Abstract
Introducing small DNA molecules (Dbait) impairs the repair of damaged chromosomes and provides a new method for enhancing the efficiency of radiotherapy in radio-resistant tumors. The radiosensitizing activity is dependent upon the efficient delivery of Dbait molecules into the tumor cells. Different strategies have been compared, to improve this key step. We developed a pipeline of assays to select the most efficient nanoparticles and administration protocols before preclinical assays: (i) molecular analyses of complexes formed with Dbait molecules, (ii) cellular tests for Dbait uptake and activity, (iii) live zebrafish embryo confocal microscopy monitoring for in vivo distribution and biological activity of the nanoparticles and (iv) tumor growth and survival measurement on mice with xenografted tumors. Two classes of nanoparticles were compared, polycationic polymers with linear or branched polyethylenimine (PEI) and covalently attached cholesterol (coDbait). The most efficient Dbait transfection was observed with linear PEI complexes, in vitro and in vivo. Doses of coDbait ten-fold higher than PEI/Dbait nanoparticles, and pretreatment with chloroquine, were required to obtain the same antitumoral effect on xenografted melanoma. However, with a 22-fold lower ‘efficacy dose/toxicity dose' ratio as compared with Dbait/PEI, coDbait was selected for clinical trials.
Keywords: siDNA, PEI, cholesterol, irradiation, antitumoral
Introduction
Increased understanding of the molecular pathological mechanisms of cancer, the advent of novel molecular tools such as synthetic small interfering RNA or plasmid DNA-based vectors and technology for their in vivo delivery, have provided an encouraging perspective for the use of nucleic acid-based cancer molecular therapies.1 The efficiency and cytotoxicity of synthetic DNA delivery systems must be further addressed to optimize nucleic acid-based therapies. We recently demonstrated that small interfering DNA molecules (siDNA) can be used to trick the cell, by triggering a false DNA damage signal that results in inhibition of DNA repair.2, 3 siDNA mimicking DNA double-strand breaks (called Dbait) were successfully used to sensitize tumors to irradiation. Dbait are 32-bp long double-stranded DNA molecules protected from exonucleases and helicases by substitution, at one end, of the 3′- and 5′-terminal nucleotide residues, with phosphorothioate nucleotides and, at the other end, by tethering the two strands with a hexaethylene glycol loop.3 Dbait administration before radiotherapy enables control of tumor growth and, in some cases, a cure. Dbait acts within the cell in a strictly dose-dependent manner and, therefore, its efficient intracellular delivery is pivotal for controlling disease.
Naked DNA does not efficiently enter the cells. To facilitate its delivery, non-viral methods using a combination of synthetic polymers and nucleic acids to form particles, called lipoplexes or polyplexes, have been developed and optimized for use with large DNA in cultured cells. It is unclear how these vectors will be effective for small DNA and clinical applications. Identifying and overcoming each hurdle along the DNA entry pathways can improve DNA delivery to the nucleus and hence, improve the overall transfection efficiency. There are three major barriers to DNA delivery: (i) low uptake across the plasma membrane, (ii) inadequate release of DNA molecules with limited stability and (iii) lack of nuclear targeting. In addition to these ‘cellular barriers', ‘tissue and systemic barriers' also exist, which include degradation, opsonization of particles by charged serum components, rapid clearing and accumulation in non-target tissues. In this study, we established a pipeline of assays from analyses of physicochemical and stability properties of complexes, imaging of cellular uptake and tissue distribution in animals, to activity assays in adapted cell culture and in vivo models. A multi-system approach is necessary to successfully complete the characterization and selection of a vector for in vivo DNA delivery. Cell cultures have been used for vector selection, based on activity assays, zebrafish early embryos for quasi-instantaneous and live imaging of cellular uptake of Dbait nanoparticles and mice for in vivo imaging of Dbait distribution in healthy or tumoral tissue and therapeutic efficacy assays. Integrating these assays provides a powerful tool for selection of the most efficient delivery system for biomolecules to optimize pre-clinical assays.
Materials and methods
Dbait and particles formation
Dbait and coDbait molecules were obtained by automated solid-phase oligonucleotide synthesis from Eurogentec (Seraing, Belgium) as previously described.2 They were purified by denaturing reversed-phase high-performance liquid chromatography and/or high-performance liquid chromatography–ion exchange. Sequence in Dbait and coDbait is: 5′-GCTGTGCCCACAACCCAGCAAACAAGCCTAGA-(H)-TCTAGGCTTGTTTGCTGGGTTGTGGGCACAGC-3′, where H is a hexaethylene-glycol linker. Sequence of the inactive siDNA are: 5′ACGCACGG-(H)-CCGTGCGT-3′ for the 8H and 5′-AGATCGCCAACACCGAACAAACGACCGTGCGT-(H)-AGATCCGAACAAACGACCCAACACCCGTGCGT-3′ for the 64ss. Some Dbait derivatives were labeled with the fluorophores Cy3 (λexcitation=540 nm; λemission=560 nm) or Cy5.5 (λexcitation=650 nm; λemission=670 nm). Linear polyethylenimine (PEI) (11 and 22 kDa) were obtained from Polyplus-Transfection (Illkirch, France) and provided as a ready-to-use solution of 300 m nitrogen concentration. Branched bPEI25kd was purchased from Sigma-Aldrich (Saint Quentin, France). Lutrol was purchased from In Cell Art (Nantes, France). Dbait, coDBait and PEI solutions (stock PEI) were diluted in 10% sucrose or 150 m NaCl (for in vivo and in vitro transfection experiments, respectively) to obtain various ratios of vector/Dbait. The ratio of PEI/Dbait (or ratio N/P) was determined according to the number of amine nitrogen for PEI and phosphate for Dbait. Typically, for 300 μl of complexes at 0.6 mg ml−1 and N/P of 6, Dbait (180 μg, 0.54 μmol of phosphate) and the desired amount of polymer solution (11.4 μl of PEI stock solution contains 0.3 μmol of amine nitrogen) were diluted into a volume of 150 μl each. Superfect/Dbait particles were prepared according to manufacturer's instructions (Qiagen, Courtaboeuf, France) in a ratio of 10 μl Superfect per μg of DNA. The vector/Dbait complexes were analyzed by agarose gel electrophoresis method. The samples (18 μl) were mixed with bromophenol blue dye (1 μl) and electrophoresed on 1.5% agarose gels in Tris-acetate-ethylene diamine tetraacetic acid buffer 1 × (40 m Tris-acetate, pH 8.3, 1 m ethylene diamine tetraacetic acid) at 100 V for 30 min. The gel was stained with ethidium bromide and the bands visualized under UV light to detect uncomplexed Dbait.
Cell culture, Dbait molecules and transfection
Studies on cells in culture were performed using the SV40-transformed fibroblasts MRC-5. Cells were grown at 37 °C in monolayer cultures in complete Dulbecco's modified Eagle medium (DMEM) (Gibco, Cergy Pontoise, France) with 10% fetal calf serum and antibiotics (100 μg ml−1 streptomycin and 100 μg ml−1 penicillin), under conditions of 100% humidity, 95% air and 5% CO2. Unless otherwise specified, transfections were performed in 1.2 ml DMEM medium without serum in 60 mm diameter plates. Transfection with jetPEI (Polyplus-transfection) was performed at an N/P ratio of 6, according to the manufacturer's instructions. Briefly, Dbait molecules were diluted in 150 m NaCl and gently mixed with an equal volume of PEI in 150 m NaCl, and added to DMEM medium without serum. CoDbait was directly added to DMEM medium without serum. Transfection of Dbait molecules was performed with Superfect reagent in 1.2 ml DMEM medium without serum (in 60 mm diameter plates) for 5 h. For electroporation, 1.2 × 106 cells were transfected with 2 μg Dbait, using the Gene Pulser II (Bio-Rad, Marnes-la-Coquette, France). At the end of the 5-h transfection (time zero), medium was replaced by complete medium and cells were grown for the indicated amount of time before analysis. Chloroquine (50 μ) was added 30 min before transfection.
DNA-dependent protein kinase (DNA-PK) activity assay
DNA-PK activity was monitored using the kit SignaTECT DNA-dependent Protein Kinase Assay System (Promega, Madison, WI). The biotinylated substrate, 50 units of DNA-Dependent Protein Kinase (Promega) and 0.25 μg (500 n) Dbait molecules were incubated for 5 min at 30 °C with (γ-32P)ATP, according to the manufacturer's instructions. The biotinylated substrate was captured on a streptavidin membrane, washed and counted in a scintillation counter. Percentage of phosphorylation was calculated by dividing the bound radioactivity by the total count of (γ-32P)ATP per sample.
Flow cytometry
Cells were transfected with different complexes with Dbait-cy3 for 5 h and directly analyzed or left to grow for 24 h. For immunofluorescence detection by flow cytometry, the cells were fixed overnight in cold 100% ethanol before immunodetection. Cells were permeabilized in 0.2% Triton X-100 for 30 min, blocked with 2% bovine serum albumin, incubated for 2 h on ice with mouse monoclonal antibodies anti γ-H2AX (Upstate Biotechnology, Temecula, CA) and revealed with secondary antibodies conjugated with Alexa-488 (Molecular Probes, Eugene, OR) for 30 min at room temperature. Cells were analyzed using a FACScalibur flow cytometer (BD Biosciences, Franklin Lakes, NJ), and data was analyzed using BD CellQuest Pro (BD Biosciences) and the free WinMDI 2.8 (Scripps Research Institute, La Jolla, CA) software. Note that permeabilization treatment used for immunostaining removed most of the Dbait, impairing accurate Dbait detection and immunofluorescence detection on the same sample.
Zebrafish husbandry, embryo collection and treatment
Zebrafish eggs were obtained from natural spawning of wild-type or transgenic (βactin:egfp-ras) fish lines. A Narishige (MN 153) micromanipulator (East Meadow, NY) fixed on a dissecting scope with epifluorescence illumination (Leica MZ16F, Mannhein, Germany) and an air injector (Eppendorf FemtoJet, Le Pecq, France) were used to perform Dbait injection at cell stage 1 K. Glass capillaries (Harvard Apparatus GC100-10, Les Ulis, France) were pulled with a KopF vertical pipette puller (KopF 720, Tujunga, CA) to make injection needles. Two to five nanoliters of Dbait solution were injected at the animal pole of embryos in cell cycle ten and immediately processed by confocal laser scanning microscopy imaging (upright Leica SP2) with × 40/0.8 NA water dipping lens objective. Imaging was performed using simultaneous 480 nm (enhanced green fluorescent protein) and 561 nm (cy3) excitation. Embryos were grown at 28.5 °C for a further 24 h post-infection. One-day-old larvae were observed under the dissecting microscope and phenotypes categorized as described in Figure 4. Chloroquine treatment before injection consisted of a 2-h incubation in embryo medium4 with 50 μ chloroquine (CQ). Dbait (coDbait)-cy3 of 50 μ was injected either alone or in combination with PEI25K (ratio N/P=9), PEI22K (ratio N/P=6), PEI11K (ratio N/P=6), Superfect (10 μl 1−1 μg−1 Dbait).
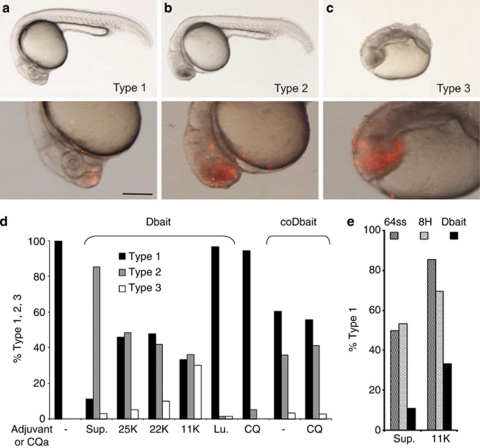
Figure 4.
Phenotypes 24 h after Dbait injection into the extracellular space of cell stage 1 K zebrafish embryos. (a–c) Lateral views anterior to the left of zebrafish embryos, 24 h after Dbait-cy3+polyethylenimine (PEI) injection (2–5 nl) at the animal pole of cell stage 1 K zebrafish embryos: top panel, bright field view; bottom panel, 2 × magnification of the head region with epifluorescence overlay showing in red Dbait-cy3, scale bar 250 μm. (a) Type 1 phenotype indistinguishable from non-injected (not shown). (b) Type 2 mild phenotype with extensive cell death in the head region. (c) Type 3 strong teratogenesis and widespread cell death. (d) Histogram showing the percentage of the three phenotypic classes depending on the adjuvant. More than 100 embryos were analyzed for each condition. Sup: Superfect; 25, 22, 11 K PEI of the corresponding size; Lu, Lutrol; CQ, chloroquine; ‘−', without adjuvant. (e) Histogram showing the percentage of the animals with type 1 phenotype after injections of nanoparticules formed with 8H, 64ss or Dbait32H and Superfect or PEI11K. The Dbait32H, and the 8H and 64ss inactive nanoparticules, were used at equivalent adjuvant concentrations.
Dbait and Irradiation treatments in mice
SK28 or U87G xenograft tumors were obtained by injecting 106 tumor cells into the flank of adult female nude mice (Charles River strain; L'arbresle, France). The animals were housed in the laboratory for at least 1 week before commencing experiments. There were five to six animals per cage, under controlled conditions of light and dark cycles (12 h:12 h), relative humidity (55%) and temperature (21 °C). Food and tap water were available ad libitum. After approximately 12 days, when the subcutaneous tumors measured 150–200 mm3, the mice were separated into homogeneous groups of at most 12 each, to receive different treatments. Irradiation was performed in a 137Cs unit (0.5 Gy min−1) with a shield designed to protect about two-thirds of the animal's body. Doses were controlled by thermoluminescence dosimetry. A total dose of 30 Gy were delivered in six sessions at intervals of three sessions of 5 Gy per week for a period of 2 weeks. Dbait and coDbait molecules were prepared in 100 μl of 10% sucrose. The Dbait mixtures were incubated for 15 min at room temperature before injection. Injections of the indicated amount of formulated Dbait were performed 5 h before each radiotherapy session. Mock-treated animals were injected with 100 μl of 10% glucose according to the protocol of the associated assays. When specified, animals received intraperitoneal injections of 100 μl of CQ (10 mg ml−1) 1 h before coDbait. Tumor size was assessed by caliper measurements every 3 days, and size was calculated using the formula (½ × length × width2). Mice were weighed and pictures of tumors were taken every week during 100 days. For ethical reasons, the animals were killed when their tumors reached 2000 mm3. The endpoint used in survival analysis was death day. For diffusion analyses, tumors were excised 5 h after one session of treatment and cryosections were labeled with To-Pro5 iodine (Invitrogen, Cergy, France) and scanned using an Odyssey scanner (ScienceTec, les Ulis, France). The Local Committee on Ethics of Animal Experimentation (Orsay, France) approved all experiments.
Statistical analysis
Descriptive analyses of the tumor responses were performed for each treatment and each tumor type. Day 1 was the day of the first treatment session. All animals were followed for at least 100 days. Median lifetime was estimated according to the Kaplan–Meier method. Tumor growth delay was calculated by subtracting the mean tumor volume quadrupling time of the control group from tumor volume quadrupling times of individual mice in each treated group. The mean tumor growth delay was calculated for each treated group, using individual measurements. We performed a Mann–Whitney U-test to assess the in vivo effect of the various treatments on the growth of xenografted tumors using statEL software (ad Science, Paris, France). The number of animals (N) and the P-value are reported in Table 2. All tests were considered significant at the P=0.05 significance level.
Results
Characterization of Dbait nanoparticles
Cholesterol-conjugated small interfering RNAs have markedly improved pharmacological properties in vitro and in vivo.5 Cholesterol has been shown to increase dendriplex uptake.6, 7 As cholesterol travels in the blood as low density lipoproteins and is taken up via receptor-mediated endocytosis by cells, we questioned if this property could be utilized for Dbait uptake. We therefore synthesized a modified Dbait molecule covalently linked to a fatty chain of cholesterol at the 5′-end nearby hexaethylene-glycol loop (called coDbait). The coDbait nanoparticle solution was very stable and did not form aggregates at concentrations as high as 80 mg ml−1.
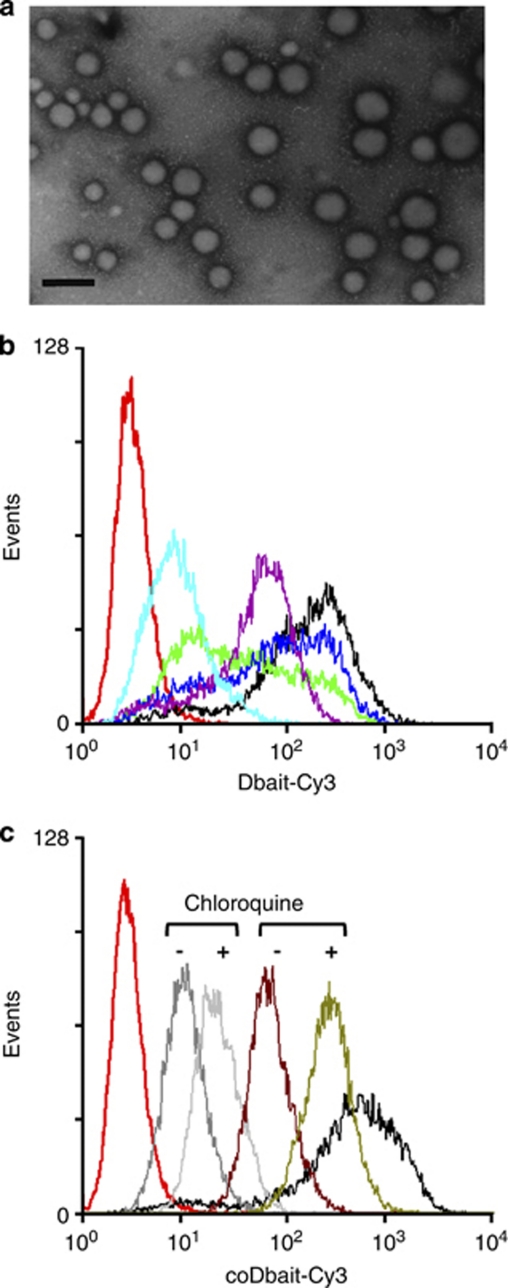
We compared the activity of coDbait to several PEI particle polyplexes formed with Dbait. PEI has been used for gene delivery with increasing popularity since its description as a transfection agent.8 PEI forms non-covalent interpolyelectrolyte complexes with DNA,8 oligonucleotides9 and RNA.10 Long PEI chains are more effective, but more cytotoxic in gene transfection.11 For each PEI vector tested, our main goal was to develop a formulation with the most homogeneous particle size distribution at the highest Dbait concentration. The diameter and surface charge of the Dbait/PEI nanoparticles were measured by dynamic laser light scattering. Using multimodal analysis, we found that branched PEI (bPEI25K) with a mean size of 25 KDa, and linear PEI with 22 KDa (PEI22K) or 11 KDa (PEI11 K) in size, formed Dbait complexes with similar properties (Supplementary Table S1). Different ratios of PEI on Dbait were tested. The lowest ratio resulting in formation of 100% complexed particles with Dbait was determined by gel shift assay (data not shown). The ratios of PEI/Dbait (N/P) 6, 6 and 9 were chosen for PEI11K, PEI22K and bPEI25K, respectively, for further study. Dbait-PEI complex particles were stable over a period of 1 h in 10% sucrose (data not shown). The highly homogenous morphology of the spherical particles in the population (sizes range from 125 to 140 nm) was confirmed by transmission electron microscopy (Figure 1a). Homogeneous complex solutions were obtained at concentrations not exceeding 60 μg ml−1. The presence of salt in the dilution buffer at a concentration exceeding 0.8 mg ml−1 or prolonged storage, induced aggregation of PEI complexes. Superfect complexes (60 μg Superfect per μg Dbait) that yielded larger and polydisperse aggregates (>2 μm) were used as positive controls. The uncharged amphiphilic copolymer Lutrol12 did not form stable interacting complexes with Dbait and was used as negative control in some experiments.
Figure 1.
Cellular uptake of formulated Dbait. (a) Microscopic analyses of Dbait complexes with polyethylenimine (PEI)11k. (b and c) Flow cytometric analyses of cellular uptake were performed 5 h after beginning treatment for various transfection conditions. (b) 1.6 μg ml−1 Dbait-cy3 (red), Dbait-cy3 with Superfect (black), bPEI25K (purple), PEI11K (green), PEI22K (blue), electroporation (light blue); (c) Dbait-cy3 with Superfect (black), 1.6 μg ml−1 coDbait-cy3 (grey), with and without chloroquine (CQ) treatment (light grey) before transfection, 21 μg ml−1 coDbait-cy3 without CQ (brown) and with CQ (green).
Cell uptake of Dbait nanoparticles
We monitored the cellular uptake of the different nanoparticles using a fluorescent cy3-modified Dbait or coDbait. The initial fluorescence of the cy3-Dbait complex was measured immediately before transfection. In PEI complexes, the Dbait fluorescence was two to three-fold lower, indicating that the compaction of the molecules with PEI might quench fluorescence (Table 1). The coDbait was also less fluorescent than the naked Dbait, indicating that cholesterol might interact with the cyanine on the same molecule. Superfect or Lutrol did not affect fluorescence. Cellular content of human transfected fibroblast cells was measured by flow cytometry analysis. The distribution of fluorescence in cells treated with naked Dbait or Dbait–Lutrol mixture was not different to the untreated control (data not shown), indicating that the Dbait molecules did not spontaneously enter into the cells. Electroporation was relatively inefficient and increasing the concentration of Dbait did not improve the transfection efficiency (Figure 1b). All polycationic polymers (PEI and Superfect) promoted efficient cellular uptake; however, linear PEIs showed a wider distribution than Dbait/Superfect or Dbait/PEIb25 K complexes (Figure 1b). Interestingly, coDbait entered cells without the help of any transfection factors, indicating that conjugation to cholesterol facilitated DNA uptake as previously observed for small interfering RNA uptake. However, the mean fluorescence intensity in cells was lower than for Dbait/PEI, and a ten-fold higher concentration of coDbait was required to reach Dbait/PEI transfection efficiency (Figure 1c and Table 1).
Table 1. Fluorescence and cellular uptake of formulated Dbait.
| Molecules/method | Dbait | Chloroquine | Fluorescencea | MCC | Cor MCC | MCC | Cor MCC |
|---|---|---|---|---|---|---|---|
| (μg ml−1) | (100 mg ml−1) | × 106 μg−1ml−1 | 5 h | 5 h | 24 h | 24 h | |
| — | — | − | 0 | 3 | ND | 3 | ND |
| — | — | + | 0 | 3 | ND | ND | ND |
| Electroporation | 1.6 | − | 6.17 | 9 | 1 | 11 | 2 |
| Dbait | 1.6 | − | 6 | 3 | 0.4 | 3 | 0.5 |
| Dbait/PEI11K | 1.6 | − | 1.92 | 26 | 14 | 13 | 7 |
| Dbait/bPEI25K | 1.6 | − | 3.04 | 52 | 17 | 58 | 19 |
| Dbait/PEI22K | 1.6 | − | 3.32 | 54 | 16 | 43 | 13 |
| Dbait/Superfect | 1.6 | − | 6.41 | 220 | 34 | 204 | 32 |
| Dbait/Superfect | 1.6 | + | 6.41 | 218 | 34 | ND | ND |
| coDbait | 1.6 | − | 3.47 | 10 | 3 | ND | ND |
| coDbait | 1.6 | + | 3.47 | 21 | 6 | ND | ND |
| coDbait | 16 | − | 3.47 | 66 | 19 | 64 | 18 |
| coDbait | 16 | + | 3.47 | 236 | 68 | 214 | 62 |
| coDbait | 32 | − | 3.47 | 145 | 42 | ND | ND |
| coDbait | 32 | + | 3.47 | 390 | 112 | ND | ND |
Abbreviations: Cor MCC, corrected cellular content FL2/fluorescence; MCC, mean cellular content FL2 value (>3 experiments); ND, not defined.
Fluorescence at FL2 value.
One limitation of the activity of transferred DNA is its retention in endosomes, which prevents it from interacting with its target or being transcribed. Inside the cell, to avoid degradation, DNA must escape from normal endosomal pathways. Therefore, the efficiency of DNA delivery is associated not only with cellular uptake, but also with destabilization and escape from endosomes.13 PEI is known to have a high buffering capacity that facilitates DNA release from endosomes and lysosomes (proton sponge hypothesis).8 Recycling cholesterol from endosomes requires systems that are poorly understood. Fusogenic agents such as CQ raise endosomal pH and inhibit sequestration of DNA in endosomes. We tested the capacity of CQ to enhance coDbait uptake by adding 50 μ to the cells 30 min before transfection. CQ increased the mean fluorescence of cells treated with cy3–coDbait by two- to four-fold (Figure 1c).
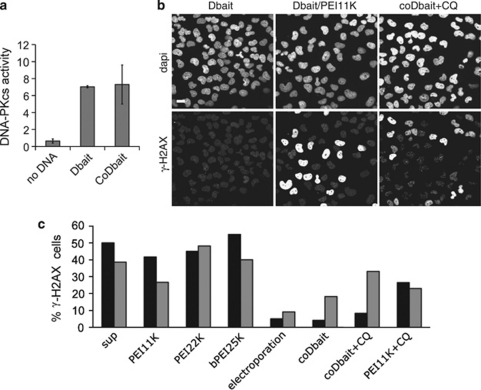
Binding of DNA-PK catalytic subunits (DNA-PKcs) kinase to Dbait molecules3 triggers its activation, which is an indicator of the amount of Dbait released in the cell.3 Addition of cholesterol did not affect the ability of Dbait to activate purified DNA-PKcs (Figure 2a). We have previously shown that the DNA-PKcs kinase activation by Dbait can be monitored by the amount of H2AX phosphorylation in the cell.3 Both Dbait/PEI and coDbait induced H2AX phosphorylation in treated cells (Figure 2b). Branched and linear PEI/Dbait complexes rapidly promoted H2AX phosphorylation (Figure 2c) that reached a maximal value 1 h after the end of transfection and persisted for 24 h. Dbait-induced kinase activity was very low after electroporation at any timepoint (Figure 2c). Transfection with ten-fold more coDbait was very inefficient and required at least 24 h to reach a maximal value of phosphorylated H2AX. Addition of CQ during transfection increased the level of activation of DNA-PKcs in coDbait-transfected cells to that observed with ten-fold less Dbait/PEI at 24 h after transfection (Figure 2c). CQ did not increase DNA-PKcs activation in Dbait/PEI-transfected cells, indicating that Dbait is efficiently released from endosomes when complexed to PEI.14 As cellular uptake of coDbait was not increased 24 h after transfection, the slow activation of DNA-PKcs indicates the slow release of coDbait from endosomes.
Figure 2.
Activity of formulated Dbait. (a) DNA-dependent protein kinase (DNA-PK) activation was measured after addition of 50 μg purified enzyme complex to no DNA, 0.25 μg Dbait or 0.25 μg coDbait. (b) Immunodetection of γ-H2AX in cells 24 h after treatment (down panels) with 1.6 μg ml−1 Dbait, 1.6 μg ml−1 Dbait/polyethylenimine (PEI)11 K, 16 μg ml−1 coDbait with CQ and corresponding cell nuclei stained with 4',6-diamidino-2-phenylindole (DAPI; upper panels). Scale bar: 20 μm. (c) Quantification of γ-H2AX, 1 h (black) and 24 h (grey) after treatment with various Dbait formulations. All transfections were performed with 1.6 μg ml−1 Dbait or 16 μg ml−1 coDbait. When indicated, CQ was added before transfection.
Cellular uptake and overall toxicity in zebrafish early embryos
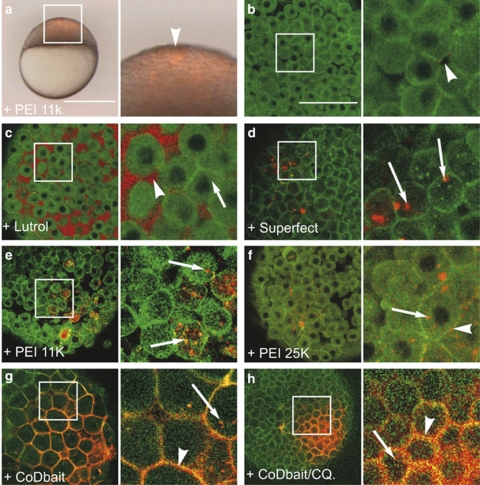
Zebrafish (Danio rerio) have recently emerged as a valuable model organism in the drug discovery processes, including target identification, disease modeling, lead discovery and compound toxicology. Their evolutionary proximity to humans, embryonic accessibility and high fecundity make zebrafish favorable for screening large numbers of small molecules in vivo.15 Analysis of Dbait uptake and its activity on cell cultures does not allow us to conclude on Dbait diffusion, cellular uptake and activity in the whole organism. To assess these activities, we injected Dbait-cy3 nanoparticles, into the intercellular space of 1000 cell stage (stage 1 K) zebrafish embryos.16 This protocol enabled in vivo observation by confocal microscopy of real-time Dbait-cy3 distribution at the cellular and subcellular level, as well as its activity on the rapidly-dividing cells of the early zebrafish embryo (Figure 3a). As shown in Figure 3b, naked Dbait-cy3 injected at the animal pole of stage 1 K embryos, rapidly diffused throughout the whole blastoderm and was no longer detected 15 min after injection. The addition of Lutrol enabled the retention of Dbait in the extracellular space around the injection point, but did not facilitate cellular uptake (Figure 3c). In the presence of Superfect or PEI, numerous fluorescent patches were observed inside the cells, indicating efficient cellular uptake (Figures 3d–f). The coDbait-cy3 demonstrated different behavior with strong and persistent staining of plasma membranes, together with patchy intracellular fluorescence (Figure 3g). Note that coDbait localization at the cellular membrane was also partially observed in mammalian cells (Supplementary Figure S1). Incubation of embryos with CQ before injection converted part of the large fluorescent patches of coDbait into a diffuse intracellular distribution (Figure 3h).
Figure 3.
Dbait injection into stage 1 K zebrafish embryos. (a–h) 2–5 nl Dbait-cy3 were injected either alone (b) or with adjuvant (c–h) indicated bottom left into the extracellular space at the animal pole of act26:egfp-ras zebrafish embryos at cell stage 1 K. (a) Bright field and epifluoresence overlay of an injected embryo at sphere stage (1 h after injection), lateral view, scale bar 250 microns, white box is a 3 × magnification in right panel, white arrowhead indicates the animal pole. (b–h) Injected embryos imaged live by confocal microscopy (Leica SP2 488 and 561 nm simultaneous excitation) from the animal pole 15–30 min after injection. Left panel, white box is a 3 × magnification in right panel (scale bar 100 μm). White arrows point to intracellular Dbait-cy3, white arrowheads point to Dbait-cy3 staining either at the cell surface (b, f, g, h) or in the extracellular space (c).
The observation of phenotypic effects 24 h after injection enabled the assessment of Dbait activity and the overall toxicity of the treatment. Dbait fluorescence was mainly detected in the head cells of larvae, 24 h after injection (Figures 4a–c), which according to the zebrafish fate map development,17 are derived from the animal pole area of pre-gastrulation embryos that received the Dbait injection. Injection of Dbait without adjuvant or combined with Lutrol (Lu) showed no effect on development (Figure 4d) correlating with the poor intracellular uptake of Dbait described above. Addition of adjuvant led to cell death in the head and, correlating with the injected volume, extensive cell death and teratogenesis might be observed. The 24 h phenotypes were categorized as described in Figure 4, allowing quantification of the toxicity of the injected mixture. For the same concentration of Dbait, clear differences appeared in terms of cell death and subsequent developmental abnormalities, depending on the adjuvant. Addition of Superfect (sup) was very toxic to embryonic cells and extensive cell death at early stages resulted in a high percentage of type 2 phenotypes. Addition of PEI (25, 22 and 11 K) proved to be also toxic to zebrafish blastomeres. Although less toxic than Dbait/PEI, coDbait injection resulted in significant cell death. Pre-incubation of embryos with CQ did not significantly increase toxicity. Altogether, early embryonic cell death and subsequent developmental abnormalities was a fast and reliable protocol for assessing the overall toxicity of Dbait±adjuvant in zebrafish embryos. Correlation of cell death with cellular uptake suggested that the anti-tumoral activity of Dbait in embryonic cells might have an important role in toxicity. Indeed, embryo cells, after injection of Dbait directly into cytoplasm, rapidly encounter mitotic catastrophe as a consequence of Dbait activity on rapidly proliferating cells (unpublished data).
We confirmed that toxicity of nanoparticules with Superfect or PEI was partly due to the Dbait activity using nanoparticules formed with inactive siDNA molecules: 8 H, a short 8-bp long double-stranded molecule and, 64ss, a single-stranded molecule corresponding to a 32-bp long Dbait with non-complementary strands. None of these molecules bind or activate DNA-PKcs.3 The toxicity of the nanoparticules with inactive siDNA was significantly lower than toxicity of nanoparticles formed with Dbait (Figure 4e).
Local and systemic toxicity in mice
To assess the relevance of toxicity data on zebrafish embryo with toxicity in mammalian, we analyzed the tolerance of nude mice skin to repeated administration of Dbait/PEI1K, Dbait/PEI22K, Dbait/bPEI25K and coDbait. The toxicity of the different formulated Dbait was analyzed after three daily subcutaneous (SC) injections. All the Dbait/PEI showed high toxicity with injections that were tolerated at 3.75 mg kg−1 (3.75 nmoles per injection), but at 5 mg kg−1, it started to trigger local inflammation associated with local necrosis that rapidly disappeared with the arrest of the treatment. Intravenous (IV) injection toxicity gave similar results; the Dbait/PEI IV injections were lethal at 3 mg kg−1 (3 nmoles per injection), with death occurring during injection, probably by blood vessel clogging. Slow injections by perfusion (0.4 μl min−1) increased tolerance up to 6 mg kg−1 Dbait/PEI (6 nmoles per injection), confirming that most of the IV toxicity is due to the local concentration at the bolus injection site. The coDbait, with or without CQ, did not show any toxicity at all doses tested (up to 640 mg kg−1; 800 nmoles per injection), irrespective of the route chosen: SC, IV bolus or IV perfusion.
Antitumoral activity in xenografted tumors
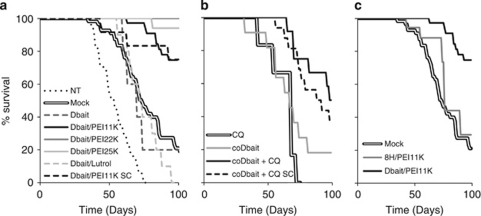
The antitumoral effect of the various Dbait formulations was tested, at doses that do not show any toxicity on healthy tissues, in combination with radiotherapy on SK28-xenografted human melanoma. Dbait/vector complexes were administered 5 h before each irradiation, using intratumoral (IT) injections. The results were consistent with the observations from previous experiments. The Dbait/PEI formulations induced tumor growth delay and tumor regression after IT injection (Table 2 and Figure 5a). The Dbait/PEI22k formulation was the most efficient. Nanoparticules formed with the inactive 8H molecule and PEI11K did not show significant effect on tumor growth, indicating that the tumor regression observed with Dbait/PEI11K is mainly due to Dbait activity (Figure 5c). The coDbait administration without CQ did not remarkably improve radiotherapy efficiency. Although radiosensitization effects have been observed with administration of a high CQ concentration,18 prophylactic doses of CQ (intraperitoneal injections of 100 μg) did not affect tumor growth in control groups, nor in groups receiving radiotherapy. In contrast, the radiosensitivity of tumors significantly increased when mice received CQ followed by coDbait 1 h later (Table 2 and Figure 5b). The activity of Dbait and coDbait in tumoral cells was confirmed by the activation of DNA-PKcs demonstrated by H2AX phosphorylation (Figure 6b). Activation of DNA-PKcs in a large number of cells within the tumor area correlated with the accumulation of small cells, with fragmented nuclei typical of apoptotic cells (Figure 6b). This result confirmed the previous observation that activation of DNA-PKcs by Dbait induced cell death in tumor cells.2, 3
Table 2. Xenografted mice survival after irradiation associated with various treatments.
| Dbait/vector complexes | Dbait conc. | Mode admin. | N mice | Median survival time | Mean TGDa | s.d. TGD | Mean % TGD | TGD P-valueb |
|---|---|---|---|---|---|---|---|---|
| Mock | — | IT | 56 | 74 | 8 | 12 | 139 | — |
| Dbait | 6 × 60 μg (3 nmol) | IT | 6 | 70 | 2 | 8 | 108 | 0.18 |
| Dbait/PEI11K | 6 × 60 μg (3 nmol) | IT | 38 | >100 | >36 | 19 | >281 | 1.06 × 10−10 |
| Dbait/PEI22K | 6 × 60 μg (3 nmol) | IT | 10 | >100 | >48 | 28 | >341 | 2.73 × 10−4 |
| Dbait/PEIb25K | 6 × 60 μg (3 nmol) | IT | 19 | >100 | >51 | 21 | >356 | 1.31 × 10−8 |
| Dbait/Lutrol | 6 × 60 μg (3 nmol) | IT | 10 | 74 | 4 | 5 | 121 | 0.57 |
| Dbait/PEI11K SC | 6 × 150 μg (7.5 nmol) | SC | 12 | >100 | >26 | 22 | >229 | 1.52 × 10−3 |
| CQ | — | IT | 6 | 68 | 7 | 6 | 134 | 0.93 |
| coDbait | 6 × 600 μg (30 nmol) | IT | 11 | 67 | 19 | 25 | 195 | 0.17 |
| coDbait+CQ | 6 × 600 μg (30 nmol) | IT | 13 | 98 | >44 | 26 | >321 | 8.66 × 10−6 |
| coDbait+CQ SC | 6 × 1.2 mg (60 nmol) | SC | 16 | 92 | >24 | 26 | >224 | 1.45 × 10−2 |
Abbreviations: CQ, chloroquine; IT, intratumoral injection; Mode admin, mode of administration; SC, subcutaneous injection; TGD, tumor growth delay;
TGD was calculated as described in Material and Methods.
Statistical analyses were performed using mock treatment as a reference.
Figure 5.
Kaplan–Meier survival representation of animals with SK28 tumors treated in six sessions over 2 weeks with different nanoparticules associated with radiotherapy (5 Gy per session). The dose of nanoparticules per session was 60 μg (3 nmol) for Dbait/adjuvant nanoparticules by intratumoral (IT) injections, 150 μg (7.5 nmol) for Dbait/polyethylenimine (PEI)11K subcutaneous (SC) injections, 600 μg (30 nmol) for coDbait IT injections and 1.2 mg (60 nmol) by coDbait SC injections. (a) Comparison of treatment efficiencies with radiotherapy associated with different nanoparticules: IT injections of 5% glucose vehicle (Mock), Dbait without adjuvant (Dbait), Dbait/PEI11K, Dbait/PEI22K, Dbait/PEI25K, Dbait/Lutrol, or SC injections of Dbait/PEI11K (Dbait/PEI11K SC). NT, non-irradiated and non-treated control. (b) Comparison of coDbait treatments and radiotherapy with (+ CQ) or without intraperitoneal injections of chloroquine. coDbait +CQ or coDbait, IT injections of coDbait; coDbait +CQ SC, SC injections of coDbait. Control cells treated by radiotherapy with CQ (CQ) showed similar survival than control receiving only radiotherapy (Mock in a). (c) Comparaison of survival of animals treated by radiotherapy and IT injections of nanoparticules formed with PEI11K and Dbait (Dbait/11K) or 8H (8H/PEI11K) molecules. Mock, irradiated tumors injected with 5% glucose vehicle.
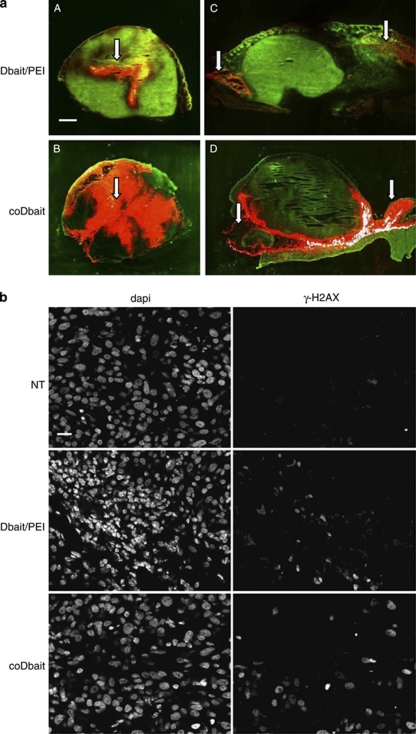
Figure 6.
Diffusion and activity in tumors. Tumors were injected with 1.6 μg Dbait-cy5.5/polyethylenimine (PEI) or 16 μg coDbait-cy5.5, excised and analyzed the following day for fluorescence distribution and DNA-PKcs activation. (a) Diffusion of fluorescent Dbait after two types of injections, intratumoral (IT) injection (A and B), or subcutaneous (SC) injections (C and D); white arrows indicate the sites of injection; scale: 2 mm. (b) DNA-PKcs activity was detected by H2AX phosphorylation (γ-H2AX) in tumors without treatment (NT) or after IT injection of Dbait/PEI or coDbait; left panels, cell nuclei stained with 4',6-diamidino-2-phenylindole (DAPI); right panels, immunodetection of γ-H2AX; scale, 20 μm.
Although drug administration by IT injection has been used in many trials, it is currently advised to avoid this route of delivery in clinical assays. We investigated how Dbait/PEI11K or coDbait could be administered SC in the area adjacent to the tumor. This route of administration has been successfully used in several clinical assays.19, 20, 21 We first compared the diffusion of the molecules in tumors treated by one IT injection or two SC injections performed at opposite sides of the tumor (Figure 6a). Fluorescent Dbait complexed to PEI11k tended to form aggregates at the site of injection and diffused progressively to the edge of the tumor. In contrast, coDbait showed a more even distribution around the injection site, whether inside the tumor or in its vicinity. Despite demonstrating a reduced diffusion to the tumor, SC injections of Dbait/PEI11K or coDbait increased tumor growth control by irradiation (Table 2 and Figure 5). Increasing the number of injection sites should allow a significant improvement to tumor growth control without the addition of local toxicity.
Discussion
In this work, we used a set of assays to guide development of administration protocols and drug formulation. These assays enabled the comparison of different formulations of Dbait before performing preclinical assays on mice. Cellular and zebrafish embryo assays were used to assess the efficiency of Dbait cellular uptake, a prerequisite step in the antitumoral drug effect, and for selection of the most appropriate protocols and formulations for preclinical studies on mammals.
Zebrafish embryo possesses a cell-autonomous mechanism that can trigger apoptosis in response to DNA damage.22, 23 We have exposed early zebrafish embryos to different Dbait containing nanoparticles that are predicted to specifically inhibit DNA repair. Overall, toxicity in the zebrafish embryo did not correlate with toxicity in mice skin or after systemic injection, but corresponded to cellular Dbait activity. Dbait/PEI (5 μ) and coDbait (50 μ)+CQ that triggered comparable DNA-PKcs activation in cell culture had similar toxic effects on zebrafish embryos (Figure 4d) and displayed significant antitumoral activity on mouse tumors (Table 2). However, although coDbait is very well tolerated by healthy tissue, Dbait/PEI proved to be highly toxic. These results suggest that toxicity in zebrafish early embryos is an indicator of tumor sensitivity rather than healthy tissue sensitivity. These observations are consistent with the sensitivity to antitumoral activity of zebrafish embryonic cells that share characteristic properties with tumor cells, including mitotic index and, biochemical and phenotypic traits.15, 24 Consistent with this hypothesis, we recently developed a method for measuring embryo cell proliferation25 and observed Dbait anti-proliferation activity by direct intracellular injection of Dbait into zebrafish blastomeres (unpublished data).
PEI polymers were the most efficient, of all adjuvant molecules tested, in forming Dbait complexes. Their use was limited, however, by their toxicity on tissues, as well as in the blood system. Local toxicity was partly overcome by slow administration (perfusion) and by administering the doses into different injection sites. The covalent combination of cholesterol and Dbait provided the most promising alternative for Dbait delivery to the cells. Indeed, the lack of toxicity within the range of tested doses suggests that this molecule might prove useful despite the high doses required for antitumoral effects. The doses of 3 and 30 nmoles per injection of Dbait/PEI11K and coDbait, respectively, tripled the delay in tumor growth delay induced by irradiation alone. The respective toxicity of both nanoparticles (LD50 of 3.7 nmoles for Dbait/PEI and >800 n for coDbait in mice) resulted in relative ratios of efficiency dose/toxicity dose of 0.8 for Dbait/PEI11K and <0.037 for coDbait, indicating that coDbait is safer and is a good candidate for local administration in clinical trials.
Acknowledgments
This work was supported by the Institut Curie, the Centre National de la Recherche (CNRS), the Muséum National d'Histoire Naturelle, Genopole (Evry), FP6 funding (BioEmergences NEST programme), ARC funding to Nadine Peyrieras, ARC post-doctoral fellowship to Benoit Maury. We thank Emmanuel Dauty for the characterization of the PEI complexes, Vincent Lemesre for technical assistance and the members of the imagery platforms at Institut Curie.
C Agrario and A Herbette are employees of DNA Therapeutics. Dr Dutreix and Dr Sun are cofounders of DNA Therapeutics, the Dbait patent holder society. Dr Sun is director of DNA Therapeutics. N Berthault, Dr Peyrieras and Dr Maury declare no potential conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cancer Gene Therapy website (http://www.nature.com/cgt)
Supplementary Material
References
- Dutreix M, Cosset JM, Sun JS. Molecular therapy in support to radiotherapy. Mutat Res. 2010;704:182–189. doi: 10.1016/j.mrrev.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Quanz M, Berthault N, Roulin C, Roy M, Herbette A, Agrario C, et al. Small-molecule drugs mimicking DNA damage: a new strategy for sensitizing tumors to radiotherapy. Clin Cancer Res. 2009;15:1308–1316. doi: 10.1158/1078-0432.CCR-08-2108. [DOI] [PubMed] [Google Scholar]
- Quanz M, Chassoux D, Berthault N, Agrario C, Sun JS, Dutreix M. Hyperactivation of DNA-PK by double-strand break mimicking molecules disorganizes DNA damage response. PLoS One. 2009;4:e6298. doi: 10.1371/journal.pone.0006298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book. University of Oregon Press: Eugene, OR, USA; 2000. [Google Scholar]
- Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- Gokhale PC, Soldatenkov V, Wang FH, Rahman A, Dritschilo A, Kasid U. Antisense raf oligodeoxyribonucleotide is protected by liposomal encapsulation and inhibits Raf-1 protein expression in vitro and in vivo: implication for gene therapy of radioresistant cancer. Gene Ther. 1997;4:1289–1299. doi: 10.1038/sj.gt.3300543. [DOI] [PubMed] [Google Scholar]
- Manoharan M. Oligonucleotide conjugates as potential antisense drugs with improved uptake, biodistribution, targeted delivery, and mechanism of action. Antisense Nucleic Acid Drug Dev. 2002;12:103–128. doi: 10.1089/108729002760070849. [DOI] [PubMed] [Google Scholar]
- Boussif O, Lezoualc'h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bologna JC, Dorn G, Natt F, Weiler J. Linear polyethylenimine as a tool for comparative studies of antisense and short double-stranded RNA oligonucleotides. Nucleosides Nucleotides Nucleic Acids. 2003;22:1729–1731. doi: 10.1081/NCN-120023124. [DOI] [PubMed] [Google Scholar]
- Aigner A, Fischer D, Merdan T, Brus C, Kissel T, Czubayko F. Delivery of unmodified bioactive ribozymes by an RNA-stabilizing polyethylenimine (LMW-PEI) efficiently down-regulates gene expression. Gene Ther. 2002;9:1700–1707. doi: 10.1038/sj.gt.3301839. [DOI] [PubMed] [Google Scholar]
- Godbey WT, Wu KK, Mikos AG. Poly(ethylenimine) and its role in gene delivery. J Control Release. 1999;60:149–160. doi: 10.1016/s0168-3659(99)00090-5. [DOI] [PubMed] [Google Scholar]
- Desigaux L, Gourden C, Bello-Roufai M, Richard P, Oudrhiri N, Lehn P, et al. Nonionic amphiphilic block copolymers promote gene transfer to the lung. Hum Gene Ther. 2005;16:821–829. doi: 10.1089/hum.2005.16.821. [DOI] [PubMed] [Google Scholar]
- El Ouahabi A, Thiry M, Pector V, Fuks R, Ruysschaert JM, Vandenbranden M. The role of endosome destabilizing activity in the gene transfer process mediated by cationic lipids. FEBS Lett. 1997;414:187–192. doi: 10.1016/s0014-5793(97)00973-3. [DOI] [PubMed] [Google Scholar]
- Murphy EA, Waring AJ, Murphy JC, Willson RC, Longmuir KJ. Development of an effective gene delivery system: a study of complexes composed of a peptide-based amphiphilic DNA compaction agent and phospholipid. Nucleic Acids Res. 2001;29:3694–3704. doi: 10.1093/nar/29.17.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler GN, Brandli AW. Simple vertebrate models for chemical genetics and drug discovery screens: lessons from zebrafish and Xenopus. Dev Dyn. 2009;238:1287–1308. doi: 10.1002/dvdy.21967. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Woo K, Shih J, Fraser SE. Fate maps of the zebrafish embryo. Curr Opin Genet Dev. 1995;5:439–443. doi: 10.1016/0959-437x(95)90046-j. [DOI] [PubMed] [Google Scholar]
- Briceno E, Calderon A, Sotelo J. Institutional experience with chloroquine as an adjuvant to the therapy for glioblastoma multiforme. Surg Neurol. 2007;67:388–391. doi: 10.1016/j.surneu.2006.08.080. [DOI] [PubMed] [Google Scholar]
- Kudo-Saito C, Schlom J, Hodge JW. Intratumoral vaccination and diversified subcutaneous/intratumoral vaccination with recombinant poxviruses encoding a tumor antigen and multiple costimulatory molecules. Clin Cancer Res. 2004;10:1090–1099. doi: 10.1158/1078-0432.ccr-03-0145. [DOI] [PubMed] [Google Scholar]
- Moreau P, Coiteux V, Hulin C, Leleu X, van de Velde H, Acharya M, et al. Prospective comparison of subcutaneous versus intravenous administration of bortezomib in patients with multiple myeloma. Haematologica. 2008;93:1908–1911. doi: 10.3324/haematol.13285. [DOI] [PubMed] [Google Scholar]
- Negrier S, Perol D, Ravaud A, Bay JO, Oudard S, Chabaud S, et al. Randomized study of intravenous versus subcutaneous interleukin-2, and IFNalpha in patients with good prognosis metastatic renal cancer. Clin Cancer Res. 2008;14:5907–5912. doi: 10.1158/1078-0432.CCR-08-0236. [DOI] [PubMed] [Google Scholar]
- Ikegami R, Rivera-Bennetts AK, Brooker DL, Yager TD. Effect of inhibitors of DNA replication on early zebrafish embryos: evidence for coordinate activation of multiple intrinsic cell-cycle checkpoints at the mid-blastula transition. Zygote. 1997;5:153–175. doi: 10.1017/s0967199400003828. [DOI] [PubMed] [Google Scholar]
- Ikegami R, Zhang J, Rivera-Bennetts AK, Yager TD. Activation of the metaphase checkpoint and an apoptosis programme in the early zebrafish embryo, by treatment with the spindle-destabilising agent nocodazole. Zygote. 1997;5:329–350. doi: 10.1017/s0967199400003919. [DOI] [PubMed] [Google Scholar]
- Peifer M, Polakis P. Wnt signaling in oncogenesis and embryogenesis--a look outside the nucleus. Science. 2000;287:1606–1609. doi: 10.1126/science.287.5458.1606. [DOI] [PubMed] [Google Scholar]
- Campana M, Maury B, Dutreix M, Peyrieras N, Sarti A. Methods toward in vivo measurement of zebrafish epithelial and deep cell proliferation. Comput Methods Programs Biomed. 2010;98:103–117. doi: 10.1016/j.cmpb.2009.08.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.