Abstract
Salinity is a major factor controlling the distribution of biota in aquatic systems, and most aquatic multicellular organisms are either adapted to life in saltwater or freshwater conditions. Consequently, the saltwater–freshwater mixing zones in coastal or estuarine areas are characterized by limited faunal and floral diversity. Although changes in diversity and decline in species richness in brackish waters is well documented in aquatic ecology, it is unknown to what extent this applies to bacterial communities. Here, we report a first detailed bacterial inventory from vertical profiles of 60 sampling stations distributed along the salinity gradient of the Baltic Sea, one of world's largest brackish water environments, generated using 454 pyrosequencing of partial (400 bp) 16S rRNA genes. Within the salinity gradient, bacterial community composition altered at broad and finer-scale phylogenetic levels. Analogous to faunal communities within brackish conditions, we identified a bacterial brackish water community comprising a diverse combination of freshwater and marine groups, along with populations unique to this environment. As water residence times in the Baltic Sea exceed 3 years, the observed bacterial community cannot be the result of mixing of fresh water and saltwater, but our study represents the first detailed description of an autochthonous brackish microbiome. In contrast to the decline in the diversity of multicellular organisms, reduced bacterial diversity at brackish conditions could not be established. It is possible that the rapid adaptation rate of bacteria has enabled a variety of lineages to fill what for higher organisms remains a challenging and relatively unoccupied ecological niche.
Keywords: brackish water, bacterioplankton, SAR11, Verrucomicrobia, bacterial diversity, 454 pyrosequencing
Introduction
Major physical and/or chemical environmental shifts, such as oxic–anoxic interfaces, strong temperature, nutrient and salinity gradients, are regions of dramatic change in community composition and richness for many organisms. The changes in community composition and richness often have consequences for the functional characteristics of whole ecosystems. Among the abiotic factors, salinity has been shown to have a particularly strong impact on the distribution patterns of species, including benthic and pelagic organisms (Remane, 1934; Wasmund et al., 1999; Cognetti and Maltagliati, 2000; Wetzel, 2001; Ysebaert et al., 2003; Ojaveer et al., 2010; Telesh and Khlebovich, 2010). As most multicellular organisms are either adapted to freshwater or marine environments, only few brackish specialists and species with broad salinity tolerance are found in the transition between marine and freshwater environments. Consequently, these brackish environments, which occur frequently in coastal and estuarine areas, are characterized by reduced diversity in faunal and floral communities (Telesh and Khlebovich, 2010). Salinity has also been suggested to be the major determinant of microbial community composition (Wu et al., 2006), exceeding the influence of temperature and pH (Lozupone and Knight, 2007). Nevertheless, it is yet unclear how indigenous bacterial communities in the transition zone between marine and freshwater systems are organized. Several studies on shifts in bacterial community composition along aquatic salinity gradients have been conducted (Murray et al., 1996; Crump et al., 1999; Cottrell and Kirchman, 2003; Kirchman et al., 2005; Kan et al., 2008). However, these are usually performed on river outlets, which are characterized by short hydrological retention times and dynamic shifts in salinity owing to tidal action and variable freshwater inputs. The hydrology of these systems limits the possibility for an autochthonous brackish microbial community to establish (Crump et al., 1999, 2004). Nevertheless, as a general trend, an increase of α-Proteobacteria and γ-Proteobacteria, and a decrease of β-Proteobacteria, with increasing salinity has been identified in different estuaries (that is, Delaware Bay (Cottrell and Kirchman, 2003), Chesapeake Bay (Bouvier and del Giorgio, 2002) or Pearl River estuary (Zhang et al., 2006)).
In contrast to most estuaries, the Baltic Sea is a relatively stable environment that provides an ideal system to investigate autochthonous brackish bacterial communities. Tidal influences are negligible, and a combination of freshwater discharge and limited water exchange with the North Sea generates far-reaching saltwater–freshwater gradients. In fact, the water retention time in the central Baltic is between 3 and 30 years (Kullenberg, 1981; Reissmann et al., 2009), which may allow brackish-adapted bacterial lineages to establish. Accordingly, previous studies succeeded at isolating bacteria with growth rate optima at salinities of 5–10 from the Baltic Sea (Rheinheimer, 1984). The recent development of high-throughput next-generation sequencing of 16S rRNA genes now allows detailed investigations of bacterial community composition with coverage far beyond previous clone libraries (Sogin et al., 2006; Andersson et al., 2009). Here, we report a comprehensive survey of the distribution of bacteria from 213 samples, generated from 60 stations along the horizontal and vertical salinity gradients of the Baltic Sea, representing the first detailed taxonomic study of an indigenous brackish water microbiome.
Materials and methods
Sampling
Water samples were obtained on a research cruise (MSM0803) of the RV Maria S Merian in June/July 2008. Conductivity, temperature, pressure, chlorophyll a fluorescence and dissolved oxygen of water samples were measured using a conductivity/temperature/depth sensor (CTD) SeaBird 911 connected to a rosette of 24 10-L bottles. Concentrations of inorganic nutrients and oxygen were analyzed as described by Grasshoff et al. (1983). Water samples for DNA analysis were filtered (0.22-μm pore-size white polycarbonate filters), and DNA was extracted according to Weinbauer et al. (2002).
PCR and 454 sequencing
Samples from the Baltic Sea (213) and two samples from Swedish freshwater lakes (the eutrophic Lake Ekoln and oligotrophic Lake Långsjön) were PCR amplified using 25 cycles with the primers Bakt_341F (CCTACGGGNGGCWGCAG) and Bakt_805R (GACTACHVGGGTATCTAATCC). The PCR primers were designed using an in-house program for broad-range 16S rRNA primer design. Bakt_341F matches perfectly to 1 081 974 of 1 124 456 bacterial sequences of ‘Good' quality score spanning Escherichia coli positions 300–400, and Bakt_805R matches perfectly to 905 196 of 1 000 727 bacterial sequences of ‘Good' quality score spanning positions 750–850 in the Ribosomal Database Project (RDP) release 10.25 (Cole et al., 2009) using the RDP ProbeMatch tool. The primers were complemented with 454 adapters and sample-specific 5-bp barcodes, designed such that two barcodes differ by at least 2 bp and homopolymers are lacking. Samples were purified using Agencourt AMPure XP (Becker Coulter, Brea, CA, USA). Amplicons were quantified with the Picogreen assay (Molecular Probes), mixed in equimolar amounts and sequenced from the reverse primer direction by LGC Genomics GmbH using Roche/454 GS FLX Titanium technology.
Sequence processing
Primer sequences were trimmed from the reads and sequences were clipped 400 bp downstream of the primer. Reads shorter than 400 bp (excluding primer) and/or containing Ns were excluded. Unique sequences were aligned and clustered into operational taxonomic units (OTUs) using the pyrosequencing pipeline at RDP (Cole et al., 2009). To co-cluster sequences ⩾97% identical, we empirically found that complete linkage clustering at the 95% level was appropriate for our data set. The complete linkage clustering at the 95% level resulted in co-clustering of 81% of the sequence pairs that were ⩾97% identical, whereas only 1.4% of co-clustered pairs were <97% identical. The most abundant sequence for each OTU was BLAST (Altschul et al., 1997) searched against a local RDP database (v. 10.18; good-quality score sequences only), and the OTU inherited the taxonomy of the best hit if this was ⩾95% identical over ⩾380 bp. If multiple best hits (of same score) were found, the taxonomy was set to the most-detailed level of taxonomy shared by the best hits. Perl scripts for sequence processing can be obtained from the authors on request. Flowgram distributions for pre-clustering and denoizing of 454 Titanium sequences were not available at the time of the analysis. During the revision process these became available, and denoizing was performed with AmpliconNoise (Quince et al., 2011). Denoized sequences were clustered into OTUs using the RDP pipeline as above and used for calculating Shannon diversity indices. All other analysis is based on the original data, as denoizing will have marginal influence here (β-diversity was highly correlated with and without noise removal; Pearson's r=0.98 for all pair-wise sample comparisons).
Calculation of diversity indices
β-Diversity (community composition differences) for principal coordinates analysis and hierarchical clustering was calculated using Spearman's rank correlations of OTU frequencies (Kuczynski et al., 2011): β-diversity=0.5+(−0.5 × ρ), where ρ is the Spearman coefficient (−1⩽ρ⩽1) of OTU frequencies for a pair of samples (not considering OTUs absent in both samples). The Spearman correlation-derived measure of β-diversity correlated well with the Bray–Curtis dissimilarity (Pearson's r=0.95 for all pair-wise sample comparisons). The correlation was lower with UniFrac (Hamady et al., 2010), with Pearson's r=0.68. When we used UniFrac in the principal coordinate analysis, projecting our OTU sequences onto the default Greengenes reference tree (DeSantis et al., 2006), principal coordinate 1 correlated most strongly with oxygen content (Spearman's ρ=0.63) and principal coordinate 2 (PC2) with salinity (Spearman's ρ=0.53). A probable explanation for this is that the pronounced changes at broad phylogenetic levels at the transition from oxic to anoxic conditions has a stronger impact on the UniFrac measure, which takes phylogenetic distances between OTUs into account. In contrast, the gradual replacement of closely related OTUs observed along the salinity gradient has a greater impact on the OTU-based measurements. Hierarchical clustering was calculated using average linkage clustering, and bootstrap approximately unbiased P-values were computed via multiscale bootstrap resampling with 100 replications using the R package Pvclust (www.cran.r-project.org), modified to allow Spearman's correlations for distance calculations. Shannon α-diversity index was calculated according to Hayek and Buzas (1996) based on original as well as denoized OTU frequencies. Calculations and statistics were carried out with the program R (www.r-project.org).
Phylogenetic analyses
The sequences identified as the 100 most abundant in our data set were aligned using the SINA webaligner (www.arb-silva.de) and imported to a SILVA 102 NR template tree (Pruesse et al., 2007) using the ARB program suite (Ludwig et al., 2004). Reference sequences were selected based on the phylogenetic affiliation of the 400 bp sequences to sequences in the SILVA 102 NR. Selected reference sequences were used to construct core trees with 1533 unambiguously aligned sequences using maximum-likelihood algorithms (RAxML, PHyML) and the general bacteria filter provided in ARB. Candidatus ‘Nitrosopumilus maritimus' (acc.CP000866) was used in all trees as root. Short sequences from our study (400 bp), positionally filtered by base frequency (50%), were added without changing the global tree topology using the ARB parsimony tool.
BLAST-based comparison with reference data sets
Marine and freshwater PCR-amplified 16S rRNA sequences (Acinas et al., 2004; Pommier et al., 2007; Shaw et al., 2008) were downloaded from the RDP (Cole et al., 2009), and shotgun 16S rRNA sequences (Rusch et al., 2007) were downloaded from CAMERA (www.camera.calit2.net). Freshwater sequences were combined with a collection of high-quality freshwater bacterial 16S rRNA sequences representing 38 globally distributed lakes (Newton et al., 2011). A total of 6992 freshwater and 7730 marine reference sequences were used in a local BLAST database.
Results and discussion
Factors structuring the bacterial community in the Baltic Sea
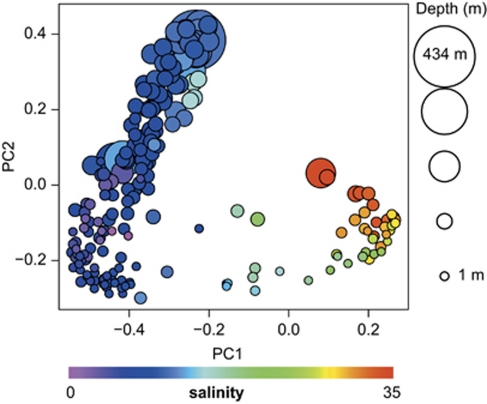
Using barcoded pyrosequencing (Andersson et al., 2008; Hamady et al., 2008), we generated on average 1052±419 (s.d.) quality-filtered clipped (trimmed) 400bp sequences per sample, spanning variable regions V3 and V4 of the 16S rRNA gene. Principal coordinates analysis separated the samples in a way that correlated well with salinity along the first principal coordinate, and depth along the second principal coordinate (Figure 1, Supplementary Table S1). As described earlier (Labrenz et al., 2007), the bacterial community composition changed drastically at the oxygen-sulfide transition zone (Supplementary Figure S1), where we observed a shift towards a Sulfurimonas sp. dominated bacterial community. In the deeper, purely sulfidic zones sulfate-reducing δ-Proteobacteria became the dominant group. This shift in bacterial community composition was not recognized where oxygen was present throughout the water column (Supplementary Figure S1). As only few samples were below the oxygen-sulfide transition, the impact of the oxic–anoxic transition on the results of the principal coordinates analysis was probably relatively low.
Figure 1.
Principle coordinates analysis of Baltic Sea bacterioplankton community composition. Pair-wise sample differences (β-diversity) were calculated by Spearman's rank-order correlations of OTU counts. Each dot is one sample and is colour-coded according to salinity and sized according to depth. PC1 (3.9% variation explained) correlated most strongly with salinity (Spearman's ρ=–0.92, P<10−16). Principal coordinate 2 (3.1% variation explained) correlated most strongly with depth (ρ=0.86, P<10−16), but also with depth-related factors, including oxygen, phosphate and light intensity (see also Supplementary Table S1).
For surface water samples (Figure 2a, ⩽10m), differences in community composition (β-diversity) correlated most strongly with differences in salinity (Spearman's ρ=0.81; Supplementary Figure S2) among the measured environmental variables (Supplementary Table S2). As salinity in the Baltic Sea is coupled to the geographic location (Figure 2a), the observed relationship could potentially also be due to spatial effects, either reflecting spatially limited microbial dispersal (Hubbell, 2001; Sloan et al., 2006) or involving other geographically correlated environmental factors (for example, substrates in run-off water from surrounding land). However, differences in salinity correlated strongly with β-diversity also when we controlled for geographic distance (partial Mantel test r=0.78, P<0.0001), whereas the correlation between β-diversity and geographic distance was lower when we controlled for differences in salinity (r=0.37, P<0.0001; Supplementary Figure S2), which indicates that salinity itself is the major structuring factor in the surface waters.
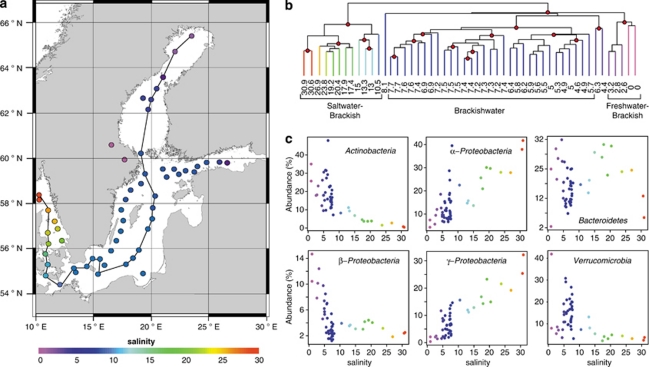
Figure 2.
Bacterial communities of surface samples along the Baltic Sea salinity gradient. (a) Map of the Baltic Sea with sampling stations colour-coded according to the measured surface water salinity. The connecting line indicates the stations displayed in Figure 3. (b) Hierarchical clustering based on bacterial community composition similarities. Nodes supported by high bootstrap values (>90%) are marked with red circles. The samples cluster in three salinity ranges: freshwater–brackish 0–3.2, brackish 4.6–7.7 and marine–brackish 10.5–30.9. Three samples do not show a clear affiliation to any cluster (8.1, 6.3 and 4.4). The samples having salinities of 6.3 and 4.4 were sampled outside Stockholm and St. Petersburg, respectively, the two largest cities in the region, and may potentially be influenced by anthropogenic emissions of nutrients and/or pollutants. (c) Relative abundance vs salinity for abundant phyla and proteobacterial classes.
The brackish bacterial community
Hierarchical clustering of the surface samples (⩽10 m) led to a separation of marine–brackish, brackish and brackish–freshwater bacterial communities with high bootstrap values (Figure 2b). A cluster consisting of samples in the salinity range 5–8 (brackish) is separated from samples in the salinity ranges 10–31 (brackish–marine) and 0–3 (freshwater–brackish), which indicates the existence of a characteristic brackish bacterial community in the central Baltic Sea. A similar pattern was previously observed for marine animals with the major shift between marine and brackish communities occurring at salinity 8–10 (Remane, 1934). Moreover, the border between the brackish–marine and the brackish cluster coincides with the Darss Sill (salinity ∼9), an important biological boundary also for phytoplankton communities (Kell, 1973; Wasmund et al., 2004; Witkowski et al., 2005).
The surface water communities along the salinity gradient of the Baltic Sea were dominated by Actinobacteria, α-Proteobacteria, β-Proteobacteria, γ-Proteobacteria, Cyanobacteria, Bacteroidetes and Verrucomicrobia (Figure 2c). At this broad phylogenetic level, the relative abundance of α-Proteobacteria and γ-Proteobacteria increased with salinity, whereas Actinobacteria and β-Proteobacteria displayed the opposite trend (Figure 2c). This is in accordance with previous studies along estuarine salinity gradients (Murray et al., 1996; Crump et al., 1999; Cottrell and Kirchman, 2003; Kirchman et al., 2005; Kan et al., 2008). Moreover, we identified a pronounced peak of the phylum Verrucomicrobia in brackish conditions (salinities 5–10; Figure 2c).
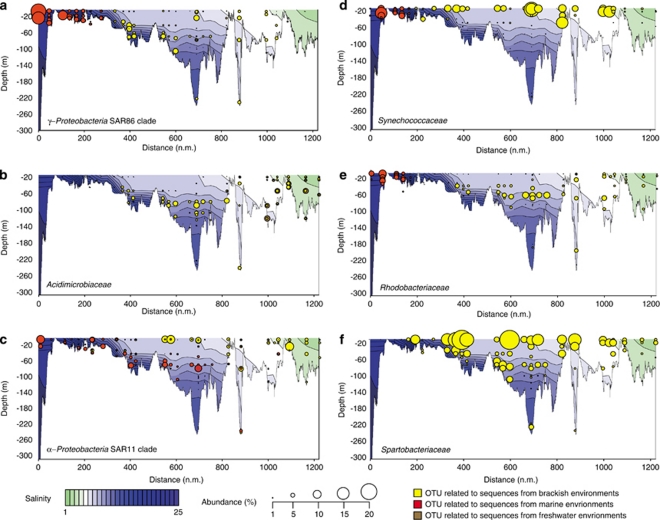
Most of the major bacterial groups in the Baltic Sea were characterized by phylogenetic shifts, where closely related populations (see Supplementary Figure 3 for a detailed phylogenetic analysis of the 100 most abundant OTUs in our study) occupied different salinity ranges, some apparently specifically adapted to the brackish environment. An OTU from the ubiquitous marine clade SAR86 (Suzuki et al., 1997) was abundant in the marine–brackish region (Figure 3a; Supplementary Figure 3 SA1), but was substituted by an SAR86 OTU closest related to a sequence previously retrieved from brackish conditions around salinity 6.5 (Shaw et al., 2008; Figure 3a; Supplementary Figure 3 SA2). Analogously, an OTU from the freshwater clade Actinobacteria acIV (within the Acidimicrobiaceae; Warnecke et al., 2004), abundant at salinities 0–5 (Figure 3b; Supplementary Figure 3 AB1), was replaced by an Actinobacteria acIV OTU, identical to previously retrieved brackish water sequences (Shaw et al., 2008) at salinities of 5–10 (Figure 3b; Supplementary Figure 3 AB2).
Figure 3.
Spatial distribution and abundance of selected OTUs along a Baltic Sea transect extending east of Gotland and into the Bay of Bothnia (see Figure 2). (a–e) Shifts in populations of (a) SAR86, (b) Acidimicrobiaceae, (c) SAR11, (d) Synechococcaceae and (e) Rhodobacteriaceae along the salinity gradient of the Baltic Sea. (f) The most abundant OTU in our study, which is related to Spartobacteriaceae, in the different depth and salinity regions.
Most OTUs occupied only a portion of the salinity spectrum, but two abundant SAR11 populations displayed partly overlapping, but contrasting distribution patterns over the complete salinity range of the Baltic Sea (Figure 3c). The OTU related to the marine Candidatus ‘Pelagibacter ubique' was abundant in the marine–brackish part of the Baltic Sea (Figure 3c, Supplementary Figure 3 SR1), whereas an OTU identical to SAR11 sequences previously retrieved from brackish environments (Kan et al., 2008; Shaw et al., 2008) dominated the brackish–freshwater transition (Figure 3c, Supplementary Figure 3 SR2). Although Candidatus ‘Pelagibacter ubique' has been shown to carry proteorhodopsin genes (Giovannoni et al., 2005; Sabehi et al., 2005), the organism followed its putative salinity optimum also at depths that are not reached by solar radiation (Figure 3c). This implies that salinity has a stronger impact than light on the distribution of Candidatus ‘Pelagibacter ubique'. In contrast, the distribution of the photosynthetic picocyanobacterium Synechococcus sp., which also exhibit phylogenetic shifts along the salinity gradient of the Baltic Sea, does not extend into deeper waters (Figure 3d, Supplementary Figure 3 CY1 and CY2). This is in line with its obligate phototropic metabolism. A different pattern was observed for Rhodobacteriaceae, which are major components of global ocean bacterioplankton (Rappé et al., 2000). Whereas various Roseobacter OTUs were abundant in the marine parts of the Baltic Sea (Figure 3e; Supplementary Figure 3 RB1), the central Baltic Sea was dominated by an OTU highly similar to Pseudorhodobacter ferrugineus (Figure 3e; Supplementary Figure 3 RB2). This OTU has highest relative abundances below 50 m in the Baltic Proper (salinity 6–10; Figure 3e), which is in accordance with the aerobic, non-photosynthetic lifestyle of P. ferrugineus (Uchino et al., 2002). Many representatives of the Roseobacter clade have also been proposed to perform photoheterotrophy in coastal zones (Labrenz et al., 2005), but as no Rhodobacteriaceae were present in the photic zone of the Baltic Sea, this function could be less significant for this bacterial family at brackish conditions.
The highly abundant phylum Verrucomicrobia (salinities 5–10; Figure 2) was, in contrast to the high diversity within other phyla, largely represented by a single dominant OTU (Figure 3f; Supplementary Figure 3 VE1), highly similar (>97%) to cloned sequences from the Baltic Sea affiliated to the Spartobacteriaceae (Supplementary Figure 4). Even though amplification biases in the PCR cannot be ruled out (Suzuki and Giovannoni, 1996, Polz and Cavanaugh, 1998), its high abundance (>10% of reads in many samples), over a range of salinities and depths, indicates that this OTU probably represents one of the most abundant organisms of the Baltic Sea in summer. Although some members of the Spartobacteriaceae have recently been identified as abundant representatives in freshwater lakes (Arnds et al., 2010), this specific OTU seems to be well adapted to the saltwater–freshwater transition zone, as indicated by its abundance maximum at brackish conditions and the lack of closely related lineages in the adjacent freshwater and marine parts of the Baltic Sea. As no close relative has yet been obtained in axenic culture (closest isolate is Chthoniobacter flavus; 88% 16S rRNA sequence identity), nothing is known about the metabolism or ecological role of this bacterium.
Bacterial α-diversity along the salinity gradient
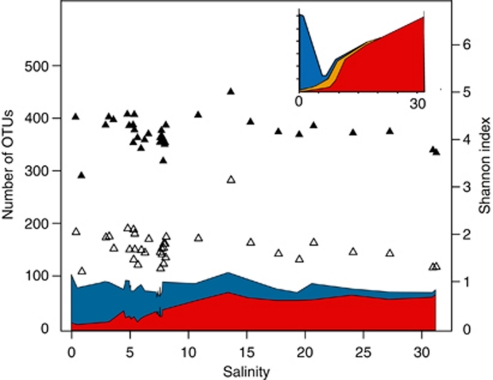
According to the Remane curve (Figure 4, inset), which describes the distribution of marine animal richness along a Baltic Sea salinity gradient, the number of marine species declines and the number of freshwater species increases with decreasing salinity (Remane, 1934). A ‘Remane relationship' has also been shown for mesozooplankton composition of the Baltic Sea (Schiewer, 2008). To test systematically whether this model is also valid for bacteria, we compared the surface water OTU sequences obtained in our study to environmental sequences from freshwater and marine environments outside of the Baltic Sea using BLAST (Altschul et al., 1997). Analogous to the situation for marine animals, the number of observed sequences matching marine sequences increased with increasing salinity, whereas the number of sequences matching freshwater sequences decreased (Figure 4). Comparable to the Remane curve, and in accordance with the clustering results above, marine bacteria are dominant in a larger salinity spectrum (8–32) than freshwater bacteria (0–8). In the Baltic Proper (salinity 6–10), roughly equal proportions of sequences matched the two reference environments, which suggests that the community is derived from these biomes to similar extents.
Figure 4.
Comparison of surface water OTU sequences obtained in our study to environmental sequences from freshwater and marine reference environments outside of the Baltic Sea. Blue and red areas indicate the number of OTUs ⩾97% identical to freshwater (blue) and marine sequences (red), respectively. Empty triangles are observed number of OTUs and black triangles are Shannon index. To account for sampling size differences, the OTUs corresponding to 800 randomly picked reads (without replacement) were used for each surface sample (samples with fewer than 800 reads were excluded from the analysis). Inset: The Remane curve (reconstructed from Remane (1934)) showing benthic fauna richness along a Baltic Sea salinity gradient. Blue area: number of freshwater species; red area: number of marine species; and yellow area: number of brackish specialist species.
The Remane curve predicts a minimum of species richness at intermediate salinity levels (Figure 4, inset), because the number of brackish specialists does not compensate for the decline in marine and freshwater diversity (Remane, 1934). However, no trends in the number of observed OTUs along the salinity gradient could be observed, nor did the Shannon diversity index change markedly (Figure 4). These α-diversity measurements did not differ significantly between salinity groups when dividing the surface samples into freshwater–brackish (salinity 0–4), brackish (salinity 4–9) and brackish–marine (salinity 10–31; Wilcoxon's rank-sum test P>0.16 for all pair-wise comparisons of groups), and were also nonsignificant when sequences had been pre-processed with AmpliconNoise for noise removal (Quince et al. (2011); P>0.09 for all comparisons). As the sum of OTUs matching to freshwater and marine sequences is relatively constant along the salinity gradient (Figure 4), the α-diversity at intermediate salinities is likely not primarily maintained by phylogenetically divergent brackish water specialists, but rather by adapted bacteria originating from marine and freshwater environments. The observed diversity likely represents actively reproducing populations, as protist grazing implies a fast turnover of picoplankton in surface waters of the Baltic Sea, similar to in other pelagic systems (Setälä and Kivi, 2003). The Baltic Sea was formed after the last glaciation (10 000–15 000 years ago) and is a, in a geological and evolutionary timescale, young sea (Feistel et al., 2008). The observed deviation from the Remane model could reflect that bacteria have been able to faster adapt to the brackish conditions of this sea than multicellular eukaryotes. Physiological differences between bacteria and multicellular organisms, which influence their possibilities to adapt to novel salinity ranges, could be a reason for these results, but also due to a more general rapid bacterial adaptation to new environments, potentially due to their shorter generation times and more dynamic genomes.
In conclusion, we have observed marked differences in the brackish bacterial community composition compared with its freshwater and marine counterparts. The brackish waters of the Baltic Sea are occupied by a diverse combination of freshwater and marine clades that appears to have adapted to the brackish conditions. We currently know very little about the function and ecology of even the most dominant members of these microbial communities, but the mapping of the present brackish sea microbiome provides the foundation for functional and genomic investigations of the important bacterial players and their specific biogeochemical roles in the brackish environment. Future studies may also reveal to what extent the different populations identified in this study are in fact composed of multiple genomically determined ecotypes specialized in more narrow salinity and depth ranges.
Acknowledgments
We thank the crew and staff of the RV Maria S Merian (MSM0803) for taking samples; Christian Stolle, Carlo Berg, Christian Meeske, Anke Gerber and Katja Becker for technical assistance; Günther Nausch for contributing chemical data; and the LGC Genomics company (formerly AGOWA) for 454 sequencing of the samples. The study was funded by the Leibniz Institute for Baltic Sea Research (IOW), the Knut and Alice Wallenberg Foundation, the Swedish Research Council (grant to SB) and the Swedish Research Council FORMAS (grant to AFA). We thank Eva Lindström, Paul Wilmes and the groups of Mary Ann Moran and Gregory Dick for constructive comments on the manuscript, and the German Science Foundation for financial support for the MSM0803 cruise.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Acinas SG, Klepac-Ceraj V, Hunt DE, Pharino C, Ceraj I, Distel DL, et al. Fine-scale phylogenetic architecture of a complex bacterial community. Nature. 2004;430:551–554. doi: 10.1038/nature02649. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson AF, Lindberg M, Jakobsson H, Backhed F, Nyren P, Engstrand L. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One. 2008;3:e2836. doi: 10.1371/journal.pone.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson AF, Riemann L, Bertilsson S. Pyrosequencing reveals contrasting seasonal dynamics of taxa within Baltic Sea bacterioplankton communities. ISME J. 2009;4:171–181. doi: 10.1038/ismej.2009.108. [DOI] [PubMed] [Google Scholar]
- Arnds J, Knittel K, Buck U, Winkel M, Amann R. Development of a 16S rRNA-targeted probe set for Verrucomicrobia and its application for fluorescence in situ hybridization in a humic lake. Syst Appl Microbiol. 2010;33:139–148. doi: 10.1016/j.syapm.2009.12.005. [DOI] [PubMed] [Google Scholar]
- Bouvier TC, del Giorgio PA. Compositional changes in free-living bacterial communities along a salinity gradient in two temperate estuaries. Limnol Oceanogr. 2002;47:453–470. [Google Scholar]
- Cognetti G, Maltagliati F. Biodiversity and adaptive mechanisms in brackish water fauna. Marine Pollut Bull. 2000;40:7–14. [Google Scholar]
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottrell MT, Kirchman DL. Contribution of major bacterial groups to bacterial biomass production (thymidine and leucine incorporation) in the Delaware estuary. Limnol Oceanogr. 2003;48:168–178. [Google Scholar]
- Crump BC, Armbrust EV, Baross JA. Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia river, its estuary, and the adjacent coastal ocean. Appl Environ Microbiol. 1999;65:3192–3204. doi: 10.1128/aem.65.7.3192-3204.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump BC, Hopkinson CS, Sogin ML, Hobbie JE. Microbial biogeography along an estuarine salinity gradient: combined influences of bacterial growth and residence time. Appl Environ Microbiol. 2004;70:1494–1505. doi: 10.1128/AEM.70.3.1494-1505.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Green genes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feistel R, Nausch G, Wasmund N. State and Evolution of the BALTIC Sea, 1952–2005: A Detailed 50-Year Survey of Meteorology and Climate, Physics, Chemistry, Biology, and Marine Environment. General Oceanography of the Baltic Sea. John Wiley & Sons Inc.: Hoboken: NJ; 2008. [Google Scholar]
- Giovannoni SJ, Bibbs L, Cho JC, Stapels MD, Desiderio R, Vergin KL, et al. Proteorhodopsin in the ubiquitous marine bacterium SAR11. Nature. 2005;438:82–85. doi: 10.1038/nature04032. [DOI] [PubMed] [Google Scholar]
- Grasshoff K, Erhardt M, Kremling KV.1983Methods of Seawater Analysis, vol. 2.Verlag Chemie: Weinheim [Google Scholar]
- Hamady M, Lozupone C, Knight R. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2010;4:17–27. doi: 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayek LAC, Buzas MA. Surveying Natural Populations. Columbia University Press: New York, NY; 1996. [Google Scholar]
- Hubbell SP. The Unified Neutral Theory of Biodiversity and Biogeography. Princeton University Press: Princeton, NJ; 2001. [Google Scholar]
- Kan J, Evans S, Chen F, Suzuki M. Novel estuarine bacterioplankton in rRNA operon libraries from the Chesapeake Bay. Aquat Microb Ecol. 2008;51:55–66. [Google Scholar]
- Kell V. Die Darβer Schwelle—eine biologische Grenze. Wiss Zeit Univ Rostock. 1973;22:617–623. [Google Scholar]
- Kirchman DL, Dittel AI, Malmstrom RR, Cottrell MT. Biogeography of major bacterial groups in the Delaware estuary. Limnol Oceanogr. 2005;50:1697–1706. [Google Scholar]
- Kuczynski J, Liu Z, Lozupone C, McDonald D, Fierer N, Knight R. Microbial community resemblance methods differ in their ability to detect biologically relevant patterns. Nat Methods. 2011;7:813–819. doi: 10.1038/nmeth.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullenberg G.1981Physical oceanographyIn: Voipio A (ed).The Baltic Sea. Elsevier Scientific Publishing Company: Amsterdam; 135–175. [Google Scholar]
- Labrenz M, Jost G, Jürgens K. Distribution of abundant prokaryotic organisms in the water column of the central Baltic Sea with an oxic–anoxic interface. Aquat Microb Ecol. 2007;46:177–190. [Google Scholar]
- Labrenz M, Lawson PA, Tindall BJ, Collins MD, Hirsch P. Roseisalinus antarcticus gen. nov., sp. nov., a novel aerobic bacteriochlorophyll a-producing α-proteobacterium isolated from hypersaline Ekho Lake, Antarctica. Int J Syst Evol Micr. 2005;55:41–47. doi: 10.1099/ijs.0.63230-0. [DOI] [PubMed] [Google Scholar]
- Lozupone CA, Knight R. Global patterns in bacterial diversity. Proc Natl Acad Sci USA. 2007;104:11436–11440. doi: 10.1073/pnas.0611525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Kumar Y, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AE, Hollibaugh JT, Orrego C. Phylogenetic compositions of bacterioplankton from two California estuaries compared by denaturing gradient gel electrophoresis of 16S rDNA fragments. Appl Environ Microbiol. 1996;62:2676–2680. doi: 10.1128/aem.62.7.2676-2680.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton RJ, Jones SE, Eiler A, McMahon KD, Bertilsson S. A guide to the natural history of freshwater lake bacteria. Microbiol Mol Biol Rev. 2011;75:14–49. doi: 10.1128/MMBR.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojaveer H, Jaanus A, Mackenzie BR, Martin G, Olenin S, Radziejewska T, et al. Status of biodiversity in the Baltic Sea. PLoS One. 2010;5:e12467. doi: 10.1371/journal.pone.0012467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polz MF, Cavanaugh CM. Bias in template-to-product ratios in multitemplate PCR. Appl Environ Microbiol. 1998;64:3724–3730. doi: 10.1128/aem.64.10.3724-3730.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier T, Canback B, Riemann L, Bostrom KH, Simu K, Lundberg P, et al. Global patterns of diversity and community structure in marine bacterioplankton. Mol Ecol. 2007;16:867–880. doi: 10.1111/j.1365-294X.2006.03189.x. [DOI] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quince C, Lanzen A, Davenport R, Turnbaugh P. Removing noise from pyrosequenced amplicons. BMC Bioinform. 2011;12:38. doi: 10.1186/1471-2105-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappé MS, Vergin K, Giovannoni SJ. Phylogenetic comparisons of a coastal bacterioplankton community with its counterparts in open ocean and freshwater systems. FEMS Microbiol Ecol. 2000;33:219–232. doi: 10.1111/j.1574-6941.2000.tb00744.x. [DOI] [PubMed] [Google Scholar]
- Remane A. Die Brackwasserfauna. Zool Anz. 1934;7:34–74. [Google Scholar]
- Reissmann J, Burchard H, Feistel R, Hagen E, Lass HU, Mohrholz V, et al. State-of-the-art review on vertical mixing in the Baltic Sea and consequences for eutrophication. Progr Oceanogr. 2009;82:47–80. [Google Scholar]
- Rheinheimer G. Bacterial ecology of the Baltic Sea. Bot Mar. 1984;27:277–299. [Google Scholar]
- Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, et al. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabehi G, Loy A, Jung KH, Partha R, Spudich JL, Isaacson T, et al. New insights into metabolic properties of marine bacteria encoding proteorhodopsins. PLoS Biol. 2005;3:e273. doi: 10.1371/journal.pbio.0030273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiewer UE.2008Ecology of Baltic Coastal Waters. Series: Ecological Studiesvol. 197.Springer: Berlin [Google Scholar]
- Setälä O, Kivi K. Planktonic ciliates in the Baltic Sea in summer: distribution, species association and estimated grazing impact. Aquat Microb Ecol. 2003;32:287–297. [Google Scholar]
- Shaw AK, Halpern AL, Beeson K, Tran B, Venter JC, Martiny JB. It's all relative: ranking the diversity of aquatic bacterial communities. Environ Microbiol. 2008;10:2200–2210. doi: 10.1111/j.1462-2920.2008.01626.x. [DOI] [PubMed] [Google Scholar]
- Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR, et al. Microbial diversity in the deep sea and the underexplored ‘rare biosphere'. Proc Natl Acad Sci USA. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan WT, Lunn M, Woodcock S, Head IM, Nee S, Curtis TP. Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environ Microbiol. 2006;8:732–740. doi: 10.1111/j.1462-2920.2005.00956.x. [DOI] [PubMed] [Google Scholar]
- Suzuki MT, Giovannoni SJ. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MT, Rappe MS, Haimberger ZW, Winfield H, Adair N, Strobel J, et al. Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl Environ Microbiol. 1997;63:983–989. doi: 10.1128/aem.63.3.983-989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telesh IV, Khlebovich VV. Principal processes within the estuarine salinity gradient: a review. Mar Pollut Bull. 2010;61:4–6. doi: 10.1016/j.marpolbul.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Uchino Y, Hamada T, Yokota A. Proposal of Pseudorhodobacter ferrugineus gen. nov., comb. nov., for a non-photosynthetic marine bacterium, Agrobacterium ferrugineum, related to the genus Rhodobacter. J Gen Appl Microbiol. 2002;48:309–319. doi: 10.2323/jgam.48.309. [DOI] [PubMed] [Google Scholar]
- Warnecke F, Amann R, Pernthaler J. Actinobacterial 16S rRNA genes from freshwater habitats cluster in four distinct lineages. Environ Microbiol. 2004;6:242–253. doi: 10.1111/j.1462-2920.2004.00561.x. [DOI] [PubMed] [Google Scholar]
- Wasmund N, Pollehne F, Postel L, Siegel H, Zettler ML. Biologische Zustandseinschätzung der Ostsee im Jahre 2003. Meereswissen Ber Warn. 2004;60:1–87. [Google Scholar]
- Wasmund N, Zalewski M, Busch S. Phytoplankton in large river plumes in the Baltic Sea. ICES J Mar Sci. 1999;56:22–32. [Google Scholar]
- Weinbauer MG, Fritz I, Wenderoth DF, Höfle MG. Simultaneous extraction from bacterioplankton of total RNA and DNA suitable for quantitative structure and function analyses. Appl Environ Microbiol. 2002;68:1082–1087. doi: 10.1128/AEM.68.3.1082-1087.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel RG. Salinity of Inland Waters. Limnology. Elsevier: San Diego, CA; 2001. [Google Scholar]
- Witkowski A, Broszinski A, Bennike O, Anczak-Kostecka B, Jensen JB, Lemke W, et al. Darss Sill as a border in the fossil record of the Baltic Sea: evidence from diatoms. Quatern Int. 2005;130:97–109. [Google Scholar]
- Wu QL, Zwart G, Schauer M, Kamst-van Agterveld MP, Hahn MW. Bacterioplankton community composition along a salinity gradient of sixteen high-mountain lakes located on the Tibetan Plateau, China. Appl Environ Microbiol. 2006;72:5478–5485. doi: 10.1128/AEM.00767-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ysebaert T, Herman PMJ, Meire P, Craeymeersch J, Verbeek H, Heip CHR. Large-scale spatial patterns in estuaries: estuarine macrobenthic communities in the Schelde estuary, NW Europe. Estuar Coast Shelf S. 2003;57:335–355. [Google Scholar]
- Zhang Y, Jiao N, Cottrell MT, Kirchman DL. Contribution of major bacterial groups to bacterial biomass production along a salinity gradient in the South China Sea. Aquat Microb Ecol. 2006;43:233–241. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.