Abstract
Hypersaline environments harbour the highest number of viruses reported for aquatic environments. In crystallizer ponds from solar salterns, haloviruses coexist with extremely halophilic Archaea and Bacteria and present a high diversity although little is known about their activity. In this work, we analyzed the viral expression in one crystallizer using a metatranscriptomic approach in which clones from a metaviromic library were immobilized in a microarray and used as probes against total mRNA extracted from the hypersaline community. This approach has two advantages: (i) it overcomes the fact that there is no straightforward, unambiguous way to extract viral mRNA from bulk mRNAs and (ii) it makes the sequencing of all mRNAs unnecessary. Transcriptomic data indicated that the halovirus assemblage was highly active at the time of sampling and the viral groups with the highest expression levels were those related to high GC content haloarchaea and Salinibacter representatives, which are minor components in the environment. Moreover, the changes in the viral expression pattern and in the numbers of free viral particles were analyzed after submitting the samples to two stress conditions: ultraviolet-radiation and dilution. Results showed that Archaea were more sensitive than Bacteria to these stress conditions. The overexpression in the predicted archaeal virus fraction raised and the total numbers of free viruses increased. Furthermore, we identified some very closely related viral clones, displaying single-nucleotide polymorphisms, which were expressed only under certain conditions. These clones could be part of very closely related virus genomes for which we propose the term ‘ecoviriotypes'.
Keywords: virus, metatranscriptome, metagenome, halovirus, haloquadratum, salinibacter
Introduction
Viruses from hypersaline environments are one of the most abundant biological entities in aquatic systems (Guixa-Boixareu et al., 1996). Although some halophages have been characterized so far by culture-dependent methods (Dyall-Smith et al., 2003, www.haloarchaea.com and references therein), little is known about their diversity and activity in natural samples (NSs). Previous studies on haloviruses in NSs using transmission electron microscopy revealed that the numbers of virus-like particles (VLPs) increased along the salinity gradient and their abundance was correlated with the numbers of cells (Guixa-Boixareu et al., 1996). Pulsed-field gel electrophoresis was also applied to analyze the size of viral genomes in hypersaline environments (Díez et al., 2000; Jiang et al., 2003; Sandaa et al., 2003; Santos et al., 2007) showing that the viral assemblage changes from low to high salinity ponds, with genome ‘populations' between 10 and 533 kb. Metagenomic analyses (Santos et al., 2007, 2010) of the CR30 crystallizer from the multipond solar salterns of Santa Pola (Alicante, Spain) revealed that the halophilic viral assemblage was highly diverse as indicated by the analyses of their sequences (referred here as halophilic viral sequences, HVSs) which were classified according to their dinucleotide frequency and GC content (Santos et al., 2010). Genomic sequences from the extremely halophilic Haloquadratum walsbyi, Salinibacter ruber (Archaea and Bacteria, respectively, which coexist with haloviruses in the crystallizer pond) and some high GC content haloarchaea, such as Halobacterium salinarum, Haloarcula marismortui and Natronomonas pharaonis, were also included in the analysis. The resulting classification scheme grouped all the sequences into five different clusters (HVS-1 to HVS-5, Figure 1a). The genomic sequences of high GC haloarchaea, S. ruber, and Hqr. walsbyi were grouped within the clusters HVS-1, HVS-2 and HVS-4, respectively, which included HVS of similar GC content and dinucleotide frequencies. According to this grouping schema, we suggested that the high GC content clusters HVS-1 and HVS-2 could correspond to viruses that might infect, respectively, high GC haloarchaea and Salinibacter representatives, while the low GC content cluster HVS-4 would include viruses infecting Haloquadratum lineages (Santos et al., 2010).
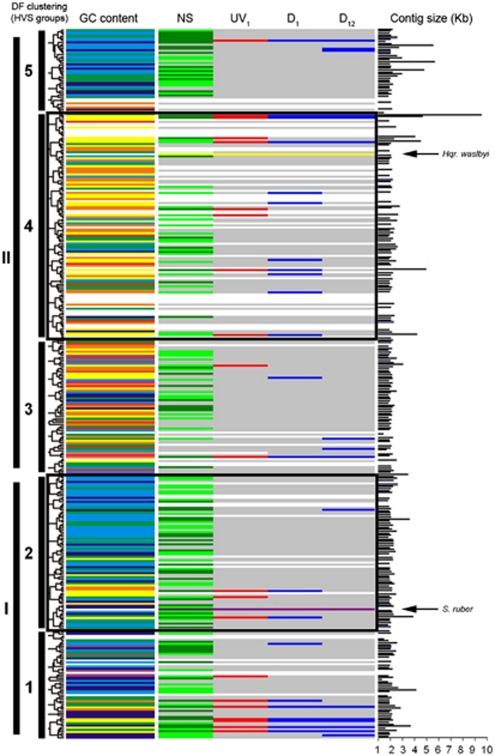
Figure 1.
Schematic representation showing the expression and overexpression of viral contigs. Clustering of HVSs is based on dinucleotide frequency analysis (DF). The GC content of each viral contig is also shown in colours (according to the rainbow spectrum, from low to high GC; for example, yellow is 45–50% GC and dark blue is 60–65%, as in Santos et al., 2010). The column labelled as NS shows the expression of viral contigs in the sample taken in May 2007, where contigs highly expressed are coloured in light green and contigs very highly expressed are indicated in dark green. Contigs marked in red in column UV1 are the contigs overexpressed 1 h after the UV-radiation treatment. Contigs marked in blue in columns D1 and D12 are the contigs overexpressed 1 and 12 h after the application of the osmotic shock. Black bars indicate the size of each contig, in kb. Arrows in panels show the position of Hqr. walsbyi and S. ruber genomes, according to their dinucleotide frequency (their GC content is also indicated by yellow and purple lines, respectively).
Once the viral ‘community' was analyzed by the above mentioned metagenomic study, we took a metatranscriptomic approach using microarray technology to detect and quantify the expression of viral genes. This technology has been applied in the field of microbial ecology to study the composition of microbial communities using ribosomal RNA (rRNA)-targeted probes (for example, Garrido et al., 2008) and the expression of selected genes (Parro et al., 2007; Moreno-Paz et al., 2010), which have provided information about the functional relevance of the diversity in a given ecosystem. In viruses, as there are no ‘universal markers', microarrays have been basically used as a tool for studies within virus families (probes designed for the genes from one viral strain can be used to study their presence or absence in related viral genomes (Willson et al., 2005; Allen and Wilson, 2006; Chou et al., 2006; Allen et al., 2006, 2007; Webster et al., 2009)) and analyses of the viral dynamics in the infant gut using metaviromic DNA as probes (Breitbart et al., 2008). Recently, Snyder et al. (2010) used microarrays designed based on CRISPR spacer sequences to identify viruses in hot springs. The use of microarrays for detecting viral expression in natural communities is, however, very scarce. To the best of our knowledge, the only examples on such an approach are the works by Kunin et al. (2008) who monitored a sludge bioreactor at three time points spanning 3 months using expression arrays constructed from predicted genes from both phage and bacterial metagenomes, and the use of microarrays to assess the diversity and activity of viruses related to infections in humans, allowing the detection of both known and novel pathogenic viral variants (Wang et al., 2002; Kistler et al., 2007).
Viral activity in natural communities has been traditionally measured using a few techniques. The counting of infected cells by transmission electron microscopy is direct evidence that viruses infect prokaryotes. Viral activity can also be measured by calculating the rate of production of viruses using different techniques: by measuring tritiated thymidine incorporation into viral DNA, by incorporation of fluorescently labelled viruses added into a sample, by inhibiting viral production and comparing with a non-inhibited control or, finally, by diluting the sample before directly counting virus production (Fuhrman, 1999; Wilhelm et al., 2002; Weinbauer, 2004). Viral decay rates can also be analyzed by several techniques, for example, incubating NSs after adding a toxic agent against the cellular fraction (Wommack and Colwell, 2000). However, these techniques consider the whole viral assemblage as a ‘black box' and do not provide information regarding the expression of different genotypes within the viral assemblage.
In this work, we have constructed microarrays containing clones from the previously characterized metavirome of the above mentioned CR30 crystallizer (Santos et al., 2010), which were used as probes for hybridization against complementary DNAs (cDNAs) obtained from total environmental RNAs. The same NSs were used for constructing the viral metagenomic libraries analyzed in Santos et al. (2010), and for the microarray construction, nucleic acid extractions and stress experiments described here. Thus, we were able to identify viral transcripts from the bulk mRNAs and ascertain which components of the viral community were active under different conditions at the time of sampling. In addition, we analyzed the viral overexpression when the NSs were submitted to two stress conditions (ultraviolet (UV)-radiation and dilution). Changes in the composition of the stressed prokaryotic communities were monitored by denaturing gradient gel electrophoresis (DGGE) and fluorescence in situ hybridization (FISH), while changes in the numbers of free viral particles were detected by SYBR-green staining. Our results indicated that the viral halophilic ‘community' that inhabits the crystallizers was highly active at the time of sampling and that stress treatments had different effects on the prokaryotic and viral assemblages.
Materials and methods
Sampling
Hypersaline water samples (named ‘NS') were collected in May 2007 and May 2009 from a crystallizer (CR30) located in the multipond solar saltern ‘Bras del Port' (Santa Pola, Alicante, Spain, 38°12′N, 0°36′W). The samples were taken a few centimetres below the surface using acid-washed polypropylene bottles. Salinity was measured in situ with a hand refractometer (Sper Scientific, Scottsdale, AZ, USA).
Microarray construction
Purification of the viral particles contained in 2 l of the NS taken in 2007 and viral DNA extraction, cloning and PCR amplification of viral inserts using vector primers were carried out as described in Santos et al. (2010). Purified PCR products were dried in a Speed Vac Concentrator (Savant, Thermo Fisher Scientific, Waltham, MA, USA), resuspended at 50–200 ng μl–1 in microSpotting Solution Plus 1 × (Arrayit Corp., Sunnyvale, CA, USA) and used for the viral microarray construction. Spotting was carried out with the MicroGrid-TAS II Arrayer (Genomic Solutions, Huntingdon, UK) at 22 °C and 50–60% relative humidity on epoxy-substrate slides (Arrayit Corp.) according to the manufacturer's instructions. PCR products of 16 S rRNA genes from Hqr. walsbyi, S. ruber and Halomonas sp. were included as hybridization controls.
Stress experiments
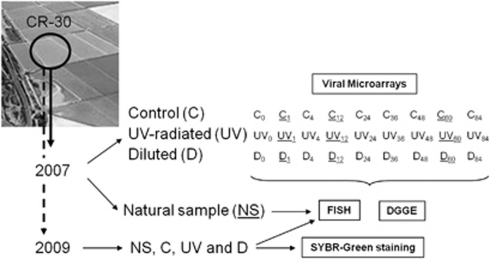
A scheme of the experimental design is shown in Figure 2. Samples were submitted to two different stress conditions: (i) UV-radiation (30-min exposure in a TELSTAR AV-100 cabin flux, operating at 40 W and λ=254 nm; ‘UV' samples) and (ii) dilution (by adding 1 volume of sterile mQ water; ‘D' samples). Each stress experiment was carried out in duplicate. Water samples not submitted to stress were used as controls (‘C'). All the samples were incubated in the laboratory and aliquots were taken for fixation and nucleic acid extraction at different times after the application of the shocks: C0, UV0 and D0 (t=0 h); C1, UV1 and D1 (t=1 h after the treatments); C4, UV4 and D4 (t=4 h); C12, UV12 and D12 (t=12 h); C24, UV24 and D24 (t=24 h); C36, UV36 and D36 (t=36 h); C48, UV48 and D48 (t=48 h); C60, UV60 and D60 (t=60 h); C84, UV84 and D84 (t=84 h). The time elapsed between the collection of NS and the beginning of the stress experiments was 2 h.
Figure 2.
Experimental setting. NSs taken in May 2007 and May 2009 were analyzed by FISH and used for stress experiments. In both cases, subsamples were submitted to UV-radiation (UV) and dilution (D) and changes in the prokaryotic communities were monitored by FISH during 84 h, comparing with not-stressed subsamples used as controls (C). Subsamples C, UV and D from May 2007 were also analyzed by DGGE in all the sampling time points. Expression and overexpression of viral genes in the NS sample and after the treatments (1, 12 and 60 h after the shocks) were analyzed only in the samples from May 2007 by using microarrays. Samples from May 2009 were also used to study the changes in the free viral fraction by SYBR-green staining of viral particles.
DAPI counts and FISH
NS, C, UV and D samples were fixed with formaldehyde (7% final concentration; Sigma-Aldrich, Steinhem, Germany). Hybridization, 4,6-diamidino-2-phenylindole (DAPI) staining and microscopy were carried out as described in Antón et al., 1999. For every sample, at least one filter was analyzed, and 500 DAPI-stained cells were counted. Probes EUB338 and Arc915 were used for counting Bacteria (Amann et al., 1990) Archaea (Stahl and Amann, 1991), respectively. Cells were visualized and counted in an epifluorescence microscope (Leica, type DM4000B, Vashaw Scientifics Inc., Norcross, GA, USA).
Nucleic acids extraction and RNA amplification
Ten ml of NS, C, UV and D samples from 2007 were filtered through 0.22 μm pore-size GV filters (Durapore, Millipore, Billerica, MA, USA) to retain cells and the viral fraction inside cells (that contains actively replicating viruses). Filters were stored at −70 °C until the extraction was carried out. Filters were cut into small pieces, mixed with 500 μl of TE (Tris-Hcl 10 m, EDTA 1 m, pH 8.0) with lysozyme (final concentration: 40 μg μl–1) and incubated for 5 min at room temperature before nucleic acid extraction with the RNeasy Mini Kit (Qiagen, Venlo, The Netherlands). An aliquot of each sample was used for electrophoresis onto 1% LE agarose gels (FMC products, Rockland, MF, USA) and PCR amplification (see below). The rest of each extract was digested with RNase-free DNase DNA-free (Ambion, Austin, TX, USA) at 37 °C for 1 h PCR amplifications of 16S rRNA genes with several amounts of digested samples (1–5 μl) were performed in order to confirm that genomic DNA was absent from the samples.
The amount and quality of each RNA preparation was checked with the 2100 Bioanalyzer (Agilent Technologies, Van Nuys, CA, USA). Aliquots of 250–350 ng of total viral RNA were amplified through a method based on T7 RNA polymerase linear amplification as previously described (Moreno-Paz and Parro, 2006) using the MessageAmp II aRNA Kit (Ambion).
PCR amplification of 16S rRNA genes
DNA from the microbial fraction in each sample was extracted as described above and used for PCR amplification of 16S rRNA genes by using universal DGGE primers for Bacteria (Muyzer et al., 1993) and Archaea (Muyzer et al., 1996) with the conditions previously reported in Muyzer et al., (1993), adding a final step at 72 °C for 30 min (Janse et al., 2004). Reaction mixtures contained 100 ng of DNA, 1.5 m MgCl2, 50 m KCl, 10 m Tris-HCl (pH 9.0), 200 μ of each dNTP, 1 U of Taq DNA polymerase (Invitrogen, Leek, The Netherlands) and 0.5 pmol of each oligonucleotide primer, in a total volume of 50 μl. To eliminate heteroduplexes, PCR products were diluted 10-fold and used as templates for a low-cycle-number reamplification using fresh reaction mixture, as described by Thompson et al (2002). To check for quality, 5 μl of the amplification products were loaded into 1% LE agarose gels (FMC).
Denaturing gradient gel electrophoresis
DGGE was carried out with a Dcode System (Bio-Rad, Hercules, CA, USA). In all the samples, 500 ng of PCR products were separated by electrophoresis at 100 V for 16 h in a linear gradient of denaturing agents from 45% to 65% (where 100% of denaturing agents was 7 M urea and 40% deionized formamide) in a 6% acrylamide-bysacrylamide (37.5:1) gel. After electrophoresis, the gels were stained with SYBR-green and the bands were cut, re-amplified, and checked again by DGGE to ascertain that they corresponded to single bands with the expected mobility compared with the NS. The nucleotide sequences of the bands were determined using the Big Dye Terminator Cycle Sequencing kit ABI PRISM 310 DNA Sequencer (Applied Biosystems, Foster City, CA, USA). Partial 16S rRNA gene sequences were compared with reference sequences at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST).
cDNA labelling and microarray hybridization
Around 2 μg of total amplified RNA (see above) were labelled by cDNA synthesis with the CyScribe cDNA direct labelling Kit, using Cy3- or Cy5-labeled dUTP (GE Healthcare, Waukesha, WI, USA) and random hexamers, following the manufacturer's instructions. Printed arrays were denatured and pre-hybridized at 42 °C in pre-hybridization buffer (saline-sodium citrate 5 × solution, 0.1 mg ml–1 herring sperm denatured-DNA, 0.1% sodium dodecyl sulphate and 0.1% bovine serum albumin) for 1 h, and then hybridized against environmental labelled cDNAs. Two types of hybridizations were carried out: single hybridizations (using NS or Leptospirillum ferrooxidans labelled cDNAs, see Supplementary Material) and double hybridizations, in which labelled cDNAs from control and stressed samples were simultaneously hybridized in a microarray (C1/UV1, C1/D1, C12/UV12, C12/D12, C60/UV60 and C60/D60). Hybridizations were performed overnight at 50 °C in HybIt hybridization solution (Arrayit Corp.). Washes were carried out with saline-sodium citrate 2 × buffer and SDS 0.1% for 5 min, saline-sodium citrate 2 × buffer for 5 min, and 0.2 × saline-sodium citrate for 5 min.
Scanning and data analysis
Hybridized arrays were scanned for Cy3 and Cy5 dyes in a GenePix 4100A Scanner (Axon Instruments Inc., Foster City, CA, USA). The scanned images were saved as 16-bit greyscale tagged image file format and analyzed by quantifying the fluorescence intensity of each spot using GenePix Pro v.6.0 software (Axon Instruments Inc.). The local background signal was subtracted automatically from the hybridization signal for each spot. Microarray hybridization results were analyzed with Genepix pro v.6.0 software (Axon Instruments Inc.) and were normalized by Global normalization using the global median of log intensity ratios (Yang et al., 2001).
SYBR-green staining of viral particles
One ml from the NS taken in May 2009 and from subsamples submitted to UV-radiation and dilution were centrifuged for 5 min at 15 700 g (Heraeus Labofuge 400R, DJB Labcare Ltd, Buckinghamshire, UK). The supernatant was fixed with 0.02 μm Anodisc 25 filters (Whatman Int. Ltd, Maidstone, UK) filtered formaldehyde (4% final concentration) for 15–30 min, at room temperature. In all, 10 μl from the fixed samples were filtered using 0.02 μm Anodisc 25 filters and stained with SYBR-green as described in Noble and Fuhrman (1998). Viral particles were then visualized and counted in an epifluorescence microscope (Leica, type DM4000B).
Results
Metatranscriptome analysis of viral assemblages in the NS
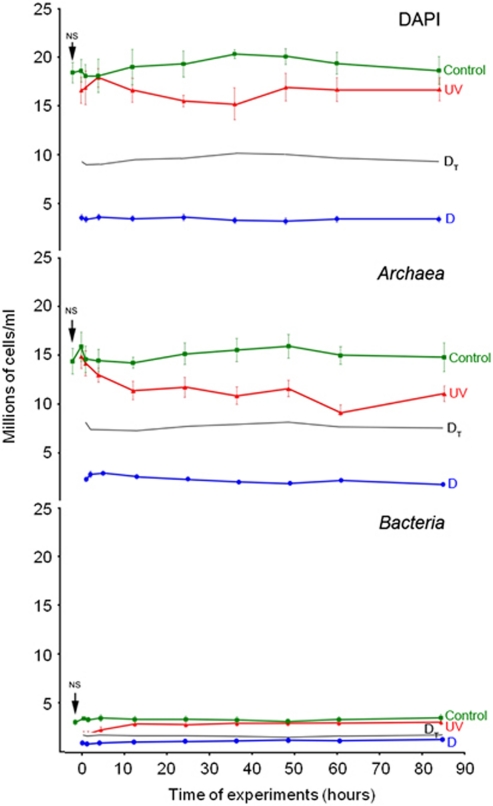
The salinity of the NS taken in May 2007 was 32% and the DAPI and FISH counts (Figure 3) were in the range of those from other crystallizer samples analyzed around the world (Rosselló-Mora et al., 2003; Elevi-Bardavid et al., 2007; Antón et al., 2008). The dominant group was Archaea, whose representatives accounted for up to 77% of the DAPI counts, while members of Bacteria accounted for up to 17.6%.
Figure 3.
Changes in the number of total cells (DAPI), Archaea and Bacteria with time after the application of the stress treatments, compared with values in the NS taken in May 2007. Green lines are the values of the control samples. In red (UV), the number of cells after the UV-radiation treatment. In blue (D), the number of cells under osmotic shock. Black lines (DT) represent the theoretical values after the dilution of the sample without considering lysis.
Total RNA from the sample was extracted, amplified, reverse-transcribed and labelled as described above and then hybridized against the viral microarray that had been constructed with the metaviromic shotgun clones from the same sample. Up to 340 spots out of 596 showed fluorescence above nine times the background fluorescence and therefore were selected for the analysis (see Supplementary Material). As the viral nucleic acids used for the microarray had been previously sequenced, assembled and annotated (Santos et al., 2010), the expression in the spots could be correlated with the expression in their corresponding contigs (Figure 1, section NS). Larger contigs were formed by a higher number of clones, thus representing viruses that were more abundant in the environment. However, the level of expression detected was not always directly related to the size of the contig being expressed (Figure 1, section contig size). In general, larger contigs showed the highest fluorescent signal, indicating a high expression level of the corresponding genes. However, some small contigs, which could be related to less abundant viruses, were also very highly expressed in the NS.
The percentage of the contigs with an expression above nine times the background fluorescence included in each group was: 65% and 69% in the HVS-1 and HVS-2 groups (which included the sequences of high GC content haloarchaea and S. ruber, respectively), 67% for HVS-3, 48% for HVS-4 (that included the sequence of Hqr. walsbyi) and 72% in the HVS-5 group. In general, the contigs that show expression are expressed along their whole length. Table 1 shows a list of the predicted open reading frames in these contigs. Putative methyltransferases, terminases, membrane and regulation proteins were expressed in all groups, as well as many hypothetical and conserved hypothetical proteins.
Table 1. List of predicted ORFs in contigs expressed in the NS.
| HVS group | Contigs | Predicted ORFs |
|---|---|---|
| HVS-1 | 022, 030, 058, 110, 141, 151, 198, 225, 246 | HP |
| 078, 209 | CHP | |
| 126, 152 | CHP, HP | |
| 008, 113 | HP, Zn-finger | |
| 012 | Zn-finger, HP, CHP | |
| 029 | RHH protein, CHP | |
| 038 | HP, terminase | |
| 042 | CHP, Zn-finger | |
| 061 | Transcription factor, HP | |
| 070 | RHH protein, integrase | |
| 096 | HP, envelope phage protein | |
| 140 | HP, transposase | |
| 178 | Replicative ATPase | |
| 193 | Integrase | |
| 208 | HP, methyltransferase | |
| HVS-2 | 014, 024, 027, 041, 065, 106, 120, 128, 138, 162, 179, 194, 200, 202, 218, 228, 242, 247 | HP |
| 055, 088, 107 | Membrane, HP | |
| 066, 090, 237 | Membrane | |
| 067, 199 | CHP | |
| 127, 129 | HP, CHP | |
| 163, 190 | HP, methyltransferase | |
| 011 | Terminase, HP | |
| 049 | Membrane, CHP, tat-signal protein | |
| 059 | Zn-finger, CHP | |
| 062 | Prohead peptidase, capsid structure | |
| 069 | HP, phosphohydrolase | |
| 072 | Membrane, CHP | |
| 104 | Terminase | |
| 123 | SsDNA-binding protein, HTH-protein, HP | |
| 213 | Phage mu-like | |
| 214 | HP, transcriptional regulator | |
| 261 | Deoxyribosyltransferase | |
| HVS-3 | 017, 031, 144, 176, 221, 226, 248 | HP |
| 171, 116 | Membrane, HP | |
| 236, 251 | Membrane | |
| 044 | MCM protein (DNA replication initiator) | |
| 060 | Replicative DNA helicase (intein endonuclease) | |
| 076 | HNH endonuclease, HP | |
| 080 | HP, ssDNA-binding protein | |
| 084 | HTH phage repressor (peptidase) | |
| 114 | LAGLIDADG endonuclease | |
| 130 | CHP | |
| 137 | Methyltransferase, phosphoadenosine reductase | |
| 150 | Ssl1-like protein (transcription factor) | |
| 227 | Hypothetical DUF protein | |
| 249 | HNH endonuclease, membrane | |
| HVS-4 | 006, 036, 047, 050, 063, 098, 118, 149 | HP |
| 005, 035, 074 | HP, CHP | |
| 001 | HP, capsid, methyltransferase | |
| 004 | HP, CHP, terminase, methyltransferase | |
| 007 | Methyltransferase, nuclease, HP | |
| 009 | HP, traslocation domain protein | |
| 019 | Terminase, HP | |
| 043 | HP, transposase | |
| 068 | Lamgl protein (glucanase) | |
| 113 | HP, homeodomain protein | |
| 125 | Membrane, HP | |
| 158 | WD40/YVTN repeat-like domain | |
| 165 | HP, mRNA autoregulator | |
| 174 | Arginine N-succinyltransferase | |
| HVS-5 | 083, 131, 132, 142, 164, 169, 173, 216, 255 | HP |
| 002, 015 | HP, CHP, membrane | |
| 026, 182 | Phage DNA primase | |
| 160, 266 | Membrane | |
| 075 | HP, membrane | |
| 081 | Hypothetical ParB nuclease-like protein | |
| 111 | Terminase | |
| 124 | Replicative DNA-dependent ATPase | |
| 167 | HP, CHP | |
| 212 | CHP |
Abbreviations: CHP, conserved hypothetical protein; DUF, domain of unknown function; HP, hypothetical protein; HTH, helix-turn-helix domain; HVS, halophilic viral sequence; NS, natural sample; ORF, open reading frame; RHH, ribbon-helix-helix domain; ssDNA, single-stranded DNA.
Metatranscritome analysis of viral assemblages under stress conditions
The viral microarrays were used for double hybridizations in which UV and D samples were compared with the C samples (Figure 1, sections UV1, D1 and D12). Changes in the prokaryotic community caused by these stress conditions were monitored by FISH and DGGE, as explained above.
Stability of the control sample
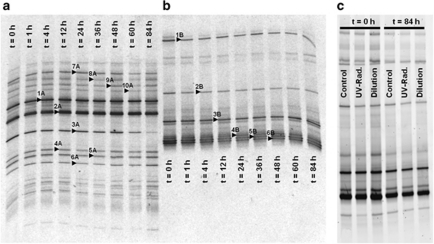
The sample used as a control was highly stable throughout the stress experiments; both DAPI and FISH counts (Figure 3, green lines) in the C sample did not change significantly with time, and differences in DAPI were no higher than 11% in all the analyzed points. Between 93% and 100% of total cell counts could be assigned by FISH to either Archaea, which always dominated the community, or Bacteria. Both archaeal and bacterial DGGE profiles (Figures 4a and b) in the sample did not show any change over the course of the experiment. Sixteen bands were excised from the gel and sequenced and, as in previous studies (Benlloch et al., 2002; Casamayor et al., 2002;Gomariz et al., unpublished), the sequences obtained matched in databases with cultured and uncultured members of Halobacteriaceae and Bacteroidetes (see Supplementary Table 1).
Figure 4.
DGGE profiles from archaeal (a) and bacterial (b) 16S rRNA gene amplification products in the samples used as controls. Each lane corresponds to different sampling times in the experiment (in hours). In (c), comparison among the bacterial assemblages in the control, diluted and UV-radiated samples in two different sampling times. Sequences 1, 2 and 10, in (a), were related to the species Hqr. walsbyi. The rest of the bands in (a) were associated to uncultured Halobacteriaceae environmental clones. Sequences from band 3 were related to the species S. ruber. The rest of the sequences in (b) were associated to uncultured Salinibacter (bands 2, 4-6) and Bacteroidetes (band 1) clones.
A double hybridization with C1 and NS showed (Supplementary Material) that during the time elapsed between the collection of the sample and 1 h after the treatments there was no overexpression of viral genes.
Changes under UV-radiation treatment
Figure 3 (red lines) shows the variation in the numbers of DAPI, Archaea and Bacteria following the UV treatment. Just after the shock there was a remarkable reduction in the number of Bacteria (43.42%), probably due to lysis, compared with the effect on the archaeal populations (6.42% reduction). However, after 1 h, Archaea started to decrease and numbers of Bacteria increased.
When 16S rRNA genes from UV samples were amplified for monitoring by DGGE changes in archaeal and bacterial diversity, the same patterns as in the control were obtained (Figure 4c), thus indicating that the ‘species' composition of the crystallizer remained stable. Further analysis would be needed to ascertain if the treatments produced changes in the microdiversity of the sample, as DGGE profiles only provide information on diversity patterns based on a partial amplification of the 16S rRNA genes.
Up to 114 spots in the array showed overexpression in the sample UV1 when compared with sample C1 while no overexpression was detected in UV12 and UV60 when compared with their corresponding controls. As with the NS, a correlation among the overexpressed spots with their corresponding contigs was established (Figure 1, section UV1). A list of contigs overexpressed after radiation with their respective open reading frames is shown in Table 2.
Table 2. Viral contigs overexpressed (+) after UV-radiation and dilution treatments, compared with expression in NS.
| Group | Contig | Predicted ORFs | Expression NS |
Overexpression (+) |
||
|---|---|---|---|---|---|---|
| UV1 | D1 | D12 | ||||
| HVS-1 | 012 | CHP, HP, Zn-finger proteins | VH | + | + | + |
| 030 | HPs | VH | + | + | P | |
| 039 | CHP, ribbon-helix protein | H | — | P | — | |
| 042 | CHP, Zn-finger proteins | VH | + | + | + | |
| 073 | HPs | —a | + | + | + | |
| 152 | CHP, HP | VH | + | + | + | |
| 193 | Integrase | VH | — | — | + | |
| 225 | HPs | H | + | + | — | |
| 250 | CHP | H | + | — | — | |
| HVS-2 | 011 | Terminase, HP | VH | P | P | — |
| 106 | HPs | VH | + | + | — | |
| 129 | CHP, HP | VH | — | — | P | |
| 199 | CHP | H | + | — | — | |
| HVS-3 | 017 | HPs | H | + | — | — |
| 060 | Replicative DNA helicase | H | — | — | + | |
| 100 | HNH endonuclease, HP | — | — | + | — | |
| 177 | Phage capsid protein, HP | — | — | — | + | |
| 226 | HP | VH | + | + | + | |
| HVS-4 | 001 | HP, methyltransferase, capsid protein | VH | + | + | P |
| 005 | CHP, HP | VH | P | + | P | |
| 006 | HPs | H | P | P | P | |
| 007 | Methyltransferase, nuclease, HP | P | P | P | — | |
| 009 | HP, translocation protein | VH | P | — | — | |
| 010 | Terminase | — | — | P | — | |
| 019 | Terminase, HP | H | P | — | — | |
| 068 | LamGL protein (glucanase) | H | — | P | — | |
| 113 | HP, homeodomain | H | + | — | — | |
| 165 | HP, mRNA autoregulator | H | — | + | — | |
| 191 | HP, membrane protein | H | — | + | — | |
| 241 | HPs | — | — | + | — | |
| HVS-5 | 021 | Integrase, HP | H | — | — | + |
| 159 | HP | — | — | — | + | |
| 167 | HP, CHP | H | + | + | + | |
Abbreviations: CHP, conserved hypothetical protein; H, contigs highly expressed in NS; HNH, conserved amino acid sequence in certain endonucleases; HP, hypothetical protein; HVS, halophilic viral sequence; ORF, open reading frame; NS, natural sample; P, the contig was partially expressed; VH, contigs very highly expressed in NS; UV, ultraviolet.
Absence of expression and/or overexpression.
In the UV1 sample, the groups showing the highest numbers of overexpressed contigs were HVS-1 and HVS-4 (15% and 12% of the contigs in each group, respectively), while in the groups HVS-2, HVS-3 and HVS-5, only 4.6%, 3.3% and 2.7% of the contigs, respectively, were overexpressed with respect to the C1 sample. As mentioned above, besides viral sequences, HVS-1 includes high GC haoarchaeal sequences and HVS-4 includes the genomic sequence of Hqr. walsbyi (Santos et al., 2010). Thus, HVS groups that displayed the strongest response to UV treatment were those containing viruses related to Archaea, which could explain the continuous decrease in the archaeal numbers observed once the stress disappeared, contrasting with the recovery in bacterial counts detected in the same period (although the initial effect of UV radiation on bacterial numbers seemed more dramatic).
Changes under dilution treatment
Dilution of the sample (Figure 3, blue lines) produced a very marked decrease in Archaea numbers. Bacteria seemed more resistant to dilution and numbers started to increase after 1 h. DGGE profiles after dilution showed the same patterns as the control and UV-radiated samples (Figure 4c).
One hour after the dilution of the sample, up to 134 spots in the microarray showed overexpression with respect to the undiluted sample. After 11 h (in D12), 42 spots (some of them different from the ones expressed at t=1 h) were overexpressed. At 60 h after the dilution of the sample, no viral overexpression could be detected. The correlation between the overexpressed spots and the corresponding contigs is shown in Figure 1, sections D1–D12, and Table 2.
The groups with the highest number of overexpressed contigs in D samples were also HVS-1 and HVS-4 (15% of contigs in each group, after 1 h; 13% and 4.5% of the contigs, respectively, after 12 h). Thus, as observed with the UV treatment, HVS groups containing archaeal sequences showed the strongest response to the dilution treatment. This, again, could be related to the significant decrease in archaeal numbers.
Changes in the viral counts of stressed samples
Numbers of free viruses in the NS and subsamples C, UV and D were analyzed by SYBR-green staining in the sample taken in May 2009. This sample had 34% salinity and contained 1.52 × 109±1.45 × 108 VLPs ml−1 and 3.46 × 107±3.25 × 106 cells ml−1. Archaea and Bacteria accounted for 79.35% and 10.31% of the DAPI counts, respectively. FISH analyses indicated that changes in the prokaryotic community because of stress treatments were similar to those detected in the experiment carried out in 2007 (see Supplementary Figure 1).
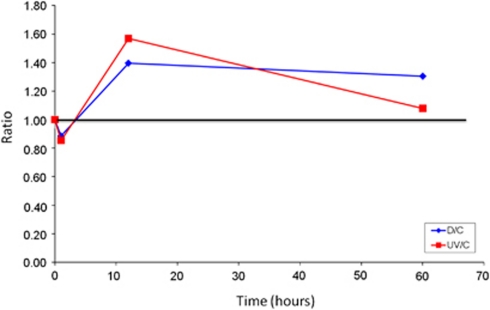
The number of VLPs present in the C sample decreased after 60 h, from 1.52 × 109 to 8.40 × 108. In both UV and D samples, a reduction in the number of VLPs was observed 1 h after the treatments, when compared with their corresponding C subsamples (Figure 5). After 12 h, the number of VLPs increased, being slightly higher in the UV sample. At 60 h after the shocks, the free VLPs in the radiated sample decreased while diluted samples maintained the levels reached previously. An explanation for the initial decrease in the number of free viruses could be that capsids were disrupted because of high radiation and osmotic shock. However, part of the viral particles (located either inside or outside the cells) might be activated because of the treatments (or the hosts made more susceptible), favouring the release of new viruses and their subsequent increase.
Figure 5.
Changes in the numbers of the viral fraction in the May 2009 sample under stress conditions (UV radiation, in red; dilution, in blue) divided by changes in the control sample.
The observed increase in free VLPs numbers was in agreement with the high levels of overexpression observed in the metatranscriptomic analyses. Indeed, both techniques indicate a more persistent effect of the dilution since viral numbers did not decrease along the treatment and viral overexpression was detected also 12 h after diluting the sample.
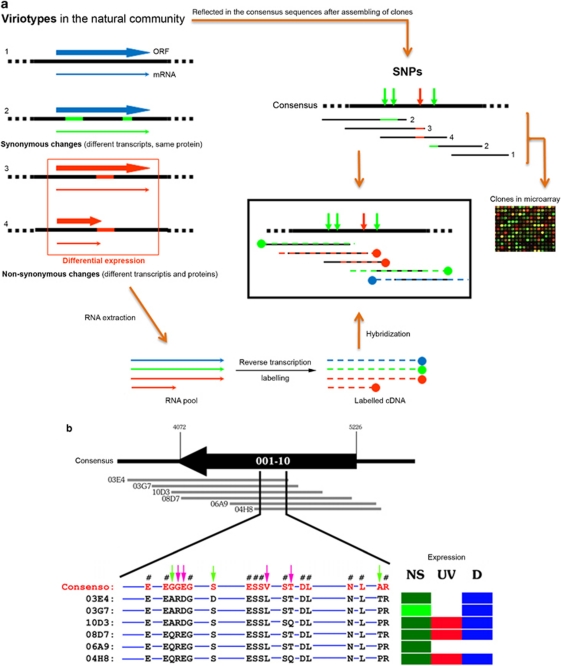
Expression of ‘ecoviriotypes'
The characterization of the CR30 metavirome (Santos et al., 2010) revealed the existence of a significant number of single-nucleotide polymorphisms (SNPs) in some viral contigs. The presence of SNPs corresponded to a very high mutation frequency (7.65 × 10−3 substitutions per nucleotide) that reflected a high microdiversity of closely related haloviruses. Around 40% of these SNPs corresponded to non-synonymous changes that could have a phenotypic effect on the corresponding viriotypes. We hypothesize that certain viriotypes of the community, because of these non-synonymous mutations, could code for transcripts varying in nucleotide composition and length (Figure 6a), which finally result in different proteins from a given homologous open reading frame. Differences in the function/efficiency (fitness) among such polymorphic proteins would determine the expression of a given viriotype under a given environmental condition. The analysis of the metatranscriptomic data could then reveal these differences in the expression of clones (that is, spots in the microarray) covering the same polymorphic open reading frames (Figure 6a). For this analysis we only considered the clones that were expressed under at least one of the assayed conditions because the absence of expression in a clone under all the conditions would indicate an absence of DNA in the microarray (if other clones in the same contig were expressed).
Figure 6.
Ecoviriotypes. (a) Theoretical scheme showing how nearly identical viruses (viriotypes) with non-synonymous changes can respond differentially to environmental changes. This differential response can be detected in the expression of the overlapping clones from a given contig. (b) Differential expression of ‘ecoviriotypes'. The overlapping clones (grey lines) that form a given contig present SNPs that can be translated in neutral–non-synonymous changes (pink arrows) and non-neutral–non-synonymous changes (green arrows). Symbol #indicates synonymous changes. Blue lines indicate 100% identity in amino acid and nucleotides among clones.
With these premises, we analyzed the differences in the expression of the 49 polymorphic contigs (an example is shown in Figure 6b). Out of the 49 contigs, 24 showed expression only in the NS while 12 were expressed and/or overexpressed in two or three analyzed conditions, thus showing differential expression (the remaining 13 contigs were not expressed under any conditions or were wholly expressed in the three conditions assayed). Up to 67% of these differentially expressed contigs had consensus sequences displaying both synonymous and non-synonymous changes. If we consider that the non-synonymous changes reflect viral proteins that could be differentially expressed (the amino acidic composition in each clone was not identical), the transcripts with a given number of SNPs would not hybridize well with their polymorphic spots (Figures 6a and b). On the basis of these observations, we suggest the term ‘ecoviriotype' to refer to these different but very closely related viral genotypes that could respond differentially to changes in the environmental parameters.
However, the remaining 33% of the contigs displaying differential expression showed only synonymous changes. In these cases, the differences in the nucleotide sequence of the clones that form a given contig could also be affecting the hybridization efficiency of the viral transcripts. These clones could not be considered ecoviriotypes because the observed differential expression would not be due to differential responses to environmental conditions, but to a differential efficiency of the hybridization between target and probes in the microarray.
Discussion
This work presents the use of microarrays as a tool to carry out metatranscriptomic analyses in viral communities. This technique makes it unnecessary to carry out the complete sequencing of all the transcripts present in the sample and the in silico search for viral messengers in the total RNA extracts.
The CR30 crystallizer showed a high viral expression at the time of sampling (60% of the metaviromic contigs were expressed) although the expression was not homogeneous for all the contigs. Groups HVS-1, HVS-2 and HVS-5 had the highest percentages of expressed contigs. Given that sequences from HVS-1 and -2 groups could correspond, respectively, to viruses infecting high GC content haloarchaea and S. ruber representatives (Santos et al., 2010), these viruses could be responsible for the lower numbers that their hosts reach in the CR30 sample compared with the high abundance of Hqr. walsbyi (Antón et al., 1999).
It is well known that Hqr. walsbyi dominates the prokaryotic community in many (but not all) crystallizers analyzed around the world (Antón et al., 1999; Bolhuis et al., 2004; Burns et al., 2004; Legault et al., 2006; Maturrano et al., 2006; Baati et al., 2008; Mutlu et al., 2008), but little is known about the reasons for this phenomenon. One could argue that, besides phage control, the dominance of Haloquadratum over Salinibacter could be due either to competition for resources, different nutrient requirements, or halocin activity, among other reasons. When archaeal populations are reduced by the stress conditions used in this work (see below), Bacteria do not seem to obtain any benefit and reach the same numbers they had in the NS. Thus, competition with Archaea for specific resources does not seem to be controlling bacterial populations.
Haloquadratum populations are considered to be highly versatile metabolically and very tolerant to high concentrations of magnesium (Cuadros-Orellana et al., 2007). In addition, it seems that different lineages of Haloquadratum harbour a large variety of surface proteins (Cuadros-Orellana et al., 2007; Rodríguez-Valera et al., 2009) that could mediate phage processes and confer resistance to viral infection in a significant proportion of their natural strains. Given the high abundance of their hosts, viruses affecting Haloquadratum representatives should be the most abundant viruses in the system, which is supported by the fact that largest contigs in the metavirome (those formed by the largest number of clones) are included in the HVS-4 group, which also contains the Hqr. walsbyi genome. However, this is not the most highly expressed (that is, active) group, a fact in agreement with the idea that ‘abundance alone does not tell how active the viruses may be in infecting hosts' (Fuhrman and Schwalbach, 2003). Although, in our case, most of the contigs probably do not correspond to very minor components of the community, given the amount of analyzed sequences, our findings somehow contrasts with the ‘Bank model' proposed by Breitbart and Rohwer (2006) that assumes that ‘only the most abundant viruses are active'. In the ‘Bank model', different hosts become dominant and their viruses move from the ‘bank' into the active fraction when the environment changes. Data reported for years from the CR30 crystallizer show that Haloquadratum members always dominate the prokaryotic community (Antón et al., 1999; Bolhuis et al., 2006). Our results indicate that their dominance could be related to resistance/evasion mechanisms to viral infection and are in agreement with the experiments carried out with marine samples by Bouvier and del Giorgio (2007), who proposed a scenario of bacterial–viral interactions where the dominant bacterial phylotypes in the system might have a lower susceptibility to viral infection because of the development of resistance or by intrinsic metabolic activity, in a density-independent form. In principle, this would contradict the density-dependent ‘kill the winner' hypothesis, in which it is assumed that ‘viruses should preferentially infect the most common hosts'. In the ‘kill the winner' model, proposed by Thingstad and Lignell (1997) and revised by Winter et al. (2010), the ‘winner' refers to the most active population in the community. In the CR30 crystallizer, Hqr. walsbyi would be the ‘winner', which would dominate the system not only because of their metabolical adaptability but also because of their survival strategies when faced with a viral attack. Once again, it seems that a semantic issue is at the heart of this discussion, because the winner can be either the dominant species (Hqr. walsbyi in our case) or the most abundant lineage within a given species (one specific Hqr. walsbyi strain, below what would be considered a ‘population'). Indeed, DGGE patterns display a diversity of phylotypes within the Haloquadratum assemblage, in agreement with the vast pan-genome reported for this species (Legault et al., 2006). Such a wide range of microdiversity has been reported for other prokaryotes inhabiting this low diversity environment, such as Halorubrum spp. isolates (Papke et al., 2004) and different S. ruber strains (Peña et al., 2010).
Electromagnetic radiation is known to inactivate prokaryotic viruses, which are particularly vulnerable because of the absence of DNA reparation mechanisms. Sunlight, particularly UV radiation, can be very important for viral inactivation in the upper water column (Suttle and Chen, 1992; Murray and Jackson, 1993), as the inactivated viruses are unable to infect sensitive hosts. Since solar salterns represent ecosystems submitted to high solar radiation, the UV treatment could give information about the strategies for the activity and survey of halophages in the crystallizers. In addition, the dilution of brines is a common phenomenon after rainfalls and it is known that decreasing the salinity of the medium increases the burst size (Dyall-Smith et al., 2003). High salinities inhibit the absorption of some haloviruses that infect Halobacterium salinarum, favouring high cell densities. However, when the salinity is reduced, viruses are stimulated to reproduce and exit cells quickly given that cells become unstable because of the change in the osmotic pressure (Dyall-Smith et al., 2003).
Our results showed that both stress conditions increased the expression of some viral groups and the number of viruses found in the sample. In addition, they affected differentially the behaviour of archaeal and bacterial populations. Putative archaeal viruses also showed more activity under stress, which in fact implies that the decrease in the archaeal populations could be due to both the treatments and the increase in the expression of archaeal viruses. In any case, neither of these factors were affecting the rank of ‘species' in the samples, at least at the level of resolution provided by the analysis of DGGE patterns.
The question now is to what extent viruses may select prokaryotic lineages within a given phylotype under the assayed conditions. Indeed, this strain-specific phage susceptibility has been previously reported for members of the species S. ruber (Peña et al., 2010) and proposed for members of the Haloquadratum assemblage (Rodríguez-Valera et al., 2009). The replacement of closely related strains by the effect of viral activity within a temporarily stable community has been recently proposed for solar salterns in San Diego (Rodriguez-Brito et al., 2010). These observations are coherent with the constant-diversity dynamics model (Rodríguez-Valera et al., 2009) according to which the diversity of prokaryotic communities is preserved by phage predation.
Acknowledgments
This work was supported by projects CGL2006-12714-CO2-01 and -02, and AYA2008-04013 of the Spanish Ministry of Science and Innovation (MICINN), which include funding from the European Union (‘FEDER') and ACOM09/155 from the Generalitat Valenciana.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Allen MJ, Forster T, Schroeder DC, Hall M, Roy D, Ghazal P, et al. Locus-specific gene expression pattern suggests a unique propagation strategy for a giant algal virus. J Virol. 2006;80:7699–7705. doi: 10.1128/JVI.00491-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen MJ, Martínez-Martínez J, Schroeder DC, Somerfield PJ, Wilson WH. Use of microarrays to assess viral diversity: from genotype to phenotype. Environ Microbiol. 2007;94:971–982. doi: 10.1111/j.1462-2920.2006.01219.x. [DOI] [PubMed] [Google Scholar]
- Allen MJ, Wilson WH. The coccolithovirus microarray: and array of uses. Brief Funct Genomic Proteomic. 2006;5:273–279. doi: 10.1093/bfgp/ell033. [DOI] [PubMed] [Google Scholar]
- Amann RI, Krumholz L, Stahl DA. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antón J, Llobet-Brossa E, Rodríguez-Valera F, Amann R. Fluorescente in situ hybridization analysis of the prokaryotic community inhabiting crystallizer ponds. Environ Microbiol. 1999;1:517–523. doi: 10.1046/j.1462-2920.1999.00065.x. [DOI] [PubMed] [Google Scholar]
- Antón J, Peña A, Santos F, Martínez-García M, Schmitt-Kopplin P, Rosselló-Mora R. Distribution, abundance and diversity of the extremely halophilic bacterium Salinibacter ruber. Saline Syst. 2008;4:15. doi: 10.1186/1746-1448-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baati H, Guermazi S, Amdouni R, Gharsallah N, Sghir A, Ammar E. Prokaryotic diversity of a Tunisian multipond solar saltern. Extremophiles. 2008;12:505–518. doi: 10.1007/s00792-008-0154-x. [DOI] [PubMed] [Google Scholar]
- Benlloch S, Lopez-Lopez A, Casamayor EO, Ovreas L, Goddard V, Daae FL, et al. Prokaryotic genetic diversity throughout the salinity gradient of a coastal solar saltern. Environ Microbiol. 2002;4:349–360. doi: 10.1046/j.1462-2920.2002.00306.x. [DOI] [PubMed] [Google Scholar]
- Bolhuis H, Palm P, Wende A, Falb M, Rampp M, Rodriguez-Valera F, et al. The genome of the square archaeon Haloquadratum walsbyi: life at the limits of water activity. BMC Genomics. 2006;4:169. doi: 10.1186/1471-2164-7-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolhuis H, Poele EM, Rodríguez-Valera F. Isolation and cultivation of Walsby's square archaeon. Environ Microbiol. 2004;6:349–360. doi: 10.1111/j.1462-2920.2004.00692.x. [DOI] [PubMed] [Google Scholar]
- Bouvier T, del Giorgio PA. Key role of selective viral-induced mortality in determining marine bacterial community composition. Environ Microbiol. 2007;9:287–297. doi: 10.1111/j.1462-2920.2006.01137.x. [DOI] [PubMed] [Google Scholar]
- Breitbart M, Haynes M, Kelley S, Angly F, Edwards RA, Felts B, et al. Viral diversity and dynamics in an infant gut. Res Microbiol. 2008;159:367–373. doi: 10.1016/j.resmic.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Breitbart M, Rohwer F. Here a virus, there a virus, everywhere the same virus. Trends Microbiol. 2006;13:278–284. doi: 10.1016/j.tim.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Burns DG, Camakaris HM, Janssen PH, Dyall-Smith ML. Cultivation of Walsby's square haloarchaeaon. FEMS Microbiol Lett. 2004;238:469–473. doi: 10.1016/j.femsle.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Casamayor EO, Massana R, Bencholl S, Ovreas L, Díez B, Goddard VJ, et al. Changes in archaeal, bacterial and eukaryal assemblages along a salinity gradient by comparison of genetic fingerprinting methods in a multipond solar saltern. Environ Microbiol. 2002;4:338–348. doi: 10.1046/j.1462-2920.2002.00297.x. [DOI] [PubMed] [Google Scholar]
- Chou CC, Lee TS, Chun-Houh C, Hsiao HY, Lin YL, Ho MS, et al. Design of microarrays probes for virus identification and detection of emerging viruses at the genus level. BMC Bio. 2006;7:232–242. doi: 10.1186/1471-2105-7-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadros-Orellana S, Martín-Cuadrado AB, Legault B, D'Auria G, Zhaxybayeva O, Papke RT, et al. Genomic plasticity in prokaryotes: the case of the square haloarchaeon. ISME J. 2007;1:235–245. doi: 10.1038/ismej.2007.35. [DOI] [PubMed] [Google Scholar]
- Díez B, Antón J, Guixa-Boixareu N, Pedrós-Alió C, Rodríguez-Valera F. Pulsed-field gel electrophoresis analysis of virus assemblages present in a hypersaline environment. Int Microbiol. 2000;3:159–164. [PubMed] [Google Scholar]
- Dyall-Smith M, Tang SL, Bath C. Haloarchaeal viruses: how diverse are they. Res Microbiol. 2003;154:309–313. doi: 10.1016/S0923-2508(03)00076-7. [DOI] [PubMed] [Google Scholar]
- Elevi-Bardavid R, Ionescu D, Oren A, Rainey FA, Hollen BJ, Bagaley DR, et al. Selective enrichment, isolation and molecular detection of Salinibacter ruber and related extremely halophilic Bacteria from hypersaline environments. Hydrobiologia. 2007;576:3–13. [Google Scholar]
- Fuhrman JA. Marine viruses and their biogeochemical and ecological effects. Nature. 1999;399:541–548. doi: 10.1038/21119. [DOI] [PubMed] [Google Scholar]
- Fuhrman JA, Schwalbach M. Viral influence of aquatic bacterial communities. Biol Bull. 2003;204:192–195. doi: 10.2307/1543557. [DOI] [PubMed] [Google Scholar]
- Garrido P, González-Toril E, García-Moyano A, Moreno-Paz M, Amils R, Parro V. An oligonucleotide prokaryotic acidophile microarray: its validation and its use to monitor seasonal variations in extreme acidic environments with total environmental RNA. Environ Microbiol. 2008;10:836–850. doi: 10.1111/j.1462-2920.2008.01477.x. [DOI] [PubMed] [Google Scholar]
- Guixa-Boixareu N, Calderón-Paz JI, Heldal M, Bratbak G, Pedrós-Alió C. Viral lysis and bacterivory as prokaryotic loss factors along a salinity gradient. Aquat Microb Ecol. 1996;11:215–227. [Google Scholar]
- Janse I, Bok J, Zwart G. A simple remedy against artifactual double bands in denaturing gradient gel electrophoresis. J Microbiol Meth. 2004;57:279–281. doi: 10.1016/j.mimet.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Jiang S, Steward G, Jellison R, Chu W, Choi S. Abundance, distribution and diversity of viruses in alkaline, hypersaline Mono Lake, California. Microb Ecol. 2003;47:9–17. doi: 10.1007/s00248-003-1023-x. [DOI] [PubMed] [Google Scholar]
- Kistler A, Avila PC, Rouskin S, Wang D, Ward T, Yagi S, et al. Pan-viral screening of respiratory tract infections in adults with and without asthma reveals unexpected human coronavirus and human rhinovirus diversity. J Infec Dis. 2007;196:817–825. doi: 10.1086/520816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin V, He S, Warnecke F, Peterson SB, Garcia-Martin H, Haynes M, et al. A bacterial metapopulation adapts locally to phage predation despite global dispersal. Genome Res. 2008;18:293–297. doi: 10.1101/gr.6835308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legault BA, López-López A, Alba-Casado JC, Doolittle WF, Bolhius H, Rodríguez-Valera F, et al. Environmental genomics of ‘Haloquadratum walsbyi' in saltern crystallizer indicates a large pool of accessory genes in an otherwise coherent species. BMC Genomics. 2006;4:171. doi: 10.1186/1471-2164-7-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maturrano L, Santos F, Rosselló-Mora R, Antón J. Microbial diversity in Maras salterns, a hypersaline environment in the Peruvian Andes. Appl Environ Microbiol. 2006;72:3887–3895. doi: 10.1128/AEM.02214-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Paz M, Gómez MJ, Arcas A, Parro V. Environmental transcriptome analysis reveals physiological differences between biofilm and planktonic modes of life of the iron oxidizing bacteria Leptospirillum spp. in their natural microbial community. BMC Genomics. 2010;11:404. doi: 10.1186/1471-2164-11-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Paz M, Parro V. Amplification of low quatity bacterial RNA for microarray studies: time-course analysis of Leptospirillum ferrooxidans under nitrogen-fixing conditions. Environ Microbiol. 2006;8:1064–1073. doi: 10.1111/j.1462-2920.2006.00998.x. [DOI] [PubMed] [Google Scholar]
- Murray A, Jackson G. Viral dynamics II: a model of the interaction of ultraviolet light and mixing processes on virus survival in seawater. Mar Ecol Prog Ser. 1993;102:105–114. [Google Scholar]
- Mutlu MB, Martínez-García M, Santos F, Peña A, Guven K, Antón J. Prokaryotic diversity in Tuz Lake, a hypersaline environment in inland Turkey. FEMS Microbiol Ecol. 2008;65:474–483. doi: 10.1111/j.1574-6941.2008.00510.x. [DOI] [PubMed] [Google Scholar]
- Muyzer G, De Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyzer G, Hottentrager S, Teske A, Wagner C.1996Denaturing gradient gel electrohpresis of PCR-amplified 16S rDNA. A new molecular approach to analyze the genetic diversity of mixed microbial communitiesIn: Akkermans ADL, van Elsas JD, de Brujin FJ (eds),Molecular Microbial Ecology Manual Kluwer Academic Publishing: Dordrecht; pp. 3.4.4.1-3.4.4.2.2. [Google Scholar]
- Noble RT, Fuhrman JA. Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat Microb Ecol. 1998;14:113–118. [Google Scholar]
- Papke RT, Koening JE, Rodríguez-Valera F, Doolittle WF. Frequent recombination in saltern population of Halorubrum. Science. 2004;306:1928–1929. doi: 10.1126/science.1103289. [DOI] [PubMed] [Google Scholar]
- Parro V, Moreno-Paz M, González-Toril E. Analysis of environmental transcriptomes by DNA microarrays. Environ Microbiol. 2007;9:453–464. doi: 10.1111/j.1462-2920.2006.01162.x. [DOI] [PubMed] [Google Scholar]
- Peña A, Teeling H, Huerta-Cepas J, Santos F, Yarza P, Brito-Echevarría J, et al. Fine-scale evolution: genomic, phenotypic and ecological differentiation in two coexisting Salinibacter ruber strains. ISME J. 2010;4:882–895. doi: 10.1038/ismej.2010.6. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Brito B, Li L, Wegley L, Furlan M, Angly F, Breitbart M, et al. Viral and microbial community dynamics in four aquatic environments. ISME J. 2010;4:739. doi: 10.1038/ismej.2010.1. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Valera F, Martín-Cuadrado AB, Rodríguez-Brito B, Pasic L, Thingstad TF, Rohwer F, et al. Explaining microbial population genomics through phage predation. Nat Rev Microbiol. 2009;7:828–836. doi: 10.1038/nrmicro2235. [DOI] [PubMed] [Google Scholar]
- Rosselló-Mora R, Lee N, Antón J, Wagner M. Substrate uptake in extremely halophilic microbial communities revealed by microautoradiography and fluorescence in situ hybridization. Extremophiles. 2003;7:409–413. doi: 10.1007/s00792-003-0336-5. [DOI] [PubMed] [Google Scholar]
- Sandaa RA, Skjoldal EF, Bratbak G. Virioplankton community structure along a salinity gradient in a solar saltern. Extremophiles. 2003;7:347–351. doi: 10.1007/s00792-003-0328-5. [DOI] [PubMed] [Google Scholar]
- Santos F, Meyerdierks A, Peña A, Rosselló-Mora R, Amann R, Antón J. Metagenomic approach to the study of halophages: the environmental halophage 1. Environ Microbiol. 2007;9:1711–1723. doi: 10.1111/j.1462-2920.2007.01289.x. [DOI] [PubMed] [Google Scholar]
- Santos F, Yarza P, Briones C, Parro V, Antón J. The metavirome of a hypersaline environment. Environ Microbiol. 2010;12:2965–2976. doi: 10.1111/j.1462-2920.2010.02273.x. [DOI] [PubMed] [Google Scholar]
- Snyder JC, Bateson MB, Lavin M, Young MJ. The use of cellular CRISPR spacer-based microarrays for detection of viruses in 1 environmental samples. Appl Environ Microbiol. 2010;76:7251. doi: 10.1128/AEM.01109-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl DA, Amann RI.1991Development and application of nucleic acid probes in bacterial systematicsStackebrandt E, Goodfellow M (eds). Nucleic Acid Techniques in Bacterial Systematics Wiley & Sons Ltd: Chichester, England; 205–248. [Google Scholar]
- Suttle CA, Chen F. Mechanisms and rates of decay of marine viruses in seawater. Appl Environ Microbiol. 1992;58:3721–3729. doi: 10.1128/aem.58.11.3721-3729.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thingstad TF, Lignell R. Theoretical models for the control of bacterial growth rate, abundance, diversity and carbon demand. Aquat Microb Ecol J. 1997;13:19–27. [Google Scholar]
- Thompson JR, Marcelino LA, Polz MF. Heteroduplexes in mixed-template amplifications: formation, consequence and elimination by ‘reconditioning PCR'. Nucl Acids Res. 2002;30:2083–2088. doi: 10.1093/nar/30.9.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Coscoy L, Zylberberg M, Avila PC, Boushey A, Ganem D, et al. Microarray-based detection and genotyping of viral pathogens. Proc Natl Acad Sci USA. 2002;99:15687–15692. doi: 10.1073/pnas.242579699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster DR, Hekele AG, Lauring AS, Fischer KF, Li H, Yino R, et al. An enhanced single base extension technique for the analysis of complex viral populations. PLoS One. 2009;410:e7453. doi: 10.1371/journal.pone.0007453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinbauer MG. Ecology of prokaryotic viruses. FEMS Microbiol Rev. 2004;28:127–181. doi: 10.1016/j.femsre.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Wilhelm SW, Brigden SM, Suttle CA. A dilution technique for the direct measurement of viral production: a comparison in stratified and tidally mixed coastal waters. Microb Ecol. 2002;43:168–173. doi: 10.1007/s00248-001-1021-9. [DOI] [PubMed] [Google Scholar]
- Willson WH, Schroeder DC, Allen MJ, Holden MT, Parkhill J, Barrell BG, et al. Complete genome sequence and lytic phase transcription profile of a Coccolithovirus. Science. 2005;309:1090–1092. doi: 10.1126/science.1113109. [DOI] [PubMed] [Google Scholar]
- Winter C, Bouvier T, Weinbauer MG, Thingstad TF. Trade-offs between competition and defense specialist among unicellular planktonic organisms: the ‘Killing the Winner' hypothesis revisited. Microb Mol Biol Rev. 2010;74:42–57. doi: 10.1128/MMBR.00034-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wommack KE, Colwell RR. Virioplankton: viruses in aquatic ecosystems. Microb Mol Biol Rev. 2000;641:69–114. doi: 10.1128/mmbr.64.1.69-114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MC, Ruan QG, Yang JJ, Eckenrode S, Wu S, McIndoe RA, et al. A statistical method for flagging weak spots improves normalization and ratio estimates in microarrays. Physiol Genomics. 2001;7:45–53. doi: 10.1152/physiolgenomics.00020.2001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.