Abstract
Self-incompatibility (SI) is a genetic system found in some hermaphrodite plants. Recognition of pollen by pistils expressing cognate specificities at two linked genes leads to rejection of self pollen and pollen from close relatives, i.e., to avoidance of self-fertilization and inbred matings, and thus increased outcrossing. These genes generally have many alleles, yet the conditions allowing the evolution of new alleles remain mysterious. Evolutionary changes are clearly necessary in both genes, since any mutation affecting only one of them would result in a nonfunctional self-compatible haplotype. Here, we study diversification at the S-locus (i.e., a stable increase in the total number of SI haplotypes in the population, through the incorporation of new SI haplotypes), both deterministically (by investigating analytically the fate of mutations in an infinite population) and by simulations of finite populations. We show that the conditions allowing diversification are far less stringent in finite populations with recurrent mutations of the pollen and pistil genes, suggesting that diversification is possible in a panmictic population. We find that new SI haplotypes emerge fastest in populations with few SI haplotypes, and we discuss some implications for empirical data on S-alleles. However, allele numbers in our simulations never reach values as high as observed in plants whose SI systems have been studied, and we suggest extensions of our models that may reconcile the theory and data.
GENES involved in recognition systems, such as the major histocompatibility complex in vertebrates (Solberg et al. 2008), mating types either in fungi (Billiard et al. 2011) or protists (Phadke and Zufall 2009), and self-incompatibility (SI) in plants (Lawrence 2000), typically show extraordinarily high levels of genetic diversity. SI is widespread in angiosperms [found in >100 families (Igic et al. 2008)] and is highly multiallelic [up to 200 SI haplotypes (Lawrence 2000)]. This system enables hermaphrodite plants to avoid selfing and mating with close relatives and is based on recognition and rejection of pollen by pistils if they express cognate SI specificities. In many species, SI specificity is controlled by a single genetic locus (the S-locus) composed of two linked genes, one expressed in pollen and the other in styles (Takayama and Isogai 2005). The maintenance of high diversity in gametophytic self-incompatibility (GSI) is easily explained by negative frequency-dependent selection, whereby individuals with a rare SI haplotype can fertilize more partners than individuals with a common SI haplotype. As a consequence, rare SI haplotypes are unlikely to be lost by drift, especially new SI haplotypes arising by mutation (Wright 1939). A major unsolved puzzle, though, is how new SI haplotypes appear, while it is a long-standing question (Lewis 1949; Fisher 1961). Some information exists about sequence changes affecting the specificity of the pollen or pistil protein, e.g., in the Solanum chacoense pistil gene (Matton et al. 1999) and the Brassica oleracea pollen gene (Chookajorn et al. 2004). These investigations showed that the substitution of just a few amino acids in the pollen or pistil sequences may be sufficient to alter specificity. Yet there is no plausible evolutionary scenario for the emergence of new SI haplotypes, because, to create a new functional SI haplotype, the two genes must both change appropriately. The large number of SI haplotypes typically found in SI species nevertheless shows that such diversification occurred repeatedly.
Three evolutionary models have so far been proposed to explain how new SI haplotypes can arise. Matton et al. (1999) proposed a verbal model on the basis of the observation in experiments with S. chacoense of a dual-specificity artificial chimeric S-RNase protein (produced by the style gene) able to reject two distinct pollen specificities. Their model considered a mutation in the style gene conferring dual specificity, recognizing a new pollen type, not yet present, as well as its initial pollen type. The pollen gene of this haplotype might then mutate to acquire the new specificity, and the ancestral pistil recognition type might be lost, resulting in a new functional SI haplotype. This scenario was criticized because the new pollen allele would transiently be rejected by two different haplotypes, placing it at a selective disadvantage when challenged against its ancestral nonmutated copy (Charlesworth 2000; Uyenoyama and Newbigin 2000), and is thus expected to be lost from the population. Furthermore, this scenario involves the unlikely occurrence of three consecutive mutations on the same haplotype (Charlesworth 2000).
A second verbal model is based on the observation that the B. oleracea pollen gene can tolerate several amino acid substitutions without affecting its binding affinity or specificity (Chookajorn et al. 2004). These authors proposed that nearly neutral standing variation exists within each functional allelic class. If, by chance, recognition became stronger among a subset of haplotypes within a class than with the other haplotypes of the same class, selection might reinforce this new interaction and reduce recognition of the remaining haplotypes in the class, eventually disrupting recognition between the two classes. This model did not specify the selective force reinforcing interactions within nascent functional classes and did not consider inbreeding depression, the main evolutionary force maintaining and driving the evolution of SI. Self-compatible haplotypes can invade SI populations (Charlesworth and Charlesworth 1979), but the conditions under which this model can generate new functional SI haplotypes are unclear.
The only population genetics model of new SI haplotypes involves self-compatible (SC) intermediates (Uyenoyama et al. 2001). SC pollen mutants expressing a new pollen specificity can invade populations and be stably maintained at intermediate frequencies along with functional SI haplotypes when inbreeding depression is high and the proportion of self-pollen is low to intermediate (Charlesworth and Charlesworth 1979; Uyenoyama et al. 2001). Long-term maintenance of pollen mutants was proposed to allow diversification, with new SI haplotypes arising through a single compensatory mutation in the pistil gene of the haplotype carrying the pollen mutant. However, the haplotype in which the pollen mutant arose is almost always excluded at equilibrium. Thus, even though new SI haplotypes eventually appear, this almost always involved loss of their ancestral haplotype, resulting in replacement of SI haplotypes rather than diversification, suggesting that an increased number of SI haplotypes is almost impossible in a panmictic population. The authors therefore suggested that diversification might occur in subdivided populations, but did not model subdivision.
We extend the analytical model of Uyenoyama et al. (2001) by studying the conditions for the evolution of new SI haplotypes and also by simulations. We first study the conditions under which diversification at the S-locus is expected to occur in an infinite population, in terms of the proportion of self-pollen deposited and the inbreeding depression, and then investigate the effect of genetic drift using simulations including recurrent mutations for new pollen and pistil specificities.
Models
We assumed GSI, i.e., pollen specificities determined by the pollen's haploid genome, and style specificities determined codominantly by the diploid genome of the style. Pollinations are compatible if the pollen specificity of the donor differs from both specificities expressed by the style of the recipient (de Nettancourt 2001). Following Uyenoyama et al. (2001), we assumed two completely linked genes, A determining the style type and B the pollen specificity. Alleles Ai and Bj segregate at the A and B genes and express specificities i and j. Functionally self-incompatible haplotypes carry identical specificities at the two genes, e.g., AiBi, and we denote haplotypes by Si (Table 1 gives our notation). Under these assumptions, a new SI haplotype AuBu requires two mutations, one in each gene.
TABLE 1.
Genotypes and associated phenotypes produced in a population composed of n − 1 SI haplotypes Si, the ancestral haplotype Sn, and the different studied mutants (the pollen-part mutant Sb and the compensated mutant Sn+1)
| Genotype | Frequency | Rejected haplotypes | Proportion of compatible pollen | Selfing rate |
| SiSj | xij | Si, Sj | Nij | 0 |
| SbSb | xbb | Sn | Nbb | sbb |
| SbSn | xbn | Sn | Nbn | sbn |
| SbSi | xbi | Sn, Si | Nbi | sbi |
| SnSi | xni | Sn, Si | Nni | 0 |
| SbSn+1 | xbn+1 | Sb, Sn, Sn+1 | Nbn+1 | 0 |
| SnSn+1 | xnn+1 | Sb, Sn, Sn+1 | Nnn+1 | 0 |
| SiSn+1 | xin+1 | Sb, Sn+1, Si | Nin+1 | 0 |
Infinite population model
In the infinite population model, the starting population had equal frequencies of n Si haplotypes (1 ≤ i ≤ n). We modeled inbreeding depression by assuming that each plant received a proportion α of self-pollen (rate of self-pollen deposition) and that selfed offspring have a probability 1 – δ of reaching maturity, relative to outcrossed offspring. The average fitness of the population was thus
| (1) |
with xuv the frequency of genotype SuSv (see Table 1) and suv its selfing rate. We sequentially introduced different mutants and followed their fate in the population. Because Uyenoyama et al. (2001) showed that diversification can proceed only through pollen-part mutants, we give only the results when the first mutation affects the pollen type.
Pollen-part mutation (first mutation):
Using Mathematica 7.0 (Wolfram Research 2008), we first investigated the fate of a mutation in the pollen component of haplotype Sn, generating haplotype Sb = AnBn+1. Following the introduction of one such mutation, the population has n + 1 distinct haplotypes: n − 1 unaffected haplotypes Si (1 ≤ i < n), the ancestral haplotype Sn, and the pollen-part mutant Sb, with respective frequencies in the pollen pool p, pn, and pb. The population has n(n − 1)/2 + n + 1 diploid genotypes, the n(n − 1)/2 SiSj genotypes (fully SI when 1 ≤ i, j ≤ n), the n SiSb genotypes (partially self-compatible when 1 ≤ i ≤ n), and the SbSb genotype (which can self-fertilize, but rejects Sn pollen). Assuming equal pollen production among diploid genotypes and given the haplotypes rejected by each genotype (Table 1), the proportions of compatible pollen received by each genotype are
| (2) |
where the double subscripts indicate the diploid genotypes (SiSj, SbSb, SbSn, …).
Hence, the selfing rates of SC genotypes are
| (3) |
The factor of 2 in the denominators of sbn and sbi arises because only half of the pollen of SC heterozygotes is self-compatible.
Using Equations 1–3, we computed the recursion equations describing the genotypic frequency change in a generation, which allowed us to investigate the evolutionary fate of all haplotypes in the population, in particular the pollen-part mutant Sb. The resulting recursion equations are given in Appendix A1. Pollen-part mutation.
Compensatory (second) mutation:
We next introduced a mutation in the pistil gene of the pollen-part mutant Sb, generating a fully functional new SI haplotype, denoted by Sn+1 = An+1Bn+1. Let pn+1 be the frequency of Sn+1 in the pollen pool. In the population, n(n + 1)/2 + n + 1 + 1 diploid genotypes can now be formed, the n(n + 1)/2 SiSj genotypes (fully SI if 1 ≤ i, j ≤ n + 1), the n partially self-compatible SiSb genotypes (1 ≤ i ≤ n), the genotype SbSn+1 (which is fully self-incompatible and rejects the three haplotypes Sn, Sb, and Sn+1), and the SbSb genotype, which can self but rejects Sn pollen. The proportions of compatible pollen for each genotype are
| (4) |
The selfing rates sbb, sbn, and sbi defined in Equation 3 remain unchanged, and the recursion equations needed to investigate the evolutionary fate of the compensated haplotype Sn+1 and its SC ancestral Sb are given in Appendix A2. Compensatory mutation.
Evolution of the system:
For diversification to occur, four conditions are clearly necessary (Uyenoyama et al. 2001):
i. The pollen-part mutant Sb must increase in frequency when rare.
ii. Sb must not fix (i.e., a functional SI system must be retained).
iii. Sn (the ancestral allele from the pollen-part mutant arises) must not be excluded by the introduction of Sb.
iv. The mutation in the pistil gene that creates the new fully functional SI haplotype, Sn+1, must increase in frequency when both Sb and Sn are present in the population.
We determined parameter values under which these conditions were jointly satisfied. Conditions i and ii were investigated using local stability analysis. For condition i, we studied invasion of a population with n Si haplotypes by Sb, and for condition ii we studied the stability of the equilibrium with Sb fixed. We could not use this approach to investigate conditions iii and iv because we found no tractable analytical solution for the equilibrium when n − 1 Si and Sb were all present in the population. We therefore used recursion equations in Appendixes A1 and A2 to numerically approximate the parameter values satisfying these conditions. For each of 5 to 10 values of the self-pollen deposition parameter (α), we varied inbreeding depression (δ) values in steps of 10−3 and followed the population for 10,000 generations. Haplotypes with frequencies <10−5 were considered lost from the population. Specifically, for condition iii we introduced Sb at low frequency (xbi = 2 × 10−5; i.e., pb = 10−5) into an initially isoplethic population (with all functional SI haplotypes equally frequent) and determined the value of δ for which Sn was not eliminated when Sb spread. For condition iv, we started with a population at equilibrium with n Si haplotypes plus Sb, as obtained under condition iii, and introduced the Sn+1 haplotype at low frequency (xin+1 = 2 × 10−5; i.e., pn+1 = 10−5) to determine the critical δ-value for Sn + 1 to increase in frequency.
Finite populations simulation
We assumed a population of N diploid individuals in which all adults died after zygote production. The initial population contained n different SI haplotypes Si = Sii in equal frequencies. We assumed a maximum of k different specificities (1 ≤ i ≤ k). The life cycle included three steps: mutation, fertilization during which selection on S-alleles during pollination plays, and viability selection through inbreeding depression.
Mutation:
The number of mutations occurring each generation (U) was randomly drawn from a Poisson distribution with parameter 4Nμ, where μ is the mutation rate per gene per generation. The factor 4 arises because mutations arise in both the pollen and the pistil genes of the 2N chromosomes in the population. U chromosomes were randomly drawn with replacement in the population. For each chromosome, the pollen or pistil gene was mutated with equal probability, and the specificity was randomly changed to one of the remaining k − 1 specificities. Hence, unlike the infinite population model, the order of mutations is not constrained.
Pollination:
Plants were assumed to produce infinite numbers of pollen grains and ovules. Hence we assume no pollen limitation. Diploid individuals were randomly drawn with replacement as maternal plants. Let us consider a plant with genotype SijSgh that expresses specificities i and g in pistil and j and h in pollen, with {g, h, i, j} ∈ . The probability for this plant to self-fertilize was
| (5) |
where α is the proportion of self pollen received, pmn is the frequency of the Smn haplotype in the population, and γij is an indicator variable such that γij = 0 if i = j (i.e., Bj pollen is rejected by Ai pistils) and γij = 1 if i ≠ j (compatible mating).
If the plant self-fertilizes, it could be fertilized by Sij with probability fij = γijγgj/(γijγgj + γghγih) or by Sgh with probability fgh = 1 − fij. Otherwise, the probability that it was fertilized by a pollen with haplotype Suv was
| (6) |
Inbreeding depression:
Finally, to form the zygote genotype, a maternal chromosome was drawn randomly with probability 1/2. Selfed seeds survived with probability 1 − δ. N individuals were generated.
Diversification, loss, or maintenance of SI:
We first simulated an initial population of n = 5 different SI haplotypes in equal frequencies and k = 20. We simulated four population sizes (N = 50, 500, 5000, and 50,000) and five mutation rates (μ = 5 × 10−7, 5 × 10−6, 5 × 10−5, 5 × 10−4, and 5 × 10−3). We defined four possible different final states of the population:
Loss of SI: Only SC haplotypes were present in the population.
Maintenance of SI: No SC haplotypes were present and the number of SI haplotypes remained stable or decreased (n ≤ 5).
Polymorphism: Both SI and SC haplotypes were present at equilibrium.
Diversification: n > 5 SI haplotypes and no SC haplotypes were present.
One hundred runs were performed for each set of parameters. The simulations were stopped when either diversification occurred (more than five SI haplotypes and no SC haplotypes in high frequencies) or all SI haplotypes were lost. Otherwise (no new SI haplotypes and/or presence of SC haplotypes at high frequencies), the system was run for 105 generations. Since we were interested in the final state of the population, and since SC and SI haplotypes could arise by mutation at any time, we then arbitrarily characterized the state of the system on the basis of haplotypes present at frequencies >0.01 (0.05 for N = 50).
Dynamics of diversification:
In the previous section we were interested only in the final evolutionary outcome. To investigate the numbers of SI haplotypes over time, and the times between successive diversification events, we performed further simulations for 106 generations with N = 5000, μ = 5 × 10−5, δ = 0.9, and α-values of 0.2 and 0.4. Runs were started at isoplethy with three SI haplotypes, and we studied three k values, 20, 100, and 200. One hundred runs were performed for each set of parameters.
To investigate the detailed dynamics of SI haplotype gains and losses, we also tracked the genotypes in the population for 150,000 generations for 20 replicates, sampling genotypic frequencies every 1000 generations, using the following approximations. Since new SI haplotypes necessarily arise through SC intermediates, their rate of gain should be approximated by the population's total frequency of SC haplotypes. Similarly, since in our model the loss of haplotypes is caused by genetic drift of low-frequency haplotypes, and all functional SI haplotypes in GSI systems have equal expected equilibrium frequencies, the rate of loss should be approximated by the average frequency of SI haplotypes.
Results
Infinite populations
Evolutionary outcomes:
Extending the stability analysis of the Uyenoyama et al. (2001) model (Appendix A3. Stability analysis) allowed us to divide the parameter space into six regions (L, M1, M2, R, D1, and D2; see Figure 1A), instead of the four originally described (Table 2). Uyenoyama et al.’s (2001) four regions were defined only on the basis of the frequency change of the pollen-part mutant Sb when rare or frequent. Our inclusion of the fate of the ancestral SI haplotype Sn splits two of the regions where Sb increases in frequency when rare, depending on whether rare Sn (compensating) mutations increase or decrease. The six resulting regions correspond to four qualitatively different evolutionary patterns for Sb, Sn, and Sn+1 haplotypes. Three evolutionary outcomes were identified by Uyenoyama et al. (2001): in region L, the SI system was lost, whereas it was maintained unaltered in regions M1 and M2, or haplotypes were replaced (region R). Our analysis identifies additional regions D1 and D2 in which new SI haplotypes evolve. We next describe these regions in detail.
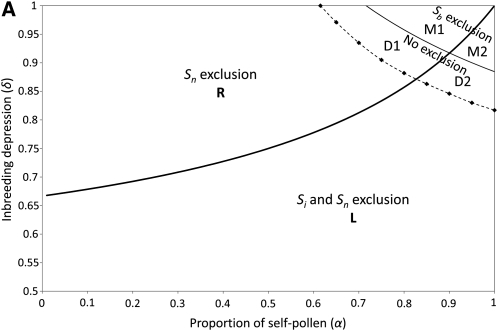
Figure 1.—
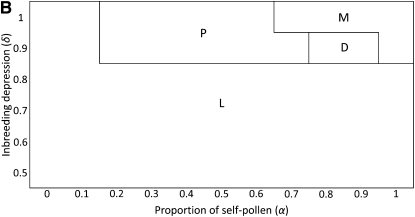
Limits of the regions where the different evolutionary outcomes are observed, for an initial number of S haplotypes n = 5. (A) In an infinite population: solid lines are the results of the stability analysis (thick line, fate of the pollen-part mutant Sb when it is frequent; thin line, evolutionary outcome when Sb is rare; see text for further details). The dashed line limit was obtained through numerical iterations and delimits the region where haplotype Sn was not excluded after the introduction of the pollen-part mutant Sb. Diamonds refer to the parameters that we investigated to approximate this limit. We define four regions according to which haplotypes are excluded when Sb is introduced at low frequency in the population. These four regions are the loss region (L), when the pollen-part mutant Sb goes to fixation; the replacement region (R), when only Sn is excluded; the maintenance regions (M1 + M2), when Sb alone is excluded; and the diversification regions (D1 + D2), when no haplotypes are excluded, and a new S haplotype can emerge. (B) In a finite population (N = 5000, μ = 5 × 10−7, k = 20): we defined four regions according to which evolutionary outcome was the most frequent in our simulations for a given set of parameters: the loss region (L, where loss of the SI system was mainly observed), the polymorphism region (P, where the populations were composed of a mix of SC and SI haplotypes), the maintenance region (M, where only SI haplotypes were observed and the number of specificities did not increase), and the diversification region (D, where no SC haplotypes were observed and the number of SI haplotypes increased).
TABLE 2.
Evolution of the frequency of Sb near fixation and of Sb, Sn and Sn+1 near exclusion, in the six described regions
| Sb frequent | Sb rare | Sn rare | Sn+1 rare | |
| L | + | + | − | − |
| R | − | + | − | + |
| M1 | − | − | + | + |
| M2 | + | − | + | + |
| D1 | − | + | + | + |
| D2 | + | + | + | + |
“+” indicates that the frequency increases. “−” indicates that the frequency decreases. Regions M1, M2, R + D1, and L + D2 correspond, respectively, to the regions E, D, P, and S described in Uyenoyama et al. (2001).
Low inbreeding depression yields outcomes in region L (unconditional increase in the frequency of Sb, to fixation excluding all functional haplotypes, with loss of SI). With high inbreeding depression and a high proportion of self-pollen deposition, regions M1 and M2 result (Sb does not increase in frequency when introduced at low initial frequency and an unaltered SI is maintained; the two regions differ in the behavior of Sb near fixation, effectively fixing in M2, but decreasing in frequency in M1). When inbreeding depression is high and self-pollen deposition low to intermediate, region R results. In this region of parameter space, Sb establishes a polymorphism along with the preexisting Si haplotypes, but this is always accompanied by the exclusion of the ancestral Sn haplotype. Subsequent introduction of Sn+1 leads to the exclusion of Sb, resulting in a fully SI population with the n − 1 initial Si haplotypes and the new haplotype Sn+1 replacing Sn, with no increase in the number of SI haplotypes.
Finally, for intermediate to high values of both δ and α, diversification is possible. Such situations yield regions D1 and D2, in which Sb also establishes a polymorphism at equilibrium (like region R), but without loss of Sn (Sb fixes in region D2, but in D1 it decreases in frequency when initially present at high frequency). The frequency of Sb has one stable equilibrium in region D1 and two equilibria, one stable and one unstable, in D2 (Table 2); the stable equilibrium in D2 has a lower Sb frequency than the unstable one. Because Sn is also maintained in regions D1 and D2, the compensatory pistil mutation Sn+1 can produce an increase the number of SI haplotypes. Indeed, the Sn+1 mutation always led to the exclusion of Sb (see condition iv in Appendix A3).
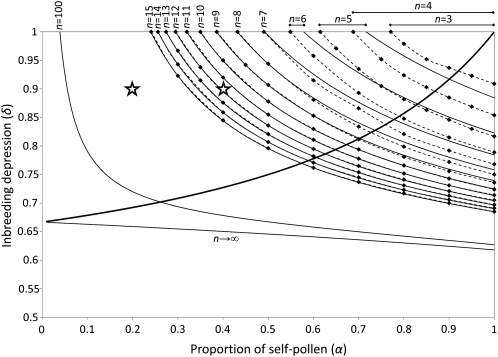
The relative sizes of the different regions depended on the initial number of SI haplotypes (n), with an increase of the M regions and a decrease of the R and D regions as n increases (Figure 2). This relates to the possibility of polymorphism for pollen-part mutation until n becomes large (Charlesworth and Charlesworth 1979).
Figure 2.—
Effect of the initial number of specificities (n) on the different evolutionary outcomes in an infinite population. Solid and dashed lines define the limits of each region (L, R, M1, M2, D1, and D2; see Figure 1 legend). The diversification region is indicated at the top for the different n’s. Note that some diversification regions are overlapping. The area of the diversification region decreased when n increased, except for n = 3 and n = 4. The diversification region was also displaced toward smaller values of δ and α when n increased. Stars indicate the values of δ and α that were chosen to investigate the diversification dynamics across 106 generations in Figure 4.
Finite population simulations
Our simulations starting with n = 5 support the analytical predictions, but genetic drift and recurrent pollen-part and compensatory pistil mutation can increase the parameter space where diversification can occur. Figure 1B shows four different evolutionary outcomes: region L, where SC haplotypes became fixed; M, where SI was maintained; a polymorphism region (P), where both SC and SI haplotypes are present; and a diversification region (D), where the final population contained more than the initial number of SI haplotypes and no SC haplotypes. Note that Figure 1B is a simplified representation of the results for N = 5000, μ = 5 × 10−7, and k = 20 (detailed results are shown in Figure 3 and supporting information, Figure S1, Figure S2, and Figure S3).
Figure 3.—
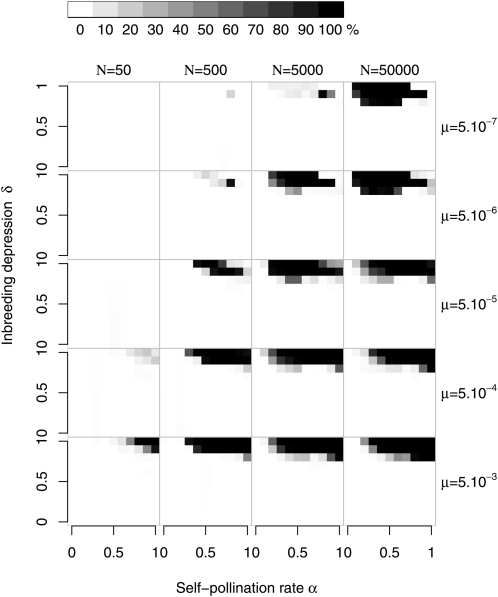
The diversification region in a finite population for different population sizes and mutation rates, with an initial number of specificities n = 5. Grid cells are figured in shades of gray that are proportional to the number of simulations where diversification was observed (n > 5 at the end of the simulation), with black squares corresponding to sets of parameters under which the evolutionary outcome was only diversification and white squares corresponding to cases under which no diversification was observed. One hundred replicate runs were performed for each combination of parameter values (population size N, mutation rate μ). k = 20 for all simulations.
Whereas the relative positions of these different zones are largely independent of the parameter values, their sizes depend strongly on the population size and mutation rate. Except when the values of μ and N were both low, region D was considerably larger than in the analytical model (Figure 3), extending over much of the R region determined for an infinite population. Indeed, for high Nμ, diversification occurred even with high inbreeding depression and self-pollen deposition (whereas, with low Nμ, the system was maintained, as in an infinite population; see Figure S1). The parameter values for which SI was lost were similar to those for an infinite population (Figure S2), except for high inbreeding depression and low self-pollen deposition where SI is lost in finite populations whereas replacement is expected in an infinite population. As expected from the fact that a compensated mutant Sn+1 always excludes its SC ancestor Sb, coexistence of SI and SC haplotypes was very rare, observed mainly with low mutation rates and intermediate N (Figure S3).
Dynamics of diversification:
With n = 3 in the initial population, there was generally loss of some SI haplotypes in the early generations, since, with such low numbers of SI haplotypes, an Sb pollen-part mutant is expected to exclude its ancestral SI haplotype Sn (see Figure 2). However, the populations rapidly returned to n > 3. For the parameter values investigated (N = 5000, μ = 5 × 10−5, δ = 0.9, and α = 0.2 and 0.4), the numbers of SI haplotypes in the populations increased from 3 to between 7 and 18 within 106 generations (Figure 4). For α = 0.2, the population had apparently reached a stationary number of haplotypes. Diversification always proceeded through pollen-part mutants followed by compensatory mutations in the pistil gene, confirming for finite populations this prediction from the analytical model (Uyenoyama et al. 2001).
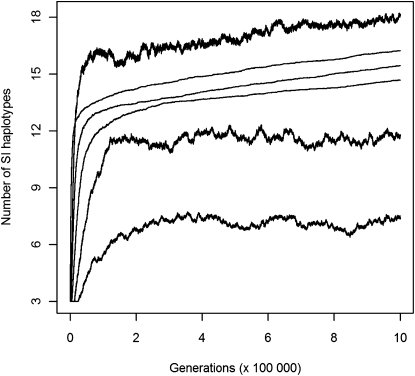
Figure 4.—
Diversification dynamics in finite populations across 106 generations. The number of SI haplotypes in the population was averaged over 100 replicates. Simulations were performed with N = 5000, μ = 5 × 10−5, δ = 0.9, the initial number of SI haplotypes n = 3, and for three different values of the maximum number of specificities (k) and two values of the proportion of self-pollen (α). Thick line, α = 0.2; thin line, α = 0.4. For a given α, curves correspond to k = 20, 100, and 200 from top to bottom.
The mean waiting time between successive diversification events (see Models for the approximations used to study this) increased with n, with two distinct phases. Haplotype numbers first increased rapidly and then suddenly either stabilized or increased much more slowly. In the initial fast diversification, the overall frequency of SC haplotypes was very high (Figure 5A), implying a high birth rate of SI haplotypes, but the overall frequency of SC haplotypes then decreased, thus slowing down diversification as n increased (Figure 5A). Interestingly, during the same time, the rate of loss of functional SI also decreased (as estimated from the increased mean frequency of SI haplotypes, see Models) (Figure 5B). This suggests that the decrease of the birth rate more than outweighs the decreased death rate and therefore that the slowdown of diversification is due to the decreased birth rate. This is consistent with the fact that the ability of pollen-part mutations to invade decreases when n is high.
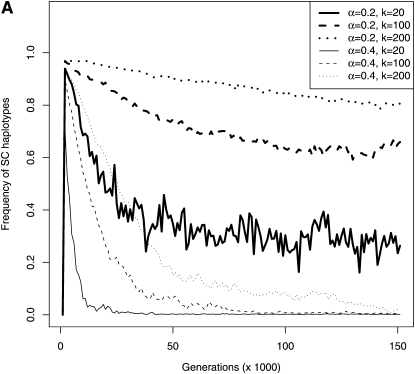
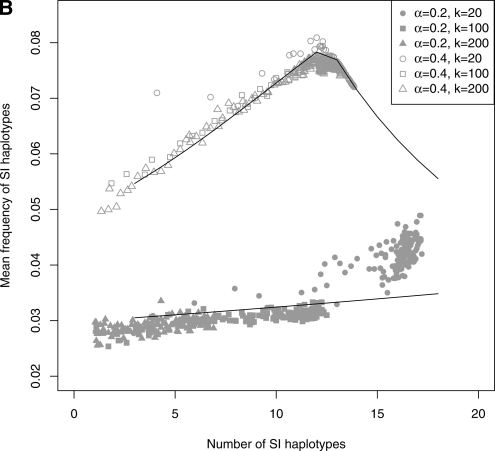
Figure 5.—
Genotypic composition of the population in the first 150,000 generations to investigate the dynamics of birth and death of SI haplotypes (genotypic frequencies sampled every 1000 generations in 20 replicates for N = 5000, μ = 5 × 10−5, and δ = 0.9). (A) Mean cumulated frequency of all SC haplotypes as a function of time. After an initial rapid increase in the total frequency of SC haplotypes, their frequency started to decrease as the number of SI haplotypes increased (see Figure 4). (B) Mean frequency of SI haplotypes as a function of their total number. Symbols are mean frequencies in simulations and lines are the expected frequencies at deterministic equilibrium in an infinite population (computed using equations in Appendix A1. Pollen-part mutation with a frequency of Sn, pn = 0 and setting n = x + 1 when x SI haplotypes were segregating within the population). For both values of α, the mean frequency of SI haplotypes increased with their total number. This is due to the fact that when the number of SI haplotypes increases, the total frequency of SC haplotypes decreases, thus leaving a larger range of frequency for SI haplotypes. When α = 0.4, the mean frequency of SI haplotypes decreased when their number was >12 because the frequency of SC haplotypes approached zero. There was no similar threshold value of the number of SI haplotypes in the case α = 0.2 because the frequency of SC haplotypes remained high.
SI haplotype diversification rate increased with decreasing k, especially during the first phase (Figure 4), which explains the lower number of SI haplotypes at a given time. We interpret this in terms of the increased probability 1/(k − 1) that compensatory mutations can occur as k decreases. However, the value of k should not affect diversification probability for given values of α and δ. In other words, we do not expect that the parameter space for diversification (Figure 3) would shrink as k increases. The detailed dynamics depend on the level of self-pollen deposition. With high self-pollen deposition (α = 0.4), the overall frequency of SC haplotypes was near zero (Figure 5A), implying a very low rate of gain of new SI haplotypes, and the frequency of SI haplotypes was high (Figure 5B), suggesting a low probability of haplotype loss by genetic drift. Diversification therefore occurred with slow haplotype turnover, with both birth and death rates that were low. For lower proportions of self-pollen (α = 0.2), however, SC haplotypes were at higher frequency (Figure 5A) but SI haplotypes were less frequent (Figure 5B); i.e., diversification occurred with fast haplotype turnover and high birth and death rates. This is consistent with the observation of more stochastic variation in the number of haplotypes in the population for the lower α-value (Figure 4) and with additional simulations with larger population size, up to N = 10,000 (data not shown), which did not detectably affect the final number of haplotypes when α = 0.4 (i.e., there was no effect of genetic drift on the dynamics of diversification) but did increase the final number of haplotypes for α = 0.2.
Discussion
Importance of genetic drift and recurrent mutations
Allelic diversification is clearly possible in an infinite, undivided population, and the region of parameter space where it can occur can be large, especially when the number of SI haplotypes in the population is fewer than six. Our simulations show that, in finite populations, diversification is generally possible under a larger set of parameter values than in an infinite population (with the exception of very small populations and/or very low mutation rates). The difference was largely dependent on the proportion of self-pollen deposition, low values of which can greatly extend the lower limit of the diversification region, so that it could cover most of the region where replacement of SI haplotypes, without diversification, is expected in an infinite population. In contrast, the critical inbreeding depression allowing diversification in finite populations never extended beyond that for an infinite population, for the parameter space we explored (even for the largest population sizes and mutation rates). Hence, while stochastic processes are generally associated with reduced genetic diversity within small, finite populations, our results show that diversification of SI halotypes behaves in the opposite way and that the forces promoting diversification can outweigh stochastic loss of SI haplotypes, which becomes strongly limited due to frequency-dependent selection under conditions when SI is favored.
The role of inbreeding depression
Our results support previous conclusions that diversification requires high inbreeding depression (Charlesworth and Charlesworth 1979; Uyenoyama 1988a,b). However, these conclusions were based on a fixed inbreeding depression parameter, ignoring its evolution as the outcrossing rate changes. Porcher and Lande (2005) showed that, for high inbreeding depression due to deleterious mutations, and high selfing rates, mutations may be efficiently purged, eventually leading to the loss of SI. Our study again treated inbreeding depression as a fixed parameter, even though clearly invasion of a population by an Sb allele may allow purging of deleterious mutations, making the conditions for diversification increasingly stringent. However, Porcher and Lande (2005, Figure 2) also showed, in single-locus models for S-alleles, that SC haplotypes can be maintained in coexistence with SI haplotypes under a wide range of parameter values, especially when the selfing rate is low or when there is a sheltered load. Moreover, our results showed that inbreeding depression must be high for diversification to occur, while the self-pollination rate may be moderate to high. This should now be added to further studies of two-gene models for S-allele evolution.
Dynamics of diversification
The observed increase in the time between successive diversification events may be due to the fact that, for a given combination of self-pollen deposition and inbreeding depression, the expected frequency of pollen-part intermediates decreases as n increases, which decreases the probability of a compensatory (pistil) mutation occurring in the intermediate haplotype before it is lost by drift. The prediction that diversification is not constant through time may explain some otherwise puzzling features of SI systems, including cases of rapid S-allele diversification, and the unexpectedly long terminal branches typically observed at SI genes.
S-allele numbers and the shape of phylogenies
The observation that all SI haplotypes in Physalis and Witheringia belong to only three ancestral lineages (Paape et al. 2008; see also Miller et al. 2008 for the genus Lycium) was interpreted by these authors in terms of rapid S-allele diversification after demographic bottlenecks (when the number of SI haplotypes will tend to be small). Further, Castric and Vekemans (2007) estimated much stronger positive selection among Brassica SI haplotypes than among the more anciently diverged SI haplotypes of Arabidopsis species. This might suggest rapid and recent diversification of Brassica SI haplotypes from just two ancestral lineages.
The parameter space where diversification is expected to occur in an infinite population shrinks rapidly with increased n, and the D region is also displaced toward lower self-pollen deposition rates and inbreeding depression. Initially rapid diversification becomes slow as SI haplotype number increases, eventually stopping almost completely, yielding a threshold value of n that allows diversification. For α = 0.2, haplotype turnover was rapid during diversification, and the overall frequency of SC haplotypes remained high when diversification ceased.
Our simulations yield many fewer SI haplotypes than are typically observed in natural populations, where the number of SI haplotypes commonly reaches 50–100 in species-wide range samples (Lawrence 2000). This is probably not due to the low effective population size assumed in our simulations, because, at least for high proportions of self-pollen (α = 0.4), the SI haplotype number maintained was apparently independent of population size (results not shown). Moreover, under the classical single-gene mutation–drift–selection theoretical framework where a single mutation gives rise to new functional SI haplotypes, only a very low mutation rate is needed to yield high equilibrium numbers of SI haplotypes in a population of 5000 plants. In such models, to yield as few as 17 SI haplotypes at equilibrium (the maximum number of SI haplotypes we observed in our simulations), an unrealistically low mutation rate of 10−20 must be assumed (Vekemans and Slatkin 1994). Thus, although our model can generate diversification, it does not fully explain S-allele polymorphism, suggesting that other factors, such as population structure, are probably needed to explain the diversity observed, as proposed by Uyenoyama et al. (2001).
Allelic genealogies of genes under balancing selection extend deep in the evolutionary past and thus can reveal ancient evolutionary processes such as the number of breeding individuals in the species as a whole across its evolutionary history or the strength of balancing selection, but these evolutionary inferences assume a constant diversification rate (e.g., Takahata 1990; Vekemans and Slatkin 1994). Our results suggest that this assumption may be violated, since the diversification rate decreases with the number of functional SI haplotypes in the population. The diversification rate will not be constant if the equilibrium number of SI haplotypes has only recently been reached and the turnover rate is slow. In contrast, if turnover is fast enough and/or haplotype number stopped increasing a sufficiently long time ago, the rate can be considered constant.
Our results are consistent with the finding that terminal branches in the phylogenies of SI haplotypes are longer than expected by models that assume that balancing selection simply extends the lengths of branches of the coalescent tree, relative to a neutral model (Takahata 1990; Uyenoyama 1997). Two main hypotheses have been proposed to explain this. First, Uyenoyama (1997) proposed the accumulation of deleterious mutations linked to individual SI haplotypes (called sheltered load); a new SI haplotype would then share linked mutations with its ancestral copy and would thus be excluded from the population. There is some evidence for existence of sheltered load (Stone 2004; Llaurens et al. 2009). Second, Schierup et al. (2001) proposed that recombination could explain long terminal branches, by creating haplotypes whose sequences are composed of fragments with different evolutionary histories, thus homogenizing sequence divergence among them. Recombination may occur among SI haplotypes in Arabidopsis (Castric et al. 2010), Petunia inflata (Wang et al. 2001), Prunus dulcis (Ortega et al. 2006), and other Solanaceae and Rosaceae (Vieira et al. 2003). Our results here suggest that long terminal branches could simply be an intrinsic property of the diversification process, through the slowing down of the S-allele diversification process as SI haplotype numbers increase. Furthermore, it is striking that such a slowing down of diversification is apparently encountered in very different mechanisms such as the appearance of mating types in fungi (May et al. 1999) and speciation (e.g., Morlon et al. 2010; Quental and Marshall 2010). We might thus speculate the intriguing possibility that such a phenomenon is a general pattern.
The mutation process
Our hypothesis for the molecular mechanism responsible for recognition between pollen and pistil is oversimplified, particularly the limited number of possible allelic states (k-allele model) and the strict recognition reactions between pollen and pistils with cognate specificities. The pollen and pistil genes differ in size and function (Takayama and Isogai 2005) and in the numbers of amino acid sites evolving under positive selection. Estimates of this number are between 13 and 31 in the pistil gene S-RNase of Solanaceae (Takebayashi et al. 2003; Savage and Miller 2006; Igic et al. 2007), vs. 23 in the pollen gene SFB in Prunus (Nunes et al. 2006). In Brassica, estimates are between 26 and 44 in the pistil gene SRK and ∼47 in the pollen-S gene SCR (Takebayashi et al. 2003; Sainudiin et al. 2005; Castric and Vekemans 2007). Mutagenesis experiments suggest that several mutations are generally required to shift specificities [at least four amino acids in S. chacoense S-RNases (Matton et al. 1999) and four amino acids in B. oleracea SCR (Chookajorn et al. 2004)]. Compensatory mutations may thus require several mutational steps, possibly making coevolution and diversification more difficult than in our model.
Now that Arabidopsis thaliana has been established as a model plant for mechanistic studies of SI (Nasrallah et al. 2002; Tsuchimatsu et al. 2010), it will be feasible to study in detail the steps required for allele diversification, at least in this plant.
Acknowledgments
We thank Xavier Vekemans, Denis Roze, Emmanuelle Porcher, and Chi Viet Tran for discussions, Camille Roux for technical help on graphics and Marcy Uyenoyama, Deborah Charlesworth, and an anonymous reviewer for their helpful comments on the manuscript.
APPENDIX
Here we detail the recursive equations for each genotype present in a population composed of n SI haplotypes and the different studied mutants. Definition of the different genotypes and their associated variables are given in Table 1.
A1. Pollen-part mutation
Genotypic frequencies in the next generation in a population composed of n − 1 SI haplotypes Si, the ancestral SI haplotype Sn, and the pollen-part mutant haplotype Sb are
where , Ng, and sg are respectively defined in Equations 1, 2, and 3 and where p, pn, and pb, the frequencies of Si, Sn, and Sb, are
A2. Compensatory mutation
In the case of a population composed of n − 1 SI haplotypes Si, the ancestral SI haplotype Sn, the pollen-part mutant haplotype Sb, and the new SI haplotype Sn+1, the genotypic frequencies in the next generation are
where , Ng, and sg are respectively defined in Equations 1, 4, and 3 and where p, pn, pb, and pn+1, the frequencies of Si, Sn, Sb, and Sn+1, are
A3. Stability analysis
Condition i: fate of haplotype Sb when rare
Local stability analysis of the equilibrium when Sb was absent (x0 = (n − 2)/n; x4 = 2/n; x1 = x2 = x3 = 0) provides the parameter space where Sb can increase in frequency when rare. The parameter space we found where Sb increases when rare was identical to that described by Uyenoyama et al. (2001, Equation A1) and is delimited by the thin solid line in Figure 1A for n = 5 and in Figure 2 for different values of n. Above this line, Sb did not increase in frequency when rare and was excluded from the population, i.e., when the self-pollen rate α and the inbreeding depression δ were high. Below this line, Sb increased in frequency and may either invade the population (resulting in the loss of SI) or be maintained at intermediate frequencies along with functional haplotypes (see condition ii below). Note that when the population contained only three SI haplotypes (n = 3), the parameter space was empty, i.e., Sb was always expected to increase in frequency, and that for n = 4, the parameter space was limited to the points α = 1 and δ = 1. Figure 2 shows that the parameter space of condition i became larger as the number of SI haplotypes segregating in the population (n) increased.
Condition ii: fate of the haplotype Sb when frequent
Local stability analysis of the equilibrium where Sb was fixed (x1 = 1; x0 = x2 = x3 = x4 = 0) gives the parameter space where Sb increased in frequency when already frequent, i.e., where it reaches fixation. We found that, for any n, Sb increased when already frequent if
This threshold corresponds to the critical inbreeding depression above which an SI haplotype can enter a SC population from a low initial frequency (Charlesworth and Charlesworth 1979, Equation 2). This threshold is shown in Figures 1a and 2 as the thick solid line and is independent from the number of SI haplotypes in the population.
Condition iii: fate of haplotype Sn when Sb was present
The parameter space where Sn was excluded when Sb was present was determined numerically and is delimited by the dashed lines in Figure 1A for n = 5 and in Figure 2 for different values of n. The parameter space for which Sb and Sn can both be maintained in the population at the same time is located between the thin solid line, condition i, and the dashed line, condition iii. Figure 1A shows that Sn was not excluded by Sb for high values of α and δ. Figure 2 shows that the parameter space for which Sn is not excluded became smaller and was displaced toward smaller values of α and δ when n increased. Practically, we were not able to distinguish between conditions i and iii when n > 10. Note that for n = 3 and n = 4, the parameter space for condition iii was large, whereas it either did not exist or was reduced to a single point for condition i.
Condition iv: fate of haplotype Sn+1 when both Sn and Sb were present
Our results showed that the introduction of the new SI haplotype Sn+1 in a population composed of Sb and Si haplotypes (including or not the ancestor Sn) always led to the exclusion of the pollen-part mutant Sb, resulting in a fully self-incompatible population including a new SI haplotype. In cases where Sn was maintained in polymorphism with the pollen-part mutant Sb, this process can thus potentially lead to an increase in the number of SI haplotypes in the population.
Literature Cited
- Billiard S., Lopez-Villavicencio M., Devier B., Hood M., Fairhead C., Giraud T., 2011. Having sex, yes, but with whom? Inferences from fungi on the evolution of anisogamy and mating types. Biol. Rev. 86: 421–442 [DOI] [PubMed] [Google Scholar]
- Castric V., Vekemans X., 2007. Evolution under strong balancing selection: how many codons determine specificity at the female self-incompatibility gene SRK in Brassicaceae. BMC Evol. Biol. 7: 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castric V., Bechsgaard J. S., Grenier S., Noureddine R., Schierup M. H., et al. , 2010. Molecular evolution within and between self-incompatibility specificities. Mol. Biol. Evol. 27: 11–20 [DOI] [PubMed] [Google Scholar]
- Charlesworth D., 2000. How can two-gene models of self-incompatibility generate new specificities? Plant Cell 12: 309–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D., Charlesworth B., 1979. Evolution and breakdown of S-allele systems. Heredity 43: 41–55 [Google Scholar]
- Chookajorn T., Kachroo A., Ripoll D., Clark A., Nasrallah J., 2004. Specificity determinants and diversification of the Brassica self-incompatibility pollen ligand. Proc. Natl. Acad. Sci. USA 101: 911–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nettancourt D., 2001. Incompatibility and Incongruity in Wild and Cultivated Plants. Springer-Verlag, Berlin [Google Scholar]
- Fisher R. A., 1961. A model for the generation of self-sterility alleles. J. Theor. Biol. 1: 411–414 [PubMed] [Google Scholar]
- Igic B., Smith W. A., Robertson K. A., Schaal B. A., Kohn J. R., 2007. Studies of self-incompatibility in wild tomatoes: I. S-allele diversity in Solanum chilense (Dun.) Reiche (Solanaceae). Heredity 99: 562. [DOI] [PubMed] [Google Scholar]
- Igic B., Lande R., Kohn J. R., 2008. Loss of self-incompatibility and its evolutionary consequences. Int. J. Plant Sci. 169: 93–104 [Google Scholar]
- Lawrence M., 2000. Population genetics of the homomorphic self-incompatibility polymorphisms in flowering plants. Ann. Bot. 85: 221–226 [Google Scholar]
- Lewis D., 1949. Structure of the incompatibility gene. Heredity 3: 339–355 [DOI] [PubMed] [Google Scholar]
- Llaurens V., Gonthier L., Billiard S., 2009. The sheltered genetic load linked to the S locus in plants: new insights from theoretical and empirical approaches in sporophytic self-incompatibility. Genetics 183: 1105–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matton D., Luu D., Xike Q., Laublin G., O’Brien M., et al. , 1999. Production of an S RNase with dual specificity suggests a novel hypothesis for the generation of new S alleles. Plant Cell 11: 2087–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May G., Shaw F., Badrane H., Vekemans X., 1999. The signature of balancing selection: fungal mating compatibility gene evolution. Proc. Natl. Acad. Sci. USA 96: 9172–9177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. S., Levin R. A., Feliciano N. M., 2008. A tale of two continents: Baker’s rule and the maintenance of self-incompatibility in Lycium (Solanaceae). Evolution 62: 1052–1065 [DOI] [PubMed] [Google Scholar]
- Morlon H., Potts M. D., Plotkin J. B., 2010. Inferring the dynamics of diversification: a coalescent approach. PLoS Biol. 8: e1000493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrallah M., Liu P., Nasrallah J., 2002. Generation of self-incompatible Arabidopsis thaliana by transfer of two S locus genes from A. lyrata. Science 297: 247–249 [DOI] [PubMed] [Google Scholar]
- Nunes M. D. S., Santos R. A. M., Ferreira S. M., Vieira J., Vieira C. P., 2006. Variability patterns and positively selected sites at the gametophytic self-incompatibility pollen SFB gene in a wild self-incompatible Prunus spinosa (Rosaceae) population. New Phytol. 172: 577–587 [DOI] [PubMed] [Google Scholar]
- Ortega E., Boskovic R. I., Sargent D. J., Tobutt K. R., 2006. Analysis of S-RNase alleles of almond (Prunus dulcis): characterization of new sequences, resolution of synonyms and evidence of intragenic recombination. Mol. Genet. Genomics 276: 577. [DOI] [PubMed] [Google Scholar]
- Paape T., Igic B., Smith S. D., Olmstead R., Bohs L., et al. , 2008. A 15-Myr-old genetic bottleneck. Mol. Biol. Evol. 25: 655–663 [DOI] [PubMed] [Google Scholar]
- Phadke S. S., Zufall R. A., 2009. Rapid diversification of mating systems in ciliates. Biol. J. Linn. Soc. 98: 187–197 [Google Scholar]
- Porcher E., Lande R., 2005. Loss of gametophytic self-incompatibility with evolution of inbreeding depression. Evolution 59: 46–60 [PubMed] [Google Scholar]
- Quental T. B., Marshall C. R., 2010. Diversity dynamics: molecular phylogenies need the fossil record. Trends Ecol. Evol. 25: 434–441 [DOI] [PubMed] [Google Scholar]
- Sainudiin R., Wong W., Yogeeswaran K., Nasrallah J., Yang Z., et al. , 2005. Detecting site-specific physicochemical selective pressures: applications to the class-I HLA of the human major histocompatibility complex and the SRK of the plant sporophytic self-incompatibility system. J. Mol. Evol. 60: 315–326 [DOI] [PubMed] [Google Scholar]
- Savage A., Miller J., 2006. Gametophytic self-incompatibility in Lycium parishii (Solanaceae): allelic diversity, genealogical structure, and patterns of molecular evolution at the S-RNase locus. Heredity 96: 434–444 [DOI] [PubMed] [Google Scholar]
- Schierup M., Mikkelsen A., Hein J., 2001. Recombination, balancing selection and phylogenies in MHC and self-incompatibility genes. Genetics 159: 1833–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solberg O. D., Mack S. J., Lancastera A. K., Single R. M., Tsai Y., et al. , 2008. Balancing selection and heterogeneity across the classical human leukocyte antigen loci: a meta-analytic review of 497 population studies. Hum. Immunol. 69: 443–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone J., 2004. Sheltered load associated with S-alleles in Solanum carolinense. Heredity 92: 335–342 [DOI] [PubMed] [Google Scholar]
- Takahata N., 1990. A simple genealogical structure of strongly balanced allelic lines and transspecies evolution of polymorphism. Proc. Natl. Acad. Sci. USA 87: 2419–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S., Isogai A., 2005. Self-incompatibility in plants. Annu. Rev. Plant Biol. 56: 467–489 [DOI] [PubMed] [Google Scholar]
- Takebayashi N., Brewer P., Newbigin E., Uyenoyama M., 2003. Patterns of variation within self-incompatibility loci. Mol. Biol. Evol. 20: 1778–1794 [DOI] [PubMed] [Google Scholar]
- Tsuchimatsu T., Suwabe K., Shimizu-Inatsugi R., Isokawa S., Pavlidis P., et al. , 2010. Evolution of self-compatibility by a mutation in the male specificity gene. Nature 464: 1342–1346 [DOI] [PubMed] [Google Scholar]
- Uyenoyama M., 1988a. On the evolution of genetic incompatibility systems 2. Initial increase of strong gametophytic self-incompatibility under partial selfing and half-sib mating. Am. Nat. 131: 700–722 [Google Scholar]
- Uyenoyama M., 1988b. On the evolution of genetic incompatibility systems 3. Introduction of weak gametophytic self-incompatibility under partial inbreeding. Theor. Popul. Biol. 34: 47–91 [DOI] [PubMed] [Google Scholar]
- Uyenoyama M., 1997. Genealogical structure among alleles regulating self-incompatibility in natural populations of flowering plants. Genetics 147: 1389–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyenoyama M., Newbigin E., 2000. Evolutionary dynamics of dual-specificity self-incompatibility alleles. Plant Cell 12: 310–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyenoyama M., Zhang Y., Newbigin E., 2001. On the origin of self-incompatibility haplotypes: transition through self-compatible intermediates. Genetics 157: 1805–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vekemans X., Slatkin M., 1994. Gene and allelic genealogies at a gametophytic self-incompatibility locus. Genetics 137: 1157–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira C., Charlesworth D., Vieira J., 2003. Evidence for rare recombination at the gametophytic self-incompatibility locus. Heredity 91: 262–267 [DOI] [PubMed] [Google Scholar]
- Wang X., Hughes A., Tsukamoto T., Ando T., Kao T., 2001. Evidence that intragenic recombination contributes to allelic diversity of the S-RNase gene at the self-incompatibility (S) locus in Petunia inflata. Plant Physiol. 125: 1012–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfram Research, 2008. Mathematica Edition: Version 7.0. Wolfram Research. http://www.wolfram.com/ [Google Scholar]
- Wright S., 1939. The distribution of self-sterility alleles in populations. Genetics 24: 538–552 [DOI] [PMC free article] [PubMed] [Google Scholar]