Several studies have demonstrated the acquisition of microbial genes by different marine cyanophages, including core photosystem-I (PSI) and photosystem-II (PSII) genes, as well as many other genes of the electron transport chain (Mann et al., 2003; Lindell et al., 2004; Millard et al., 2004; Sullivan et al., 2005; Sharon et al., 2009; Alperovitch et al., 2011).
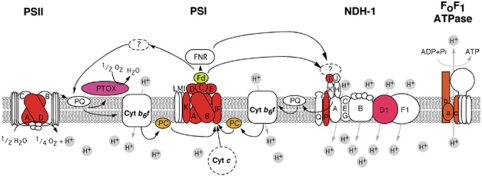
A unique search strategy on the Global Ocean Survey (GOS) metagenomes (Rusch et al., 2007) in combination with marine virome and microbiome pyrosequencing-based data sets (Dinsdale et al., 2008a, 2008b), has recently revealed previously undetected microbial oxygenic photosynthesis and electron transport protein genes within genomes of uncultured marine viral communities (the VirMic project (Sharon et al., 2011), http://www.cs.technion.ac.il/~itaish/VirMic/). Among them are new viral electron transport protein genes, ndhI and ndhD, from the cyanobacterial type I NAD(P)H dehydrogenase (NDH-1) complex (Figure 1). In cyanobacteria these complexes participate in different tasks such as CO2 uptake, respiration and cyclic electron flow around PSI (for review, see (Battchikova et al., 2011)). On the basis of homology it was suggested that the viral proteins found in the VirMic project participate in cyclic electron flow and respiration during viral infection, probably enhancing the production of ATP required for protein synthesis (Sharon et al., 2011). Thus, cyanophages might additionally improve their fitness by tuning the electron transfer routes to optimise phage reproduction, especially in conditions of moderate heat and strong light stresses, which inhibit PSII activity.
Figure 1.
Schematic draft of cyclic electron transfer around PSI and the cyanobacterial equivalent of chlororespiration pathways in phage-infected-cyanobacterial membranes. In colour are proteins predicted to be encoded by cyanophage genomes (based on the VirMic Project). It is important to note that the proteins predicted to be encoded in cyanophage genomes do not all co-occur in any single phage. Proteins predicted to be present on a single phage or scaffold are coloured with the same colour. NDH-1 stands for the cyanobacterial type I NAD(P)H dehydrogenase complex; PTOX for plastid terminal plastoquinone oxidase; PQ for plastoquinone; PC for plastocyanin; Fd for ferredoxin; FNR for ferredoxin: NADP(H) oxireductase. The ellipse labeled with a question mark on top of the NDH-1 complex indicates unknown subunits responsible for NAD(P)H oxidation. The colour reproduction of this figure is available on the html full text version of the manuscript.
Recenly, two novel small subunits of the cyanobacterial NDH-1 complex, NdhP and NdhQ, were identified via biochemical studies in Thermosynechococcus elongatus. Homology searches with the ndhP gene revealed the presence of ndhP genes in different cyanophage PSI gene cassettes (Nowaczyk et al., 2011). These gene cassettes were already known to include another NDH-1 gene, the ndhI gene (Alperovitch et al., 2011). Using blastX searches with the new ndhP gene on the NCBI environmental samples data set (env_nr) and also using the VirMic website, we have detected different viral scaffolds from the GOS data set that include, in addition to the ndhP gene, unique gene combinations related to the electron transport chain. Among them, are scaffolds that include genes coding for plastid terminal plastoquinone oxidase (PTOX), ndhI and different viral PSI genes (data not shown). A model of oxygenic photosynthetic electron transport chain is shown in Figure 1, highlighting in colour those membrane proteins, the homologues of which have been found to be encoded by phage genomes. We have included different known viral-encoded proteins involved in electron transfer processes, such as the cyanophage-encoded plastocyanins and ferredoxins (Lindell et al., 2004; Sullivan et al., 2005) and PTOX (Weigele et al., 2007). Interestingly, and related to the electron transport chain issue, we have identified via the VirMic project a GOS cyanophage clone (JCVI_SCAF_1096628171668) that includes three genes encoding for subunits a, b and c from the Fo membrane part of the FoF1 ATPase complex. It should be noted, as a word of caution that all of our predictions are based on metagenomic analyses and homology searches and are therefore prone to errors such as wrong annotations, different functions within the same cluster and so on.
Cyanophages hijack the host's translation and transcription systems (Rohwer and Thurber, 2009). In addition, phage-harboured microbial genes differ in sequence and can even introduce novel fusion genes such as the recently found PSI J-F subunits viral fusion gene (Sharon et al., 2009). The phages' fast rate of evolution and their ability to carry hosts' genes enable them to constantly change, both at the sequence level and at the components level, the respiratory chain and the photosynthetic electron transfer chain. Marine cyanophages probably mix and modify the components belonging to these systems, linking complexes that otherwise tend to be separated under optimal growth of uninfected cells. As shown in Figure 1, we hypothesise that the viral-encoded proteins modify the photosynthetic electron transfer chain such that the cyclic electron flow around PSI is favoured over the linear one. This would result in preferential production of ATP (and less NADPH), which is likely to be crucial for efficient reproduction of cyanophages. This idea needs be tested in vivo by replacing different combinations of cyanobacterial genes with the ones found in cyanophages. We would expect that future marine environmental sequencing projects would reveal that other components of the photosynthetic and respiratory chains are carried by marine cyanophages.
Marine bacteria have an important role in global biogeochemical processes (Falkowski et al., 2008). Phages infecting these bacteria would likely have an impact on these processes as well (Danovaro et al., 2011). However, as previously suggested by Rohwer and Thurber (2009), we would expect phages to contribute to biogeochemical processes not only indirectly by predation of bacteria and the subsequent effects on bacterial populations (such as the ‘kill the winner' hypothesis and ‘viral-shunt'), but also to have a direct effect on these processes by constantly changing and reshuffling genes that have a part in these processes. Although phages carry these genes and manipulate them probably for their own fitness, the resulting effect on global biogeochemical processes may be profound. This is indicated by preliminary evidence that some cellular processes (for example, photosynthesis (Lindell et al., 2005; Clokie et al., 2006)) do not cease but in fact continue while the bacterial cells are under viral attack, with the phage-encoded genes being expressed in higher copy numbers compared with the hosts' intrinsic genes. The growing repertoire of phage-harbored bacterial metabolic genes and the variety of infected hosts suggests that marine phages are major, but still underrated, players in processes of global importance.
References
- Alperovitch A, Sharon I, Rohwer F, Aro EM, Glaser F, Milo R, et al. Reconstructing a puzzle: existence of cyanophages containing both photosystem-I & photosystem-II gene suites inferred from oceanic matagenomic datasets. Environ Microbiol. 2011;13:24–32. doi: 10.1111/j.1462-2920.2010.02304.x. [DOI] [PubMed] [Google Scholar]
- Battchikova N, Elsenhut M, Aro EM.2011Cyanobacterial NDH-1 complexes: novel insights and remaining puzzles Biochim Biophys Actadoi: 10.1016/j.bbabio.2010.10.017 [DOI] [PubMed]
- Clokie MR, Shan J, Bailey S, Jia Y, Krisch HM, West S, et al. Transcription of a ‘photosynthetic' T4-type phage during infection of a marine cyanobacterium. Environ Microbiol. 2006;8:827–835. doi: 10.1111/j.1462-2920.2005.00969.x. [DOI] [PubMed] [Google Scholar]
- Danovaro R, Corinaldesi C, Dell'anno A, Fuhrman JA, Middelburg JJ, Noble RT, et al. 2011Marine viruses and global climate change FEMS Microbiol Revdoi: 10.1111/j.1574-6976.2010.00258.x [DOI] [PubMed]
- Dinsdale EA, Edwards RA, Hall D, Angly F, Breitbart M, Brulc JM, et al. Functional metagenomic profiling of nine biomes. Nature. 2008a;452:629–632. doi: 10.1038/nature06810. [DOI] [PubMed] [Google Scholar]
- Dinsdale EA, Pantos O, Smriga S, Edwards RA, Angly F, Wegley L, et al. Microbial ecology of four coral atolls in the northern Line Islands. PLoS ONE. 2008b;3:e1584. doi: 10.1371/journal.pone.0001584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkowski PG, Fenchel T, Delong EF. The microbial engines that drive Earth's biogeochemical cycles. Science. 2008;320:1034–1039. doi: 10.1126/science.1153213. [DOI] [PubMed] [Google Scholar]
- Lindell D, Jaffe JD, Johnson ZI, Church GM, Chisholm SW. Photosynthesis genes in marine viruses yield proteins during host infection. Nature. 2005;438:86–89. doi: 10.1038/nature04111. [DOI] [PubMed] [Google Scholar]
- Lindell D, Sullivan MB, Johnson ZI, Tolonen AC, Rohwer F, Chisholm SW. Transfer of photosynthesis genes to and from Prochlorococcus viruses. Proc Natl Acad Sci USA. 2004;101:11013–11018. doi: 10.1073/pnas.0401526101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann NH, Cook A, Millard A, Bailey S, Clokie M. Bacterial photosynthesis genes in a virus. Nature. 2003;424:741. doi: 10.1038/424741a. [DOI] [PubMed] [Google Scholar]
- Millard A, Clokie MRJ, Shub DA, Mann NH. Genetic organization of the psbAD region in phages infecting marine Synechococcus strains. Proc Natl Acad Sci USA. 2004;101:11007–11012. doi: 10.1073/pnas.0401478101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowaczyk MM, Wulfhorst H, Ryan CM, Souda P, Zhang H, Cramer WA, et al. NdhP and NdhQ: two novel small subunits of the cyanobacterial NDH-1 Complex. Biochemistry. 2011;50:1121–1124. doi: 10.1021/bi102044b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohwer F, Thurber RV. Viruses manipulate the marine environment. Nature. 2009;459:207–212. doi: 10.1038/nature08060. [DOI] [PubMed] [Google Scholar]
- Rusch DB, Halpern AL, Heidelberg KB, Sutton G, Williamson SJ, Yooseph S, et al. The Sorcerer II Global Ocean Sampling expedition: I, the northwest Atlantic through the eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon I, Alperovitch A, Rohwer F, Haynes M, Glaser F, Atamna-Ismaeel N, et al. Photosystem-I gene cassettes are present in marine virus genomes. Nature. 2009;461:258–262. doi: 10.1038/nature08284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon I, Battchikova N, Aro EM, Giglione C, Meinnel T, Glaser F, et al. 2011Comparative metagenomics of microbial traits within oceanic viral communities ISME Jdoi: 10.1038/ismej.2011.2 [DOI] [PMC free article] [PubMed]
- Sullivan MB, Coleman ML, Weigele P, Rohwer F, Chisholm SW. Three Prochlorococcus cyanophage genomes: Signature features and ecological interpretations. PLoS Biol. 2005;3:e144. doi: 10.1371/journal.pbio.0030144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigele PR, Pope WH, Pedulla ML, Houtz JM, Smith AL, Conway JF, et al. Genomic and structural analysis of Syn9, a cyanophage infecting marine Prochlorococcus and Synechococcus. Environ Microbiol. 2007;9:1675–1695. doi: 10.1111/j.1462-2920.2007.01285.x. [DOI] [PubMed] [Google Scholar]