Abstract
Mammalian Peptidoglycan Recognition Proteins (PGRPs), similar to antimicrobial lectins, bind to bacterial cell wall and kill bacteria through an unknown mechanism. We show that PGRPs enter Gram-positive cell wall at the site of daughter cell separation during cell division. In Bacillus subtilis PGRPs activate the CssR-CssS two-component system that detects and disposes of misfolded proteins exported out of bacterial cells. This activation results in membrane depolarization, cessation of intracellular peptidoglycan, protein, RNA, and DNA synthesis, and production of hydroxyl radicals, which are responsible for bacterial death. PGRPs also bind to the outer membrane in Escherichia coli and activate functionally homologous CpxA-CpxR two-component system, which results in bacterial death. We excluded other potential bactericidal mechanisms (inhibition of extracellular peptidoglycan synthesis, hydrolysis of peptidoglycan, and membrane permeabilization). Thus we reveal a novel mechanism of bacterial killing by innate immunity proteins that bind to cell wall or outer membrane and exploit bacterial stress defense response to kill bacteria.
Multicellular eukaryotes have several mechanisms to kill bacteria, including oxidative killing by phagocytic cells, complement, and antimicrobial peptides present in phagocytic cells, on body surfaces, secretions, and tissue fluids. The mechanisms of bacterial killing by these antibacterial systems are well established. Eukaryotes also produce antibacterial proteins, such as antibacterial lectins1,2 and Peptidoglycan Recognition Proteins (PGRPs or PGLYRPs)3,4, but the mechanism of bacterial killing by these large proteins is completely unknown and likely different than killing by small antibacterial peptides5. To reveal the mechanism of bacterial killing by these proteins, we studied bacterial killing by PGRPs.
PGRPs are innate immunity proteins that are conserved from insects to mammals, recognize bacterial peptidoglycan, and function in antibacterial immunity. Mammals have four PGRPs, PGLYRP-1, -2, -3, and -4 (initially named PGRP-S, -L, -Iα, and -Iβ)6,7. PGLYRP-1, PGLYRP-3, and PGLYRP-4 are all directly bactericidal8–12, whereas PGLYRP-2 is an N-acetylmuramoyl-L-alanine amidase that hydrolyzes peptidoglycan13,14. PGLYRP-1 is found mainly in polymorphonuclear leukocytes’ granules, PGLYRP-3 and PGLYRP-4 are found in the skin, eyes, salivary glands, tongue, throat, esophagus, stomach, and intestine, and PGLYRP-2 is expressed mainly in the liver and is secreted into blood3,4.
Peptidoglycan is a polymer of β(1–4)-linked N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc), crosslinked by short peptides containing L- and D-amino acids. It is a unique component of the cell wall of virtually all bacteria and is essential for the structural integrity of bacteria, their growth, and survival. For this reason peptidoglycan is also a frequent target of antibacterial products, such as antibiotics (produced by bacteria and fungi) or animal innate immunity proteins. In mammals peptidoglycan is recognized by Nod-like receptors, PGRPs, CD14, Toll-like receptor-2, mannose binding lectin, RegIIIγ C-type lectin, and lysozyme4.
PGRPs are specific for the MurNAc-pentapeptide fragment of peptidoglycan3,4. It was hypothesized that PGRPs kill bacteria by inhibiting peptidoglycan synthesis at one of the two extracellular steps (transglycosylation and transpeptidation). This hypothesis was based on our previous observation that bactericidal PGRPs bind to peptidoglycan but do not permeabilize cytoplasmic membranes5,10,12, and on the structural analysis of interaction of PGRPs with peptidoglycan fragments15 showing that their binding to PGLYRP-3 should prevent formation of cross-links between peptidoglycan’s peptide stems in the growing cell wall.
Here we reveal a novel mechanism of bacterial killing in which PGRPs bind to the cell wall or outer membrane and activate bacterial protein-sensing two-component systems that kill bacteria by inducing exaggerated stress response.
RESULTS
PGRPs inhibit an intracellular step in peptidoglycan synthesis
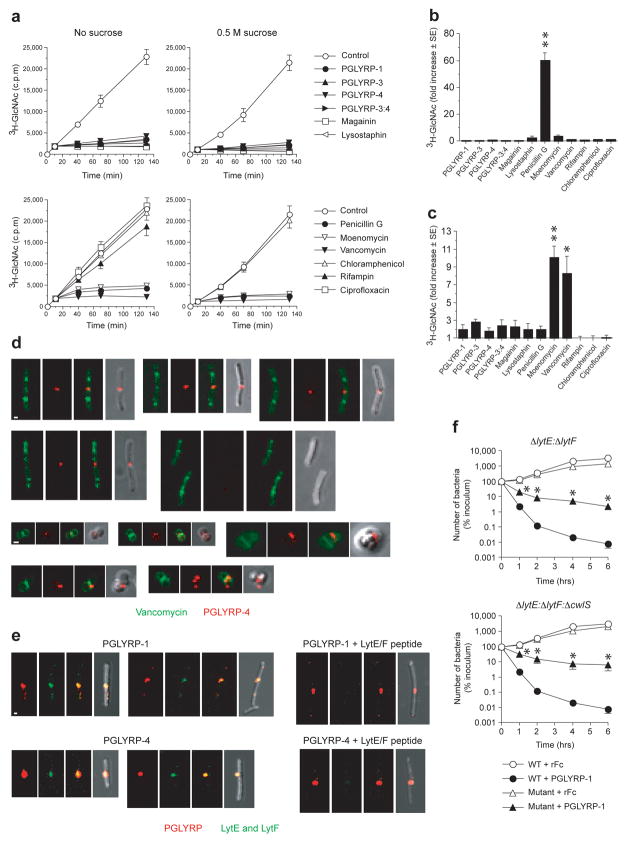
We first tested whether PGRPs inhibit peptidoglycan synthesis, because previous studies predicted that PGRPs inhibit transpeptidation10,12,15. We followed the rate of total peptidoglycan biosynthesis in Staphylococcus aureus by measuring incorporation of 3H-GlcNAc into the polymeric insoluble cell wall peptidoglycan, which was rapidly and completely inhibited by PGLYRP-1, PGLYRP-3, PGLYRP-4, and PGLYRP-3:4, as it also was by known peptidoglycan synthesis inhibitors, penicillin G, moenomycin, and vancomycin, and also by magainin and lysostaphin (Fig. 1a). Magainin rapidly permeabilizes the cytoplasmic membrane and immediately stops all metabolic reactions in the cell16,17, whereas lysostaphin rapidly hydrolyzes the pentaglycine bridge in S. aureus peptidoglycan, which causes osmotic lysis of bacteria and also stops all metabolic reactions. These results demonstrate that bactericidal PGRPs cause rapid inhibition of peptidoglycan synthesis.
Figure 1. PGRPs inhibit intracellular step in peptidoglycan synthesis and localize to the newly formed cell separation site.
(a) PGRPs inhibit peptidoglycan synthesis in S. aureus measured by incorporation of 3H-GlcNAc into insoluble peptidoglycan; 0.5 M sucrose prevents osmotic lysis; means ± SEM of 3 – 4 experiments. (b) PGRPs do not inhibit transpeptidation in S. aureus measured by incorporation of 3H-GlcNAc into secreted uncrosslinked polymeric peptidoglycan. (c) PGRPs do not inhibit transglycosylation in S. aureus measured as accumulation of 3H-GlcNAc-lipid II in cell membrane; means ± SEM of 3–4 (b) or 4–5 (c) experiments; *, P<0.03; **, P<0.0002 (t-test). (d) PGRPs localize to the newly formed cell separation site and not to the sites of peptidoglycan synthesis in B. subtilis (top rows) or S. aureus (bottom rows); different stages of cell division shown; each panel from left to right shows green (BODIPY-FL-vancomycin), red (Alexa Fluor 594-PGLYRP-4), overlay red and green, and overlay of red and dark field view. (e) PGLYRP-1 or PGLYRP-4 (red) co-localize with LytE and LytF (green) in B. subtilis and LytE/LytF peptide inhibits detection LytE and LytF; each panel from left to right shows red (PGRP), green (LytE and LytF), overlay red and green (yellow), and overlay of red and green and dark field view; bar = 1 μm. (f) LytE- and LytF-generated cell separation sites are required for efficient killing of B. subtilis by PGRPs, shown by lower killing of ΔlytE:ΔlytF or ΔlytE: ΔlytF: ΔcwlS mutants than of WT bacteria by PGLYRP-1; means ± SEM of 3 experiments; *, P<0.05 versus WT (t-test).
We then tested whether PGRPs inhibit the two extracellular steps of peptidoglycan synthesis, transglycosylation and transpeptidation. Penicillin G, as expected, inhibited transpeptidation and induced high incorporation of 3H-GlcNAc into newly-synthesized secreted polymeric uncrosslinked peptidoglycan, whereas PGRPs (at concentrations that were bactericidal and inhibited peptidoglycan synthesis), as well as other antibiotics that have other mechanisms of action, and also magainin and lysostaphin, did not inhibit transpeptidation (Fig. 1b). These results indicate that PGRPs do not inhibit transpeptidation, and thus must inhibit an earlier step in peptidoglycan synthesis.
We then tested whether PGRPs inhibit transglycosylation, which precedes transpeptidation in peptidoglycan synthesis. As expected, vancomycin or moenomycin induced accumulation of 3H-GlcNAc in the cell membrane lipid fraction containing lipid II in S. aureus (the membrane-bound undecaprenyl-phosphate-GlcNAc-MurNAc-peptide, Fig. 1c). However, PGRPs, similar to other antibiotics, magainin, and lysostaphin did not induce accumulation of lipid II in the cell membrane (Fig. 1c). These results indicate that PGRPs do not inhibit transglycosylation, and thus must inhibit an earlier intracellular step in peptidoglycan synthesis.
PGRPs localize to cell separation sites, which is required for killing
To selectively and directly inhibit an intracellular step in peptidoglycan synthesis PGRPs would have to enter the cytoplasm. Therefore, we next determined the localization of PGRPs in bacterial cells. Alexa Fluor 594-labeled PGLYRP-4 (red) exclusively localized at the sites of newly-formed daughter cell separation in Bacillus subtilis. Dual-labeling clearly showed that PGLYRP-4 did not co-localize with BODIPY-FL-labeled vancomycin (green); the latter localized at the sites of new peptidoglycan synthesis, which is primarily at the synthesis of new septa (Fig. 1d). Cells that still synthesize new septa and have not started to separate labeled only with vancomycin and did not bind PGLYRP-4. Similar results were obtained with S. aureus: vancomycin labeled the sites of new septa synthesis, and PGLYRP-4 labeled cell-separation sites, although because of the smaller size of S. aureus cells, perpendicular formation of new septa, and tendency of S. aureus to grow in clusters, there was smaller spatial separation of PGLYRP-4 and vancomycin (Fig. 1d). Similar results were obtained in B. subtilis with unlabeled PGLYRP-1 and PGLYRP-4 detected with antibody to V5 tag, and also in another unrelated Gram-positive bacterium, Listeria monocytogenes (Fig. 1e and Supplementary Fig. 1a, b).
Cell separation after cell division is carried out by dedicated peptidoglycan hydrolases, which in B. subtilis are LytE, LytF, and CwlS18, and whose expression is limited to the cell separation sites19. PGLYRP-1 and PGLYRP-4 completely co-localized with LytE and LytF (Fig. 1e), which further demonstrates preferential localization of PGRPs to the cell separation sites.
To determine whether localization of PGRPs to the cell separation sites is important for bacterial killing, we tested PGRP-mediated killing of B. subtilis ΔlytE:ΔlytF and ΔlytE:ΔlytF:ΔcwlS mutants18, whose cells do not separate after cell division, because they lack LytE, LytF, and CwlS peptidoglycan hydrolases. Both ΔlytE:ΔlytF and ΔlytE:ΔlytF:ΔcwlS mutants were significantly less killed by PGLYRP-1 than the wild type (WT) strain (Fig. 1f). These mutants also did not show specific staining with Alexa Fluor 594-labeled PGLYRP-4 (Supplementary Fig. 2a, b), which demonstrates that cell-separating LytE and LytF enzymes are required for efficient PGRP binding to bacteria and bacterial killing. This effect is selective for LytE and LytF, because deficiency in two other peptidoglycan hydrolytic enzymes, LytC and LytD, which have different specificity and function as autolytic but not cell-separating enzymes18–20, has no effect on bacterial sensitivity to PGRP-induced killing (Supplementary Fig. 3).
Killing of ΔlytE:ΔlytF and ΔlytE:ΔlytF:ΔcwlS mutants by other bactericidal agents (aminoglycoside antibiotics and pore-forming peptide magainin) was similar to the killing of WT bacteria (Supplementary Fig. 2c, d). These results indicate that other mechanisms of bacterial killing that are not dependent on the entry of the killing compound through the cell separation site are intact in these mutants.
Neither PGLYRP-4 nor PGLYRP-1 was detected intracellularly in B. subtilis, S. aureus, or L. monocytogenes when merged stacks or individual scans from confocal microscope of PGRP-stained bacteria were analyzed (Fig. 1d, e and Supplementary Fig. 1). Our results thus show that PGRPs do not enter the cytoplasm and stay in the cell separation sites bound to the cell wall in the vicinity of the cytoplasmic membrane. These results further support our biochemical data showing that PGRPs do not inhibit any of the extracellular steps in peptidoglycan synthesis, because at the time the cells begin to separate peptidoglycan synthesis at the separation sites has been completed.
Thus, our results indicate that in Gram-positive bacteria PGRPs gain access to the inner cell wall and cell membrane at the sites of peptidoglycan hydrolysis where daughter cells separate, and trigger their lethal effect from this extracellular site without entering the cytoplasm.
PGRPs inhibit protein, RNA, and DNA synthesis
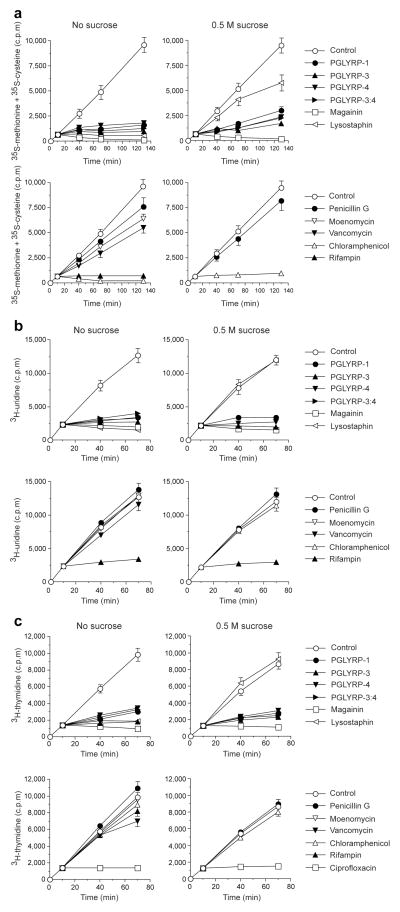
We next tested whether PGRPs cause a general inhibition of biosynthetic reactions in bacteria. PGLYRP-1, PGLYRP-3, PGLYRP-4, and PGLYRP-3:4 all rapidly and completely inhibited protein, RNA, and DNA synthesis, as shown by the inhibition of incorporation of 35S-methionine and 35S-cysteine, 3H-uridine, and 3H-thymidine into proteins, RNA, and DNA, respectively (Fig. 2a – c), at the same concentrations and with the same kinetics as they inhibited peptidoglycan synthesis (Fig. 1a). Titrating the inhibitory concentrations of PGRPs from 25 to 300 μg ml−1, revealed the same minimal concentrations for bactericidal effect and inhibition of all four biosynthetic processes (peptidoglycan, protein, RNA, and DNA).
Figure 2. PGRPs inhibit protein (a), RNA (b), and DNA (c) synthesis.
(a) Incorporation 35S-methionine and 35S-cysteine into cellular proteins, (b) 3H-uridine into RNA, and (c) 3H-thymidine into DNA in S. aureus treated with PGRPs, magainin, lysostaphin, antibiotics, or BSA (control) added at 10 min is shown; 0.5 M sucrose was used to prevent osmotic lysis; means ± SEM of 3–4 experiments.
Similar rapid and complete inhibition of peptidoglycan, protein, RNA, and DNA synthesis (Figs. 1a, 2a – c) was also obtained with a membrane-permeabilizing peptide, magainin (due to the loss of membrane integrity), and with a peptidoglycan-lytic enzyme, lysostaphin (due to the loss of peptidoglycan, which results in immediate osmotic lysis).
To differentiate between the effects due to osmotic lysis from the loss of peptidoglycan integrity and the effects due to membrane permeabilization, we performed similar experiments on inhibition of peptidoglycan, protein, RNA, and DNA synthesis in the hyperosmotic medium (with 0.5 M sucrose), which prevents osmotic lysis after the loss of peptidoglycan integrity, but has no effect on the consequences of membrane permeabilization. As expected, 0.5 M sucrose prevented lysostaphin-, but not magainin-induced inhibition of protein, RNA, and DNA synthesis, confirming that the inhibitory effect of lysostaphin is due to osmotic lysis of bacteria (Fig. 2a – c). Inhibition of protein, RNA, and DNA synthesis by PGLYRP-1, PGLYRP-3, PGLYRP-4, and PGLYRP-3:4 was not prevented in 0.5 M sucrose, which differentiates these PGRPs from lysostaphin and suggests that they do not kill bacteria by rapidly hydrolyzing peptidoglycan and do not induce osmotic lysis of bacteria.
PGRPs do not permeabilize cell membranes
Permeabilization of bacterial cell membranes by PGRPs would explain their rapid and simultaneous inhibition of all biosynthetic reactions that was not prevented by hyperosmotic medium (and thus resembled the effect of membrane-permeabilizing peptides, such as magainin). We considered this possibility in detail in Supplementary Information, where we show that PGRPs do not permeabilize bacterial cell membranes over the period of 6 h, despite rapid killing that exceeded 95% in 2 – 4 h and was not prevented by 0.5 M sucrose (Supplementary Fig. 4). Thus, our results so far demonstrate that the mechanism of bactericidal activity of PGRPs is different from the mechanism of bactericidal activity of antibiotics that inhibit peptidoglycan, protein, RNA, or DNA synthesis, and is also different from membrane-permeabilizing peptides and from enzymes that rapidly hydrolyze bacterial cell wall.
Bactericidal PGRPs do not hydrolyze peptidoglycan
Another possible mechanism of bacterial killing by PGRPs is enzymatic digestion of peptidoglycan by PGRPs. However, we show in Supplementary Information that bactericidal PGRPs do not hydrolyze insoluble S. aureus or B. subtilis peptidoglycan, uncrosslinked soluble polymeric peptidoglycan, synthetic peptidoglycan fragments, or heat-killed S. aureus and B. subtilis bacteria (Supplementary Fig. 5). Thus, these PGRPs do not have amidase, carboxypeptidase, or any other peptidoglycan-hydrolytic activity, and, therefore, these activities cannot be responsible for their bactericidal activity. We also excluded the possibility that activation of autolytic enzymes is responsible for PGRPs-induced killing of bacteria (Supplementary Figs. 3 and 5).
PGRPs induce membrane depolarization and •OH production
We next considered whether a loss of membrane potential is responsible for inhibition of intracellular biosynthetic reactions and killing of bacteria by PGRPs, because all these reactions require energy from ATP, whose production is largely dependent on ATP synthase driven by the proton gradient maintained by the membrane potential.
Indeed, PGRPs at bactericidal concentrations induced rapid and sustained membrane depolarization in B. subtilis (Fig. 3a), measured by flow cytometry with a membrane potential-sensitive fluorescent probe DiBAC4(3)21. Membrane depolarization was accompanied by intracellular production of toxic hydroxyl radicals (•OH), which are part of a stress-response in bacteria (Fig. 3b). All bactericidal PGRPs also induced membrane depolarization and high •OH production in S. aureus (data not shown).
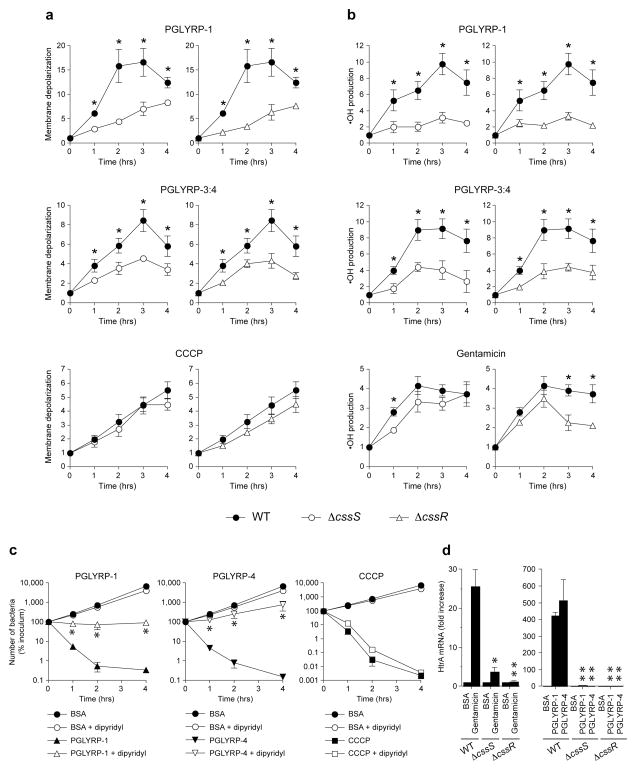
Figure 3. PGRPs induce membrane depolarization, •OH formation, and HtrA expression through the CssR-CssS two-component system, and •OH is responsible for PGRP-induced killing.
(a) Membrane depolarization or (b) •OH production in WT B. subtilis (closed symbols) or ΔcssS or ΔcssR mutants (open symbols) incubated with the indicated PGRPs, or CCCP, or gentamicin (positive controls), or BSA (negative control) measured by flow cytometry with fluorescent probes DiBAC4(3) (a) or HPF (b) and expressed as ratios of mean fluorescence intensity of PGRP-, CCCP-, or gentamicin-treated to BSA-treated bacteria are shown. (c) •OH-dependent killing of WT B. subtilis incubated with the indicated PGRPs, 20 μM CCCP, or BSA as a control without or with 300 μM dipyridyl measured by colony counts is shown. (d) HtrA mRNA expression in WT B. subtilis or ΔcssS or ΔcssR mutants incubated with PGLYRP-1, PGLYRP-4, or BSA (100 μg ml−1), or gentamicin (5 μg ml−1) for 30 min measured by qRT-PCR is shown. The results are means of 3 to 6 experiments ± SEM; *, P≤ 0.05; **, P≤ 0.001 (t-test) versus WT (a, b, d) or versus no dipyridyl (c).
To determine whether •OH production is responsible for PGRP-induced bacterial killing and how •OH is generated, we then tested the effect of dipyridyl and thiourea on PGRP-induced •OH production and bacterial killing. Dipyridyl is a membrane-permeable selective Fe2+ chelator that inhibits Fenton reaction-mediated •OH production, whereas thiourea is a •OH scavenger21. Both dipyridyl and thiourea significantly inhibited PGRP-induced bacterial killing (Fig. 3c and Supplementary Fig. 6). Effective inhibition of intracellular •OH production by dipyridyl was confirmed using •OH-specific fluorescent probe (Supplementary Fig. 7a). These results demonstrate that •OH are generated through Fenton reaction and are required for PGRP-induced bacterial killing.
We then tested the contribution of membrane depolarization to generation of •OH and bacterial killing. Chemical membrane potential decoupler, carbonyl cyanide 3-chlorophenylhydrazone (CCCP), as expected, induced membrane depolarization and killed bacteria, and the killing was not inhibited by dipyridyl (Fig. 3c), indicating •OH-independent killing by CCCP (further discussed in Supplementary Information).
PGRPs kill B. subtilis through CssR-CssS two-component system
We then tested whether the CssR-CssS two-component system in B. subtilis22 is involved in PGRPs-induced membrane depolarization, •OH production, and bacterial killing, because functionally homologous CpxA-CpxR two-component system in Escherichia coli detects misfolded proteins in antibiotic-treated bacteria and is responsible for antibiotic-induced membrane depolarization, stress response, •OH production, and killing21,23.
PGRPs-induced membrane depolarization and •OH production were significantly reduced in both ΔcssS and ΔcssR mutants compared to isogenic WT B. subtilis (Fig. 3a, b). Membrane depolarization by CCCP was similar in WT and ΔcssS and ΔcssR mutants, indicating that these mutants do not have an inherent defect in maintaining membrane potential and that the effect of PGRPs is selectively mediated by CssS and CssR. CCCP was bactericidal to both WT and mutant strains at the same concentration that induced membrane depolarization (Fig. 3c), indicating that this extent of membrane depolarization is sufficient to kill bacteria. •OH production induced by gentamicin (Fig. 3b), a control antibiotic that was previously shown to induce OH production in E. coli and S. aureus21,23, was also reduced in ΔcssS and ΔcssR mutants, although to a lower extent than the PGRPs-induced effect. These results indicate that both membrane depolarization and •OH production induced by PGRPs is mediated through the CssR-CssS two-component system.
PGRPs also caused rapid high-level induction of htrA mRNA (HtrA is membrane-bound protease directly regulated by CssR22) in WT B. subtilis, but not in ΔcssS and ΔcssR mutants (Fig. 3d). These results demonstrate direct CssS- and CssR-dependent activation of CssR-CssS two-component system by PGRPs and indicate that PGRPs activate the regulator (the transcription factor CssR) through the sensor (CssS), because PGRP-induced htrA transcription is inhibited in the ΔcssS mutant.
To test whether the CssR-CssS two-component system is responsible for bactericidal activity of PGRPs, we then compared the sensitivity to PGRPs-induced killing of ΔcssS and ΔcssR mutants and isogenic WT B. subtilis. Whereas WT B. subtilis was readily killed by all four PGRPs (PGLYRP-1, PGLYRP-3, PGLYRP-4, and PGLYRP-3:4), ΔcssS and ΔcssR mutants were 10 to 1000 times less sensitive to killing by all four PGRP proteins (Fig. 4a). Both WT and all mutant bacteria had similar growth rates and cell separation in the presence of a control protein (BSA). These results indicate that the CssR-CssS two-component system in B. subtilis is required for bacterial killing by all four PGRP proteins.
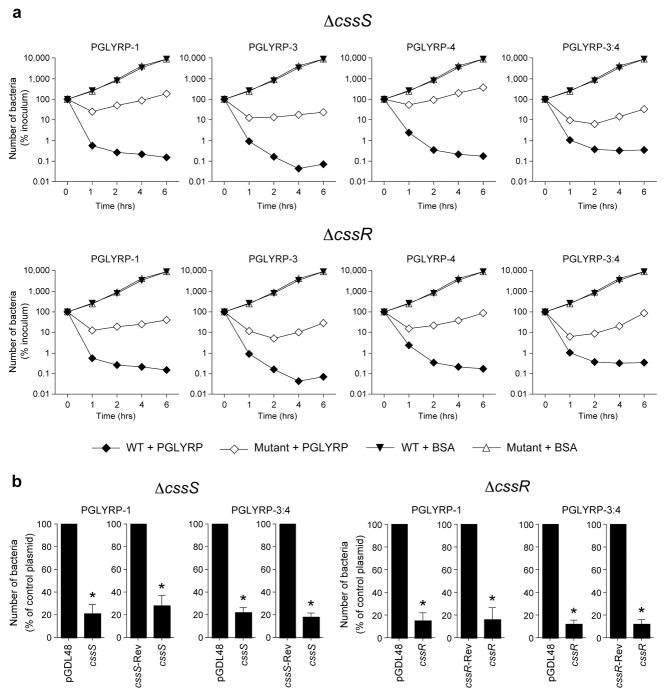
Figure 4. PGRPs kill B. subtilis through the CssR-CssS two-component system.
(a) Killing of WT B. subtilis (closed symbols) or ΔcssS or ΔcssR mutants (open symbols) incubated with the indicated PGRPs (100 μg ml−1) or BSA (control) measured by colony counts; means of 3 experiments; the average SEM were 5 – 19% and are not shown; all mutants had significantly reduced sensitivity to killing by all PGRPs (P<0.05 versus WT, t-test). (b) Complementation of PGRP-induced bacterial killing by cssS and cssR inΔcssS or ΔcssR mutants transfected with an empty vector (pGDL48) or pGDL48 containing cssS in forward orientation (cssS), cssS in reverse orientation (cssS-Rev), cssR in forward orientation (cssR), or cssR in reverse orientation (cssR-Rev) and incubated with PGLYRP-1 or PGLYRP-3:4 (100 μg ml−1) for 4 h; the results are % of surviving bacteria in cssS- or cssR-transfected groups compared to groups transfected with control plasmids (empty vector or reverse sequence); means of 3 experiments ± SEM; *, P<0.05 versus control plasmid (t-test).
To confirm the role of the CssR-CssS two-component system in PGRP killing we performed complementation experiments. ΔcssS and ΔcssR mutants complemented with cssS-or cssR-expressing pGDL48 plasmids were significantly more sensitive to killing by PGRP proteins than the same mutants transfected with an empty vector or with the same genes inserted in a reverse orientation (Fig. 4b). The role of the CssR-CssS two-component system in bacterial killing is further supported by lower sensitivity to PGRP killing of ΔhtrA mutant, which is deficient in HtrA protease regulated by the CssR-CssS system (Supplementary Fig. 8). These results confirm that the CssR-CssS two-component system is required for the bactericidal effect of PGRP proteins.
PGRPs kill E. coli through CpxA-CpxR two-component system
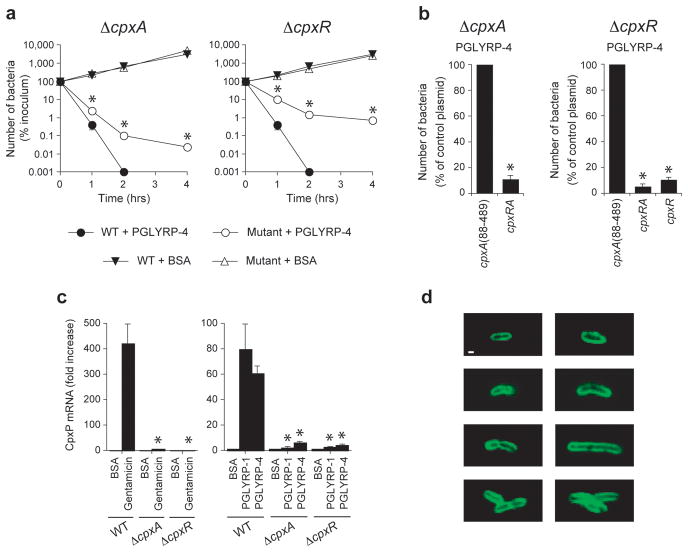
To determine whether PGRPs also kill Gram-negative bacteria through the protein-sensing two-component systems, we compared the sensitivity to PGRPs of WT E. coli and its isogenic ΔcpxA and ΔcpxR mutants. E. coli CpxA-CpxR is a functional homolog of B. subtilis CssR-CssS two-component system – they both sense extracytoplasmic misfolded proteins and induce membrane depolarization, •OH production, and stress response22,23, this paper. WT E. coli bacteria were readily killed by PGRPs (as expected), whereas ΔcpxA and ΔcpxR mutants were significantly more resistant to PGRP killing (Fig. 5a). These results indicate that both CpxA and CpxR are required for efficient killing of E. coli by PGRP proteins.
Figure 5. PGRPs kill E. coli through the CpxA-CpxR two-component system and bind to the entire outer membrane.
(a) Killing of WT E. coli or ΔcpxA or ΔcpxR mutants incubated with 50 μg ml−1 PGLYRP-4 or BSA (control); means ± SEM of 3 experiments (SEM were smaller than the size of the symbols); *, P<0.02 (t-test) mutants versus WT; similar results were obtained with PGLYRP-1 (not shown). (b) Complementation of PGRP-induced bacterial killing by cpxA and cpxR in ΔcpxA or ΔcpxR mutants transfected with a control plasmid cpxA(88–489) (containing periplasmic loop of cpxA), or cpxRA (plasmid expressing both CpxR and CpxA), or cpxR (plasmid expressing CpxR only); the results are % of surviving bacteria at 2 h in cpxRA- or cpxR-transfected groups compared to groups transfected with the control plasmid, cpxA(88–489); means of 3 experiments ± SEM; *, P<0.002 versus control plasmid (t-test). (c) CpxP mRNA expression in WT E. coli or ΔcpxA or ΔcpxR mutants incubated with PGLYRP-1, PGLYRP-4, or BSA (100 μg ml−1), or gentamicin (5 μg ml−1) for 60 min measured by qRT-PCR; means of 3 experiments ± SEM; *, P<0.005 (t-test) mutants versus WT. (d) Binding of PGLYRP-1 to the entire outer membrane in WT E. coli detected by confocal microscopy with FITC-mAbs to V5; merged stacks of representative single cells, dividing cells, and cells separating after division are shown; there was no cytoplasmic staining when individual scans were analyzed and similar results were obtained with PGLYRP-4 (not shown); bar = 1 μm.
To confirm the role of the CpxA-CpxR two-component system in PGRP killing we performed complementation experiments. ΔcpxA and ΔcpxR mutants complemented with ΔcpxA and ΔcpxR-expressing plasmids were significantly more sensitive to killing by PGRP than the same mutants transfected with a control plasmid cpxA(88–489) containing only the periplasmic loop of cpxA (Fig. 5b). These results confirm that the CpxA-CpxR two-component system is required for the bactericidal effect of PGRP proteins.
To test direct activation of the CpxA-CpxR two-component system by PGRPs, we then determined whether PGRPs increase the expression of cpxP, which is a stress-response gene directly regulated by CpxR23,24. PGLYRP-1 and PGLYRP-4 (as well as gentamicin, used as a positive control) caused rapid high-level induction of cpxP mRNA in WT E. coli, but not in ΔcpxA and ΔcpxR mutants (Fig. 5c). These results demonstrate CpxA- and CpxR-dependent activation of the CpxA-CpxR two-component system by PGRPs and indicate that PGRPs activate the regulator (the transcription factor CpxR) through the sensor (CpxA), because PGRP-induced cpxP transcription is abolished in the ΔcpxA mutant.
To address how PGRPs access the CpxA-CpxR two-component system, we demonstrated by immunofluorescence and confocal microscopy that PGRPs bind to the entire outer membrane at all stages of growth in E. coli, but do not enter the cytoplasm (Fig. 5d). These results indicate that in Gram-negative bacteria PGRPs bind to the outer membrane, where they can be sensed by the CpxA-CpxR two-component system, consistent with the ability of the E. coli CpxA-CpxR to sense proteins in the outer membrane25.
DISCUSSION
Our results reveal a previously unknown mechanism of bacterial killing by mammalian innate immunity proteins (PGRPs). In Gram-positive bacteria, PGRPs enter the cell wall at the site of daughter cell separation during cell division, bind to cell wall peptidoglycan in the vicinity of cell membrane, and activate the bacterial CssR-CssS two component system, which triggers bacterial killing by inducing membrane depolarization and •OH production in the cytoplasm (Fig. 6). This process is accompanied by cessation of all major intracellular biosynthetic reactions, likely because of the lack of membrane-potential-dependent generation of energy. PGRPs kill Gram-negative bacteria by binding to their outer membrane and activating the functionally homologous CpxA-CpxR two-component system. CssR-CssS and CpxA-CpxR two-component systems are designed to detect and dispose of extracellular misfolded bacterial proteins at the cell membrane-cell wall interface, after these proteins have been exported from the cell22,23, and in Gram-negative bacteria CpxA-CpxR also detects proteins in the outer membrane25.
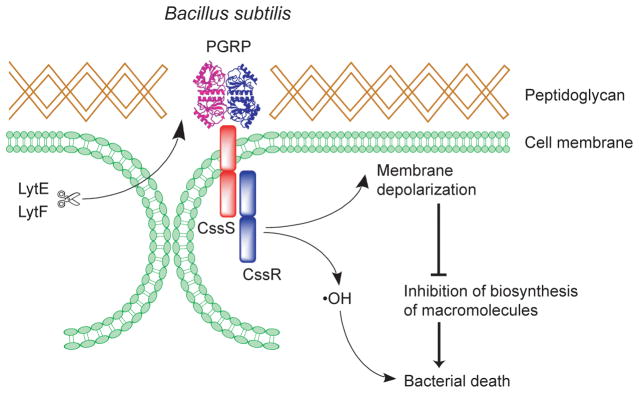
Figure 6. Mammalian PGRPs exploit bacterial defense mechanism to kill bacteria.
In Gram-positive bacteria PGRPs bind to peptidoglycan in bacterial cell wall at the cell separation sites (the sites of hydrolysis by LytE and LytF, which in B. subtilis are the enzymes that separate daughter cells after cell division). PGRPs then activate CssR-CssS two-component system that senses extracytoplasmic misfolded proteins. CssR-CssS triggers membrane depolarization and •OH production, which results in inhibition of macromolecule synthesis and bacterial death.
This could be a general mechanism of bacterial killing by other antibacterial proteins that bind to the cell wall or outer membrane, but do not enter the cytosol, such as peptidoglycan-binding bactericidal RegIIIγ C-type lectin1, galectins2, other cell-wall-binding lectins, or even lysozyme, which has an antibacterial effect that is independent of its peptidoglycan-hydrolyzing activity26.
Bacteria have many two-component systems, which consist of a sensor and a regulator and enable bacteria to quickly react to changing conditions in the environment. The CssR-CssS and CpxA-CpxR two-component systems of B. subtilis and E. coli are functional homologs and similar two-component systems are likely present in most, if not all, bacteria22,23. These systems are designed to respond to stress, which induces misfolding of bacterial proteins. These misfolded proteins are exported from the cell, detected by these two-component systems, and disposed of by associated proteases, such as HtrA in B. subtilis22. CssR-CssS responds only to membrane-translocated extracytoplasmic (but not cytoplasmic) misfolded or aggregated proteins, both self and foreign, irrespective of the cause that leads to the protein accumulation near the bacterial membrane27,28.
Binding of PGRPs to peptidoglycan induces a structural change in PGRPs that locks peptidoglycan in the protein’s bindings groove14, which makes the binding irreversible and accounts for persistence of peptidoglycan-bound PGRPs in the cell wall. PGRPs also have a hydrophobic region on the opposite side from the peptidoglycan-binding groove, and thus they are either sensed by CssR-CssS as misfolded proteins (which usually have exposed hydrophobic regions) or aggregated proteins (since misfolded proteins often aggregate and binding to peptidoglycan induces PGRP aggregation29).
The protein-sensing two-component systems not only trigger removal of the misfolded proteins, but also signal into the cell to activate a repair and defense response, and induce membrane depolarization and production of toxic •OH23. If stress is sustained or its level is too high, bacteria die. Perhaps such a suicidal mechanism evolved as advantageous to the entire bacterial population, because bacteria that are damaged beyond their ability to repair the damage would promptly die and not use up scarce resources. Thus mammalian PGRPs exploit this bacterial defense/suicidal mechanism to kill bacteria.
Because Gram-positive bacteria have a thick cell wall, to activate the two-component system in the cytoplasmic membrane PGRPs need to traverse the cell wall and bind to peptidoglycan near the cell membrane. This is accomplished at the sites of separation of newly formed daughter cells, which in B. subtilis is accomplished by dedicated peptidoglycan hydrolases, LytE, LytF, and CwlS18,19. PGRPs co-localize with these enzymes at the cell-separation sites, which correspond to the digestion sites of these enzymes observed by electron microscopy30,31 (discussed in Supplementary Information).
The initial interaction of PGRPs with Gram-negative bacteria is different than with Gram-positive bacteria and involves the outer membrane. Mammalian PGRPs have dual specificity and activity that involves both peptidoglycan-binding groove and additional binding sites outside peptidoglycan-binding groove29,32. PGRPs’ hydrophobic region on the opposite face of the molecule from the peptidoglycan-binding groove may be involved in binding additional ligands33, such as hydrophobic structures in the outer membrane of Gram-negative bacteria or the two-component systems. Thus, in addition to peptidoglycan, mammalian PGRPs also bind LPS11,34 (the main component of Gram-negative outer membrane) and exogenous LPS inhibits killing of Gram-negative bacteria by PGRPs11,12 in a similar way as exogenous peptidoglycan inhibits killing of Gram-positive bacteria by PGRPs10. We now show that PGRPs bind to the entire outer membrane in E. coli, which induces activation of the CpxA-CpxR two-component system and bacterial killing. This proposed killing model is consistent with the previously demonstrated ability of CpxA-CpxR to detect proteins in the outer membrane25, although it is not clear exactly how outer-membrane-bound PGRPs interact with CpxA-CpxR. Thus, although the initial steps of interaction of PGRPs with Gram-positive and Gram-negative bacteria are different, the killing mechanism of PGRPs is common for all bacteria and depends on the activation of the protein-sensing two-component systems.
These two-component systems may become targets for development of new antibacterial therapies, for example by designing peptides modeled on PGRPs that would be bactericidal, non-immunogenic, less expensive to manufacture than full-length proteins, and active against antibiotic-resistant bacteria (because full-length PGRPs kill multi-drug-resistant bacteria10,12).
METHODS
Materials
All antibacterial compounds were used at or above minimum bactericidal concentrations. Human PGLYRP-1, PGLYRP-3, PGLYRP-4, and PGLYRP-3:4 (PGLYRP-3:PGLYRP-4 heterodimer), control proteins, enzymes, antibiotics, bacterial strains, mutants, and plasmids are described in Supplementary Information.
Peptidoglycan synthesis
The rate of peptidoglycan biosynthesis in exponentially growing S. aureus was assayed using a three-step procedure that in the same aliquot measures total inhibition of peptidoglycan synthesis and determines whether peptidoglycan synthesis is inhibited at the transpeptidation step, transglycosylation step, or any of the earlier cytoplasmic steps. Total inhibition of peptidoglycan synthesis (at any step) was manifested by a decrease in 3H-GlcNAc incorporation into insoluble cell wall peptidoglycan. Inhibition of transpeptidation was manifested as inhibition of 3H-GlcNAc incorporation into insoluble cell wall peptidoglycan and simultaneous secretion into the medium of newly synthesized (3H-GlcNAc-labeled) polymeric uncrosslinked peptidoglycan. Inhibition of transglycosylation was manifested as inhibition of 3H-GlcNAc incorporation into insoluble cell wall peptidoglycan and simultaneous accumulation of 3H-GlcNAc-labeled undecaprenyl-phosphate-GlcNAc-MurNAc-peptide (lipid II) in the cell membrane (see Supplementary Information).
Protein, RNA, and DNA synthesis
The rate of protein, RNA, and DNA biosynthesis in S. aureus was measured by following incorporation of 35S-methionine + 35S-cysteine into proteins, 3H-uridine into RNA, or 3H-thymidine into DNA, as described in Supplementary Information. Our assays were selectively sensitive to appropriate antibiotics, i.e., only chloramphenicol and rifampin inhibited protein synthesis (rifampin inhibits protein synthesis because RNA and protein synthesis are coupled in bacteria), only rifampin inhibited RNA synthesis, and only ciprofloxacin inhibited DNA synthesis (Fig. 2a – c).
Bactericidal assay
Bactericidal assays were performed on exponentially growing bacteria as described previously10,12 in the same medium as for peptidoglycan synthesis, without or with 0.525 M sucrose (which prevents osmotic lysis). All differences in bacterial numbers >1 log10 were statistically significant at P<0.05 determined by the t-test.
Localization of PGRPs, vancomycin, LytE, and LytF in bacterial cells
Localization of PGLYRP-4 or BSA (negative control) labeled with Alexa Fluor 594 (red), or unlabeled PGLYRP-1 and PGLYRP-4 (detected with anti-V5 mAbs labeled with Alexa Fluor 594 or FITC), and of vancomycin labeled with BODIPY-FL (green, used to visualize the sites of peptidoglycan synthesis) was determined in exponentially growing B. subtilis, S. aureus, L. monocytogenes, or E. coli using confocal microscopy as described in Supplementary Information. Co-localization of PGRPs with LytE and LytF in B. subtilis was determined by confocal microscopy with anti-V5 (PGRPs) and anti-LytE and LytF antibodies as described in Supplementary Information.
Membrane permeabilization
Membrane permeabilization in S. aureus and B. subtilis was studied as described previously10 using membrane-nonpermeable DNA-binding fluorescent dye SYTOX-Green (2 μM, Molecular Probes) under the same conditions as for the bactericidal assays without or with 0.525 M sucrose at 37°C.
Hydrolysis of peptidoglycan and bacteriolysis
Hydrolysis of insoluble peptidoglycan was followed spectrophotometrically, hydrolysis of biotin-labeled soluble uncrosslinked peptidoglycan was determined by Western blotting, hydrolysis of synthetic peptidoglycan fragments was studied by mass spectrometry, and bacteriolytic activity was measured spectrophotometrically, as described in Supplementary Information.
Membrane depolarization and •OH production
Membrane depolarization and •OH production were measured by flow cytometry with fluorescent probes, DiBAC4(3) (bis-(1,3-dibutylbarbituric acid)trimethine oxanol, from InVitrogen, 2 μg ml−1, 15-min staining) and HPF (3′(p-hydroxyphenyl) fluorescein from Molecular Probes, 5 μM present during incubation), respectively, as previously described21,35, which are standard methods for these measurements in bacteria. Briefly, B. subtilis 168 or S. aureus (5 × 106 ml−1) were incubated at 37°C under the same conditions as for bactericidal assays with PGRPs, negative control (BSA or recombinant mouse albumin), or positive controls (CCCP, carbonyl cyanide 3-chlorophenylhydrazone from Sigma, 10 μM, for membrane depolarization, or gentamicin, 5–15 μg ml−1, for •OH production). At 0, 1, 2, 3, and 4 h, bacterial fluorescence was measured using Miltenyi MACSQuant flow cytometer. Increases in DiBAC4(3) and HPF fluorescence reflect membrane depolarization and intracellular •OH production, respectively. The data were collected as mean fluorescence intensity and presented as mean ratios of treated to BSA control fluorescence from 3 to 6 experiments, and the significance of differences was determined by t-test.
Gene expression
Bacteria were treated with PGRPs, control proteins, or antibiotics as described above. RNA was isolated using RiboPure-Bacteria kit from Ambion, and the amounts of mRNA were measured using quantitative reverse transcription real-time PCR (qRT-PCR) as described in Supplementary Information.
Complementation of ΔcssS, Delta;cssR, ΔhtrA, ΔcpxA, and ΔcpxR mutants
cssS, cssR and htrA were amplified from B. subtilis DNA and sub-cloned into pGDL48 low-copy expression plasmid36 in the forward or reverse (negative control) orientation. The entire cpxRA operon, cpxR, or cpxA(88–489) periplasmic loop (negative control) were amplified from E. coli and sub-cloned into the pCR2.1 vector. The plasmids were used for complementation of B. subtilisΔcssS, ΔcssR and ΔhtrA mutants or E. coli ΔcpxA and ΔcpxR mutants, respectively, in bactericidal assays with PGRP proteins as described in Supplementary Information.
Supplementary Material
Acknowledgments
We are grateful to J. M. van Dijl, O. P. Kuipers, V. P. Kontinen, and their associates J. Zweers and S. Holsappel (University of Groningen, Groningen, Netherlands, and National Institute of Health and Welfare, Helsinki, Finland); to J. Sekiguchi (Shinshu University, Nagano, Japan); and to S. J. Foster (University of Sheffield, Sheffield, UK) for providing B. subtilis mutants; to J. J. Collins and M. A. Kohanski (Boston University, Boston, MA) for providing E. coli mutants; to J. M. van Dijl and his associates T. Kouwen and M. Sibbald (University of Groningen, Groningen, Netherlands) for pGDL48 plasmid and its sequence; to M. Wang for analyzing samples by mass spectrometry; and to Huvepharma, Inc for providing moenomycin. This work was supported by USPHS Grants from NIH AI073290 and AI028797 to R.D. and D.G., and GM061761 to G.-J.B.
Footnotes
AUTHOR CONTRIBUTIONS
D.R.K., M.W, D.G., and R.D. designed the experiments, D.R.K., M.W, and R.D. performed the experiments, L.-H.L. and D.R.K. obtained and purified the proteins, G.-J.B. synthesized muramyl peptides, and R.D. wrote the manuscript.
References
- 1.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stowell SR, et al. Innate immune lectins kill bacteria expressing blood group antigen. Nature Med. 2010;16:295–301. doi: 10.1038/nm.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dziarski R, Gupta D. The peptidoglycan recognition proteins (PGRPs) Genome Biol. 2006;7:232.1–13. doi: 10.1186/gb-2006-7-8-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Royet J, Dziarski R. Peptidoglycan Recognition Proteins: pleiotropic sensors and effectors of antimicrobial defenses. Nature Rev Microbiol. 2007;5:264–277. doi: 10.1038/nrmicro1620. [DOI] [PubMed] [Google Scholar]
- 5.Dziarski R, Gupta D. Mammalian PGRPs: novel antibacterial proteins. Cell Microbiol. 2006;8:1056–1069. doi: 10.1111/j.1462-5822.2006.00726.x. [DOI] [PubMed] [Google Scholar]
- 6.Kang D, Liu G, Lundstrom A, Gelius E, Steiner H. A peptidoglycan recognition protein in innate immunity conserved from insects to humans. Proc Natl Acad Sci USA. 1998;95:10078–10082. doi: 10.1073/pnas.95.17.10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu C, Xu Z, Gupta D, Dziarski R. Peptidoglycan recognition proteins: a novel family of four human innate immunity pattern recognition molecules. J Biol Chem. 2001;276:34686–34694. doi: 10.1074/jbc.M105566200. [DOI] [PubMed] [Google Scholar]
- 8.Tydell CC, Yount N, Tran D, Yuan J, Selsted M. Isolation, characterization, and antimicrobial properties of bovine oligosaccharide-binding protein. J Biol Chem. 2002;277:19658–19664. doi: 10.1074/jbc.M200659200. [DOI] [PubMed] [Google Scholar]
- 9.Dziarski R, Platt KA, Gelius E, Steiner H, Gupta D. Defect in neutrophil killing and increased susceptibility to infection with non-pathogenic Gram-positive bacteria in peptidoglycan recognition protein-S (PGRP-S)-deficient mice. Blood. 2003;102:689–697. doi: 10.1182/blood-2002-12-3853. [DOI] [PubMed] [Google Scholar]
- 10.Lu X, et al. Peptidoglycan recognition proteins are a new class of human bactericidal proteins. J Biol Chem. 2006;281:5895–5907. doi: 10.1074/jbc.M511631200. [DOI] [PubMed] [Google Scholar]
- 11.Tydell CC, Yuan J, Tran P, Selsted ME. Bovine peptidoglycan recognition protein-S: antimicrobial activity, localization, secretion, and binding properties. J Immunol. 2006;176:1154–1162. doi: 10.4049/jimmunol.176.2.1154. [DOI] [PubMed] [Google Scholar]
- 12.Wang M, et al. Human peptidoglycan recognition proteins require zinc to kill both Gram-positive and Gram-negative bacteria and are synergistic with antibacterial peptides. J Immunol. 2007;178:3116–3125. doi: 10.4049/jimmunol.178.5.3116. [DOI] [PubMed] [Google Scholar]
- 13.Gelius E, Persson C, Karlsson J, Steiner H. A mammalian peptidoglycan recognition protein with N-acetylmuramoyl-L-alanine amidase activity. Biochem Biophys Res Commun. 2003;306:988–994. doi: 10.1016/s0006-291x(03)01096-9. [DOI] [PubMed] [Google Scholar]
- 14.Wang ZM, et al. Human peptidoglycan recognition protein-L is an N-acetylmuramoyl-L-alanine amidase. J Biol Chem. 2003;278:49044–49052. doi: 10.1074/jbc.M307758200. [DOI] [PubMed] [Google Scholar]
- 15.Cho S, et al. Structural insights into the bactericidal mechanism of human peptidoglycan recognition proteins. Proc Natl Acad Sci USA. 2007;104:8761–8766. doi: 10.1073/pnas.0701453104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hancock REW, Sahl HG. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nature Biotech. 2006;24:1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 17.Peschel A, Sahl HG. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nature Rev Microbiol. 2006;4:529–536. doi: 10.1038/nrmicro1441. [DOI] [PubMed] [Google Scholar]
- 18.Fukushima T, et al. A new D,L-endopeptidase gene product, YojL (renamed CwlS), plays a role in cell separation with LytE and LytF in Bacillus subtilis. J Bacteriol. 2006;188:5541–5550. doi: 10.1128/JB.00188-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamamoto H, Kurosawa S, Sekiguchi J. Localization of the vegetative cell wall hydrolases LytC, LytE, and LytF on the Bacillus subtilis cell surface and stability of these enzymes to cell wall-bound or extracellular proteases. J Bacteriol. 2003;185:6666–6677. doi: 10.1128/JB.185.22.6666-6677.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blackman SA, Smith TJ, Foster SJ. The role of autolysins during vegetative growth of Bacillus subtilis 168. Microbiology. 1998;144:73–82. doi: 10.1099/00221287-144-1-73. [DOI] [PubMed] [Google Scholar]
- 21.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 22.Hyyryläinen HL, et al. A novel two-component regulatory system in Bacillus subtilis for the survival of severe secretion stress. Mol Microbiol. 2001;41:1159–1172. doi: 10.1046/j.1365-2958.2001.02576.x. [DOI] [PubMed] [Google Scholar]
- 23.Kohanski MA, Dwyer DJ, Wierzbowski J, Cottarel G, Collins JJ. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell. 2008;135:679–690. doi: 10.1016/j.cell.2008.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DiGiuseppe PA, Silhavy TJ. Signal detection and target gene induction by the CpxRA two-component system. J Bacteriol. 2003;185:2432–2440. doi: 10.1128/JB.185.8.2432-2440.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Danese PN, Snyder WB, Cosma CL, Davis LJ, Silhavy TJ. The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, DegP. Genes Dev. 1995;9:387–398. doi: 10.1101/gad.9.4.387. [DOI] [PubMed] [Google Scholar]
- 26.Nash JA, Ballard TN, Weaver TE, Akinbi HT. The peptidoglycan-degrading property of lysozyme is not required for bactericidal activity in vivo. J Immunol. 2006;177:519–526. doi: 10.4049/jimmunol.177.1.519. [DOI] [PubMed] [Google Scholar]
- 27.Darmon E, et al. A novel class of heat and secretion stress-responsive genes is controlled by the autoregulated CssRS two-component system of Bacillus subtilis. J Bacteriol. 2002;184:5661–5671. doi: 10.1128/JB.184.20.5661-5671.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Westers H, et al. The CssRS two-component regulatory system controls a general secretion stress response in Bacillus subtilis. FEBS J. 2006;273:3816–3827. doi: 10.1111/j.1742-4658.2006.05389.x. [DOI] [PubMed] [Google Scholar]
- 29.Lim JH, et al. Structural basis for preferential recognition of diaminopimelic acid-type peptidoglycan by a subset of peptidoglycan recognition proteins. J Biol Chem. 2006;281:8286–8295. doi: 10.1074/jbc.M513030200. [DOI] [PubMed] [Google Scholar]
- 30.Touhami A, Jericho MH, Beveridge TJ. Atomic force microscopy of cell growth and division in Staphylococcus aureus. J Bacteriol. 2004;186:3286–3295. doi: 10.1128/JB.186.11.3286-3295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamada S, et al. An autolysin ring associated with cell separation of Staphylococcus aureus. J Bacteriol. 1996;178:1565–1571. doi: 10.1128/jb.178.6.1565-1571.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang CI, Chelliah Y, Borek D, Mengin-Lecreulx D, Deisenhofer J. Structure of tracheal cytotoxin in complex with a heterodimeric pattern-recognition receptor. Science. 2006;311:1761–1764. doi: 10.1126/science.1123056. [DOI] [PubMed] [Google Scholar]
- 33.Kim MS, Byun M, Oh BH. Crystal structure of peptidoglycan recognition protein LB from Drosophila melanogaster. Nat Immunol. 2003;4:787–793. doi: 10.1038/ni952. [DOI] [PubMed] [Google Scholar]
- 34.Liu C, Gelius E, Liu G, Steiner H, Dziarski R. Mammalian peptidoglycan recognition protein binds peptidoglycan with high affinity, is expressed in neutrophils, and inhibits bacterial growth. J Biol Chem. 2000;275:24490–24499. doi: 10.1074/jbc.M001239200. [DOI] [PubMed] [Google Scholar]
- 35.Dwyer DJ, Kohanski MA, Hayete B, Collins JJ. Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol Syst Biol. 2007;3:91. doi: 10.1038/msb4100135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meijer WJ, et al. The endogenous Bacillus subtilis (natto) plasmids pTA1015 and pTA1040 contain signal peptidase-encoding genes: identification of a new structural module on cryptic plasmids. Mol Microbiol. 1995;17:621–631. doi: 10.1111/j.1365-2958.1995.mmi_17040621.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.