotted using antibodies against SUMO-1 or SUMO-2/ 3. Using this approach no signal was detected using either antibody (data not shown).
Effect of altering RGS17 expression on cell proliferation and migration
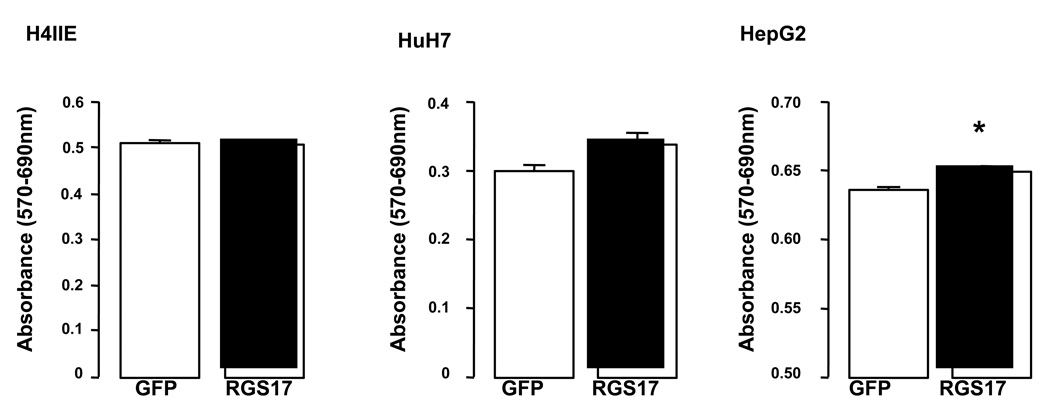
Over-expressing RGS17 in rat (H4IIE) and human (HuH-7) HCC cells did not significantly affect proliferation as measured using an MTT assay (Figure 4A). Conversely, over-expressing RGS17 in human HepG2 cells led to a modest, though significant increase in proliferation (Figure 4A, p<0.05). These data led us to consider whether over-expressing RGS17 in cells with high(er) endogenous RGS17 mRNA levels may be less effective in regulating cell function. To test this stable transfections of HuH-7 cells using RGS17 shRNA, or scrambled sequence, were performed and RGS17 silencing efficiency confirmed (Figure 5 A and B). Using these stably transfected cells proliferation in RGS17 shRNA-expressing HuH-7 cells was significantly inhibited compared to scrambled shRNA sequence (Figure 5C). To determine the role of RGS17 in invasion/ migration, studies were performed using stably transfected (RGS17 shRNA) HuH-7 cells in a migration assay. This approach demonstrated inhibiting RGS17 expression using an shRNA significantly inhibited migration (Figure 5D).
Figure 4. Effect of over-expressing RGS17 on cell proliferation.
RGS17 was over-expressed in rat (H4IIE) and human (HuH-7 and HepG2) HCC cell lines and proliferation measured using an MTT assay. *p<0.05 RGS17 expressing versus control (GFP), n = 12 independent experiments.
Figure 5. shRNA efficiently decreases RGS17 expression in human HuH7 cells and inhibits migration.
A. HuH-7 cells transfected with RGS17-GFP fusion construct alone (left panel) or with RGS17-GFP fusion construct and shRNA cassette designed against RGS17 (right panel). B. Representative RT-PCR analysis of RGS17 expression HuH-7 cells transfected with control plasmid vector (C1 and C2) or plasmid vector containing shRNA sequence against RGS17. After antibiotic selection two clones were selected (Clone 1 and 2; CL1 and CL2). β2microglobulin was used as a loading control (βGlob; Lower panel). C. RGS 17 expression was decreased by stable transfection of HuH-7 cells with RGS17 shRNA (Clones 1 and 2; CL1/ CL2), and proliferation measured using an MTT assay. *p<0.05 RGS17 shRNA versus control (scrambled; Scr) shRNA, n = 12 independent experiments. D. RGS 17 expression was decreased by stable transfection of HuH-7 cells with RGS17 shRNA and cell migration measured. #p<0.01 RGS17 shRNA versus scrambled (scr) shRNA, n = 6 independent experiments.
RGS17 associates with Giα and Gqα-subunits in HCC cells
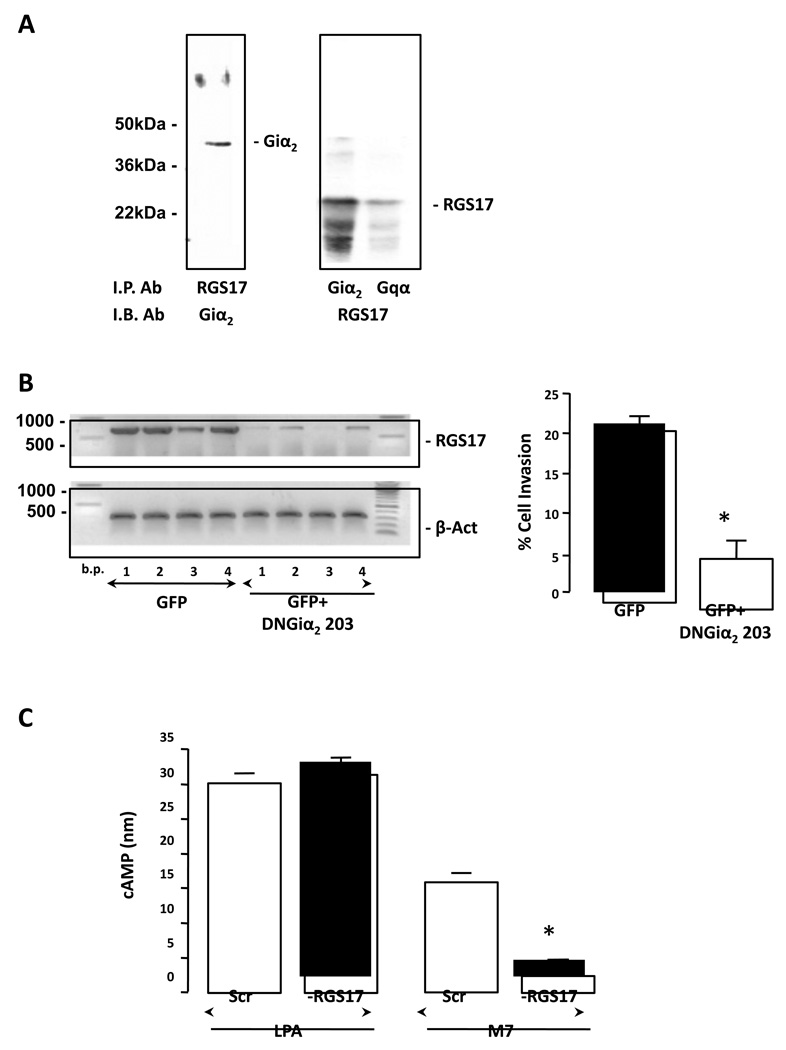
To identify specific G-protein subunit interaction with RGS17, RGS17 was over-expressed in H4IIE and HuH-7 cells and immunoprecipitation with anti-Giα1–2 or anti-Gqαwas performed. The resulting immunoprecipitates were resolved by SDS-PAGE, immunoblotted, and probed with anti-RGS17. This approach demonstrated RGS17 co-immunoprecipitated with Giα1–2 and Gqα (Figure 6A). These data were confirmed by immunoprecipitation using anti-RGS17, and immunoblot using anti-Giα1–2 or anti-Gqα (Figure 6A).
Figure 6. RGS17 protein associates with Giα and Gqα subunits, and regulates cell migration and cAMP production.
A. RGS17 was over-expressed in HuH-7 cells, immunoprecipitated with anti-Giα or Gqα and probed with anti-RGS17 antibody (left panel). Specificity of effect was confirmed by immunoprecipitating with anti-RGS17 and probing with anti-Giα. B. HuH-7 cells were transduced to over-express GFP (GFP) or dominant negative (DN)Giα2 (GFP+DNGiα2 203). Representative RT-PCR analysis (left panel) of RGS17 mRNA expression in 4 different clones (1–4) expressing either GFP or GFP+(DN)Giα2 203. Cells were subsequently analyzed for cell migration (lower panel). *p<0.01 GFP versus GFP+(DN)Giα2 203T mutant. n = 6 independent experiments. C. RGS17 expression was stably inhibited using an RGS17 shRNA, and cells treated with LPA (10µg/ ml) or mastoparan M7 (M7; 10µM), and intracellular cAMP levels measured. *p<0.01 scrambled shRNA (Scr) versus RGS17 shRNA. n = 12 independent experiments.
To confirm that RGS17 interacts with Giα1–2, a functional DNGiα2-protein-GFP construct (or GFP only; control) was over-expressed in HuH-7 cells using an adenoviral vector. This DNGiα2 (Gly203->Thr; G203T) is unable to form a functional complex with GTP, yet retains the ability to associate with GPCRs at the inner plasma membrane surface [32]. Based on data reported by Clark and Traynor, this non-functional G-protein should cause a reduction in abundance of interacting RGS protein(s) [33]. Using this approach a highly significant decrease in RGS17 mRNA expression was evidenced in HuH-7 cells expressing DN-Giα2 (Figure 6B). Subsequently, cells transfected to over-express DNGiα2, demonstrated significantly decreased migration compared to control (GFP only) (Figure 6B, p<0.01, GFP+DNGiα2 versus GFP only).
RGS Expression determines cAMP levels in response to G-protein activity
HuH-7 cells stably transfected with RGS17 shRNA were treated with LPA (10µg/ ml) or mastoparan M7 (M7 (10µM)). These studies demonstrated inhibition of RGS17 expression did not significantly alter cAMP in response to LPA (Figure 6C). However, inhibiting basal RGS17 expression dramatically reduced cAMP levels compared to control cells in response to M7 (Figure 6C).
DISCUSSION
G-protein-dependent signaling is critical to normal hepatocyte function, liver development, hepatic regeneration, and HCC. In recent years increasing attention has been focused on regulators of G-protein signaling (RGS) as potential targets for therapeutic intervention. However, the role of RGS outside of the nervous system remains poorly defined. Data herein identify elevated RGS17 mRNA expression in human and animal models of HCC versus non-tumorigenic liver. Furthermore, the increased levels of RGS17 in HCC tissue is maintained in immortalized hepatoma cell lines (H4IIE, HepG2 and HuH-7). These data support recent findings describing altered RGS17 mRNA expression in lung, prostate, and breast tumors [34, 35]. Further, we report that altering RGS17 expression in human HuH-7 HCC cells has a pronounced effect on mitogenesis and cell migration, hallmark properties of tumorigenesis.
The analysis of human HCC samples demonstrated increased RGS17 mRNA expression in 5 of 7 samples analyzed. Of note, in those patients in which increased RGS17 mRNA was detected, all of the patients presented with underlying macronodular cirrhosis or steatosis (Table 1). Conversely, in the samples analyzed where RGS17 mRNA was decreased or unchanged, tumors arose in the setting of micronodular cirrhosis or ostensibly normal underlying hepatic architecture. The varied nature of the risk factors and underlying disease state(s) in which HCC most commonly arise likely make the sample size for this analysis too small to be conclusive. However, it may be of importance that the onset and progression of cirrhosis and steatosis are characterized by dramatic changes in hepatic immune status [36–39]. This raises the possibility that changes in hepatic immune response/ signaling may play a significant role on modulating RGS17 expression, and affect hepatic tumor progression. While the change in hepatic immune status/ signaling may prove to be important in regulating RGS17 expression, other possibilities cannot be discounted. Of note, hepatic cirrhosis is characterized by the activation and replication of hepatic stellate cells to a transformed, myofibroblastic phenotype [36, 37]. This in turn raises the possibility that the differences in RGS17 mRNA expression measured may actually be due to the comparison of HCC to diseased/ cirrhotic liver, as opposed to “healthy” liver. Unfortunately the lack of specificity and / or sensitivity of antibodies commercially available to detect RGS17 made it impossible to determine the specific cell populations within which changes in RGS17 mRNA/ protein occurred. As such, increasing population size analyzed, coupled with specific cell population isolation and analysis, is likely to be necessary in order to definitively describe RGS expression/ function in the human HCC disease, and underlying hepatic disease states.
Despite employing numerous commercially available antibodies, and analysis of increasing amounts of total cell protein lysates (up to 150µg total cell lysate), we were also unable to detect RGS17 protein by immunoblot. Given the catalytic nature of RGS physiologic function it is maybe of little surprise that the protein exists at (relatively) low abundance within the cell. The predominant function of RGS17 is to accelerate GTP hydrolysis by the Gα subunits via binding and releasing from the Gα-GTP (GDP) complex, and subsequent deactivation of the G-protein [15, 17]. Immunodetection of many RGS proteins is a commonly recognized problem described in the literature [34, 35, 40]. Furthermore, promiscuity of interaction between members of the RGS family and heterotrimeric G-protein counterparts make it highly likely that RGS expression is tightly regulated within the cell and may explain why, despite comparative abundance of RGS17 mRNA over-expression, corresponding protein could not be detected by immunoblot. That is, over-expression of RGS17 by adenoviral infection at low doses led to dramatic increases in mRNA expression, yet yielded undetectable/ low protein levels. Rather, increasing the intensity of transduction levels were required to detect protein translation to detectable levels in the cell lines studied in vitro. Furthermore, as might be predicted, heterologous RGS17 expression at a given infection intensity was inversely proportional to endogenous RGS17 level of the respective cell line; being the highest in HepG2 and lowest in HuH-7.
An analysis of RGS17 regulation in hepatoma cell lines did not demonstrate a role for miRNAs in regulating RGS17 mRNA via 3'UTR. This finding in HCC cells is in contradiction with data published by Sun et al., in lung cancer cells who report mir-182 to be involved in regulating RGS17 expression via two conserved sites located in the 3' UTR region [35]. In this report the investigators demonstrate the level of mir-182 exhibits a reverse correlation with RGS17 expression. Conversely, our microarray expression data using two miRNA sets, in non - transformed rat hepatocytes and the H4IIE HCC cell line, demonstrate the opposite; a n ≈10 fold increase in mir-182 expression being detected in H4IIE cells compared to hepatocytes (data not shown). At present it is uncertain as to why such dramatic discrepancies exist, although it is tempting to speculate that it may be due to both the origin of the cells forming the tumor, as well as species differences. In contrast to our observations with miRNAs, employing inhibitors to block protein degradation via the proteasome demonstrated increased RGS17 accumulation within cells, especially at lower levels of infection. Collectively, these data suggest that the regulation of RGS17 in cells of hepatic origin occurs preferentially at the protein, as opposed to mRNA, level.
Given the high degree of regulation in RGS 17 expression, it was somewhat surprising that heterologous over-expression of RGS17 (by adenovirus) had little, or no, effect on different HCC cell lines in terms of proliferation; a slight, though significant, increase in proliferation being observed only in the HepG2 cell line. One possible explanation might be that, within the confines of the high degree of regulation that exists to define protein expression, the endogenous RGS17 protein level in the hepatoma cell lines is close to “saturation”, such that there exists only a limited physiological effect following further over-expression. Indeed, further support of such an effect may be evidenced by the significantly decreased proliferation rate of RGS17 deficient cells, an effect further emphasizing a role for RGS17 in hepatoma progression.
In addition to proliferation, altering RGS17 expression had a pronounced effect on migration. Previous studies have indentified a significant role for Gi-proteins during cell invasion, hepatic regeneration, and HCC progression [7, 10, 11, 41]. These reports identify the importance of Gi/ Gs-proteins in regulating cAMP-PKA signaling, and the effect of changes brought about by aberrant Giα expression/ signaling. Analysis of Gi/ Gs expression and function in HCC demonstrate increased Giα-protein expression in human and animal models of HCC [10, 11]. Using parallel in vitro models of HCC, pharmacological stimulation of Giα signaling led MAPK activation and cell proliferation, effects observed in other cell lines but not in isolated, cultured hepatocytes [10, 11, 42, 43].
Gi and Gq proteins can be coupled to the same GPCR within the same cell type [44], so both G-proteins can be activated via the same extracellular stimuli. Such cooperative Gi/ Gq activation would lead to more efficient signal transmission to MAPK signaling cascade than would occur via either individually [45]. In light of these findings, the interaction of RGS17 with both Giα and Gqα subunits reported herein may prove highly significant in mediating such activation.
The role of cAMP-PKA signaling in mitogenesis is somewhat ambiguous, depending on both cell type and cAMP levels. In theory, increases in intracellular cAMP should lead to a pro-mitogenic response via PKA-cAMP responsive element binding protein (CREB) signaling. However, we have previously reported sustained increases in intracellular cAMP leads to induction of antimitogenic inducible cAMP early repressor (ICER) expression [30]. Similarly, prolonged cAMP accumulation followed by PKA activation can exert an antimitogenic effect via PKA-mediated inhibition of Raf kinase activity [46]. This raises the possibility that RGS17 fits within this network by balancing cAMP-PKA and MAPK signaling pathways such that RGS17 acts in favor of the cAMP-PKA arm.
In conclusion, the data presented in our study demonstrate elevated RGS17 mRNA expression in HCC tumors/ cells, in both human and animal disease models. While basal protein expression is not detectable in these systems (by immunoblot), the use of alternative approaches demonstrates that RGS17 protein expression is under close regulation at the protein level, predominantly by protein sequestration and degradation. Furthermore, manipulation of RGS17 expression, either over-expression or inhibition, significantly affects cellular physiology, including migration, proliferation, and the physical-functional interaction with Giα2. Given the central role that Gi-proteins play in normal hepatic function, liver regeneration, and HCC progression, our data support an equally important likely role for changes in RGS expression/ function during these events. As such, despite the difficulties associated with measuring RGS expression at the protein level, future studies are required to elucidate the precise mechanism of RGS17 interaction during cell migration, the involvement of RGS17 in malignant tumor transformation, and the potential role of RGS17 during hepatic tumorigenesis in animal models and human liver disease states.
ACKNOWLEDGEMENTS
This work was funded in part by grants from the NIH (Grant #AA014891 (LWS) & AA016858 (IHM)).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Eugene Sokolov, Email: evgueni.sokolov@carolinashealthcare.org.
David A. Iannitti, Email: david.iannitti@carolinashealthcare.org.
Laura W. Schrum, Email: laura.schrum@carolinashealthcare.org.
Iain H. McKillop, Email: iain.mckillop@carolinashealthcare.org.
REFERENCES
- 1.Ahuja S, Smith SO. Trends Pharmacol Sci. 2009;30(9):494–502. doi: 10.1016/j.tips.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 2.Gurevich VV, Gurevich EV. Mol Pharmacol. 2008;74(2):312–316. doi: 10.1124/mol.108.049015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smrcka AV. Cell Mol Life Sci. 2008;65(14):2191–2214. doi: 10.1007/s00018-008-8006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feldman RD, Gros R. Life Sci. 2007;81(4):267–271. doi: 10.1016/j.lfs.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 5.Rozengurt E. J Cell Physiol. 2007;213(3):589–602. doi: 10.1002/jcp.21246. [DOI] [PubMed] [Google Scholar]
- 6.Woehler A, Ponimaskin EG. Curr Mol Pharmacol. 2009;2(3):237–248. doi: 10.2174/1874467210902030237. [DOI] [PubMed] [Google Scholar]
- 7.Diehl AM, Yang SQ, Wolfgang D, Wand G. J Clin Invest. 1992;89(6):1706–1712. doi: 10.1172/JCI115771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawai Y, Arinze IJ. Mol Cell Endocrinol. 1993;90(2):203–209. doi: 10.1016/0303-7207(93)90153-b. [DOI] [PubMed] [Google Scholar]
- 9.Yagami T, Kirita S, Matsushita A, Kawasaki K, Mizushima Y. Biochim Biophys Acta. 1994;1222(1):81–87. doi: 10.1016/0167-4889(94)90027-2. [DOI] [PubMed] [Google Scholar]
- 10.McKillop IH, Wu Y, Cahill PA, Sitzmann JV. J Cell Physiol. 1998;175(3):295–304. doi: 10.1002/(SICI)1097-4652(199806)175:3<295::AID-JCP7>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt CM, McKillop IH, Cahill PA, Sitzmann JV. Hepatology. 1997;26(5):1189–1194. doi: 10.1002/hep.510260516. [DOI] [PubMed] [Google Scholar]
- 12.Sethakorn N, Yau DM, Dulin NO. Cell Signal. 2010;22(9):1274–1281. doi: 10.1016/j.cellsig.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sjogren B, Neubig RR. Mol Pharmacol. 2010;78(4):550–557. doi: 10.1124/mol.110.065219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bos JL, Rehmann H, Wittinghofer A. Cell. 2007;129(5):865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 15.Ross EM. Curr Biol. 2008;18(17):R777–R783. doi: 10.1016/j.cub.2008.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sjogren B, Blazer LL, Neubig RR. Prog Mol Biol Transl Sci. 2010;91:81–119. doi: 10.1016/S1877-1173(10)91004-1. [DOI] [PubMed] [Google Scholar]
- 17.Willars GB. Semin Cell Dev Biol. 2006;17(3):363–376. doi: 10.1016/j.semcdb.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 18.Xie GX, Palmer PP. J Mol Biol. 2007;366(2):349–365. doi: 10.1016/j.jmb.2006.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Vries L, Zheng B, Fischer T, Elenko E, Farquhar MG. Annu Rev Pharmacol Toxicol. 2000;40:235–271. doi: 10.1146/annurev.pharmtox.40.1.235. [DOI] [PubMed] [Google Scholar]
- 20.Kurrasch DM, Huang J, Wilkie TM, Repa JJ. Methods Enzymol. 2004;389:3–15. doi: 10.1016/S0076-6879(04)89001-3. [DOI] [PubMed] [Google Scholar]
- 21.Burns ME, Wensel TG. Neuron. 2003;38(6):853–856. doi: 10.1016/s0896-6273(03)00361-1. [DOI] [PubMed] [Google Scholar]
- 22.Larminie C, Murdock P, Walhin JP, Duckworth M, Blumer KJ, Scheideler MA, Garnier M. Brain Res Mol Brain Res. 2004;122(1):24–34. doi: 10.1016/j.molbrainres.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 23.Huang J, Pashkov V, Kurrasch DM, Yu K, Gold SJ, Wilkie TM. Comp Hepatol. 2006;5:8. doi: 10.1186/1476-5926-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang Y, Li C, Guzman VM, Chang WW, Evinger AJ, 3rd, Sao D, Woodward DF. FEBS J. 2005;272(3):791–799. doi: 10.1111/j.1742-4658.2004.04516.x. [DOI] [PubMed] [Google Scholar]
- 25.Mittmann C, Schuler C, Chung CH, Hoppner G, Nose M, Kehrl JH, Wieland T. Naunyn Schmiedebergs Arch Pharmacol. 2001;363(4):456–463. doi: 10.1007/s002100000376. [DOI] [PubMed] [Google Scholar]
- 26.Bonifaz V, Shan Y, Lambrecht RW, Donohue SE, Moschenross D, Bonkovsky HL. Liver Int. 2009;29(3):366–373. doi: 10.1111/j.1478-3231.2008.01833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brandon-Warner E, Sugg JA, Schrum LW, McKillop IH. Cancer Lett. 2010;291(1):120–129. doi: 10.1016/j.canlet.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt CM, McKillop IH, Cahill PA, Sitzmann JV. Eur J Gastroenterol Hepatol. 1999;11(12):1393–1399. doi: 10.1097/00042737-199912000-00009. [DOI] [PubMed] [Google Scholar]
- 29.Sun H, Liu GT. Cancer Lett. 2006;236(2):239–249. doi: 10.1016/j.canlet.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 30.Kovach SJ, Price JA, Shaw CM, Theodorakis NG, McKillop IH. J Cell Physiol. 2006;206(2):411–419. doi: 10.1002/jcp.20474. [DOI] [PubMed] [Google Scholar]
- 31.Rodriguez-Munoz M, Bermudez D, Sanchez-Blazquez P, Garzon J. Neuropsychopharmacology. 2007;32(4):842–850. doi: 10.1038/sj.npp.1301184. [DOI] [PubMed] [Google Scholar]
- 32.Inoue S, Hoshino S, Kukimoto I, Ui M, Katada T. J Biochem. 1995;118(3):650–657. doi: 10.1093/oxfordjournals.jbchem.a124959. [DOI] [PubMed] [Google Scholar]
- 33.Clark MJ, Traynor JR. Methods Enzymol. 2004;389:155–169. doi: 10.1016/S0076-6879(04)89010-4. [DOI] [PubMed] [Google Scholar]
- 34.James MA, Lu Y, Liu Y, Vikis HG, You M. Cancer Res. 2009;69(5):2108–2116. doi: 10.1158/0008-5472.CAN-08-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun Y, Fang R, Li C, Li L, Li F, Ye X, Chen H. Biochem Biophys Res Commun. 2010;396(2):501–507. doi: 10.1016/j.bbrc.2010.04.127. [DOI] [PubMed] [Google Scholar]
- 36.McKillop IH, Schrum LW. Semin Liver Dis. 2009;29(2):222–232. doi: 10.1055/s-0029-1214377. [DOI] [PubMed] [Google Scholar]
- 37.Wasmuth HE, Tacke F, Trautwein C. Semin Liver Dis. 2010;30(3):215–225. doi: 10.1055/s-0030-1255351. [DOI] [PubMed] [Google Scholar]
- 38.Corazza N, Badmann A, Lauer C. Semin Immunopathol. 2009;31(2):267–277. doi: 10.1007/s00281-009-0168-1. [DOI] [PubMed] [Google Scholar]
- 39.Tacke F, Luedde T, Trautwein C. Clin Rev Allergy Immunol. 2009;36(1):4–12. doi: 10.1007/s12016-008-8091-0. [DOI] [PubMed] [Google Scholar]
- 40.Ghavami A, Hunt RA, Olsen MA, Zhang J, Smith DL, Kalgaonkar S, Rahman Z, Young KH. Cell Signal. 2004;16(6):711–721. doi: 10.1016/j.cellsig.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 41.Hurst JH, Henkel PA, Brown AL, Hooks SB. Cell Signal. 2008;20(2):381–389. doi: 10.1016/j.cellsig.2007.10.026. [DOI] [PubMed] [Google Scholar]
- 42.Castoria G, Migliaccio A, D'Amato L, Di Stasio R, Ciociola A, Lombardi M, Bilancio A, Di Domenico M, de Falco A, Auricchio F. Front Biosci. 2008;13:1318–1327. doi: 10.2741/2764. [DOI] [PubMed] [Google Scholar]
- 43.Fehrenbacher N, Bar-Sagi D, Philips M. Mol Oncol. 2009;3(4):297–307. doi: 10.1016/j.molonc.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gudermann T, Kalkbrenner F, Dippel E, Laugwitz KL, Schultz G. Adv Second Messenger Phosphoprotein Res. 1997;31:253–262. doi: 10.1016/s1040-7952(97)80023-7. [DOI] [PubMed] [Google Scholar]
- 45.Blaukat A, Barac A, Cross MJ, Offermanns S, Dikic I. Mol Cell Biol. 2000;20(18):6837–6848. doi: 10.1128/mcb.20.18.6837-6848.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gudermann T, Grosse R, Schultz G. Naunyn Schmiedebergs Arch Pharmacol. 2000;361(4):345–362. doi: 10.1007/s002109900208. [DOI] [PubMed] [Google Scholar]