Abstract
Proteorhodopsin (PR) is a photoprotein that functions as a light-driven proton pump in diverse marine Bacteria and Archaea. Recent studies have suggested that PR may enhance both growth rate and yield in some flavobacteria when grown under nutrient-limiting conditions in the light. The direct involvement of PR, and the metabolic details enabling light-stimulated growth, however, remain uncertain. Here, we surveyed transcriptional and growth responses of a PR-containing marine flavobacterium during carbon-limited growth in the light and the dark. As previously reported (Gómez-Consarnau et al., 2007), Dokdonia strain MED134 exhibited light-enhanced growth rates and cell yields under low carbon growth conditions. Inhibition of retinal biosynthesis abolished the light-stimulated growth response, supporting a direct role for retinal-bound PR in light-enhanced growth. Among protein-coding transcripts, both PR and retinal biosynthetic enzymes showed significant upregulation in the light. Other light-associated proteins, including bacterial cryptochrome and DNA photolyase, were also expressed at significantly higher levels in the light. Membrane transporters for Na+/phosphate and Na+/alanine symporters, and the Na+-translocating NADH-quinone oxidoreductase (NQR) linked electron transport chain, were also significantly upregulated in the light. Culture experiments using a specific inhibitor of Na+-translocating NQR indicated that sodium pumping via NQR is a critical metabolic process in the light-stimulated growth of MED134. In total, the results suggested the importance of both the PR-enabled, light-driven proton gradient, as well as the generation of a Na+ ion gradient, as essential components for light-enhanced growth in these flavobacteria.
Keywords: flavobacteria, marine, photoheterotrophy, proteorhodopsin, transcriptomics
Introduction
Some prokaryotes possess proteins that interact with light, and convert it into energy for growth or into sensory information. One class of energy-harvesting photoproteins called rhodopsins consist of single, membrane-embedded protein covalently bound to the chromophore retinal (a light-sensitive pigment) (Spudich and Jung, 2005). Ten years ago, a bacterial rhodopsin, proteorhodopsin (PR), was discovered through metagenomic analyses of marine bacterioplankton genome fragments (reviewed by DeLong and Béjà, 2010). Béjà et al. (2000) found that an uncultivated marine SAR86 clade member in gammaproteobacteria contained a bacteriorhodopsin-like gene, dubbed PR. Further, the marine SAR86-derived PR functioned as a light-driven proton pump, when the photoprotein was expressed in Escherichia coli. PRs were subsequently detected in many other marine bacteria, some of which appeared to be ‘tuned' to absorb specific wavelengths of light associated with their habitat of origin (Béjà et al., 2001). Additional studies have found PR genes in a diverse array of abundant marine bacterial and archaeal clades (Giovannoni et al., 2005; Brown and Jung, 2006; Frigaard et al., 2006; McCarren and DeLong, 2007). On the basis of genomic surveys, a large fraction of naturally occurring marine bacterioplankton in oceanic surface seawaters appear to contain the PR gene (de la Torre et al., 2003; Sabehi et al., 2005; Moran and Miller, 2007; DeLong, 2009). Interestingly, chromophore biosynthetic genes including a carotenoid biosynthetic gene cluster, and a novel blh gene encoding a 15,15′-β-carotene dioxygenase that cleaves β-carotene to yield retinal, were found linked to the PR gene in some microorganisms (Sabehi et al., 2005). Martinez et al. (2007) demonstrated that the expression of the entire PR photosystem (genetically linked PR and retinal biosynthetic genes) in E. coli can result in proton-pumping activity in light, and that the resulting proton motive force can be used for adenosine triphosphate (ATP) synthesis. Furthermore, PR in recombinant E. coli can generate a light-driven proton motive force sufficient to increase the rate of flagellar rotation, providing estimates for energy flux through the photosystem (Walter et al., 2007).
PR-containing marine bacterial isolates have been recently cultured from a variety of marine environments. These isolates include members of SAR11 (alphaproteobacteria), OM43 (betaproteobacteria) and SAR 92 (gammaproteobacteria) clades, as well as members of the Bacteroidetes and Vibrionaceae (Giovannoni et al., 2005; Frigaard et al., 2006; McCarren and DeLong, 2007; Stingl et al., 2007; González et al., 2008). Laboratory experiments examining light-stimulated growth in some of these isolates, however, have proven equivocal. Some studies could detect no significant light enhancement of either growth rates or cell yields in PR-containing isolates (Giovannoni et al., 2005; Stingl et al., 2007). However, light-enhanced growth rates and cell yields were reported in one PR-containing marine flavobacterium, Dokdonia sp. MED134 (Gómez-Consarnau et al., 2007). Additionally, microcosm studies suggested that some of marine flavobacteria and SAR11 populations exhibited enhanced expression of the PR gene in the presence of light (Lami et al., 2009). As well, Gómez-Consarnau et al. (2010) demonstrated the enhanced long-term survival of PR-containing Vibrio cells in the light, but not in darkness. Nevertheless, the specific metabolic processes that facilitate PR-enhanced growth or survival are not yet well understood.
To better characterize the photophysiology of PR-containing Flavobacteria, we performed transcriptomic analyses targeting total RNA extracted from MED134 exposed to light or in the dark. Transcriptional profiles derived from cultures incubated in the light and dark were analyzed, and these results were used to further direct laboratory experiments using different growth substrates and inhibitors. The effect of light on growth at various carbon concentrations, and the effect of retinal biosynthesis inhibitors on light-enhanced growth, were explored. In addition, the effects of sodium-translocating respiratory chain inhibitors on light-stimulated growth were also examined. The combined results from both gene expression studies and physiological experiments were used to develop a model that incorporates some of the important features of photoheterotrophic growth observed in Dokdonia strain MED 134.
Materials and methods
Strain and culture conditions
PR-containing marine flavobacterium, Dokdonia sp. MED134, was isolated from surface seawater in Northwest Mediterranean Sea (Gómez-Consarnau et al., 2007). This strain was kindly provided to us by Jarone Pinhassi (University of Kalmar, Sweden). MED134 was grown in artificial seawater (ASW) (35 practical salinity units, prepared from Sea Salts; Sigma, St. Louis, MO, USA) containing a low concentration of dissolved organic carbon (DOC) (0.05 m C). ASW was filter-sterilized through 0.2 μm-pore-size filter system (Nalgene, Rochester, NY, USA) and autoclaved. Then 250 ml aliquots of ASW (containing a background concentration of 0.05 m C of DOC), were partially supplemented with full strength medium (FSM) (0.5 g of peptone (Bacto Pepton, BD) and 0.1 g of yeast extract (Bacto Yeast Extract, BD, Franklin Lakes, NJ, USA) per 100 ml of ASW), to yield final DOC concentrations of 0.14 and 0.39 m C, respectively. All media were also supplemented with 225 μ of NH4Cl and 44.7 μ of Na2HPO4·12H2O, to avoid inorganic nitrogen and phosphate limitation. DOC concentrations were measured using the high temperature combustion method on the total organic analyzer TOC-V (Shimadzu, Kyoto, Japan) with platinized aluminum catalyst. The bacteria were initially grown in ASW enriched to 1.1 m C, washed in ASW and then diluted into three different ASW media, each containing a different DOC concentration (0.05, 0.14 and 0.39 m C). Cultures were incubated at 22 °C under continuous white light (approximately 150 μmol of photons m−2 s−1) or in the darkness.
To determine bacterial cell density, cultures were filtered with pre-blackened Isopore membrane filter (pore size, 0.22 μm; Millipore, Billerica, MA, USA). Bacterial cells on the filter were stained with SYBR Green I (1:100 dilution; Molecular Probes, Eugene, OR, USA) for 15 min, and counted under an epifluorescence microscope (Axioskop 2, Zeiss, Thornwood, NY, USA). All culture experiments were performed in triplicate.
Cultivation for transcriptomic analyses
MED134 was grown in 900 ml of ASW enriched to 0.14 m C at 22 °C in the darkness for the first 2 days. At this time, 400 ml of culture was filtered onto a pore-size 0.22-μm Durapore membrane filter (25 mm diameter, Millipore), yielding the D2 sample. The remaining culture was split in two 250 ml flasks that were incubated again at 22 °C under the continuous white light (approximately 150 μmol of photons m−2 s−1) or in the darkness. After 2 more days, the cultures were filtered onto Durapore membrane filters (Millipore), yielding samples L2 (light conditions) and D4 (dark conditions), respectively. All filtered samples were immediately placed into screw-cap tubes containing 1 ml of RNAlater (Ambion, Austin, TX, USA) and stored at −80 °C until RNA extraction.
Total RNA extraction and ribosomal RNA (rRNA) subtraction
Total RNA was extracted from the filter samples using a modification of the mirVana miRNA isolation kit (Ambion) as described previously (Shi et al., 2009; McCarren et al., 2010). Briefly, filter samples were thawed on ice, and the RNAlater surrounding each filter was removed and discarded. The filters were immersed in Lysis/Binding buffer (Ambion) and mixed to lyse attached cells. Total RNA was extracted from the lysate according to the manufacturer's protocol. Remaining genomic DNA in RNA extraction was removed using a TURBO DNA-free kit (Ambion).
Bulk DNA was extracted as previously described (Shi et al., 2009). Briefly, cells were lysed with lysozyme and proteinase K solution. Then the genomic DNA was extracted with phenol-chloroform-isoamyl alcohol and precipitated with ethanol.
16S and 23S rRNAs were removed by the subtractive hybridization as described by Stewart et al. (2010). Ribonucleotide probes targeting 16S and 23S rRNA genes were generated from the bulk DNA extracted from MED134. Templates for probe generation were first prepared by PCR using Herculase II Fusion DNA Polymerase (Stratagene, La Jolla, CA, USA) and strain-specific primers flanking nearly the full length of the bacterial 16S and 23S rRNA genes, with reverse primers modified to contain the T7 RNA polymerase promoter sequence (Supplementary Table S1). Biotinylated antisense rRNA probes were generated by in vitro transcription with T7 RNA polymerase, ATP, GTP, CTP, UTP, biotin-11-CTP, biotin-16-UTP (Roche, Branford, CT, USA). Biotinylated rRNA probes were hybridized to complimentary rRNA molecules in total RNA sample. Then biotinylated double-stranded rRNA was removed from the sample by hybridization to Streptavidin-coated magnetic beads (New England Biolabs, Ipswich, MA, USA). The subtraction efficiency was evaluated by monitoring the removal of 16S and 23S peaks from total RNA profiles using a 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA).
RNA amplification, complementary DNA (cDNA) synthesis and pyrosequencing
The rRNA-subtracted RNA (10–15 ng) was amplified using the MessageAmp II-Bacteria kit (Ambion) as described previously (Shi et al., 2009; McCarren et al., 2010). In brief, total RNA was polyadenylated using E. coli poly(A) polymerase. Polyadenylated RNA was converted to double-stranded cDNA via reverse transcription primed with an oligo(dT) primer containing a promoter sequence for T7 RNA polymerase and a recognition site for the restriction enzyme BpmI (T7-BpmI-(dT)16VN) (Supplementary Table S1). cDNA was transcribed in vitro at 37 °C for 12 h, yielding large quantities (40–60 μg) of single-stranded antisense RNA. The SuperScript double-stranded cDNA synthesis kit (Invitrogen, Carlsbad, CA, USA) was used to convert antisense RNA to double-stranded cDNA, which was then digested with BpmI to remove poly(A) tails. Before pyrosequencing, poly(A)-removed cDNA was purified using the AMPure kit (Beckman Coulter Genomics, Danvers, MA, USA). Purified cDNA was used for the generation of single-stranded DNA libraries according to established protocols (454 Life Sciences, Roche). The resulting cDNA libraries were then pyrosequenced on the 454 FLX platform (Roche). All the cDNA sequences generated in this study have been submitted to the NCBI short read archive under accession number SRA029329.
Analyses of pyrosequence data
rRNA and transfer RNA (tRNA) reads were identified using BLASTN against rRNA and tRNA sequences in MED134 genome data, which are deposited in GenBank under accession no. AAMZ00000000 (Gómez-Consarnau et al., 2007). Reads producing alignments with bit scores >50 were considered as rRNA and tRNA sequences. Protein-encoding cDNAs (from mRNA) were identified using BLASTX against peptide sequences collected from MED134 genome data (bit score ⩾50). Small RNAs were analyzed using the Rfam (version 10.0) website (http://rfam.sanger.ac.uk/). Rfam is a collection of non-coding RNA families, each represented by multiple sequence alignments, consensus secondary structures and covariance models, including 1446 families in January 2010 (Gardner et al., 2009). Finally, in order to identify MED134-specific small RNA that might not be represented in Rfam, we assembled a database of intergenic regions in the genome of MED134 longer than 100 bp (total 992 sequences), which might encode putative small RNAs. Reads with matches to the intergenic regions database (bit score ⩾50) were considered small RNA reads.
L2/D4 ratios were calculated based on read number of each cDNA, which was normalized by total number of protein-encoding reads in each sample. The statistical significance of the change observed between cultures in light and dark (L2 and D4) for each cDNA was determined based on false-discovery rate method (q-value ⩽0.05) (Benjamini and Hochberg, 1995; Storey and Tibshirani, 2003). Clustering analyses of transcriptomics datasets were performed in GenePattern (Reich et al., 2006), using hierarchical clustering (Eisen et al., 1998) by Pearson correlations for both rows and columns, using pairwise complete-linkage.
Culture experiments with specific inhibitors
To confirm the importance of retinal-bound PR for the light-stimulated growth, we performed culture experiments with 2-(4-methylphenoxy)triethylamine hydrochloride (MPTA). MPTA is known to prevent lycopene cyclization in retinal biosynthesis pathway (Cunningham et al., 1994; Armstrong, 1999). First, we cultured MED134 on Marine Agar 2216 (Difco Laboratories, Detroit, MI, USA) amended with MPTA at a final concentration of 300 μ and confirmed the effect of MPTA against strain MED134 based on the inhibition of β-carotene synthesis as indicated by colony pigmentation. Next, MED134 was grown in ASW slightly enriched with FSM (0.14 m C) and amended with MPTA. MPTA was dissolved in methanol and added to ASW at a final concentration of 100 μ. The same volume of methanol was added to negative control cultures without MPTA. These cultures were incubated at 22 °C under continuous white light (approximately 150 μmol of photons m−2 s−1) or in the darkness. The cultures were performed in triplicate. Bacterial cell density was measured every 2 days by the direct counting method with epifluorescence microscope described above. Additionally, colony-forming units (c.f.u.) were also monitored after spread 100 μl of cultures on Marine Agar 2216 (Difco) and incubation at 22 °C for 48 h. MPTA was a generous gift of Francis X (Buddy) Cunningham (University of Maryland, MD, USA).
To determine the importance of sodium pumping in light-driven growth of PR-containing marine flavobacteria, MED134 was grown in ASW with 2-n-heptyl-4-hydroxyquinoline N-oxide (HQNO) (Enzo Life Sciences, Plymouth Meeting, PA, USA). HQNO is known to be a specific inhibitor of the electron-transport-linked Na+-translocating NADH-quinone oxidoreductase (NQR) enzyme complex (Tokuda and Unemoto, 1982; Häse and Mekalanos, 1999). MED134 was incubated in ASW enriched with -alanine (0.70 m C), supplemented with trace element solution (Futamata et al., 2009) and amended with HQNO at a final concentration of 10 μ. -alanine was selected as carbon source for the bacterial cultivation, as transcriptomic analyses demonstrated significant over representation of Na+/alanine symporters in the presence of light (see Results and Discussion section). HQNO was prepared in ethanol. The same volume of ethanol was added to negative control cultures without HQNO. These cultures were incubated at 22 °C under continuous white light or in the dark. The cultures were performed in triplicate. Bacterial cell densities were measured every 2 days by the direct count and plate count method described above.
Results and Discussion
Cultivation in light and darkness
Dokdonia sp
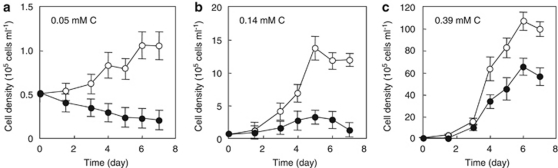
MED134 exposed to light reached a maximal abundance of 1.1 × 105 cells ml−1 in unamended ASW (DOC, 0.05 m C), 1.4 × 106 cellsml−1 in ASW enriched with 0.14 m C and 1.1 × 107 cells ml−1 in ASW enriched to 0.39 m C (Figure 1). In contrast, dark-incubated cultures remained below 5.0 × 104 cells ml−1 in unenriched ASW. In nutrient-enriched ASW media, MED134 grew moderately in the dark, but the cell yields were much lower compared with cultures grown in the light. Light–dark ratios of cell yields ranged from 1.6 to 4.6 at the peak of the growth curves. Growth rates in ASW containing 0.14 m C were 0.69 day−1 in the illuminated culture (logarithmic growth phase, 1.5 to 5 day), and 0.44 day−1 in the dark cultures (logarithmic growth phase, 1.5 to 4 day). Growth rates in ASW containing 0.39 m C were 1.17 day−1 in the light, and 1.01 day−1 in the dark (logarithmic growth phase, 0 to 4 day). These results show the considerable influence of PR on growth rate at low carbon concentrations, and its lesser influence at higher carbon concentrations. These findings confirm the previous work of Gómez-Consarnau et al. (2007), which showed that light has a definite positive impact on the growth of the PR-containing flavobacteria grown in low carbon conditions.
Figure 1.
Growth of MED134 incubated in the light or in the dark. MED134 was grown in unenriched ASW (0.05 m C) (a), in ASW enriched to 0.14 m C (b) and in ASW enriched to 0.39 m C (c). The cultures were incubated under continuous white light (○), or in the darkness (•). Error bars denote s.d. for triplicate cultures.
Transcriptome experiments
For transcriptomic analyses, MED134 was grown in ASW enriched to 0.14 m C. Bacteria grew to 1.0 × 105 cells ml−1 for first 2 days in the dark (D2). After 2 more days, MED134 reached 3.2 × 105 cells ml−1 in light (L2), whereas bacteria incubated in darkness remained 1.2 × 105 cells ml−1 (D4).
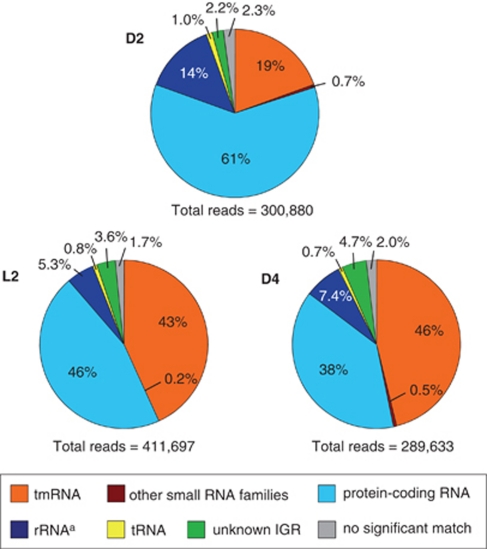
cDNAs synthesized from the RNA samples were pyrosequenced on the Roche 454 FLX platform, yielding ≈400 000 reads per sample. cDNA derived from intergenic regions accounted for 21% to 51% of the total cDNA reads (Figure 2). Remarkably, most of reads derived from intergenic regions in all samples corresponded to a single gene (399 bp) encoding transfer-messenger RNA (Supplementary Table S2). Transfer-messenger RNA is small RNA that employs both tRNA-like and mRNA-like properties as it rescues stalled ribosomes during nutrient shortage (Gillet and Felden, 2001; Moore and Sauer, 2007; Keiler, 2008). As the proportions of transfer-messenger RNA further increased with incubation time, the high percentage of transfer-messenger RNA is likely to be due to the carbon-limiting growth conditions used in this experiment. Protein-encoding transcripts, identified by comparison with the annotated MED134 genome (December 2009 GenBank version), represented 38% to 61% of total cDNA reads (Figure 2). Genes with a significant change in L2/D4 ratio (q-value ⩽0.05) were considered differentially expressed in the light versus the dark. Using these criteria, 601 genes in 2944 annotated protein-encoding genes were found to be differentially expressed. Specifically, 312 genes were upregulated in the light, whereas 289 genes exhibited downregulation in the light.
Figure 2.
Inventory of RNAs from cultures in the microbial transcriptomic datasets. MED 134 was first incubated in ASW enriched to 0.14 m C in the dark for first 2 days (D2). Then culture was split in two flasks, with one incubated in the light (L2), and the other in the dark (D4), for 2 more days. Numbers in the pie charts represent the percentage of total cDNA reads in each transcriptomic dataset. aSubtraction of 16S and 23S rRNAs were performed after total RNA extraction.
PR and retinal biosynthetic enzymes
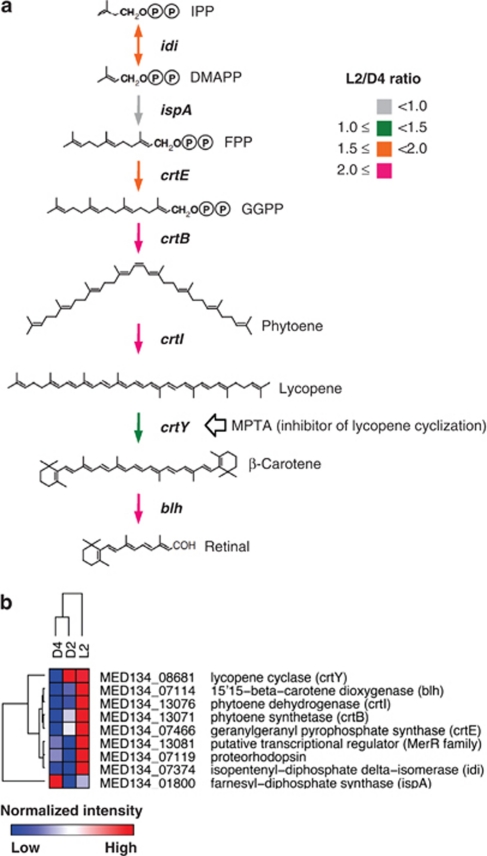
Previous genomic analysis of MED134 revealed the presence of genes encoding PR, and crtEBIY-encoding enzymes needed to synthesize β-carotene from farnesyl diphosphate (Gómez-Consarnau et al., 2007). Further, a gene (blh) encoding an enzyme that converts β-carotene to retinal has been also found next to the PR gene on the genome of MED134. Our transcriptomic survey revealed that the L2/D4 ratios for the PR gene, crtEBIY and blh were elevated (Figure 3a). In particular, statistical significance tests based on q-values showed that the PR, crtE and crtI genes were significantly upregulated in the culture exposed to light (Table 1). In addition, hierarchical clustering of transcript abundances clustered D2 and D4 together, to the exclusion of L2 (Figure 3b). This clustering pattern reflects the differential response of this PR-containing flavobacterium to light. Gómez-Consarnau et al. (2007) demonstrated by reverse transcriptase-PCR that MED134 had a higher expression of PR gene in the light than in the dark. Lami et al. (2009) also reported that marine flavobacteria and SAR11 in natural costal seawaters displayed significant high expression of PR gene in the presence of light. Our results extend these previous reports and indicate that transcription of the entire PR photosystem is upregulated in the presence of light in this flavobacterium.
Figure 3.
Transcriptomic analyses of proteorhodopsin and retinal biosynthetic genes. (a) Retinal biosynthetic pathway. The colors indicate the L2/D4 ratio of retinal biosynthetic enzymes. The ratio was calculated based on abundance of reads for each specific gene, normalized by the total number of protein-encoding reads for each sample. (b) Cluster analysis of the relative abundance of PR and retinal biosynthetic enzymes. Hierarchical clustering was performed based on the number of cDNA reads, normalized to the total number of protein-encoding cDNAs in each sample, using a Pearson correlation. The heat map shows relative difference of transcript abundance in each sample (red indicates high Pearson correlation; white indicates intermediate; blue indicates low). The numbers of cDNA reads are presented in Table 1. IPP, isopentenyl pyrophosphate; DMAPP, dimethylallyl pyrophosphate; FPP, farnesyl pyrophosphate; GGPP, geranylgeranyl pyrophosphate.
Table 1. Read numbers and L2/D4 ratios of protein-encoding cDNA associated with light-stimulated growth.
| Name | Locus tag | Size (aa) |
Read number |
L2/D4 ratio | q-Value | ||
|---|---|---|---|---|---|---|---|
| D2 | L2 | D4 | |||||
| Opsin | |||||||
| Proteorhodopsin | MED134_07119 | 247 | 1 | 33 | 5 | 3.92 | 0.0129 |
| Retinal biosynthetic enzymes | |||||||
| Isopentenyl-diphosphate -isomerase (idi) | MED134_07374 | 172 | 17 | 36 | 11 | 1.94 | 0.1568 |
| Farnesyl-diphosphate synthase (ispA) | MED134_01800 | 325 | 27 | 30 | 21 | 0.85 | 0.7416 |
| Geranylgeranyl pyrophosphate synthase (crtE) | MED134_07466 | 324 | 53 | 77 | 23 | 1.99 | 0.0160 |
| Phytoene synthetase (crtB) | MED134_13071 | 279 | 20 | 34 | 8 | 2.53 | 0.0660 |
| Phytoene dehydrogenase (crtI) | MED134_13076 | 486 | 37 | 86 | 20 | 2.56 | 0.0005 |
| Lycopene cyclase (crtY) | MED134_08681 | 403 | 10 | 10 | 4 | 1.49 | 0.7585 |
| 15,15′-b-carotene dioxygenase (blh) | MED134_07114 | 287 | 2 | 4 | 1 | 2.38 | 0.8027 |
| Transcriptional regulator, MerR family | MED134_13081 | 309 | 9 | 22 | 7 | 1.87 | 0.3648 |
| Transporters | |||||||
| Na+/alanine and glycine symporter | MED134_02355 | 561 | 90 | 84 | 27 | 1.85 | 0.0241 |
| Na+/alanine and glycine symporter | MED134_14567 | 509 | 64 | 52 | 13 | 2.38 | 0.0243 |
| Na+/phosphate symporter | MED134_11180 | 745 | 81 | 64 | 15 | 2.54 | 0.0049 |
| Electron transport chain | |||||||
| Na+-translocating NADH quinone oxidoreductase (nqrA) | MED134_00295 | 448 | 713 | 1308 | 518 | 1.50 | <0.0001 |
| Na+-translocating NADH quinone oxidoreductase (nqrB) | MED134_00300 | 400 | 406 | 753 | 268 | 1.67 | <0.0001 |
| Na+-translocating NADH quinone oxidoreductase (nqrC) | MED134_00305 | 248 | 101 | 181 | 56 | 1.92 | 0.0001 |
| Na+-translocating NADH quinone oxidoreductase (nqrD) | MED134_00310 | 215 | 93 | 147 | 34 | 2.57 | <0.0001 |
| Na+-translocating NADH quinone oxidoreductase (nqrE) | MED134_00315 | 228 | 73 | 97 | 38 | 1.52 | 0.1115 |
| Na+-translocating NADH quinone oxidoreductase (nqrF) | MED134_00320 | 435 | 250 | 485 | 165 | 1.75 | <0.0001 |
Abbreviation: cDNA, complementary DNA. Statistical significance between light and dark cultures was measured based on q-value (false-discovery rate method). The features with q-values ⩽0.05 are significant (Storey and Tibshirani, 2003), which are in bold.
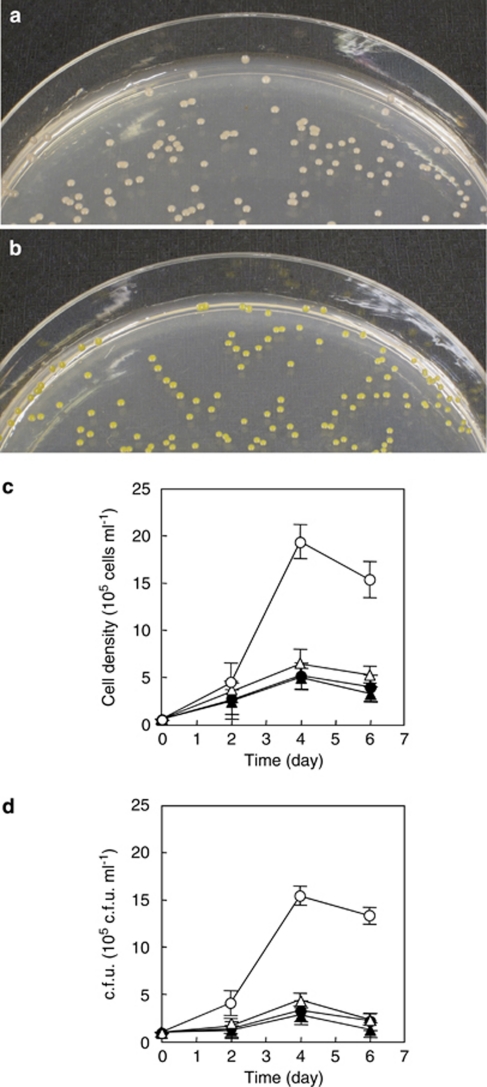
Previous evidence supporting the role of PR in light-stimulated growth in strain MED134 was the observation that only light corresponding to the wavelengths absorbed by PR elicited growth enhancement (Gómez-Consarnau et al., 2007). To provide additional data supporting the role of PR in light-stimulated growth in strain MED134, we performed culture experiments with MPTA, a specific inhibitor of lycopene cyclization in the retinal biosynthetic pathway (Cunningham et al., 1994; Armstrong, 1999; see Figure 3a). First, MED134 was grown on agar plates with and without MPTA. In the presence of MPTA, MED134 produced light pink colonies, whereas yellow colonies were observed on agar plates without MPTA (Figures 4a and b). In general, it is known that bacterial cells accumulating β-carotene display yellow or orange colonies, whereas light pink colonies indicate accumulation of lycopene (Cunningham et al., 1994; Armstrong, 1999). Our results indicate that MPTA effectively prevented β-carotene generation, the precursor for retinal, in strain MED134. Next, MED134 was grown in ASW enriched to 0.14 m C and amended with MPTA. When strain MED134 was incubated in ASW with MPTA, bacteria grew moderately to 6.3 × 105cells ml−1 and 4.3 × 105 c.f.u. ml−1 in the presence of light (Figures 4c and d). In contrast, MED134 incubated in ASW without MPTA in the light produced significantly higher yields (1.9 × 106 cells ml−1 and 1.5 × 106 c.f.u. ml−1) than those in ASW with MPTA. In culture experiments in the dark with or without MPTA, bacteria grew similarly to approximately 5.0 × 105 cells ml−1 (3.0 × 105 c.f.u. ml−1), equivalent to the results with MPTA in the light. The findings suggest that PR bound to retinal has a critical role in the light-stimulated growth of the PR-containing marine flavobacteria.
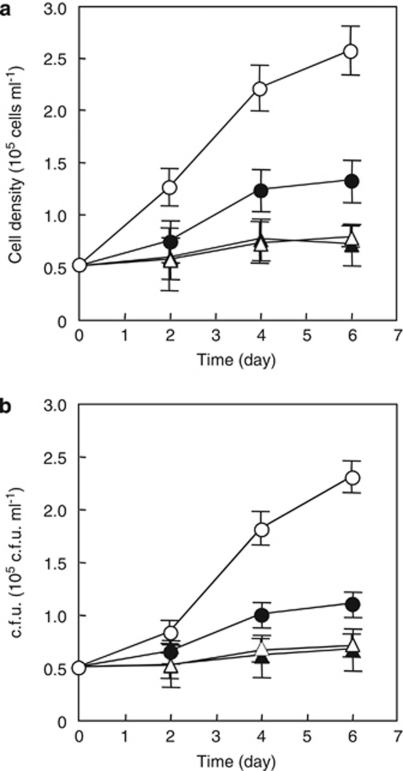
Figure 4.
Colony image and growth of MED134 in culture experiments with a specific inhibitor of lycopene cyclization, MPTA. (a, b) Colony pigmentation of MED134 on Marine Agar 2216 (Difco) plate amended with MPTA (a) and without MPTA (b). (c, d) Microbial cell density in enriched ASW (0.14 m C) amended with MPTA in culture exposed to light (▵) and in the dark (▴), and in the enriched ASW without MPTA in culture exposed to light (○) and in the dark (•). Bacterial cells were counted by epifluorescence microscopy (c) and plate count method (d).
ATP synthetase
The genome of MED134 harbors genes encoding membrane-embedded ATP synthetase, a multi-unit enzyme consisting of two large complexes. In this study, large numbers of transcripts from ATP synthetase were identified; however, there was no significant difference in ATP synthetase transcript abundance in the light and dark cultures (Supplementary Table S3).
Light sensors
MED134 contains several gene homologs of membrane sensors known to respond to light. González et al. (2008) reported that MED134 contains genes encoding bacterial cryptochrome and several DNA photolyase/cryptochromes that belong to different gene families (DASH family, (6–4) photolyase family and class I photolyase). Similar genes encoding these light sensor proteins were also identified in the genome sequence of Polaribacter sp. MED152, another marine flavobacterium (González et al., 2008). Further, MED134 has PAS (period clock protein, aryl hydrocarbon receptor and single-minded protein) and GAF (cGMP phosphodiesterase, adenylyl cyclase and FhlA) domains, known to be common components of phytochromes that detect red and far-red light (Taylor and Zhulin, 1999; Anantharaman et al., 2001). PAS domains are able to respond to oxygen levels, redox potential and light, whereas GAF domains work as phototransducers. Another gene associated with light sensing contains the BLUF (blue light sensing using FAD) domain, which specifically responds to blue light (Gomelsky and Klug, 2002). The BLUF domain is also found in the genome of related Polaribacter sp. MED152 (González et al., 2008). In addition to light sensors, several histidine kinases, which might have important roles for secondary transduction and response regulation, are contained in the genome of MED134.
In this study, we found significant upregulation of bacterial cryptochrome and two putative DNA photolyase/cryptochrome genes under light (L2) versus dark (D4) conditions (Table 2). In contrast, genes encoding PAS, GAF and BLUF domains exhibited no significant difference between light and dark cultures (Supplementary Figure S1). Of the secondary transduction enzymes, one histidine kinase (MED134_10396) showed very high expression ratio (L2/D4 ratio, 18.3) and significant upregulation in light (Table 2). This histidine kinase may therefore be involved in controlling gene expression in response to light.
Table 2. Read number and L2/D4 ratio of cDNA-encoding domains and peptides with a role in light absorption and response.
| Name | Locus tag | Size (aa) |
Read number |
L2/D4 ratio | q-Value | ||
|---|---|---|---|---|---|---|---|
| D2 | L2 | D4 | |||||
| Bacterial cryptochrome, DASH family | MED134_10201 | 432 | 9 | 29 | 5 | 3.45 | 0.0341 |
| DNA photolyase/cryptochrome, (6-4) photolyase family | MED134_10206 | 511 | 5 | 22 | 2 | 6.54 | 0.0149 |
| DNA photolyase/cryptochrome, (6-4) photolyase family | MED134_10211 | 494 | 3 | 24 | 3 | 4.75 | 0.0244 |
| DNA photolyase/cryptochrome, class I photolyase | MED134_14266 | 436 | 37 | 38 | 12 | 1.88 | 0.1700 |
| PAS domain | MED134_02435 | 1200 | 87 | 66 | 37 | 1.06 | 0.9091 |
| GAF domain | MED134_01075 | 151 | 53 | 56 | 37 | 0.90 | 0.8120 |
| Phytochrome region | MED134_02440 | 749 | 27 | 32 | 9 | 2.11 | 0.1574 |
| BLUF domain | MED134_02460 | 338 | 38 | 18 | 11 | 0.97 | 0.9371 |
| Multi-sensor hybrid His kinase | MED134_01400 | 741 | 109 | 116 | 37 | 1.86 | 0.0058 |
| Two-component system sensor His kinase | MED134_07876 | 182 | 11 | 12 | 12 | 0.59 | 0.4031 |
| Sensory transduction His kinase | MED134_10396 | 163 | 3 | 123 | 4 | 18.3 | <0.0001 |
| Sensory transduction His kinase | MED134_06794 | 657 | 48 | 54 | 31 | 1.04 | 0.9371 |
| Sensory transduction His kinase | MED134_07881 | 470 | 42 | 29 | 15 | 1.15 | 0.8630 |
Statistical significance between light and dark cultures was measured based on q-value (false-discovery rate method). The features with q-values ⩽0.05 (in bold) are considered significant (Storey and Tibshirani, 2003).
Central metabolic pathways
The main energy generating metabolic pathways of MED134 identified by genome analyses were glycolysis, the pentose phosphate cycle, and the tricarboxylic acid (TCA) cycle (Gómez-Consarnau et al., 2007). Several genes encoding enzymes working in the central metabolic pathways were significantly induced in culture exposed to light (Supplementary Table S4). In particular, genes encoding fructose-bisphosphatase (fbp) in glycolysis, glucoce-6-phosphate dehydrogenase (zwf) and phosphogluconate dehydrogenase (gnd) in pentose phosphate cycle, and succinate dehydrogenase (sdhABC) and fumarate hydratase (fumC) in TCA cycle exhibited significant upregulation in the light. In addition, hierarchical clustering of transcript abundances clustered D2 and L2 together, to the exclusion of D4 (Supplementary Figure S2). This clustering pattern may reflect the increased levels of carbon available in the D2 culture, and the additional energy source (light) available in the L2 culture, relative to the D4 culture. Hence, the expression pattern of enzymes of the central metabolic pathways may reflect the differential nutrient and energy availability between the different treatments.
MED134 also harbors genes encoding pyruvate carboxylase (pycA) and phosphoenolpyruvate (PEP) carboxylase (ppc) that function in anaplerotic metabolism and that are associated with carbon fixation (González et al., 2008). PycA generates oxaloacetate from bicarbonate and pyruvate, whereas PEP carboxylase synthesizes oxaloacetate from bicarbonate and PEP (Attwood and Wallace, 2002; Izui et al., 2004; Jitrapakdee et al., 2008). The transcript abundance of pycA and PEP carboxylase were significantly downregulated in the light, compared with that in the dark (Supplementary Table S5). In contrast, SulP-type bicarbonate transporter and carbonic anhydrase, which is known to interconvert CO2 and bicarbonate, were not significantly different between L2 and D4. Although Polaribacter sp. MED152 has been reported to fix more bicarbonate in the light than in the darkness (González et al., 2008), our findings indicate that in MED134 CO2 incorporation via pycA and PEP carboxylase may be equally important for anaplerotic carbon replenishment under both dark and light growth conditions.
Transporters and electron transport chain
Membrane transporters have critical roles in uptake of essential nutrients and minerals. The genome of MED134 contains relatively low numbers of genes encoding membrane transporters, compared with other marine bacteria. Transcriptomic analyses revealed that several of them exhibited significant upregulation in the presence of light (Supplementary Table S6). In particular, two predicted Na+/alanine symporters (MED134_02355, MED134_14567) were significantly upregulated in the light (Table 1 and Supplementary Figure S3). Further, a Na+/phosphate symporter (MED134_11180) also exhibited significant upregulation in the light. Transcriptome analyses also indicated significant upregulation of Na+-translocating NQR, sdhABC, and cytochrome c oxidase in the light (Table 1, Supplementary Table S7, and Supplementary Figure S4). These results suggested the potential importance of the sodium ion gradient for transport functions in the light, and indirect light-stimulation of sodium pump activities via the light-driven proton gradient (see Supplementary Figure S5).
To test the importance of sodium ion exchange in light-stimulated physiology of MED134, we performed growth experiments with HQNO, a specific inhibitor of Na+-translocating NQR (Tokuda and Unemoto, 1982; Häse and Mekalanos, 1999). MED134 was grown in ASW with and without HQNO. For these experiments, -alanine was chosen as carbon source, because of the high expression levels of Na+/alanine symporter we observed in cultures grown in the light. In cultures without HQNO, MED134 increased to 2.6 × 105 cells ml−1 (2.3 × 105 c.f.u. ml−1) in the light and 1.3 × 105 cells ml−1 (1.1 × 105 c.f.u. ml−1) in the dark (Figure 5) when grown on -alanine. Cell yields in cultures incubated with HQNO in the light were about three times less than those grown in the absence of inhibitor. In contrast, for cultures grown in the dark, cell yields decreased by about 1/2 in the presence of the inhibitor, although the amount of growth on alanine was much lower. These findings indicate that the Na+-translocating NQR has a critical role in sodium pumping in light-stimulated growth, and that active PR photophysiology greatly enhances the ability of these flavobacteria to grow on DL alanine.
Figure 5.
Growth of MED134 in cultures incubated with HQNO, a specific inhibitor of Na+-translocating NQR. MED134 was grown in ASW enriched with -alanine (0.7 m C). Cultures were incubated in the ASW amended with HQNO in light (▵) or dark (▴) and in the ASW without HQNO in light (○) or dark (•). Bacterial cell densities were determined by epifluorescence microscopy (a) and plate count method (b).
Conclusion
In this study, we characterized gene expression patterns associated with the higher growth rates and cell yields observed in flavobacterium strain MED134 in carbon-limited media, when grown in the light. Among protein-encoding transcripts, a number of genes were upregulated in the light, including PR, retinal biosynthetic enzymes, and several predicted light sensors (Figure 6). Previous studies had suggested the involvement of PR in light-enhanced growth, because the action spectra for this response generally matched the PR absorption spectrum. Experiments with MPTA, a specific inhibitor of an enzyme in retinal biosynthetic pathway, confirmed the involvement of PR in the observed light-enhanced growth. MED134 cultures grown with MPTA exhibited much lower cell yields than those without MPTA, when both were grown in the presence of light, while MPTA had no effect on MED134 grown in the dark. Proteins involved in some central metabolic pathways, including fbp, zwf, gnd, sdhABC and fumC, also had relatively higher expression in light than dark, suggesting an increased requirement for these enzymes during active cell growth. Transcripts for membrane-embedded ATP synthase, and pycA and PEP carboxylase were well represented in both the light and dark grown cultures. These results likely reflect a central and essential requirement for ATP synthetase and the two carboxylases, in both the light and the dark.
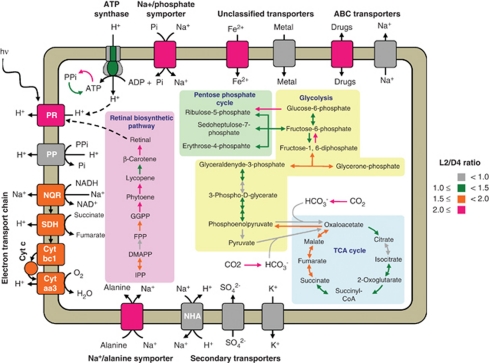
Figure 6.
Model of light-stimulated transcriptional responses in MED134. Proton-pumping processes and retinal biosynthetic pathway are shown in left, whereas central metabolic pathways, such as glycolysis, pentose phosphate cycle, and TCA cycle, are in right. The color of arrows and membrane proteins indicate the value of L2/D4 ratio. The ratio is calculated based on abundance of reads for each gene normalized by total number of protein-encoding reads for each sample. PR, proteorhodopsin; PP, pyrophosphatase; SDH, succinate dehydrogenase; Cyt, cytochrome oxidase; NHA, Na+/H+ antiporter.
Of the membrane transporters, Na+/phosphate symporter and Na+/alanine symporter exhibited particularly significant upregulation in cultures exposed to light. The Na+-translocating NQR linked electron transport chain also exhibited significantly greater expression levels in the light. The gene expression levels of other sodium pumps however, including the Na+/H+ antiporter and the Na+ efflux pump, did not appear significantly different between the L2 and D4 treatments (Figure 6). These results indicate the potential importance of these symporters, coupled with the central role of NQR in driving energy-requiring nutrient transport via its maintenance of the sodium ion gradient. Culture experiments using a specific inhibitor of Na+-translocating NQR, HQNO, also suggested the importance of the sodium ion gradient for MED134 growth, and indicated that sodium pumping via Na+-translocating NQR is critical metabolic process for the light-stimulated growth of PR-containing marine flavobacteria, in addition to proton pumping via retinal-bound PR. In total, our findings support a direct role for retinal-bound PR in light-enhanced growth in Dokdonia strain MED134. Furthermore, an important role for H+/Na+ ion exchange, and transport processes that utilize energy derived from the sodium ion gradient, appear particularly important for the photoheterotrophic growth in this flavobacterium strain.
Acknowledgments
We thank Jarone Pinhassi for providing strain MED134 and Francis X (Buddy) Cunningham for providing MPTA. We are also grateful to Jamie W Becker for measuring DOC in media, Frank J Stewart for helping with the rRNA subtraction, Yanmei Shi for helping small RNA analyses and Rachel Barry for work in preparing samples for pyrosequencing. This work was supported by a grant from the Gordon and Betty Moore Foundation (EFD), the Office of Science (BER), US Department of Energy (EFD) and NSF Science and Technology Center Award EF0424599. This work is a contribution of the Center for Microbial Oceanography: Research and Education (C-MORE). H Kimura was supported by Postdoctoral Fellowships for Research Abroad of Japan Society for the Promotion of Science (JSPS).
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Anantharaman V, Koonin EV, Aravind L. Regulatory potential, phyletic distribution and evolution of ancient, intracellular small-molecule-binding domains. J Mol Biol. 2001;307:1271–1292. doi: 10.1006/jmbi.2001.4508. [DOI] [PubMed] [Google Scholar]
- Armstrong G.1999Carotenoid genetics and biochemistryIn: Cane DE (ed.).Isoprenoids Including Carotenoids and Steroids vol. 2Elsevier: Amsterdam, The Netherlands; 321–352. [Google Scholar]
- Attwood PV, Wallace JC. Chemical and catalytic mechanisms of carboxyl transfer reactions in biotin-dependent enzymes. Acc Chem Res. 2002;35:113–120. doi: 10.1021/ar000049+. [DOI] [PubMed] [Google Scholar]
- Béjà O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP, et al. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science. 2000;289:1902–1906. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- Béjà O, Spudich EN, Spudich JL, Leclerc M, DeLong EF. Proteorhodopsin phototrophy in the ocean. Nature. 2001;411:786–789. doi: 10.1038/35081051. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- Brown LS, Jung K-H. Bacteriorhodopsin-like proteins of eubacteria and fungi: the extent of conservation of the haloarchaeal proton-pumping mechanism. Photochem Photobiol Sci. 2006;5:538–546. doi: 10.1039/b514537f. [DOI] [PubMed] [Google Scholar]
- Cunningham FX, Sun Z, Chamovitz D, Hirschberg J, Gantt E. Molecular structure and enzymatic function of lycopene cyclase from the cyanobacterium Synechococcus sp strain PCC7942. Plant Cell. 1994;6:1107–1121. doi: 10.1105/tpc.6.8.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre JR, Christianson LM, Béjà O, Suzuki MT, Karl DM, Heidelberg J, et al. Proteorhodopsin genes are distributed among divergent marine bacterial taxa. Proc Natl Acad Sci USA. 2003;100:12830–12835. doi: 10.1073/pnas.2133554100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong EF. The microbial ocean from genomes to biomes. Nature. 2009;459:200–206. doi: 10.1038/nature08059. [DOI] [PubMed] [Google Scholar]
- DeLong EF, Béjà O. Light-driven proton pump proteorhodopsin enhances bacterial survival during tough times. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigaard N-U, Martinez A, Mincer TJ, DeLong EF. Proteorhodopsin lateral gene transfer between marine planktonic Bacteria and Archaea. Nature. 2006;439:847–850. doi: 10.1038/nature04435. [DOI] [PubMed] [Google Scholar]
- Futamata H, Kaiya S, Sugawara M, Hiraishi A. Phylogenetic and transcriptional analyses of a tetrachloroethene-dechlorinating ‘Dehalococcoides' enrichment culture TUT2264 and its reductive-dehalogenase genes. Microbes Environ. 2009;24:330–337. doi: 10.1264/jsme2.me09133. [DOI] [PubMed] [Google Scholar]
- Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, et al. Rfam: updates to the RNA families database. Nucleic Acids Res. 2009;37:D136–D140. doi: 10.1093/nar/gkn766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet R, Felden B. Emerging views on tmRNA-mediated protein tagging and ribosome rescue. Mol Microbiol. 2001;42:879–885. doi: 10.1046/j.1365-2958.2001.02701.x. [DOI] [PubMed] [Google Scholar]
- Giovannoni SJ, Bibbs L, Cho J-C, Stapels MD, Desiderio R, Vergin KL, et al. Proteorhodopsin in the ubiquitous marine bacterium SAR11. Nature. 2005;438:82–85. doi: 10.1038/nature04032. [DOI] [PubMed] [Google Scholar]
- Gomelsky M, Klug G. BLUF: a novel FAD-binding domain involved in sensory transduction in microorganisms. Trends Biochem Sci. 2002;27:497–500. doi: 10.1016/s0968-0004(02)02181-3. [DOI] [PubMed] [Google Scholar]
- Gómez-Consarnau L, Akram N, Lindell K, Pedersen A, Neutze R, Milton DL, et al. 2010Proteorhodopsin phototrophy promotes survival of marine bacteria during starvation PLoS Biol 8e1000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Consarnau L, González JM, Coll-Lladó M, Gourdon P, Pascher T, Neutze R, et al. Light stimulates growth of proteorhodopsin-containing marine Flavobacteria. Nature. 2007;445:210–213. doi: 10.1038/nature05381. [DOI] [PubMed] [Google Scholar]
- González JM, Fernández-Gómez B, Fernàndez-Guerra A, Gómez-Consarnau L, Sánchez O, Coll-Lladó M, et al. Genome analysis of the proteorhodopsin-containing marine bacterium Polaribacter sp. MED152 (Flavobacteria) Proc Natl Acad Sci USA. 2008;105:8724–8729. doi: 10.1073/pnas.0712027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häse CC, Mekalanos JJ. Effects of changes in membrane sodium flux on virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci USA. 1999;96:3183–3187. doi: 10.1073/pnas.96.6.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izui K, Matsumura H, Furumoto T, Kai Y. Phosphoenolpyruvate carboxylase: a new era of structural biology. Annu Rev Plant Biol. 2004;55:69–84. doi: 10.1146/annurev.arplant.55.031903.141619. [DOI] [PubMed] [Google Scholar]
- Jitrapakdee S, St Maurice M, Rayment I, Cleland WW, Wallace JC, Attwood PV. Structure, mechanism and regulation of pyruvate carboxylase. Biochem J. 2008;413:369–387. doi: 10.1042/BJ20080709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler KC. Biology of trans-translation. Annu Rev Microbiol. 2008;62:133–151. doi: 10.1146/annurev.micro.62.081307.162948. [DOI] [PubMed] [Google Scholar]
- Lami R, Cottrell MT, Campbell BJ, Kirchman DL. Light-dependent growth and proteorhodopsin expression by Flavobacteria and SAR11 in experiments with Delaware coastal waters. Environ Microbiol. 2009;11:3201–3209. doi: 10.1111/j.1462-2920.2009.02028.x. [DOI] [PubMed] [Google Scholar]
- Martinez A, Bradley AS, Waldbauer JR, Summons RE, DeLong EF. Proteorhodopsin photosystem gene expression enables photophosphorylation in a heterologous host. Proc Natl Acad Sci USA. 2007;104:5590–5595. doi: 10.1073/pnas.0611470104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarren J, Becker JW, Repeta DJ, Shi Y, Young CR, Malmstrom RR, et al. Microbial community transcriptomes reveal microbes and metabolic pathways associated with dissolved organic matter turnover in the sea. Proc Natl Acad Sci USA. 2010;107:16420–16427. doi: 10.1073/pnas.1010732107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarren J, DeLong EF. Proteorhodopsin photosystem gene clusters exhibit co-evolutionary trends and shared ancestry among diverse marine microbial phyla. Environ Microbiol. 2007;9:846–858. doi: 10.1111/j.1462-2920.2006.01203.x. [DOI] [PubMed] [Google Scholar]
- Moore SD, Sauer RT. The tmRNA system for translational surveillance and ribosome rescue. Annu Rev Biochem. 2007;76:101–124. doi: 10.1146/annurev.biochem.75.103004.142733. [DOI] [PubMed] [Google Scholar]
- Moran MA, Miller WL. Resourceful heterotrophs make the most of light in the coastal ocean. Nat Rev Microbiol. 2007;5:792–800. doi: 10.1038/nrmicro1746. [DOI] [PubMed] [Google Scholar]
- Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- Sabehi G, Loy A, Jung KH, Partha R, Spudich JL, Isaacson T, et al. New insights into metabolic properties of marine bacteria encoding proteorhodopsins. PLoS Biol. 2005;3:e273. doi: 10.1371/journal.pbio.0030273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Tyson GW, DeLong EF. Metatranscriptomics reveals unique microbial small RNAs in the ocean's water column. Nature. 2009;459:266–269. doi: 10.1038/nature08055. [DOI] [PubMed] [Google Scholar]
- Spudich JL, Jung KH.2005Microbial rhodopsin: phylogenetic and functional diversityIn: Briggs WR, Spudich JL (eds).Handbook of Photosensory Receptors Wiley-VCH Verlag GmbH & Co KGaA: Weinheim, Germany; 1–24. [Google Scholar]
- Stewart FJ, Ottesen EA, DeLong EF. Development and quantitative analyses of a universal rRNA-subtraction protocol for microbial metatranscriptomics. ISME J. 2010;4:896–907. doi: 10.1038/ismej.2010.18. [DOI] [PubMed] [Google Scholar]
- Stingl U, Desiderio RA, Cho J-C, Vergin KL, Giovannoni SJ. The SAR92 clade: an abundant coastal clade of culturable marine bacteria possessing proteorhodopsin. Appl Environ Microbiol. 2007;73:2290–2296. doi: 10.1128/AEM.02559-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BL, Zhulin IB. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuda H, Unemoto T. Characterization of the respiration-dependent Na+ pump in the marine bacterium Vibrio alginolyticus. J Biol Chem. 1982;257:10007–10014. [PubMed] [Google Scholar]
- Walter JM, Greenfield D, Bustamante C, Liphardt J. Light-powering Escherichia coli with proteorhodopsin. Proc Natl Acad Sci USA. 2007;104:2408–2412. doi: 10.1073/pnas.0611035104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.