Abstract
Experimental analysis of gut microbial communities and their interactions with vertebrate hosts is conducted predominantly in domesticated animals that have been maintained in laboratory facilities for many generations. These animal models are useful for studying coevolved relationships between host and microbiota only if the microbial communities that occur in animals in lab facilities are representative of those that occur in nature. We performed 16S rRNA gene sequence-based comparisons of gut bacterial communities in zebrafish collected recently from their natural habitat and those reared for generations in lab facilities in different geographic locations. Patterns of gut microbiota structure in domesticated zebrafish varied across different lab facilities in correlation with historical connections between those facilities. However, gut microbiota membership in domesticated and recently caught zebrafish was strikingly similar, with a shared core gut microbiota. The zebrafish intestinal habitat therefore selects for specific bacterial taxa despite radical differences in host provenance and domestication status.
Keywords: bacteria, Danio rerio, fish, gastrointestinal tract, microbiome, pyrosequencing
Introduction
Early stages of vertebrate development typically occur in the protected confines of the chorion, an environment devoid of microorganisms. Upon leaving this germ-free environment at birth, vertebrates are exposed to the microorganisms present in their respective local environment. The external surfaces of the vertebrate body are subsequently colonized with microbes, with the majority of these microbial residents assembling into dense gastrointestinal tract communities (gut microbiota). Understanding how host-associated microbiotas assemble requires the use of model systems that reflect natural host community establishment and that allow for the rigorous experimental analysis of the microbiota. Our knowledge of how gut microbial communities assemble and interact with vertebrate hosts is largely derived from a few laboratory model species including mice, rats, and zebrafish (Bäckhed et al., 2005; Cheesman and Guillemin, 2007; Camp et al., 2009). However, the use of lab-reared animals to study these complex and subtle host-microbiota interactions is appropriate only if those interactions in the lab are representative of the interactions that occur in nature. Laboratory animals are usually reared in large enclosed facilities, where they have been domesticated over the course of many generations after their wild ancestors were originally collected from their respective natural habitat. If gut microbial communities are strongly shaped by the composition of the microbial community present in the local environment, then this temporal and spatial separation of domesticated lab animals from their natural habitat could result in significant differences in gut microbial community composition compared with wild hosts. This would also be predicted to result in significant variation in gut microbiota composition in animals raised in different lab facilities with distinct husbandry practices and histories. In contrast, if gut microbial community composition is strongly shaped by selective pressures that occur within the host gut habitat, then the microbial communities that assemble in the intestines of wild hosts and those maintained for generations in different lab facilities should be similar and perhaps share a core microbiota.
The zebrafish (Danio rerio; superorder Ostariophysi, order Cypriniformes) is an omnivorous freshwater teleost fish indigenous to the inland waters of Pakistan, India, Bangladesh, Nepal and Burma (Engeszer et al., 2007). Over the last 40 years, the zebrafish has emerged as a pre-eminent vertebrate model organism for biomedical research. Although zebrafish had long been circulated in the global pet trade, it was Dr George Streisinger at the University of Oregon who brought zebrafish into the laboratory setting in the late 1960s to develop the forward genetic techniques that would ultimately establish zebrafish as a robust research model (Grunwald and Eisen, 2002). Within a typical modern zebrafish laboratory, zebrafish of different genetic backgrounds are maintained in an indoor recirculating or flow-through aquaculture system under constant temperature and light cycle conditions, and fed combinations of artificial and live diets (Westerfield, 2000; Lawrence, 2007). Although significant differences exist between husbandry practices in different zebrafish aquaculture facilities, their potential impact on zebrafish biology has not been adequately examined.
Although the zebrafish has been used extensively to study vertebrate development and physiology, it has only recently been established as a model for studying host-microbiota interactions. We have developed gnotobiotic husbandry methods for the zebrafish, and used them to reveal host responses to the gut microbiota including effects on innate immunity, nutrient metabolism, and intestinal epithelial differentiation and renewal (Rawls et al., 2004, 2006; Bates et al., 2006, 2007; Cheesman and Guillemin, 2007; Cheesman et al., 2011; Kanther and Rawls, 2010). Preliminary insights into the membership of the zebrafish gut microbiota have been provided by sequencing libraries of bacterial 16S rRNA genes amplified from pooled intestinal samples from zebrafish reared in laboratory aquaculture facilities (Rawls et al., 2004, 2006; Bates et al., 2006; Brugman et al., 2009). These results indicate that the zebrafish gut microbiota is numerically dominated at all stages of the zebrafish life cycle by members of the bacterial phylum Proteobacteria, with the phyla Firmicutes and Fusobacteria also prevalent during larval and adult stages respectively. However, it is unknown if the gut microbiota of domesticated lab-reared zebrafish are similar to zebrafish collected from their natural habitat, nor how the composition of the zebrafish gut microbiota varies between zebrafish from different aquaculture facilities. Moreover, all previous 16S rRNA gene sequence-based surveys of the zebrafish gut microbiota have been limited to clone library analysis of only a few hundred sequences per sample, thereby only identifying the most abundant bacterial taxa. Here, we show that the gut microbiota of laboratory-reared zebrafish is similar in composition to that of zebrafish collected recently from their natural habitat. Furthermore, we demonstrate that the zebrafish gut microbiota varies among fish from different geographically separated facilities, and that this variation is explained in part by the historical connections between particular zebrafish facilities. Finally, we identify shared bacterial members of the gut microbiotas of recently caught zebrafish and domesticated zebrafish in different locations, which might comprise a zebrafish core gut microbiota.
Materials and methods
Zebrafish husbandry
All zebrafish experiments were conducted in conformity with the Public Health Service Policy on Humane Care and Use of Laboratory Animals using standard protocols approved by the Institutional Animal Care and Use Committees of the University of North Carolina at Chapel Hill, the University of Oregon, Washington University and the University of Washington (Westerfield, 2000). Wild zebrafish were collected from Shutunga River in Mathabhanga subdivision of Cooch Behar district in the Indian state of West Bengal, housed in a stand-alone static tank for ∼28 days (Deepak Nopany, Asian Exports, Calcutta, India), then transported to the University of Washington and housed in a quarantine facility in a stand-alone static tank filled with fresh reverse osmosis-purified water conditioned with Instant Ocean Sea Salt (Aquarium Systems, Mentor, OH, USA) for 4 days before sample acquisition.
DNA isolation
Zebrafish were euthanized with an overdose of MS222 (Sigma-Aldrich, St Louis, MO, USA) and exterior surfaces swabbed with 100% ethanol before dissection of the whole intestine using sterile instruments. Excised intestines were combined in 2.0 ml screw-cap tubes with 0.5 mm Zirconia/silica beads (Biospec Products, Bartlesville, OK, USA), 800 μl 120 m Na-phosphate buffer (pH 8.0) and 400 μl of lysis solution containing 10% sodium dodecyl sulfate, 0.5 M Tris-HCl (pH 8.0) and 0.1 M NaCl. Samples were homogenized in a Mini-Beadbeater (Biospec Products) for 5 min on high speed. The supernatant was transferred to new tubes and lysozyme (Roche, Indianapolis, IN, USA) was added to a final concentration of 10 mg ml−1 followed by incubation at 42 °C for 30 min. Ammonium acetate (7.5 ) was added (2:5 ratio of ammonium acetate to supernatant) and samples were incubated at −20 °C for 5 min. Samples were centrifuged for 5 min at 12 000 g and the supernatant was transferred to new tubes. DNA was precipitated with room-temperature isopropyl alcohol and pelleted by centrifugation at 12 000 g for 30 min at 4 °C. Pellets were washed with −20 °C 70% ethanol and air dried for 30 min before resuspension in 100 μl nuclease free water. Subsequent analysis of pooled samples was conducted with DNA mixtures containing equivalent amounts of DNA from the represented individual samples.
Terminal restriction fragment length polymorphism (T-RFLP) analysis of 16S rRNA gene amplicons
PCR was performed using primers 27f (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492r (5′-TACGGYTACCTTGTTACGACTT-3′). The forward primer only was labeled with a phosphamide dye (D4-PA, Sigma-Aldrich). The 50 μl reactions were carried out in triplicate using 100 ng DNA template, 5 μl of 10 × HotStart buffer (Novagen, Gibbstown, NJ, USA), 5 μl of 25 m MgCl2, 5 μl of 8 m dNTP, 1 μl of each primer and 0.5 μl of 10U μl−1 HotStart Taq (Novagen). Reaction temperatures and times were 95 °C for 10 min, 30 cycles of 94 °C for 30 s, 58 °C for 30 s, 72 °C for 30 s and 72°C for 10 min. The triplicate reactions were combined using a QIAquick PCR Purification Kit (Qiagen, Valencia, CA, USA), eluted in 30 μl, and quantified with a Nanodrop ND-1000 Spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA).
PCR products were digested in 50 μl reactions containing 300 ng of purified DNA, 0.5 μl of 100 × bovine serum albumin, 5 μl of 10 × Buffer 2 (New England Biolabs, Ipswich, MA, USA) and 10U of restriction enzyme HhaI (New England Biolabs). Samples were digested overnight at 37° and inactivated for 20 min at 65 °C. The restriction products were ethanol precipitated and pellets were resuspended in 60 μl of nuclease free water. A volume of 7.5 μl of digested DNA (∼37.5 ng) was combined with 0.5 μl of size standard 600 (Beckman-Coulter, Brea, CA, USA) and 32 μl of sample loading solution (Beckman-Coulter) and submitted in duplicate to the University of Oregon DNA Sequencing and Genomics Facility for capillary analysis on a CEQ8000 Genetic Analysis System (Beckman-Coulter). Raw data was analyzed using the CEQ8000 genetic analysis system software (Beckman-Coulter) set to default settings. Terminal restriction fragment (TRF) length in nucleotides and TRF peak area were exported to Microsoft Excel. TRF peak data for fragments less than 57 nucleotides in length were removed. The square root of each peak height was calculated and the adjusted data was analyzed by hierarchical clustering of the Pearson coefficients of the T-RFLP profile of each sample using Gene Cluster 3.0 (Eisen et al., 1998). The resulting hierarchical tree was drawn using Java TreeView version 1.1.5r2 (Saldanha, 2004). T-RFLP peak data was also imported into QIIME 1.2.0 (Caporaso et al., 2010) using the ‘trflp_file_to_otu_table.py' command. A mapping file was manually generated to annotate the samples with their metadata. The resulting mapping file and operational taxonomy unit (OTU) table were then used to generate non-phylogenetic diversity metrics, including a matrix of Pearson distances (1–Pearson coefficient) between all individual samples. This Pearson distance matrix was used to generate an unweighted principal coordinates analysis (PCoA) plot using the ‘make_3d_plots.py' command and Kinemage. The average Pearson distance was calculated for each sample relative to the other samples from the same location versus samples from each different location. These individual averages were then averaged across groups to generate a matrix of average Pearson distances within and between groups.
Clone library sequencing of 16S rRNA gene amplicons
DNA isolated from pooled samples D.rerio.India.1, D.rerio.UW.1, D.rerio.UNC.1, D.rerio.ZIRC.1 and D.rerio.UO.1 was used to generate 16S rRNA clone libraries, which were then sequenced at the Washington University Genome Sequencing Center in BigDye Terminator reactions using our established methods (Rawls et al., 2006). DNA extraction of negative control water samples did not yield detectable 16S rRNA PCR products or colonies. The 16S rRNA gene sequences were edited and assembled into consensus sequences using PHRED and PHRAP in XplorSeq software (Frank, 2008). These new sequences were combined with our published sequences from samples D.rerio.WU.1, D.rerio.WU.2, and M.musculus (Rawls et al., 2006) and other published sequence data sets (see Table 1). Contiguous sequences with at least 700 bp >Q20 were aligned using the NAST server (DeSantis et al., 2006), and putative chimeras were identified and excluded using Bellerophon version 3 implemented at the Greengenes website (http://greengenes.lbl.gov; match length to core set sequence threshold of 500 bp and window size of 200 bp; Supplementary Table S1). New non-chimeric 16S rRNA gene sequences derived from samples D.rerio.India.1, D.rerio.UW.1, D.rerio.UNC.1, D.rerio.ZIRC.1 and D.rerio.UO.1 were submitted to GenBank under accession numbers HM778178-HM780469. All 16S rRNA clone library sequences used in this study (Supplementary Table S1) were added by parsimony to a local ARB database containing the Greengenes core set (http://greengenes.lbl.gov/Download/Sequence_Data/Arb_databases/greengenes.arb.gz; updated 23-May-2007), and inserted sequences grouping with chloroplast sequences were removed. Sequences used in this study were incorporated into an ARB neighbor joining (NJ) tree using Olsen correction and lanemaskPH filter. The resulting NJ tree was analyzed using the Fast Unifrac tool (http://bmf2.colorado.edu/fastunifrac) (Hamady et al., 2010; Lozupone et al., 2010). Clone library sequences were taxonomically classified with the RDP-II Naïve Bayesian Classifier version 2.2 (Wang et al., 2007) using an 80% confidence threshold.
Table 1. Description of samples used in this study.
| Sample name | Common and scientific name of host | Environment | Location | Water habitata | Sample site | Strain | No.b | Age | Collection date | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| D.rerio.India.1 | Zebrafish (Danio rerio, superorder Ostariophysi, order Cypriniformes) | Recently-caught | River Shutunga, West Bengal, India | F | Whole intestinal contents | — | 20 | Adult | 11-Nov-2006 | This study |
| D.rerio.UW.1 | Zebrafish (Danio rerio, superorder Ostariophysi, order Cypriniformes) | Conventional lab aquaculture | University of Washington, Seattle, WA, USA | F | Whole intestinal contents | AB-wp | 4 | Adult (12 months) | 13-Nov-2006 | This study |
| D.rerio.UNC.1 | Zebrafish (Danio rerio, superorder Ostariophysi, order Cypriniformes) | Conventional lab aquaculture | University of North Carolina, Chapel Hill, NC, USA | F | Whole intestinal contents | SJA | 6 | Adult (7 months) | 5-Dec-2008 | This study |
| D.rerio.WU.1 | Zebrafish (Danio rerio, superorder Ostariophysi, order Cypriniformes) | Conventional lab aquaculture | Washington University, St Louis, MO, USA | F | Whole intestinal contents | C32 | 9 | Adult (8 months) | 2-May-2005 | Rawls et al., 2006 |
| D.rerio.WU.2 | Zebrafish (Danio rerio, superorder Ostariophysi, order Cypriniformes) | Conventional lab aquaculture | Washington University, St Louis, MO, USA | F | Whole intestinal contents | C32 | 9 | Adult (9 months) | 2-May-2005 | Rawls et al., 2006 |
| D.rerio.ZIRC.1 | Zebrafish (Danio rerio, superorder Ostariophysi, order Cypriniformes) | Conventional lab aquaculture | Zebrafish Intl. Resource Center, Eugene, OR, USA | F | Whole intestinal contents | AB | 9 | Adult (9 months) | 2-May-2005 | This study |
| D.rerio.UO.1 | Zebrafish (Danio rerio, superorder Ostariophysi, order Cypriniformes) | Conventional lab aquaculture | University of Oregon, Eugene, OR, USA | F | Whole intestinal contents | AB | 9 | Adult (7 months) | 2-May-2005 | This study |
| P.fulvidraco | Yellow catfish (Pelteobagrus fulvidraco, superorder Ostariophysi, order Siluriformes) | Wild | Niushan Lake, Hubei, China | F | Whole intestinal contents | — | 50 | Adult | — | Wu et al., 2010 |
| A.nigricans | Whitecheek surgeonfish (Acanthurus nigricans, superorder Acanthopterygii, order Perciformes) | Wild | Palmyra Atoll, USA | M | Distal intestinal contents | — | 6 | Adult | — | Smriga et al., 2010 |
| L.bohar | Two-spotted red snapper (Lutjanus bohar, superorder Acanthopterygii, order Perciformes) | Wild | Palmyra Atoll, USA | M | Distal intestinal contents | — | 5 | Adult | — | Smriga et al., 2010 |
| C.sordidus | Daisy parrotfish (Chlorurus sordidus, superorder Acanthopterygii, order Perciformes) | Wild | Palmyra Atoll, USA | M | Feces | — | 5 | Adult | — | Smriga et al., 2010 |
| C.aceratus | Blackfin icefish (Chaenocephalus aceratus, superorder Acanthopterygii, order Perciformes) | Wild | Dallmann Bay, Antarctica | M | Foregut wall | — | 1 | Adult | — | Ward et al., 2009 |
| N.coriiceps | Black rockcod (Notothenia coriiceps, superorder Acanthopterygii, order Perciformes) | Wild | Dallmann Bay, Antarctica | M | Foregut wall | — | 1 | Adult | — | Ward et al., 2009 |
| T.niphobles | Grass puffer (Takifugu niphobles, superorder Acanthopterygii, order Tetraodontiformes) | Wild | Shizuoka, Japan | M | Whole intestinal contents | — | 5 | Adult | — | Shiina et al., 2006 |
| M.musculus | Mouse (Mus musculus) | Conventional lab housing | Washington University, St Louis, MO, USA | N/A | Cecum | Swiss-Webster | 3 | Adult | — | Rawls et al., 2006 |
| H.sapiens | Human (Homo sapiens) | Wild | — | N/A | Feces | — | 1 | Adult | — | Ley et al., 2008 |
Abbreviations: F, freshwater; M, marine.
The fish was collected from freshwater or marine habitats.
The number of individual animals included in each sample set.
Phylogenetic analysis
Clone library sequences and cultured clone sequences isolated from the zebrafish intestine in this and previous studies (GenBank accession numbers HM778163-HM778168) (Rawls et al., 2006) that were classified as members of the phylum Fusobacteria (Figure 3) or the genus Edwardsiella (for Supplementary Figure S2) were selected for further phylogenetic analysis. Novel and reference sequences selected from the ARB-SILVA database (Version 100; 2009) and Genbank were aligned using the SINA aligner (http://www.arb-silva.de), and manually evaluated in MacClade 4.06 (Maddison and Maddison, 2000). Sequences were then imported into ARB (Kumar et al., 2006) where the majority of sequences were assigned to the phylum Fusobacteria or the genus Edwardsiella based on their position after ‘parsimony insertion' into the ARB database dendrogram, omitting hypervariable portions of the rRNA gene using a filter based on the Lane mask (Lane, 1991). Maximum Likelihood (ML) analysis was performed with RAxML-VI-HPC v2.2. using a GTRCAT model of evolution (Stamatakis et al., 2008) in the CIPRES portal (http://www.phylo.org/sub_sections/portal/). Bootstrap resampling (1000 replicates) was used to test the robustness of inferred topologies.
Pyrosequencing of barcoded 16S rRNA gene amplicons
Three D. rerio pooled intestinal samples (D.rerio.India.1, D.rerio.UW.1, D.rerio.UNC.1) were analyzed by massively parallel barcoded-pyrosequencing. A fragment of the 16S rRNA gene (∼330 bp), spanning the V1 and V2 hypervariable regions, was PCR amplified from each sample. The universal Bacterial primers 27F and 338RII were modified by adding ligation adaptors and/or MID barcodes (sample identification sequences) to the 5′- ends (Supplementary Table S5). PCR was performed using a high fidelity polymerase (Phusion Hot Start, Finnzymes, Lafayette, CO, USA), 50 °C annealing temperature, 1500 ng template in 400 μl volume (split between 8 tubes) and 25 cycles. Amplicons, purified and concentrated to 50 μl using the Promega Wizard SV PCR Clean-Up System (Promega, Madison, WI, USA), quantified using a Qubit fluorometer (Invitrogen, Carlsbad, CA, USA) and standardized to 100 ng μl−1, were used as templates for emulsion PCR using the emPCR kit II (Roche). DNA was sequenced using a Genome Sequencer FLX (Roche) and the GS-LR70 kit (Roche) by the Environmental Genomics Core Facility (University of South Carolina) on LR70 plates following Roche standard protocol. FASTA-formatted sequences and corresponding quality scores (QC) were extracted from the .sff data file using the GS Amplicon software package (Roche).
All data preprocessing, OTU-based analysis, pylotype analysis and hypothesis testing was performed using modules implemented in the Mothur software platform (Schloss et al., 2009). Sequences were binned by sample of origin using the unique barcodes, which were removed before downstream analyses. Sequence length and quality were evaluated for each read; sequences were culled if the length was <200 bp and >280 bp, the average SFF quality score was <30, they contained any ambiguous base calls (N's), or did not match the primer or one of the used barcode sequences. The data set was simplified by using the ‘unique.seqs' command to generate a non-redundant (unique) set of sequences. Unique sequences were aligned using the ‘align.seqs' command and an adaptation of the Bacterial SILVA SEED database as a template (available at: http://www.mothur.org/wiki/Alignment_database). Sequences were denoised using the ‘pre.cluster' command. This command applies a pseudo-single linkage algorithm with the goal of removing sequences that are likely due to pyrosequencing errors (Huse et al., 2010). A total of 1534 potential chimeric sequences were detected and removed using the ‘chimera.slayer' command.
Aligned sequences were clustered into OTUs defined by 97% similarity using the average neighbor algorithm. Rarefaction curves were plotted for each sample and an unweighted UniFrac dendrogram (Lozupone et al., 2010) was generated using the UniFrac module implemented in Mothur. Rank-abundance curves (Whittaker plots) were generated using custom Perl scripts. All community diversity parameters (Shannon-Weaver, Chao1, and Simpson's) were calculated as described in the Mothur software manual. Pyrosequences were taxonomically classified by the RDP-II Naïve Bayesian Classifier version 2.2 using a 60% confidence threshold. Sequences that could not be classified to at least the kingdom level were excluded from subsequent diversity analyses. Venn diagrams and heatmap figures were generated using custom Perl scripts. Pyrosequence data sets are available through the NCBI/EBI/DDBJ Short Read Archive (accession number ERP000213).
Results
Intestinal bacterial communities in domesticated zebrafish are similar to recently caught zebrafish
We tested whether the intestinal microbiotas of domesticated zebrafish were significantly different from zebrafish collected recently from their natural habitat. To assess the intestinal microbiotas of zebrafish that have been domesticated in lab facilities for generations, we extracted genomic DNA from intestinal contents of adult zebrafish sampled from five lab aquaculture facilities that are all derived from the original University of Oregon facility (Figure 1, Table 1). To assess the intestinal microbiota of zebrafish that have only recently been collected from their natural habitat, we extracted genomic DNA from intestinal contents of adult zebrafish that were collected from the Shutunga River, West Bengal, India and then housed in quarantine aquaculture facilities for ∼32 days (‘recently caught' zebrafish; Table 1) (Engeszer et al., 2008). We first compared intestinal bacterial diversity in individual domesticated and recently caught zebrafish using 16S rRNA gene T-RFLP analysis. Hierarchical cluster and PCoA analysis of 16S rRNA T-RFLP profiles revealed that the gut bacterial communities in individual recently caught and domesticated zebrafish have similar community structures (Supplementary Figure S1A,B), raising the possibility that intestinal bacterial communities in recently caught zebrafish are not markedly different from domesticated zebrafish. Furthermore, T-RFLP analysis of pooled intestinal samples from each location provided an approximate representation of the component members of the pool (Supplementary Figure S1A), suggesting that sample pooling may overcome interindividual variation among animals in a given location.
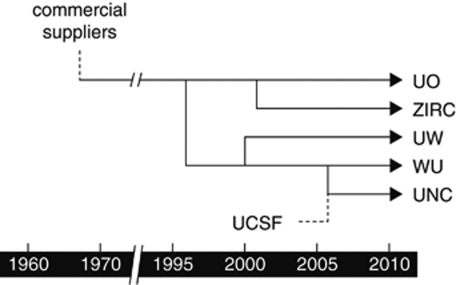
Figure 1.
Relationship between sampled zebrafish aquaculture facilities. The original University of Oregon (UO) facility was seeded by zebrafish acquired from commercial suppliers in the late 1960s (Grunwald and Eisen, 2002). Zebrafish from the UO facility was subsequently used to seed the Washington University (WU) facility in 1996 and the Zebrafish International Resource Center (ZIRC) in 2001. In 2000, zebrafish from the WU facility was used to seed a facility at the University of Texas at Austin, which was subsequently moved to the University of Washington (UW) facility in 2005. The University of North Carolina (UNC) facility was seeded in 2006 by zebrafish from the WU facility as well as from a facility at the University of California at San Francisco (UCSF) that has no historical connection to the UO facility.
As T-RFLP analysis reveals sequence diversity at only a small number of sites in the 16S rRNA gene and does not identify the specific bacterial types that are shared or distinct between different locations, we next generated 16S rRNA clone libraries from pooled intestinal samples from each group of animals, and sequenced them using Sanger chemistry (3721 clones in total; Table 1, Supplementary Table S1). To permit taxon-based assessments of diversity and coverage, we binned these sequences into OTUs defined by 97% pairwise sequence identity. Comparison of OTUs derived from zebrafish intestines revealed that the Chao1 richness and Shannon–Weaver diversity estimates of the intestinal microbiotas from domesticated and recently caught zebrafish were relatively similar (Supplementary Table S2), indicating that host provenance does not have a strong influence on the overall richness or diversity of the zebrafish gut bacterial community.
To permit phylogenetic comparisons of intestinal bacterial communities within the intestines of zebrafish sampled from different locations, we analyzed our 16S rRNA gene sequence data sets using the UniFrac metric (Hamady et al., 2010). We first supplemented our zebrafish intestinal 16S rRNA clone sequences with several additional 16S rRNA clone libraries from the intestines of human, mouse and seven other wild teleost fishes including five marine fish from order Perciformes (Acanthurus nigricans, Lutjanus bohar, Chlorurus sordidus, Chaenocephalus aceratus, Notothenia coriiceps), one marine pufferfish from order Tetraodontiformes (Takifugu niphobles) and one freshwater yellow catfish from order Siluriformes (Pelteobagrus fulvidraco; see Table 1). An unweighted UniFrac tree of the resulting set of 5217 16S rRNA gene sequences revealed several distinct clusters based on bacterial community membership (Figure 2a). The human and mouse libraries clustered together separately from all fish libraries, denoting distinct differences between the composition of the gut bacterial communities in fish and mammals (Supplementary Table S3). Another distinct cluster was formed by the five marine Perciformes fish included in the analysis, with two Antarctic fish and three fish from Palmyra Atoll forming distinct subclusters (Figure 2a and Table 1). The largest major cluster in the UniFrac tree was comprised of all the zebrafish samples plus the yellow catfish (P. fulvidraco), the only other freshwater fish included in our analysis.
Figure 2.
The 16S rRNA gene sequencing reveals the relationship and membership of intestinal microbiotas of zebrafish from different locations. (a) Unweighted UniFrac tree comparing 5217 16S rRNA clone library sequences from the gut microbiotas of adult zebrafish, other teleost fish species, mouse and human (see Table 1). The distance P value for this entire UniFrac tree (the probability that there are more unique branches than expected by chance, using 1000 iterations) was found to be <0.002, assigning high condence to the overall structure of the tree. (b) Unweighted UniFrac tree of 17 763 16S rRNA pyrosequences spanning the V1–V2 hypervariable regions derived from the gut microbiotas of recently caught (India.1) and domesticated (UNC.1 and UW.1) zebrafish. Scale bars indicate distance between the samples in UniFrac units. The shape at the end of each branch indicates host superorder (triangles: freshwater Ostariophysi fish, circles: marine Acanthopterygii fish, squares: mammalian reference samples) with color indicating host order as shown in the key (Perciformes fish from Antarctica and Palmyra Atoll are labeled separately). (c) The relative abundance of bacterial classes observed in these data sets is represented in heatmaps below each tree. Bacterial classes are grouped by phylum: Proteobacteria (Pr), Firmicutes (Fi), Bacteroidetes (Ba), Chloroflexi (Ch), Fusobacteria (Fu), Actinobacteria (At), Spirochetes (Sp), Deferribacteres (De), Acidobacteria (Ai), Nitrospira (Ni), Planctomycetes (Pl), Verrucomicrobia (Ve), Lentisphaerae (Le), and Deinococcus-Thermus (DT)(see also Supplementary Table S3). Communities are clustered using PCoA of unweighted (d) and weighted (e) UniFrac distance matrices. The gray halos encircle the cluster of freshwater Ostariophysi fish. The percentage of the variation explained by the plotted principal coordinates is indicated on the axes.
To further compare the composition of the gut microbiotas in zebrafish and other fishes, we subjected these 16S rRNA gene sequences to PCoA. PCoA plots derived from both unweighted (an assessment of community composition) and weighted (an assessment of community structure) algorithms showed that all domesticated zebrafish samples clustered together closely with recently caught zebrafish and wild yellow catfish (P. fulvidraco), establishing a high degree of similarity in composition and structure of these gut bacterial communities (Figures 2d and e).
To provide perspective on the observed relationships between gut microbiotas from different zebrafish populations, we compared the taxonomy of intestinal bacterial communities in recently caught and domesticated zebrafish to those of other teleost fish species. We classified 16S rRNA clone library sequences using the RDP-II Naïve Bayesian Classifier (Supplementary Table S3), and plotted the relative frequency of bacterial classes against the UniFrac tree (Figure 2). Several bacterial classes were observed in only a subset of zebrafish libraries (for example, α-, β-, δ-Proteobacteria, Actinobacteria and Planctomycetacia), potentially due to variation in gut microbiota composition between zebrafish from different locations and/or the limited sampling depth provided by clone libraries. Strikingly, two bacterial classes, γ-Proteobacteria and Fusobacteria, appeared consistently in the gut microbiotas of zebrafish and the other fish species (Figure 2), suggesting that members of these bacterial classes are especially well adapted to conditions in the fish intestine or their surrounding aquatic environment. The bacterial genera within these classes in recently caught zebrafish were also common in domesticated zebrafish (that is, Aeromonas spp., Pseudomonas spp., Plesiomonas spp., Vibrio spp., Shewanella spp. and Cetobacterium spp.), however Pseudomonas spp. were enriched in recently caught zebrafish (18% of clones) compared with domesticated zebrafish (0–2% of clones). Although γ-Proteobacteria were detected in the intestines of each fish species in our analysis, the γ-Proteobacteria genera observed varied between fish. For example, Aeromonas and Plesiomonas spp. were common in the intestines of freshwater fish from superorder Ostariophysi, but undetected in marine fish from superorder Acanthopterygii (P<0.001 and P<0.05, respectively). In contrast, Vibrio spp. were significantly enriched in the intestines of Acanthopterygii compared with Ostariophysi fish (P<0.05).
The Fusobacteria are underrepresented in the public 16S rRNA gene databases, therefore we compared the Fusobacteria 16S rRNA gene sequences from fish intestines to known type strains by generating a phylogenetic tree (Figure 3). Many of the Fusobacteria sequences detected in the intestines of zebrafish and other fishes were closely related to Cetobacterium somerae, whereas others had no close homolog in the public databases. These results establish that phylogenetically diverse teleost fishes host a diversity of Fusobacteria, many of which fall outside of known Fusobacteria clades.
Figure 3.
The 16S rRNA gene sequence-based phylogenetic analysis of Fusobacteria within the intestinal microbiotas of zebrafish and other fishes. Maximum likelihood tree showing the phylogenetic relationship between Fusobacteria 16S rRNA gene sequences from uncultured and cultured bacteria derived from the intestinal contents of zebrafish (red text) and other teleost fishes (blue text), and those from other sources available in the public databases (black text). Branches representing multiple identical sequences are indicated with the number of sequences in parentheses. Bootstrap support (⩾50%) is shown as results from 1000 bootstrap replicates. The scale bar indicates 0.05% estimated sequence divergence.
Gut bacterial community structure varies among domesticated zebrafish in geographically separate lab aquaculture facilities
We next sought to determine whether gut microbiota composition varies significantly between domesticated zebrafish raised in different lab aquaculture facilities. We used the T-RFLP profiles to assess individual variation in gut microbial communities within and between the recently caught and domesticated populations. On average, fish from the same location had more similar profiles (lower Pearson distance values) to each other than to fish from different locations (Supplementary Figure S1C). However, the differences between recently caught fish and domesticated fish populations were not greater than differences between domesticated fish from different aquaculture facilities. This was also evident when the Pearson distance matrix from the individual T-RFLP profiles was used to generate an unweighted PCoA plot (Supplementary Figure S1B); individual fish from the same location clustered together, with the recently caught fish lying midway along both of the first and second principle coordinate axes. Although T-RFLP profiles of individual fish from the same location clustered together in the PCoA analysis, the interindividual variation within each location overlapped considerably with that of other locations (Supplementary Figure S1B). This suggests shared characteristics in the gut microbial communities of zebrafish sampled from different locations. The basis of interindividual variation remains unclear, but gender did not appear to be a strong determinant of zebrafish gut microbiota composition, as male and female individuals were interspersed throughout the tree (Supplementary Figure S1A).
The PCoA plots from 16S rRNA T-RFLP profiles from individual fish samples (Supplementary Figure S1B) and clone library sequences from pooled fish samples (Figure 2d) revealed strikingly similar relationships between different locations (Figures 1 and 2a, d and e). For example, zebrafish from the University of Oregon clustered together with animals from the neighboring Zebrafish International Resource Center in Eugene OR, which was seeded by fish from the UO facility in 2000. Similarly, samples from a facility at Washington University clustered with samples from a facility at the University of Washington, which was originally derived from the WU facility in 2000. These results suggest that gut bacterial community structure can be explained in part by the historical connections between specific zebrafish facilities.
Deep sequencing suggests a core microbiota in the zebrafish intestine
Although our analysis of 16S rRNA clone library sequences revealed novel patterns of variation and consistency between different zebrafish populations and other fish species, we speculated that these patterns were likely to be underestimated due to the limited depth of coverage provided by clone libraries. We therefore subjected pooled samples from recently caught zebrafish (India.1) and domesticated zebrafish from two lab aquaculture facilities (UW.1 and UNC.1) to Roche GS-FLX pyrosequencing to assess the effect of domestication on the zebrafish gut microbiota in greater depth. The improved depth of coverage provided by the resulting set of 17 763 high quality 16S rRNA gene sequences revealed many new bacterial phylotypes present at low abundance that were not detected in the respective clone libraries (Supplementary Table S3). Taxon-based assessments of these pyrosequencing data sets revealed that each library contained at least 178 OTUs (defined by 97% pairwise sequence identity; Figure 4a), greatly exceeding the number of OTUs detected in the respective clone library data sets (Supplementary Table S2). A UPGMA tree of the three pyrosequencing data sets revealed a similar relationship between the three samples as observed in the clone library data set, with the India.1 and UNC.1 samples clustering away from the UW.1 library (Figure 2b). All three samples were dominated by Proteobacteria, although the relative abundance of other phyla varied between samples (for example, abundance of Fusobacteria in UNC sample, and Firmicutes and Actinobacteria in the UW sample; Figure 4d). The bacterial classes present in the three samples were highly similar, in some cases revealing shared membership that was not apparent in the respective clone libraries (for example, Bacilli, Clostridia, Flavobacteria and Actinobacteria classes; Figure 2c). We detected 525 OTUs across all three samples, however many were observed in only one pyrosequencing data set and only 21 of these OTUs were detected in all three data sets (Figure 4b). By plotting the ranked abundance of all 525 OTUs according to their occurrence in the three samples, we found that the 21 OTUs common to all three samples tended to be highly abundant whereas OTUs observed in only one or two samples tended to be relatively rare (Figure 4c). RDP Classifier analysis revealed that the 21 OTUs common to all three libraries were comprised of 12 genera within the γ-Proteobacteria, β-Proteobacteria, Fusobacteria, Bacilli, Flavobacteria and Actinobacteria classes, with Aeromonas and Shewanella appearing as the most frequent genera (Supplementary Table S4). As discussed below, these shared constituents of domesticated and recently caught zebrafish intestinal microbiotas might constitute a ‘core microbiota' of the zebrafish intestine.
Figure 4.
Deep sequencing of 16S rRNA genes reveals a core intestinal microbiota shared among recently caught and domesticated zebrafish. (a) Rarefaction curves of 16S rRNA gene pyrosequences spanning the V1–V2 region from pooled intestinal samples collected from recently caught zebrafish (India; sample D.rerio.India.1; 5582 sequences) or domesticated zebrafish raised in aquaculture facilities at the University of North Carolina (UNC; sample D.rerio.UNC.1; 9357 sequences) or the University of Washington (UW; sample D.rerio.UW.1; 2824 sequences). Sequences are binned into OTUs using a pairwise sequence similarity threshold of 97%. (b) Venn diagram showing the distribution of all 525 OTUs (97%) identified in the combined 17 763 16S rRNA gene pyrosequences from India, UNC and UW, revealing a shared community of 21 OTUs found in all three locations. (c) Rank abundance plot showing the OTUs (97%) within each category of the Venn diagram in panel b ranked according to their abundance in the combined 17 763 sequence data set. The similarity between the rank abundance plots of all 525 OTUs (black line) and the 21 shared OTUs (green line) reveals that the OTUs found in all three locations include the most abundant OTUs in any location. (d) Pie charts showing the relative abundance of bacterial phyla in the intestinal microbiotas of zebrafish from India, UNC, and UW (see Supplementary Table S3), as well as the 21 OTUs shared between all three locations, which may comprise a ‘core' zebrafish gut microbiota (see Supplementary Table S4).
Discussion
One of the long-term goals for efforts such as the Human Microbiome Project is to develop effective strategies for manipulating gut microbial communities to promote and sustain the health of human hosts (Peterson et al., 2009). To achieve this goal, we must first understand the principles governing microbial community assembly and maintenance within the intestine. An important prerequisite for understanding these principles is the development of robust model systems for the study of host-microbiota interactions. Here we have demonstrated that domesticated lab-reared zebrafish develop a gut microbiota similar to that of zebrafish collected recently from their natural habitat, but that the domesticated zebrafish gut microbiota covaries according to the historical connections of the respective lab facility. Furthermore, using pyrosequencing we have identified potential members of a core zebrafish gut microbiota, an important step toward establishing zebrafish as a powerful model for host-microbiota studies.
Our results disclose substantial interlocation variation between the gut microbiotas of adult zebrafish raised in difference lab aquaculture facilities (Figures 2, 4, and Supplementary Figure S1). Interlocation variation of gut microbiota composition is not unique to zebrafish, as marked interlocation variation has also been observed between lab-reared mice (Alexander et al., 2006; Ivanov et al., 2009; Friswell et al., 2010). Intriguingly, the relationships between the composition of zebrafish gut bacterial communities in different aquaculture facilities (Figure 2a) were reminiscent of the historical seeding relationships between these facilities (Figure 1). The causes of these observed patterns of interlocation variation remain unknown, but could include differences in housing infrastructure, water chemistry, diet composition, feeding schedule, history of antibiotic use, and the spectra of infectious microorganisms and viruses in different locations. Interlocation variation could also be in part due to genetic variation between zebrafish maintained in different lab facilities. All of the lab-reared domesticated zebrafish lines sampled in this study are derived from the same AB line established at the University of Oregon or one of its derivatives (Table 1). However the sampled lines of domesticated zebrafish are not inbred and retain significant genetic polymorphisms (Rawls et al., 2003; Guryev et al., 2006). Therefore bottleneck effects and genetic drift within different lab facilities could contribute to the observed interindividual variation in gut microbiota composition.
Our observations of interlocation variation in the zebrafish intestinal microbiota have important implications for researchers investigating aspects of zebrafish biology that could be influenced by the microbial environment (for example, immunology, nutrition, gastrointestinal development and physiology). If a given zebrafish phenotype is sensitive to the composition of the local microbial community, then differences in microbiota composition across aquaculture facilities could result in phenotypic variation and reduced experimental reproducibility. These potential complications of interlocation variation in microbiota composition could be mitigated by establishing defined mixtures of culturable bacterial types to inoculate zebrafish in different facilities, similar to the altered Schaedler flora used in rodent husbandry (Dewhirst et al., 1999).
In addition to patterns of interlocation variation, our culture-independent 16S rRNA gene data sets provided an unprecedented opportunity to define bacterial types that are broadly shared among zebrafish in different locations. Our 16S rRNA clone library sequences identified members of the γ-Proteobacteria and Fusobacteria classes as common members of the gut microbiota in adult zebrafish raised in different locations as well as in other fish species (Figure 2). The nature of our study required that we limited our UniFrac analysis to those fish species for which complete (that is, non-dereplicated) 16S rRNA clone libraries were available (Table 1), however, these same bacterial classes have been also observed in the intestinal microbiotas of other teleost fishes in culture-independent and culture-based surveys (Huber et al., 2004; Romero and Navarrete, 2006; Kim et al., 2007; Tsuchiya et al., 2008; Merrifield et al., 2009; Navarrete et al., 2009, 2010). This suggests that these specific bacterial groups are especially well adapted for the environment within the fish intestine, despite large evolutionary and geographic distances between their fish hosts.
Our phylogenetic analysis revealed a diverse set of Fusobacteria sequences isolated from the intestines of zebrafish and other fishes, most of which were closely related to Cetobacterium somerae cultured previously from human feces (Finegold et al., 2003) (Figure 3). C. somerae (initially named Bacteroides type A) is a microaerotolerant, non-spore-forming, rod-shaped, vitamin B12 (cobalamin) producing Fusobacterium that has been shown to be indigenous to the digestive tract of multiple freshwater fish species that do not require dietary supplements of vitamin B12 (Sugita et al., 1991; Tsuchiya et al., 2008). C. somerae was not detected in the digestive tract of two freshwater fish species, which show deficiency symptoms when fed vitamin B12-depleted diets (Sugita et al., 1991), suggesting that C. somerae may be involved in determining the vitamin B12 requirements of freshwater fish.
Although the same bacterial classes predominated the gut microbiotas of diverse teleost fishes, the variation in gut microbiota composition across fishes produced two major clusters in our UniFrac analysis: marine fish from superorder Acanthopterygii order Perciformes, and freshwater fish from superorder Ostariophysi (Figure 2a). Intriguingly, the Perciformes cluster included two distinct subclusters consisting of fish collected from geographically separate marine habitats in Antarctica and Palmyra Atoll. The relationship between gut bacterial community membership in fish hosts therefore matches their respective phylogenetic relationships, despite differences in geographic location and domestication status. The respective marine and freshwater habitats of these Perciformes and Ostariophysi fish could contribute to the observed differences in gut microbiota composition. However, the gut microbiota of another marine fish from superorder Acanthopterygii (T. niphobles) was distinctly different from the other marine fish (Figure 2a), suggesting that the gut microbiota in fish is shaped by factors other than water salinity alone. These observations complement a recent large-scale comparison of mammalian species (Ley et al., 2008), collectively establishing host phylogeny as a major determinant of gut bacterial diversity in fish as well as mammals.
This report is the first to compare the intestinal microbiotas of recently caught and domesticated zebrafish. Our results complement a limited number of previous 16S rRNA gene sequence-based comparisons of gut microbiotas in wild and domesticated animals. Analysis of wild and domesticated mice (Wilson et al., 2006), turkeys (Scupham et al., 2008), parrots (Xenoulis et al., 2010), fruitflies (Cox and Gilmore, 2007) and hydra (Fraune and Bosch, 2007) indicate that members of the same species tend to possess gut bacterial communities of similar taxonomic composition at the phylum or class level regardless of domestication status, with some differences between wild and domestic individuals emerging at shallower phylogenetic resolution. These previous studies have revealed varying effects of domestication on gut bacterial diversity, with increased diversity in wild mice (Wilson et al., 2006) and fruitflies (Cox and Gilmore, 2007) compared with domesticated controls, and decreased diversity in wild versus domesticated parrots (Xenoulis et al., 2010). Our clone library sequencing and pyrosequencing of the zebrafish intestinal microbiota revealed variation between recently caught and domesticated zebrafish, however, the scale of these variations were no larger than those observed between or within different zebrafish lab facilities (Figures 2, 4, and Supplementary Figure S1). Moreover, the bacterial taxa that dominated the intestines of recently caught zebrafish were largely the same as those dominating the intestines of domesticated zebrafish. One notable exception was the genus Edwardsiella, which includes the freshwater fish pathogens E. tarda and E. ictaluri (Plumb, 1999; Pressley et al., 2005; Petrie-Hanson et al., 2007). Edwardsiella spp. were detected as rare members of the gut microbiotas of recently caught zebrafish (1.24% of all sequences, all closely related to E. ictaluri) and wild yellow catfish (3.08% of all sequences, all closely related to E. tarda; Supplementary Figure S2), but did not appear in any of the clone sequences or pyrosequences derived from domesticated zebrafish (Supplementary Table S3). This raises the possibility that these Edwardsiella spp. are natural members of the zebrafish and yellow catfish microbiotas, but have been effectively excluded from zebrafish lab aquaculture facilities.
Taken together, these results indicate that the membership and structure of intestinal bacterial communities in domesticated zebrafish are strikingly similar to those collected recently from their natural habitat. The recently caught zebrafish analyzed here (India.1 samples) were collected from the wild and then housed temporarily in quarantined aquaculture facilities for a total of 32 days before sample acquisition. As these recently caught zebrafish were never exposed to the microbiota of domesticated zebrafish, there are two potential explanations for the similarity between their intestinal microbiotas. One possibility is that the gut microbiota of wild zebrafish is significantly different from domesticated zebrafish, and that the capture, transport and husbandry of wild zebrafish causes a rapid, reproducible and long-lasting change to the microbiota that was observed in all zebrafish analyzed here. The other possibility is that wild zebrafish in their natural habitat and zebrafish that have been maintained over decades of domestication acquire a common gut bacterial community. In support of this model, we observed minimal differences between the intestinal bacterial communities of recently caught and domesticated zebrafish, and yellow catfish sampled directly from their natural habitat (Figure 2). This suggests that shared features of the intestinal habitat in these freshwater Ostariophysi fish select for specific bacterial taxa, resulting in similar gut bacterial communities despite radical differences in host provenance and domestication status. We speculate that these shared features could include evolutionarily conserved aspects of digestive tract anatomy, physiology, and immunity, as well as preferred salinity levels in the surrounding water. These results also suggest that lab-reared domesticated zebrafish can serve as a valid model system for investigating coevolved host-microbe relationships that occur in their natural habitat.
To our knowledge, this report comprises the first published implementation of second-generation sequencing technology to assess bacterial diversity within the intestine of a teleost fish. The improved depth of coverage provided by 16S rRNA gene pyrosequencing revealed that a core set of bacterial genera (a core microbiota) are present in domesticated as well as recently caught zebrafish despite salient differences in their life histories and local environments. The concept of a core gut microbiota has been explored in the context of mammalian hosts (Turnbaugh et al., 2009, 2010; Qin et al., 2010), and our data indicate that these concepts may also apply to bony fishes. In zebrafish as well as humans, the mechanisms and selective pressures that produce a core gut microbiota remain unresolved. We previously observed that colonization of germ-free zebrafish larvae with a Firmicutes-dominated microbiota harvested from the intestines of conventionally raised mice, results in enrichment of γ-Proteobacteria within the recipient zebrafish gut (Rawls et al., 2006). These enriched γ-Proteobacteria consisted of genera not normally found in the intestinal microbiotas of conventionally raised zebrafish. Therefore the appearance of specific genera within the zebrafish core gut microbiota may be due in part to distinct selective pressures within the host gut habitat (for example, selection of γ-Proteobacteria in general), but may also be due to the types of γ-Proteobacteria present in their surrounding freshwater habitat that are available to colonize zebrafish hosts. Our results underscore the need to identify the selective pressures governing microbial community assembly within the intestinal habitat of different host species. This information will facilitate the development of safe and effective methods for manipulating gut microbiota composition to promote the health of humans and other animals.
Acknowledgments
We are grateful to Chad Trent and Rose Gaudreau for valuable technical support; and to Jeffrey Gordon, Julie Toplin, Roger Volker and Brendan Bohannan for helpful discussions and intellectual contributions. This work was funded by grants from the NIH (DK075549 to KG; DK081426 and DK073695 to JFR; HD22486 provided support for the Oregon Zebrafish Facility; RR012546 provided support for the Zebrafish International Resource Center), the National Science Foundation (IOB 0541733 to DMP), the University of North Carolina at Chapel Hill, the Harvard University Center for the Environment, a Harvard Digestive Disease Center Pilot and Feasibility grant to CMC, a Rubicon award from the Netherlands Organization for Scientific Research to GR, a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Disease Award to KG, and a Pew Scholars Program in the Biomedical Sciences Award to JFR.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Genbank accession numbers: HM778163-HM778168, HM778178-HM780469.
Short Read Archive accession number: ERP000213.
Supplementary Material
References
- Alexander DA, Orcutt RP, Henry JC, Baker J, Jr, Bissahoyo AC, Threadgill DW. Quantitative PCR assays for mouse enteric flora reveal strain-dependent differences in composition that are influenced by the microenvironment. Mamm Genome. 2006;17:1093–1104. doi: 10.1007/s00335-006-0063-1. [DOI] [PubMed] [Google Scholar]
- Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- Bates JM, Akerlund J, Mittge E, Guillemin K. Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell Host Microbe. 2007;2:371–382. doi: 10.1016/j.chom.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates JM, Mittge E, Kuhlman J, Baden KN, Cheesman SE, Guillemin K. Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation. Dev Biol. 2006;297:374–386. doi: 10.1016/j.ydbio.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Brugman S, Liu KY, Lindenbergh-Kortleve D, Samsom JN, Furuta GT, Renshaw SA, et al. Oxazolone-induced enterocolitis in zebrafish depends on the composition of the intestinal microbiota. Gastroenterology. 2009;137:1757–1767. doi: 10.1053/j.gastro.2009.07.069. [DOI] [PubMed] [Google Scholar]
- Camp JG, Kanther M, Semova I, Rawls JF. Patterns and scales in gastrointestinal microbial ecology. Gastroenterology. 2009;136:1989–2002. doi: 10.1053/j.gastro.2009.02.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheesman SE, Guillemin K. We know you are in there: conversing with the indigenous gut microbiota. Res Microbiol. 2007;158:2–9. doi: 10.1016/j.resmic.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Cheesman SE, Neal JT, Mittge E, Seredick BM, Guillemin K. Microbes and Health Sackler Colloquium: Epithelial cell proliferation in the developing zebrafish intestine is regulated by the Wnt pathway and microbial signaling via Myd88. Proc Natl Acad Sci USA. 2011;108 Suppl 1:4570–4577. doi: 10.1073/pnas.1000072107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox CR, Gilmore MS. Native microbial colonization of Drosophila melanogaster and its use as a model of Enterococcus faecalis pathogenesis. Infect Immun. 2007;75:1565–1576. doi: 10.1128/IAI.01496-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Jr, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM, et al. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006;34:W394–W399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst FE, Chien CC, Paster BJ, Ericson RL, Orcutt RP, Schauer DB, et al. Phylogeny of the defined murine microbiota: altered Schaedler flora. Appl Environ Microbiol. 1999;65:3287–3292. doi: 10.1128/aem.65.8.3287-3292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engeszer RE, Patterson LB, Rao AA, Parichy DM. Zebrafish in the wild: a review of natural history and new notes from the field. Zebrafish. 2007;4:21–40. doi: 10.1089/zeb.2006.9997. [DOI] [PubMed] [Google Scholar]
- Engeszer RE, Wang G, Ryan MJ, Parichy DM. Sex-specific perceptual spaces for a vertebrate basal social aggregative behavior. Proc Natl Acad Sci USA. 2008;105:929–933. doi: 10.1073/pnas.0708778105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finegold SM, Vaisanen ML, Molitoris DR, Tomzynski TJ, Song Y, Liu C, et al. Cetobacterium somerae sp. nov. from human feces and emended description of the genus Cetobacterium. Syst Appl Microbiol. 2003;26:177–181. doi: 10.1078/072320203322346010. [DOI] [PubMed] [Google Scholar]
- Frank DN. XplorSeq: a software environment for integrated management and phylogenetic analysis of metagenomic sequence data. BMC Bioinformatics. 2008;9:420. doi: 10.1186/1471-2105-9-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraune S, Bosch TC. Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc Natl Acad Sci USA. 2007;104:13146–13151. doi: 10.1073/pnas.0703375104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friswell MK, Gika H, Stratford IJ, Theodoridis G, Telfer B, Wilson ID, et al. Site and strain-specific variation in gut microbiota profiles and metabolism in experimental mice. PLoS One. 2010;5:e8584. doi: 10.1371/journal.pone.0008584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunwald DJ, Eisen JS. Headwaters of the zebrafish-emergence of a new model vertebrate. Nat Rev Genet. 2002;3:717–724. doi: 10.1038/nrg892. [DOI] [PubMed] [Google Scholar]
- Guryev V, Koudijs MJ, Berezikov E, Johnson SL, Plasterk RH, van Eeden FJ, et al. Genetic variation in the zebrafish. Genome Res. 2006;16:491–497. doi: 10.1101/gr.4791006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M, Lozupone C, Knight R. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2010;4:17–27. doi: 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber I, Spanggaard B, Appel KF, Rossen L, Nielsen T, Gram L. Phylogenetic analysis and in situ identification of the intestinal microbial community of rainbow trout (Oncorhynchus mykiss, Walbaum) J Appl Microbiol. 2004;96:117–132. doi: 10.1046/j.1365-2672.2003.02109.x. [DOI] [PubMed] [Google Scholar]
- Huse SM, Welch DM, Morrison HG, Sogin ML. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol. 2010;12:1889–1898. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanther M, Rawls JF. Host-microbe interactions in the developing zebrafish. Curr Opin Immunol. 2010;22:10–19. doi: 10.1016/j.coi.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Brunt J, Austin B. Microbial diversity of intestinal contents and mucus in rainbow trout (Oncorhynchus mykiss) J Appl Microbiol. 2007;102:1654–1664. doi: 10.1111/j.1365-2672.2006.03185.x. [DOI] [PubMed] [Google Scholar]
- Kumar Y, Westram R, Kipfer P, Meier H, Ludwig W. Evaluation of sequence alignments and oligonucleotide probes with respect to three-dimensional structure of ribosomal RNA using ARB software package. BMC Bioinformatics. 2006;7:240. doi: 10.1186/1471-2105-7-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DJ.199116S/23S rRNA sequencingIn: Stackebrandt E (ed),Nucleic Acid Techniques in Bacterial Systematics John Wiley & Sons: New York; 115–175. [Google Scholar]
- Lawrence C. The husbandry of zebrafish (Danio rerio): a review. Aquaculture. 2007;269:1–20. [Google Scholar]
- Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. UniFrac: an effective distance metric for microbial community comparison. ISME J. 2010;5:169–172. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison D, Maddison W. MacClade 4: analysis phylogeny and character evolution. Sinauer Associates: Sunderland, MA, USA; 2000. [Google Scholar]
- Merrifield DL, Burnard D, Bradley G, Davies SJ, Baker RTM. Microbial community diversity associated with the intestinal mucosa of farmed rainbow trout (Oncoryhnchus mykiss Walbaum) Aquacult Res. 2009;40:1064–1072. [Google Scholar]
- Navarrete P, Espejo RT, Romero J. Molecular analysis of microbiota along the digestive tract of juvenile Atlantic salmon (Salmo salar L) Microb Ecol. 2009;57:550–561. doi: 10.1007/s00248-008-9448-x. [DOI] [PubMed] [Google Scholar]
- Navarrete P, Magne F, Mardones P, Riveros M, Opazo R, Suau A, et al. Molecular analysis of intestinal microbiota of rainbow trout (Oncorhynchus mykiss) FEMS Microbiol Ecol. 2010;71:148–156. doi: 10.1111/j.1574-6941.2009.00769.x. [DOI] [PubMed] [Google Scholar]
- Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, et al. The NIH Human Microbiome Project. Genome Res. 2009;19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie-Hanson L, Romano CL, Mackey RB, Khosravi P, Hohn CM, Boyle CR. Evaluation of zebrafish Danio rerio as a model for Enteric Septicemia of Catfish (ESC) J Aquat Animal Health. 2007;19:151–158. doi: 10.1577/H06-026.1. [DOI] [PubMed] [Google Scholar]
- Plumb JA.1999Edwardsiella septicaemiasIn: Woo PTK, Bruno DW (eds),Fish Diseases and Disorders: Viral, Bacterial, and Fungal Infections CAB International: Wallingford, UK; 479–521. [Google Scholar]
- Pressley ME, Phelan PE, III, Witten PE, Mellon MT, Kim CH. Pathogenesis and inflammatory response to Edwardsiella tarda infection in the zebrafish. Dev Comp Immunol. 2005;29:501–513. doi: 10.1016/j.dci.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls JF, Frieda MR, McAdow AR, Gross JP, Clayton CM, Heyen CK, et al. Coupled mutagenesis screens and genetic mapping in zebrafish. Genetics. 2003;163:997–1009. doi: 10.1093/genetics/163.3.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls JF, Mahowald MA, Ley RE, Gordon JI. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell. 2006;127:423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls JF, Samuel BS, Gordon JI. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc Natl Acad Sci USA. 2004;101:4596–4601. doi: 10.1073/pnas.0400706101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero J, Navarrete P. 16S rDNA-based analysis of dominant bacterial populations associated with early life stages of coho salmon (Oncorhynchus kisutch) Microb Ecol. 2006;51:422–430. doi: 10.1007/s00248-006-9037-9. [DOI] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview - extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scupham AJ, Patton TG, Bent E, Bayles DO. Comparison of the cecal microbiota of domestic and wild turkeys. Microb Ecol. 2008;56:322–331. doi: 10.1007/s00248-007-9349-4. [DOI] [PubMed] [Google Scholar]
- Shiina A, Itoi S, Washio S, Sugita H. Molecular identification of intestinal microflora in Takifugu niphobles. Comp Biochem Phys D. 2006;1:128–132. doi: 10.1016/j.cbd.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Smriga S, Sandin SA, Azam F. Abundance, diversity, and activity of microbial assemblages associated with coral reef fish guts and feces. FEMS Microbiol Ecol. 2010;73:31–42. doi: 10.1111/j.1574-6941.2010.00879.x. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Sugita H, Miyajima C, Deguchi Y. The vitamin B12-producing ability of the intestinal microflora of freshwater fish. Aquaculture. 1991;92:267–276. [Google Scholar]
- Tsuchiya C, Sakata T, Sugita H. Novel ecological niche of Cetobacterium somerae, an anaerobic bacterium in the intestinal tracts of freshwater fish. Lett Appl Microbiol. 2008;46:43–48. doi: 10.1111/j.1472-765X.2007.02258.x. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Quince C, Faith JJ, McHardy AC, Yatsunenko T, Niazi F, et al. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc Natl Acad Sci USA. 2010;107:7503–7508. doi: 10.1073/pnas.1002355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward NL, Steven B, Penn K, Methe BA, Detrich WH., III Characterization of the intestinal microbiota of two Antarctic notothenioid fish species. Extremophiles. 2009;13:679–685. doi: 10.1007/s00792-009-0252-4. [DOI] [PubMed] [Google Scholar]
- Westerfield M.2000The Zebrafish Book. A guide for the laboratory use of zebrafish (Danio rerio)4th edn.University of Oregon Press: Eugene, OR [Google Scholar]
- Wilson KH, Brown RS, Andersen GL, Tsang J, Sartor B. Comparison of fecal biota from specific pathogen free and feral mice. Anaerobe. 2006;12:249–253. doi: 10.1016/j.anaerobe.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Wu S, Gao T, Zheng Y, Wang W, Cheng Y, Wang G. Microbial diversity of intestinal contents and mucus in yellow catfish (Pelteobagrus fulvidraco) Aquaculture. 2010;303:1–7. [Google Scholar]
- Xenoulis PG, Gray PL, Brightsmith D, Palculict B, Hoppes S, Steiner JM, et al. Molecular characterization of the cloacal microbiota of wild and captive parrots. Vet Microbiol. 2010;146:320–325. doi: 10.1016/j.vetmic.2010.05.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.