Abstract
Morphogenesis is an important component of animal development. Genetic redundancy has been proposed to be common among morphogenesis genes, posing a challenge to the genetic dissection of morphogenesis mechanisms. Genetic redundancy is more generally a challenge in biology, as large proportions of the genes in diverse organisms have no apparent loss of function phenotypes. Here, we present a screen designed to uncover redundant and partially redundant genes that function in an example of morphogenesis, gastrulation in Caenorhabditis elegans. We performed an RNA interference (RNAi) enhancer screen in a gastrulation-sensitized double-mutant background, targeting genes likely to be expressed in gastrulating cells or their neighbors. Secondary screening identified 16 new genes whose functions contribute to normal gastrulation in a nonsensitized background. We observed that for most new genes found, the closest known homologs were multiple other C. elegans genes, suggesting that some may have derived from rounds of recent gene duplication events. We predict that such genes are more likely than single copy genes to comprise redundant or partially redundant gene families. We explored this prediction for one gene that we identified and confirmed that this gene and five close relatives, which encode predicted substrate recognition subunits (SRSs) for a CUL-2 ubiquitin ligase, do indeed function partially redundantly with each other in gastrulation. Our results implicate new genes in C. elegans gastrulation, and they show that an RNAi-based enhancer screen in C. elegans can be used as an efficient means to identify important but redundant or partially redundant developmental genes.
MORPHOGENESIS involves cell and tissue movements, including the movements of gastrulation and neurulation in animal embryos. Identifying the genes that control morphogenesis in animal systems has been a long-standing challenge (Wieschaus 1997). Genes involved in morphogenesis may evade genetic screens for at least two reasons. First, some genes controlling morphogenesis encode widely pleiotropic proteins such as actin and myosin (Kiehart et al. 1990). These genes may be missed in screens for morphogenesis genes because loss of function can result in arrested development before morphogenesis begins. Second, other genes may have functions that are too subtle to be identified in forward screens, for example, genes that function redundantly or partially redundantly.
Redundancy among mechanisms that underlie morphogenesis has been called a “well-recognized aspect of development” (Newman and Comper 1990). In his Nobel Lecture, Eric Wieschaus concluded that classic Drosophila screens failed to identify many morphogenesis genes and proposed as a result that the control of cell form that underlies morphogenesis may be unusually susceptible to genetic redundancy (Wieschaus 1997). Redundancy is a challenge that biologists face increasingly, as large proportions of genes in diverse systems have been found to perform important functions as members of redundant gene groups and, as a result, are often missed in genetic screens (Johnsen and Baillie 1997; Rutherford 2000; Gu et al. 2003; Felix and Wagner 2008). We recognize that two distinct forms of genetic redundancy exist: homologous redundancy, in which homologous proteins can substitute for each other, and nonhomologous redundancy, in which proteins that do not resemble each other can substitute for each other, for example, by affecting distinct, contributing cellular mechanisms (Jorgensen and Mango 2002; Gu 2003).
Despite this challenge, some key genes that function in morphogenesis have been identified by standard forward screens and by a variety of elegant modifications of such screens (Metzger and Krasnow 1999; Beitel and Krasnow 2000; Starz-Gaiano and Montell 2004; Zohn et al. 2005; Maybeck and Roper 2009; Ellertsdóttir et al. 2010; Rochlin et al. 2010; Szabo-Rogers et al. 2010). C. elegans is a valuable model system for contributing to this effort, because genetics and RNA interference (RNAi) allow one to simultaneously disrupt the functions of multiple genes in modifier screens (Labbé et al. 2006; O’Rourke et al. 2007; Dorfman et al. 2009). Genetic modifier screens have identified genes with redundant roles in C. elegans vulval and pharyngeal morphogenesis (Fay and Yochem 2007). To our knowledge, RNAi modifier screens have not yet been used to find genes controlling morphogenesis or to specifically seek redundant and partially redundant groups of genes. The ability to observe directly the individual cells participating in morphogenesis in transparent C. elegans embryos in vivo (Chisholm and Hardin 2005; Nance et al. 2005) makes it possible to detect even subtle defects. Detecting subtle defects may be important for identifying partially redundant genes.
Gastrulation is a key morphogenetic event, a cellular reorganization that occurs in diverse metazoans. Gastrulation involves the internalization of cells that give rise to mesoderm, endoderm, and germline, leaving these cells enclosed by ectoderm. In C. elegans, gastrulation begins with the internalization of two endodermal precursor cells, Ea and Ep, from the ventral face of the embryo. These two cells are the first cells of the embryo to introduce in their cell cycles a gap phase, during which they internalize (Edgar and McGhee 1988). Six neighboring cells, including the germline precursor (P4), three of the four granddaughters of the MS founder cell, and two great great granddaughters of the AB founder cell move into the space that the internalizing E cells leave behind, completing envelopment of the Ea and Ep cells (Lee and Goldstein 2003). Sixty-four other cells internalize after the endoderm precursors, leading to roughly half of the embryonic cells ending up in the interior of the embryo (Sulston et al. 1983; Nance and Priess 2002; Harrell and Goldstein 2011).
C. elegans gastrulation requires properly specified cell fates and involves cell polarization, control of motor activity, regulation of adhesion, and mechanistic links from cell fate specification to cell movements. One genetic requirement for C. elegans gastrulation is a class of genes controlling cell fate specification. The endodermal GATA factor END-3 and genes regulating its expression in the endodermal lineage are required for timely gastrulation (Bowerman et al. 1992; Thorpe et al. 1997; Maduro et al. 2005; Lee et al. 2006). Gastrulation in C. elegans also depends on genes encoding PAR polarity proteins: loss of PAR-3 or PAR-6 in somatic cells results in Ea and Ep failing to internalize on schedule (Nance and Priess 2002). These cells normally accumulate a nonmuscle myosin heavy chain protein in their apical cortex, and this accumulation requires apical PAR proteins, which localize to contact-free surfaces via a RhoGAP-mediated exclusion of PAR-6 from other surfaces (Nance and Priess 2002; Nance et al. 2003; Anderson et al. 2008). Basolaterally localized adhesion proteins also function in apical myosin localization (Grana et al. 2010). A WD repeat protein, GAD-1, (gastrulation defective), is required to delay entry into mitosis during a period of apical myosin accumulation and is required for cell internalization (Knight and Wood 1998; Nance and Priess 2002; Lee et al. 2006). Gastrulation additionally depends on a Wnt-Frizzled signaling pathway that activates the apical myosin in Ea and Ep (Lee et al. 2006). These results have led to a model in which myosin enriches at the apical, contact-free cell cortex of endodermal precursors, and activation of myosin results in an actomyosin-dependent constriction of the apical surface of these cells, driving movement of the cells to the embryo interior (see Rohrschneider and Nance 2009; Sawyer et al. 2010 for review). Consistent with this model, F-actin and actin regulators also function in gastrulation (Severson et al. 2002; Karabinos et al. 2003; Lee and Goldstein 2003; Roh-Johnson and Goldstein 2009). Several of the genes identified to date are thought to contribute partially redundantly, as strong loss of function of genes including end-3, par-3, par-6, and genes of the Wnt pathway results in only a delay of E cell internalization (Nance et al. 2003; Lee et al. 2006).
We hypothesized that many of the genes that play direct or indirect roles in normal gastrulation remain to be identified. A screen aimed specifically at identifying C. elegans gastrulation genes has not been reported previously. Here, we report a novel screening strategy for identifying genes with roles in C. elegans gastrulation. We have constructed a double mutant worm strain to serve as a sensitized background for an enhancer screen. We found that feeding these worms bacterially produced double-stranded RNAs (dsRNAs) targeting genes involved in gastrulation succeeded in producing synthetic lethality. We exploited this sensitized background together with two published microarray analyses (Robertson et al. 2004; Baugh et al. 2005) to screen for enhancers of the sensitized background among genes likely to be expressed in gastrulating cells and/or their neighbors before or near the time that gastrulation occurs. In secondary screens, we determined which of the genes we identified as enhancers were required for normal gastrulation in a nonsensitized background. Our screens identified 16 new genes with nonredundant or partially redundant functions in C. elegans gastrulation, as well as some new genes for which we only found redundant roles in gastrulation. We validated our screening method, showing that most of the genes we identified would not have been found as efficiently by a traditional RNAi feeding screen. Our screens identified several genes whose closest relatives by sequence similarity were multiple other C. elegans genes. We predict that these genes are more likely to function redundantly or partially redundantly than single copy genes. We tested this hypothesis for one gene we identified, which encodes a predicted substrate recognition subunit (SRS) for an E3 ubiquitin ligase. We showed that this gene and several similar C. elegans genes do indeed comprise a redundant gene set required for normal gastrulation, and at least some of their protein products can bind the E3 ubiquitin complex subunits CUL-2 and elongin C. Our results identify a set of genes that will be valuable for further study of morphogenesis mechanisms in C. elegans gastrulation. Moreover, they suggest that a C. elegans modifier screen using RNAi in a sensitized background can effectively assign functions to redundant gene families that are traditionally difficult to identify genetically.
Materials and Methods
Strains and worm maintenance
Nematodes were cultured and handled as described (Brenner 1974). Experiments were performed using the following strains: wild-type N2 (Bristol), JJ1317 zuIs3 [end-1::GFP], EU452 mom-5(zu193) unc-13(e1091)/hT2I; +/hT2[bli-4(e937)let-?(h661)]III, MT4434 ced-5(n1812), MT4417 ced-5(n1812);dpy-20(e1282), RB1331 end-3(ok1448), GR1373 eri-1(mg366), VC271 end-1(ok558) (backcrossed five times), RB2454 apy-1(ok3393), RB2550 ugt-23(ok3541), GH403 glo-3(kx94), GH383 glo-3(zu446), FX03627 gad-3/b0222.9(tm3627) (backcrossed five times), FX00278 tbx-11(tm278), FX02295 sdz-19(tm2295), FX01239 sdz-31(tm1239), FX01226 vet-6(tm1226), FX01378 sdz-22(tm1378), FX01169 sdz-28(tm1169), FX04187 c10a4.5(tm4187), ET099 ekEx19 [Pcul-2::CUL-2::FLAG::cul-2 3′UTR; pRF4], and LP77 end-3(ok1448); ced-5(n1812). LP77 was constructed by crossing end-3(ok1448) males with ced-5(n1812) hermaphrodites. end-3(ok1448) is a deletion of ∼700 bp (WormBase Release WS215 at www.wormbase.org). All strains were maintained at 20°.
RNAi screening and quantification of embryonic lethality
RNAi by feeding was performed at 20° according to a standard protocol, starting with L4 larvae moved every 12 hr to fresh RNAi plates (Timmins and Fire 1998; Kamath et al. 2001). Feeding strains were obtained from a dsRNA feeding library from Medical Research Council Geneservice (Kamath and Ahringer 2003). F1 embryos and larvae were counted at least 24 hr later. Plates from a 12-hr period were counted only if lethality for a positive control, par-6 RNAi, was >80% for all genetic backgrounds involved. A negative control, gfp RNAi, was used to determine the baseline worm strain lethality fraction (W). We accounted for background worm strain lethality, defining a worm strain adjusted lethality (L) by the equation L = (1 − W)*R, where R is the raw lethality resulting from a given dsRNA fed to that worm strain. Enhancement of lethality was calculated as the difference between the adjusted lethalities (for example, L for ced-5; end-3 minus L for N2). Comparisons between worm strains were only done between corresponding 12-hr plates within the same experiment. For statistical analysis, experimental pairs were repeated in triplicate. A two-tailed Student’s t-test with two-sample unequal variance (heteroscedastic) could then be assessed between the enhancement of lethality for a given bacterial strain to the enhancement of lethality of the negative control vector, L4440 expressing dsGFP.
Templates for in vitro transcription were generated by a two-step PCR from wild-type genomic DNA. Primers for the first step included 20 bases matching the target sequence and 15 bases of the T7 promoter sequence. The resulting PCR product was purified using a PCR purification kit (Qiagen) according to the manufacturer’s recommendations. This product was used as a template for a second PCR using primers containing the full-length T7 promoter sequence. One to two micrograms of the product was then gel purified and used as a template in an in vitro transcription reaction using the T7 RiboMAX Express RNAi System (Promega) according to the manufacturer’s recommendations. The integrity of the dsRNA was assessed by gel electrophoresis, and the concentration was determined by spectrophotometry. dsRNA was injected at a concentration of 100 ng/ml into hermaphrodites using a Narishige injection apparatus, a Parker Instruments Picospritzer II, and a Nikon Eclipse TE300 microscope. dsRNA was stored in two volumes of 100% ethanol at either −20° or −80°.
Microscopy and differential interference contrast imaging
For live imaging, C. elegans embryos were mounted on poly-L-lysine coated coverslips, supported by a 2–3% agarose pad. Four-dimensional (4D) differential interference contrast (DIC) microscopy was carried out using a Diagnostic Instruments SPOT2 camera mounted on a Nikon Eclipse 800 microscope. Images were acquired at 1- to 2-µm optical sections every 1 or 1.5 min during embryogenesis and analyzed with Metamorph v.6.3r5 (Molecular Devices). Imaging was performed at 20°–23° for all strains.
Sequence alignment and phylogenetic tree construction
Amino acid sequences for the genes identified in this screen and C. elegans zyg-11, along with C. briggsae, human and mouse zyg-11 homologs were aligned using CLUSTALW and MUSCLE (Chenna et al. 2003; Edgar 2004). While clear regions of conservation were identified among these sequences, both algorithms produced generally poor alignments among all sequences. The alignments were trimmed to the conserved regions and the C. briggsae sequences were excluded. To be included in the conserved sequence alignment, we required that at least two-thirds of taxa have an aligned base. We used ProTest to determine the best model for amino acid evolution, which was JTT+ G (Abascal et al. 2005). We then constructed both maximum likelihood and maximum parsimony trees for the complete sequences and the trimmed conserved sequences (Guindon and Gascuel 2003; Kumar et al. 2008). A total of 1000 and 500 bootstraps were performed for each algorithm, respectively. Generally, the trees were congruent regardless of algorithm or sequence used. The bootstrap support, however, was best with the trimmed conserved sequence.
Comparative BLAST+ analysis
We used BLAST+ to test the hypothesis that the genes identified by the screen were enriched for genes with paralogs (Camacho et al. 2009). We wrote a computer program (supporting information, File S1) to automate BLAST+ of a gene set vs. the entire C. elegans genome, and National Center for Biotechnology Information (NCBI)’s nonredundant protein (nr) database. BLAST+ result files were then analyzed to determine how many times a gene in our query file hit a gene in the C. elegans genome or in a database of all nematode sequences. Results were then analyzed using JMP (ver. 8; SAS, Cary, NC) and Matlab (MathWorks, Natick, MA). We used a rank sum test to determine significance. We removed duplicates of Wormbase Gene IDs. The top hit (the self-hit) was removed from our count. This method does not exclude hits to multiple isoforms produced from the same gene, although we inspected BLAST results for our 29 genes and found that most hits were to products of distinct genes, which we consider to be potential paralogs.
Comparative sequence analysis
We compared the newly identified gene set to the Conserved Domains Database (CCD; Marchler-Bauer et al. 2009) and filtered our trimmed alignment by similarity. No one residue was conserved across all data, but several potential motifs became apparent between 50 and 90% stringency.
Immunostaining and confocal microscopy
F58D2.1 polyclonal antibodies were generated from rabbits expressing a 100-aa polypeptide from amino acids 198–297 RFIDCSRTMMSVELLEYLLKTHRNLQGVIATMTKSDSDIYDDARALNVATFDSTVRALTYFLKANKVFENGHTITKIDDFIAADSSRILNIRPCMEIIIK (Strategic Diagnostics). A total of 80 ml of rabbit antisera was affinity purified to an endpoint titer of 0.72 ng/ml. Embryos were immunostained for F58D2.1 (1:1000) as described (Tenlen et al. 2008) and imaged using a Zeiss LSM510 confocal microscope with Laser Scanning Microscopy software. Images were further processed with Metamorph and Adobe Photoshop.
Protein interaction experiments
Full-length cDNA clones of zyg-11, gadr-6/F47G4.2, and gadr-5/Y71A12B.17 were cloned into pCMV-Tag2 vector (Stratagene) to produce FLAG-fusion constructs; cul-2-Myc was cloned into pEGFP-N1 vector (Clontech), from which the GFP sequence was removed; and the HA-ELC-1/pEGFP-N1 construct was previously described (Starostina et al. 2007). Immunoprecipitation experiments from transient transfection of HEK293T cells were performed as described (Starostina et al. 2010), using anti-FLAG (M2; Sigma) antibody for the immunoprecipitation and anti-FLAG (M2), anti-HA.11 (Covance), and anti–CUL-2 (Feng et al. 1999) for Western blots. Affinity purification coupled to LC-MS/MS to identify CUL-2::FLAG-associated proteins utilized strains ET099 (expressing Pcul-2::CUL-2::FLAG) and N2, and was performed as previously described (Starostina et al. 2010).
Results
Identifying end-3(ok1448) as a sensitized background
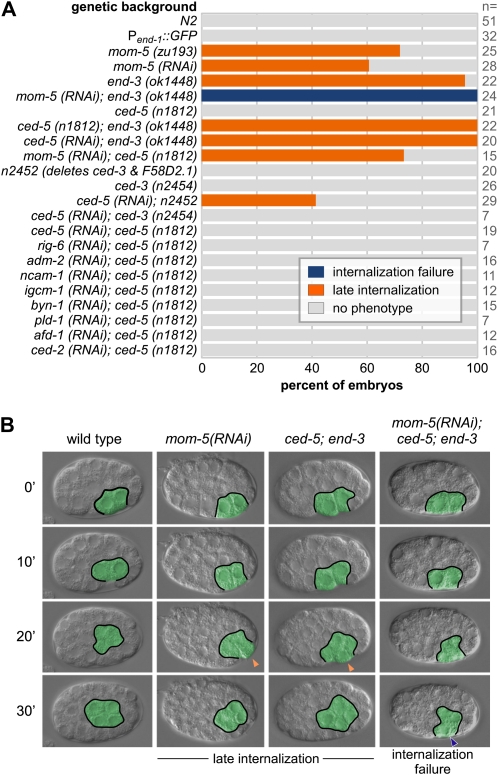
To begin to identify a sensitized background for a gastrulation screen, we sought a mutant with a subtle gastrulation defect, which might be enhanced by feeding a dsRNA, targeting another gene with a role in gastrulation (Figure 1 and Figure S1). Loss of function of either a cell fate regulator end-3 (endodermal GATA factor) or a member of the Wnt signaling pathway mom-5 (Frizzled) can result in a subtle gastrulation defect in which the Ea and Ep cells fail to internalize; however, one cell cycle later, their daughter cells internalize as four E cells (the 4E stage) (Maduro et al. 2005; Lee et al. 2006). We quantified these subtle gastrulation defects in an allele with a large deletion in end-3, end-3(ok1448). In 95% of these embryos, Ea and Ep divided on the surface and became internalized at the 4E stage (Figure 1). The strong mom-5(zu193) (Rocheleau et al. 1997) allele produced similar results, with cells internalizing late at the 4E stage in 72% of embryos (Lee et al. 2006 and Figure 1). Injection of mom-5 dsRNA into wild-type worms nearly phenocopied the mom-5(zu193) allele, with cells internalizing late at the 4E stage in 61% of embryos (Figure 1). These results confirmed that the gastrulation defects in these backgrounds are subtle, but highly penetrant.
Figure 1 .
Enhancement of subtle gastrulation defects. (A) Gastrulation defects in various mutants and/or from injected dsRNAs. (B) Four-dimensional (4D) DIC microscopy of four backgrounds with time on the left from MSa/p cell division. E lineage cells are outlined and pseudocolored in green. Late internalization at the 4E stage (orange arrowheads) and internalization failure (blue arrowhead) are indicated. “No phenotype” indicates that endodermal precursors became internalized at the 2E stage, as in wild-type embryos. Scale: C. elegans embryos are ∼50 µm long.
We discovered that targeting mom-5 and end-3 together by injecting mom-5 dsRNA into end-3(ok1448) worms resulted in a stronger and more penetrant defect than either single treatment: in all embryos, neither Ea/Ep nor their daughter cells internalized (Figure 1). This strongly synergistic effect suggests that these genes may contribute to gastrulation redundantly. The result also suggested that either of these genes might be exploited as a basis for a sensitized background to screen, ideally in a viable mutant background, for enhancement of embryonic lethality, a readily scorable phenotype. end-3 loss-of-function mutants generally produce viable embryos (Maduro et al. 2005), with only 6% embryonic lethality in end-3(ok1448) (Figure S2). Loss-of-function mutants of mom-5 resulted in embryonic lethality (Rocheleau et al. 1997), but feeding mom-5 dsRNA to wild-type animals produced a much weaker defect, with only 4% of embryos failing to hatch (Figure S3), suggesting that RNAi by feeding for mom-5 might be a means to generate partial loss of function. We fed mom-5 dsRNA to end-3(ok1448) worms and found that 24% of embryos failed to hatch, a mild but readily detectable and significant synergistic effect (P = 0.027, Student’s t-test). This result suggested that by feeding dsRNAs to end-3(ok1448) and wild-type animals in parallel, followed by quantification of embryonic lethality, an RNAi feeding screen could be carried out.
Developing a doubly sensitized background
We next determined whether other mutants can produce enhanced gastrulation defects and possibly be used to generate a more sensitized background. ced-5, which encodes a DOCK180-like guanine exchange factor for Rac (Wu and Horvitz 1998) and hmr-1, which encodes a classical cadherin (Costa et al. 1998), function redundantly in C. elegans gastrulation (G. Shemer, unpublished data). hmr-1 also contributes redundantly with sax-7, which encodes an L1CAM (Grana et al. 2010). We confirmed that Ea/Ep internalize successfully in a likely null allele of ced-5, n1812 (Wu and Horvitz 1998) (Figure 1). However, gastrulation is often delayed, with the E cells internalizing as four cells, in double hmr-1; ced-5 embryos (G. Shemer, unpublished data). RNAi to other putative adhesion genes (rig-6, ncam-1, igcm-1, and byn-1) or other genes did not similarly enhance ced-5(n1812) (Figure 1). This result suggests that ced-5(n1812) sensitizes worms to depletion of specific genes, but does not overly sensitize them to depletion of all similar genes.
We next examined whether the two useful backgrounds above might be combined to create a doubly sensitized strain. We constructed a ced-5(n1812);end-3(ok1448) double mutant, and found that it had only 6% embryonic lethality, similar to the lethality of the single alleles (Figure S2), consistent with ced-5 and end-3 being in the same pathway and/or each being redundant with one or more other pathways. We reasoned that this low level of background lethality would facilitate detecting enhancement of lethality in an RNAi feeding screen, and that including both mutations in the screening background might enable more genes to be identified in the screen than including only one or the other, particularly if multiple, partially redundant mechanisms contribute to gastrulation, as has been predicted for morphogenesis more generally (Newman and Comper 1990; Wieschaus 1997). We found that the double mutant could be maintained as homozygotes, and that it retained the ability to be enhanced by feeding mom-5 dsRNA, as expected (Figure 2). Therefore, this strain was selected as our background to screen by RNAi for new genes with possible roles in gastrulation. After screening, we confirmed the value of the double mutant, which identified some enhancers that failed to significantly enhance single mutant backgrounds (see below).
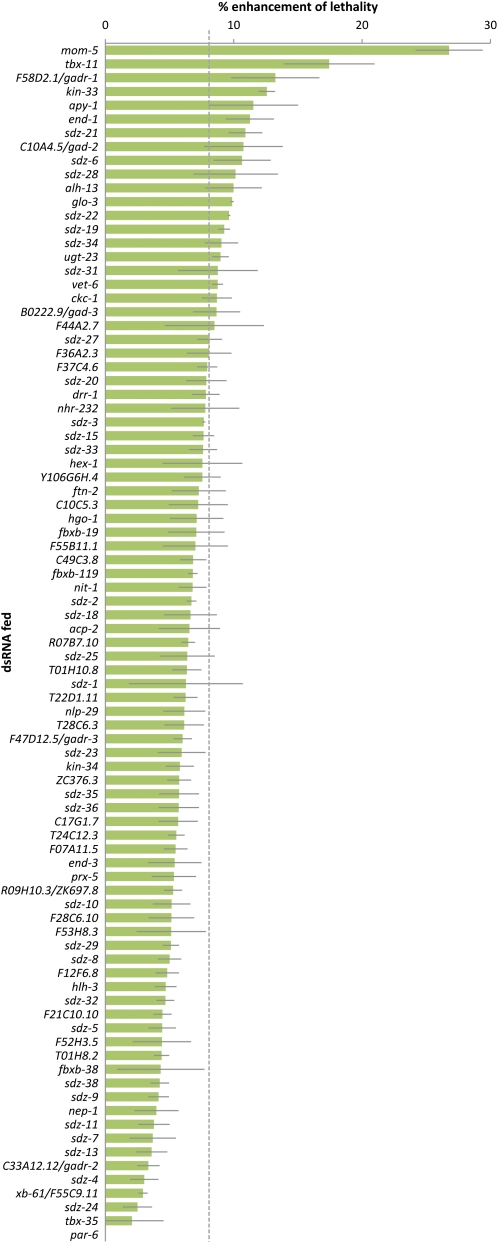
Figure 2 .
Primary screen feeding dsRNAs targeting sdz genes into the gastrulation-sensitized background ced-5(n1812); end-3(ok1448). Percentage of enhancement of lethality (see Material and Methods for calculation) is shown for each dsRNA that was fed three times or more. Dashed line indicates threshold of 8% enhancement of lethality. Error bars indicate 1 SE.
Identification of enhancers of the sensitized background among genes likely to be expressed in or near gastrulating cells
Our results above suggested that we would need to carefully quantify the degree of embryonic lethality for each treatment to identify enhancers. Therefore, to focus our effort, we selected a set of genes to screen through, making use of two previously published data sets that are likely to be enriched for genes expressed in the endodermal lineage or in their close neighbors from the MS lineage before or during gastrulation. First, the results of a published microarray expression experiment using precisely timed embryos (Baugh et al. 2005) were reordered for us by L. R. Baugh (personal communication) to identify those genes whose mRNA abundances were higher in wild-type embryos than in mex-3(zu155); skn-1(RNAi). Embryos of this background generally lack properly specified E and MS lineages at the time when Ea and Ep would normally internalize, and, as expected, early endodermally expressed mRNAs fail to accumulate (Baugh et al. 2005). We narrowed this list by the following criteria. First, we included only those genes for which mRNA abundance rose by the time that Ea/Ep cell internalization occurred, using the microarray expression profiles of known endodermal genes to choose the relevant timepoints, 23–101 min after the four-cell stage. Second, we required mRNA abundances to be higher in wild-type embryos than in mex-3(zu155); skn-1(RNAi) at these time points. Third, we also required mRNA abundances to be lower at these time points in wild-type embryos than in pie-1(zu154); pal-1(RNAi), a background where twice as many E and MS lineages form. The other list we used was a set of 50 genes identified in a microarray experiment designed to find early embryonic downstream targets of skn-1, called sdz (skn-1–dependent zygotic) genes, several of which are transcriptionally active in only MS and E descendents (Robertson et al. 2004). For convenience, we refer to both sets together as sdz genes, although skn-1 dependence has not been validated for all of the genes included. Among these two sets, 112 clones existed in an RNAi feeding library (Kamath et al. 2001).
To determine the ability of knockdown of these 112 genes to enhance the gastrulation-sensitized strain, we fed 112 corresponding bacterial RNAi feeding strains to the ced-5; end-3 worm strain and to N2 wild-type worms in parallel for 48 hr. We assessed the resulting embryonic lethality by counting unhatched embryos and hatched worms at least 24 hr after removing adults (see Materials and Methods). After the first round of feeding, we repeated the 70 with the strongest apparent enhancement of lethality twice more (Figure 2). We found 22 genes that enhanced above a threshold that we chose, 8% enhancement of lethality. These 22 genes included end-1, which is already known to function redundantly with end-3 in the E lineage as gastrulation begins (Maduro et al. 2005), confirming the effectiveness of the screening method.
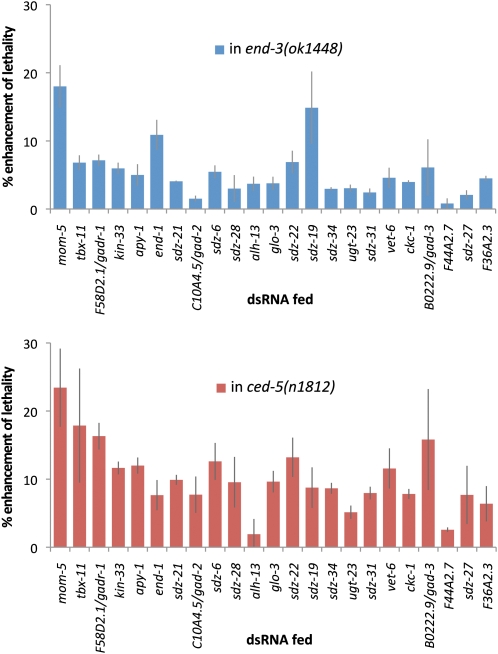
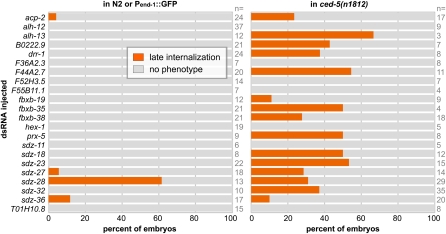
Before secondary screening, we tested whether screening in the double mutant background had increased screening efficiency as predicted, by addressing whether synergy with ced-5, end-3, or both was responsible for the enhancements in lethality. We fed dsRNAs, targeting the 22 genes identified, as well as the positive control mom-5, into the ced-5 and end-3 mutants separately (Figure 3 and Figure S4). We found that 15 genes enhanced significantly only in ced-5, and none enhanced only in the end-3 background. Three genes enhanced both ced-5 and end-3 backgrounds, including end-1 and mom-5. There were three genes that enhanced the double mutant but did not significantly enhance either of the single mutants. These results suggest that the double mutant served as a more efficient sensitized background than either single mutant. Furthermore, these results begin to suggest a structure to the redundancy, which we plan to explore more fully in the future using null mutants.
Figure 3 .
Enhancement of embryonic lethality into separate components of the sensitized background. Enhancement of lethality in ced-5(n1812) over wild type (top) and in end-3(ok1448) over wild type (bottom). Error bars indicate 1 SE.
Secondary screening implicates 16 new genes in gastrulation
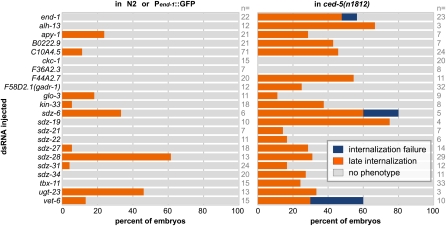
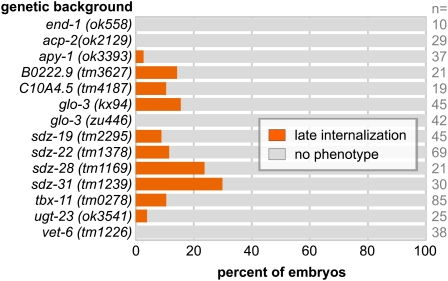
To identify which of these 22 genes were required for the normal pattern of gastrulation, we conducted a series of secondary screens. First, we injected dsRNAs, targeting each gene into the endodermal GFP reporter strain JJ1317 zuIs3 [end-1::GFP] (we refer to this as Pend-1::GFP), and we filmed gastrulation in resulting embryos by 4D DIC microscopy (Thomas et al. 1996). We also injected each dsRNA into ced-5(n1812), to more fully determine the proportion of genes that affect gastrulation in this background. For many of the genes identified in our primary screen (20/22), including end-1, injection of dsRNA into ced-5(n1812) resulted in gastrulation defects (Figure 4). The number of enhancers of ced-5 found by dsRNA injection here and by dsRNA feeding above might reflect an especially effective sensitization for gastrulation genes by ced-5(n1812), or a role for ced-5 in parallel to a large number of genes, or a combination of these possibilities. We also considered that ced-5(n1812) might have overly sensitized the primary screen, revealing genes with only marginal roles in gastrulation, roles that could not be confirmed in a nonsensitized background. This appeared to not be the case: First, for 10 of these genes, we found that injection of dsRNA resulted in gastrulation defects in at least some embryos even in the nonsensitized strain Pend-1::GFP (Figure 4). Second, to examine possible stronger loss of function and to confirm our RNAi results with true mutants, we also filmed by 4D DIC microscopy mutants that were available for 12 of the 22 genes identified in the primary screen. For 10 of these 12 genes, we found that gastrulation defects occurred in some of the filmed mutant embryos (Figure 5). Most of these genes were named previously on the basis of their sequence or as sdz genes. One of the genes, glo-3, is a novel gene that is expressed specifically in endoderm progenitors as early as the 2E cell stage (Rabbitts et al. 2008). Two of the genes were not previously named; we refer to C10A4.5 and B0222.9 as gad-2 and gad-3, respectively, for their gastrulation defective phenotypes.
Figure 4 .
Genes from ced-5; end-3 gastrulation-sensitized screen tested for roles in gastrulation by dsRNA injection. “No phenotype” indicates that endodermal precursors became internalized at the 2E stage, as in wild-type embryos.
Figure 5 .
Gastrulation defects in mutants. “No phenotype” indicates that endodermal precursors became internalized at the 2E stage, as in wild-type embryos. glo-3(zu446) is a nonnull allele (Rabbitts et al. 2008).
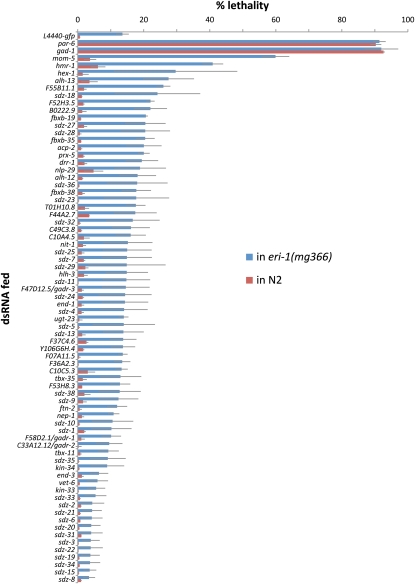
Because our starting list of 112 genes might already be enriched for genes involved in gastrulation, we further tested the value of our enhancer screen strategy by comparing it to a more commonly used method, a screen for embryonic lethality in eri-1(mg366), a background with increased effectiveness of RNAi (Kennedy et al. 2004). We fed bacterially expressed dsRNAs targeting the 70 candidate genes we had screened in triplicate in ced-5; end-3 into eri-1(mg366) and wild-type worms and quantified the degree of embryonic lethality (Figure 6). Among the 22 genes with the most penetrant embryonic lethality in the eri-1 background, 6 had been identified using ced-5; end-3. For the remaining 16, we injected dsRNAs into Pend-1::GFP animals and filmed resulting embryos by 4D DIC microscopy, quantifying gastrulation defects in these as before. This identified just 2 more genes with a very low penetrance, nonredundant role in gastrulation, and 8 more genes with a redundant role in gastrulation (Figure 7).
Figure 6 .
Embryonic lethality in an RNAi-sensitized background, eri-1(mg366), and wild type. Error bars indicate 1 SE.
Figure 7 .
Genes from eri-1 RNAi-sensitized screen tested for roles in gastrulation by dsRNA injection. “No phenotype” indicates that endodermal precursors became internalized at the 2E stage, as in wild-type embryos. Results from six genes (alh-13, B0222.9, F36A2.3, F44A2.7, sdz-27, and sdz-28) from Figure 4 are shown again here, as these genes were identified in both the ced-5; end-3 gastrulation-sensitized screen and the eri-1 RNAi-sensitized screen.
In total, 10 out of the top 22 hits from our ced-5; end-3 screen were new gastrulation genes with nonredundant phenotypes, and 14 out of 22 after examining mutants, whereas only 4 of the top 22 hits from our eri-1 screen were new gastrulation genes with nonredundant phenotypes. We view the higher efficiency of the ced-5; end-3 screen as well as the identification of unique genes in this screen as validating its value as a screening method.
These methods implicated a total of 29 new genes in successful and timely gastrulation in C. elegans. Mutants or RNAi knockdown of 16 of these genes result in gastrulation defects in some embryos even in a nonsensitized background. Interestingly, end-1 was not implicated in gastrulation by RNAi of end-1 in wild-type embryos nor by an end-1 deletion allele, suggesting that an earlier report of a role for end-1 based on a larger deletion, wDf4, is likely explained by deletion of end-3 as well in wDf4 (Maduro et al. 2005; Lee et al. 2006). Six of the 23 genes we identified had quite low penetrance effects on gastrulation, and higher penetrance in ced-5(n1812), and 13 could only be implicated in combination with ced-5(n1812), suggesting that many of these genes may act redundantly or partially redundantly in gastrulation, or in separate processes that make indirect contributions to normal gastrulation.
Several of the newly identified genes’ closest homologs are other C. elegans genes
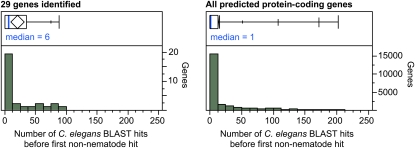
We performed BLAST searches on each of the genes we identified and discovered that for many of these genes (20/29), the closest known sequence as judged by BLAST score in the NCBI nr database as of September 2010 was another gene in the C. elegans genome. For a large proportion of the genes (18/29), multiple other C. elegans genes had higher BLAST scores than did any nonnematode genes, suggesting to us that many may belong to related groups of genes that may have arisen from rounds of gene duplication events within the nematode lineage, or represent a large set of convergently evolved genes. Since C. elegans has a compact genome with mostly single copy genes (Woollard 2005), our screen appeared to have enriched for such genes. Consistent with this, determining the number of BLAST hits from C. elegans with greater similarity by BLAST score than any nonnematode gene resulted in a median of six hits for our group of 29 genes, and a median of one hit for all 20,331 C. elegans predicted protein-coding genes (Figure 8).
Figure 8 .
Number of C. elegans BLAST hits with greater similarity by BLAST score than the first nonnematode hit, for the 29 new genes identified here (left) and for all C. elegans predicted protein-coding genes. Histograms are shown along with box-and-whisker plots at top, with boxes representing the 25–75% quartile ranges (0–35.5 for the 29 genes identified and 0–13 for all genes). Medians are marked in blue.
C. elegans gene families deriving from recent gene duplications are more likely to function redundantly than are single copy genes (Conant and Wagner 2003), and we speculate that this might be true for sets of similar genes deriving from less recent duplications or from convergent evolution as well. Given this, the subtle defects and low penetrance of many of the genes we identified, and our finding of several genes that we could implicate only in sensitized backgrounds, we hypothesized that our screening method was successful in uncovering genes that function redundantly or partially redundantly in C. elegans gastrulation. We tested this hypothesis directly for one gene family below.
gadr-1 is a redundant gastrulation gene that is expressed as gastrulation begins
One of the most penetrant enhancers of our double mutant background that we found was F58D2.1 (Figure 2). F58D2.1 appeared to act synergistically with ced-5 in gastrulation: targeting F58D2.1 and ced-5 together, by injecting F58D2.1 dsRNA into ced-5(n1812) worms, resulted in 25% of embryos failing in Ea/Ep internalization, whereas neither single treatment produced this result (Figures 1 and 4). On the basis of this result and others below, we name F58D2.1, gadr-1 (gastrulation defective, redundant).
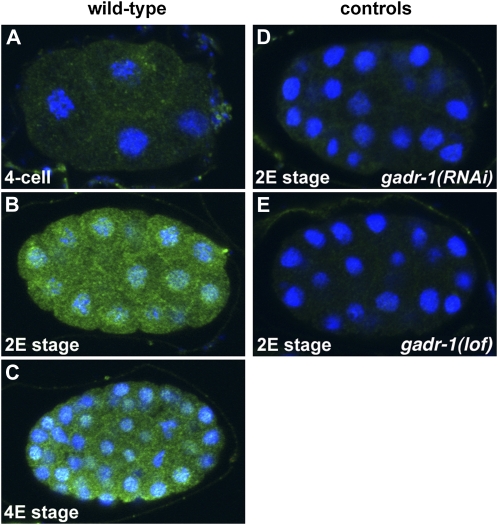
Microarray experiments on staged embryos (Baugh et al. 2005) demonstrated that gadr-1 transcript abundance increased near the time that gastrulation begins—soon after end-1 transcripts, which are first detected in the E cell by in situ hybridization (Zhu et al. 1997), and before elt-2 transcripts, which are first detected in Ea and Ep just after gastrulation begins (Fukushige et al. 1998). To determine when and where the GADR-1 protein accumulates, we generated an affinity-purified rabbit antibody to a 100-amino-acid part of the protein (see Materials and Methods) and used it to immunostain embryos. The timing of GADR-1 protein accumulation was consistent with the microarray results and with our proposed role in gastrulation: GADR-1 immunoreactivity became strong near the time of endodermal internalization. Staining was eliminated by gadr-1 RNAi or by a deletion allele, n2452, which is a 17-kb deletion that removes all or parts of seven genes including most of gadr-1 (Shaham et al. 1999) and the entire antigen sequence. GADR-1 localized to both nuclei and cytoplasm of all cells, with a small amount of enrichment near cell–cell boundaries (Figure 9). In support of our hypothesis from RNAi experiments that gadr-1 functions redundantly in gastrulation, the n2452 deletion allele produced gastrulation defects only in combination with ced-5(RNAi), and not alone (Figure 1). We conclude that gadr-1 functions redundantly in gastrulation and that it encodes a nuclear and cytoplasmic protein that is first expressed in all cells near the time that gastrulation begins.
Figure 9 .
GADR-1 protein localization. The wild-type embryos shown were imaged from the same slide under the same conditions, after staining with anti–GADR-1 (green) and DAPI to mark nuclei (blue). GADR-1 levels are low at the four-cell stage (A) and have risen by gastrulation (B and C). Staining is reduced by gadr-1 RNAi (D) and by n2452, an allele with a deletion of seven genes including part of gadr-1 (E). Scale: C. elegans embryos are ∼50 µm long.
GADR-1 and paralogs resemble substrate recognition subunits for a CUL-2 ubiquitin ligase complex
A search for similar genes by BLAST identified the predicted GADR-1 protein as belonging to a large and diverse group of C. elegans proteins that includes ZYG-11, an SRS for a CUL-2 ubiquitin ligase complex (Vasudevan et al. 2007) and ZEEL-1, a related protein implicated in reproductive incompatibility between populations (Seidel et al. 2008). By BLAST of the predicted GADR-1 protein sequence, 23 predicted C. elegans proteins had lower E values than any nonnematode sequence in the nr database, suggesting that these genes may have arisen from rounds of gene duplication within the nematodes or that they result from convergent sequence evolution. Sequence similarity among F58D2.1 and paralogs appears to be driven by a small set of residues corresponding to leucine-rich repeats (LRRs) and several uncharacterized motifs. We performed a comparative sequence analysis of the newly identified genes and zyg-11 family members. Using the Conserved Domains Database, we noticed that all genes analyzed including the mammalian zyg11 genes had at least one leucine-rich repeat-like motif (canonically, LxxLxLxxN/CxL). While most of these protein sequences are highly divergent, the strong similarity within these specific motifs in the newly identified genes suggests that these motifs are evolutionarily and functionally conserved.
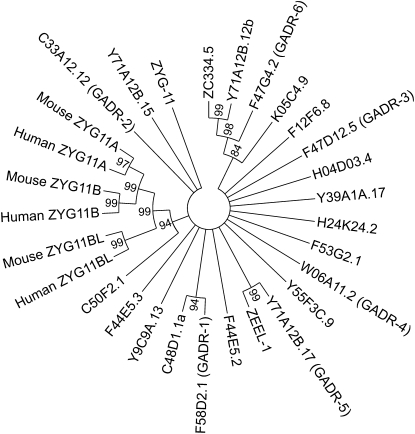
We used phylogenetic and comparative genomic analysis to reveal the evolutionary history of the newly identified genes relative to C. elegans zyg-11 and human and mouse ZYG-11 homologs. These highly diverged amino acid sequences resolved poorly, producing a star phylogeny with the exception of several sets of genes within C. elegans and the mammalian ZYG-11 gene family (Figure 10). Outside of the mammalian clade, which resolved as expected (Vasudevan et al. 2007), only three clades formed monophyletic groups (Figure 10) with significant bootstrap support using both the maximum likelihood (ML) and maximum parsimony (MP) methods.
Figure 10 .
Phylogenetic relationship of the newly identified genes, related C. elegans genes, and mammalian ZYG11 genes. We used both maximum likelihood and maximum parsimony to produce phylogenies of the newly identified genes, C. elegans zyg-11, and human and mouse zyg-11 homologs.
GADR-1 paralogs can bind ubiquitin ligase complex components CUL-2/cullin and ELC-1/Elongin C
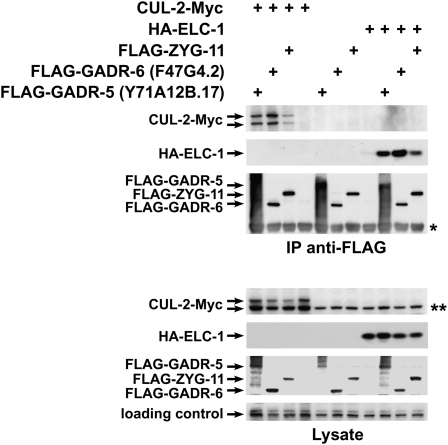
The observation that the GADR-1 and paralogs have some sequence similarity to ZYG-11 suggested that these proteins may similarly function as SRSs in CUL-2 complexes. Affinity purifications coupled to liquid chromatography and tandem mass spectrometry (LC-MS/MS) were used to identify proteins that physically associate with CUL-2::FLAG in vivo. In two separate samples, GADR-6/F47G4.2 was identified in affinity purifications from lysates of animals expressing CUL-2::FLAG. The number of peptides of GADR-6 identified by LC-MS/MS in the two samples (9 and 11 peptides) was comparable to the number of peptides observed for known SRSs (Vasudevan et al. 2007; Starostina et al. 2010): FEM-1, 24 and 32 peptides; ZER-1, 19 and 29; ZYG-11, 9 and 11; LRR-1, 3 and 5; VHL-1, 0 and 0; and ZIF-1, 0 and 0. GADR-1 to -5 were not identified in the affinity purifications. However, in separate affinity purifications that only analyzed the 85–110 kDa region of CUL-2::FLAG-associated proteins resolved on SDS–PAGE gels, GADR-5/Y71A12B.17 was identified by a single peptide (while GADR-6 was identified with 4 peptides; ZYG-11, 8 peptides; and ZER-1, 12 peptides); none of these proteins were identified from the comparable region of the control affinity purification (from wild-type animals not expressing CUL-2::FLAG).
To further probe whether GADR-5 and GADR-6 might function as SRSs, we asked whether they could interact with CUL-2 and the CUL-2 complex adaptor protein Elongin C/ELC-1 when ectopically expressed in HEK293T human cells. We observed that CUL-2 and ELC-1 co-immunoprecipitated with GADR-5 and GADR-6 at a level comparable to that observed with ZYG-11 immunoprecipitation (Figure 11). Therefore, GADR-5 and GADR-6 are likely candidates to be SRSs for CUL-2 ubiquitin ligase complexes. The failure to detect other GADR-1 paralogs in affinity purifications of CUL-2::FLAG may be due to the limitations of the analysis, as the affinity purification coupled to LC-MS/MS approach also failed to identify the previously identified SRSs VHL-1 and ZIF-1 (Derenzo et al. 2003; Mehta et al. 2009). We conclude that at least some GADR-1 paralogs can bind ubiquitin ligase complex components CUL-2 and ELC-1. RNAi targeting cul-2 or elc-1 resulted in defects before gastrulation as expected (Kipreos 2005), which precluded us from determining directly whether these complex members function in gastrulation (data not shown).
Figure 11 .
Two GADR proteins, GADR-5/Y71A12B.17 and GADR-6/F47G4.2, physically interact with both CUL-2 and ELC-1 when coexpressed in human cells. FLAG-tagged GADR-5, GADR-6, and ZYG-11 were coexpressed in HEK293T cells with CUL-2–Myc or HA–ELC-1 as noted by (+) symbols above the lanes. Anti-FLAG immunoprecipitations (IP) and lysates were analyzed by Western blot using anti-FLAG, anti-HA, or anti–CUL-2 antibodies. A cross-reacting band serves as a loading control. Note that both GADR-5 and GADR-6 bind CUL-2 and ELC-1 analogous to the known substrate recognition subunit ZYG-11. The smearing and additional lower bands for FLAG–GADR-5 presumably arise from partial degradation of the protein in HEK293T cells. * denotes the heavy chain of IgG used in the IP; ** marks nonspecific band (which comigrates with lower band of CUL-2 in the first four samples).
gadr-1 and paralogs function redundantly with each other in gastrulation
We hypothesized that gadr-1 might function redundantly in gastrulation with one or more genes that had sequence similarity. To identify such genes, we injected dsRNA, targeting the nine closest relatives of gadr-1 by BLAST into both ced-5(n1812) and Pend-1::GFP worms. We found that most of these could enhance ced-5(n1812), but none produced gastrulation defects in the nonsensitized background, Pend-1::GFP, suggesting that all of these genes might act redundantly, as gadr-1 does (Table S1). Indeed, one of these genes, C48D1.1, is also deleted by the n2452 deletion allele described above. This result implies that if gadr-1 contributes redundantly to gastrulation with some of the related genes, deleting just this pair was not sufficient to reveal a gastrulation defect.
We pursued our hypothesis of redundancy by pooled injection of dsRNAs with the other related genes. Both C48D1.1 and F53G2.1 had frequent cell division defects before gastrulation in ced-5(n1812) and were not pursued further. Injection of pooled dsRNAs targeting six remaining genes (the six with the most penetrant effects on gastrulation in ced-5(n1812)) into N2 worms resulted in 49% penetrant gastrulation defects in Ea/Ep cell internalization (Table S2). This result confirms that some or all of these six related genes function redundantly with each other in one or more processes that directly or indirectly affect gastrulation.
To elucidate whether some play more significant roles than others in gastrulation, we used a strategy of injecting all combinations of five of the six pooled dsRNAs, then omitting the one that gave the least penetrant gastrulation defects in a following round using pools of four dsRNAs, and reiterating this pattern until we had narrowed down to just a pair of genes with the most penetrant effects. We found that decreasing the number of genes decreased the penetrance of the phenotypes at nearly every step, without any genes emerging as especially major contributors (Table S2). This result suggests that these genes function partially redundantly in an additive manner with one another. We conclude that each of these genes [which we call gadr-2 (C33A12.12), gadr-3 (F47D12.5), gadr-4 (W06A11.2), gadr-5 (Y71A12B.17), and gadr-6 (F47G4.2)] acts redundantly with ced-5 in gastrulation and that all or most of them act redundantly with each other in gastrulation. Our results indicate that our strategy for identifying new gastrulation genes can successfully identify redundant players, including sets of related genes that function redundantly with each other.
Discussion
Redundancy has been proposed to be a well-recognized aspect of morphogenesis, making gene discovery a challenge (Newman and Comper 1990; Wieschaus 1997). We decided to address this challenge using classical genetics and RNAi while looking for new genes acting in C. elegans gastrulation. In this article, we have described an enhancer screen to find new C. elegans gastrulation genes, the first RNAi modifier screen for morphogenesis genes in C. elegans. We find that there is indeed developmental redundancy both between similar genes and between genes that are unrelated by sequence—homologous and nonhomologous redundancy (Jorgensen and Mango 2002). We also found that several genes that play a role in C. elegans gastrulation belong to groups of related genes, some of which may represent gene families deriving from gene duplication events in the nematodes. We predicted that such genes may be more likely than single copy genes to function redundantly or partially redundantly, and we confirmed this for one set of six related genes, gadr-1 to -6, which encode predicted substrate recognition subunits for a CUL-2 ubiquitin ligase. Our results demonstrate that screening by RNAi in a sensitized background is a viable method for tackling redundancy and that it can even identify redundant, closely related genes, traditionally thought of as difficult to identify genetically.
Using RNAi to screen for genes involved in morphogenetic processes
Many C. elegans biologists have taken advantage of the ease of RNAi to compile relatively quickly a list of genes involved in a process of interest (reviewed in Jorgensen and Mango 2002 and Boutros and Ahringer 2008). With speed and ease of methodology comes the drawback of variable and sometimes ineffective RNAi, especially when using feeding RNAi as opposed to RNAi by injection. Even with these drawbacks, an RNAi screen can be valuable in tackling redundancy and studying somewhat genetically refractory developmental processes.
Often, suppressor screens (Labbé et al. 2006; O’Rourke et al. 2007; Dorfman et al. 2009; reviewed in Boutros and Ahringer 2008) have been utilized to discover new genes that function in early developmental processes. The ability to screen for survivors starting from a conditional lethal strain is rapid and convenient. To screen for enhancers efficiently, one must be able to recognize quickly the enhanced phenotypes. In our case, we sensitized our worms using mutations known to affect gastrulation, and we used embryonic lethality as a first test for enhancement. We then used 4D microscopy to examine the initiation of gastrulation, internalization of the E cells, specifically.
One goal of our screen was to identify new genes whose functions are required for normal gastrulation. Although this succeeded, limitations exist in the screen that we have carried out. Filming embryos revealed many low penetrance gastrulation genes, and it is possible that we may have missed other genes whose loss of function in wild-type embryos may produce similar defects, but that would have been missed if they did not significantly increase lethality of the sensitized background used in our primary screen. We also did not explore defects in developmental processes other than Ea/Ep internalization. Defects in later morphogenesis or other processes could be a separate cause of enhancement of lethality from our primary screen. We started with a candidate set of zygotic genes, introducing the possibility that we have missed some important maternal genes. We expect that the genes we have identified may include genes that affect gastrulation directly or indirectly. At least two are expressed in Ea and Ep, and one in the early E and MS lineages (Table 1), suggesting that these might have more spatially restricted roles than is likely for gadr-1, which we have shown is expressed near the time of gastrulation, but in all cells. The sdz gene set is likely to be enriched for genes expressed specifically in the E and/or MS lineages (Robertson et al. 2004). The genes we have identified probably represent only a proportion of all genes that function in gastrulation, although what proportion is difficult to estimate at this stage.
Table 1.
Genes identified in this study and their predicted products
| New gad genes | Predicted products |
| acp-2 | Acid phosphatase |
| apy-1 | Apyrase |
| gad-2 (C10A4.5) | Transmembrane protein |
| gad-3 (B0222.9) | Xanthine dehydrogenase |
| glo-3 | Gut granule/lysosome formation, expressed in early E lineage |
| kin-33 | Kinase |
| sdz-6 | Unknown |
| sdz-19 | Unknown |
| sdz-22 | Transthyretin family |
| sdz-27 | Unknown |
| sdz-28 | BTB/POZ domain, MATH domain |
| sdz-31 | Unknown, expressed in early E and MS lineages |
| sdz-36 | Unknown |
| tbx-11 | T-box transcription factor |
| ugt-23 | UDP-glucuronosyl transferase |
| vet-6 | Very early transcript, contains a spectrin repeat |
| New gadr genes | |
| alh-13 | Dehydrogenase/reductase |
| drr-1 | Dietary restriction response |
| F44A2.7 | Unknown |
| fbxb-19 | F-box protein |
| fbxb-35 | F-box protein |
| fbxb-38 | F-box protein |
| gadr-1 (F58D2.1) | ZYG-11-like protein, possible SRS for a CUL-2 ubiquitin ligase |
| gadr-2 (C33A12.12) | ZYG-11-like protein, possible SRS for a CUL-2 ubiquitin ligase |
| gadr-3 (F47D12.5) | ZYG-11-like protein, possible SRS for a CUL-2 ubiquitin ligase |
| gadr-4 (W06A11.2) | ZYG-11-like protein, possible SRS for a CUL-2 ubiquitin ligase |
| gadr-5 (Y71A12B.17) | ZYG-11-like protein, possible SRS for a CUL-2 ubiquitin ligase |
| gadr-6 (F47G4.2) | ZYG-11-like protein, possible SRS for a CUL-2 ubiquitin ligase |
| prx-5 | Peroxisome import |
| sdz-18 | BTB/POZ domain, MATH domain |
| sdz-21 | Unknown |
| sdz-23 | Transmembrane, EGF domain, expressed in early E lineage |
| sdz-32 | Unknown |
| sdz-34 | Predicted E3 ubiquitin ligase |
Listed are 16 new gad genes (gastrulation-defective: genes whose loss of function results in gastrulation phenotypes) and 18 new gadr genes (gastrulation defective, redundant: genes whose loss of function only results in gastrulation phenotypes in combination with loss of function of other genes). Pend-1::GFP expression was examined for a small number of embryos after RNAi of all gad genes except gad-3, sdz-19, sdz-22, tbx-11, and was seen to be low or absent for 1/1 acp-2(RNAi) embryo, 1/1 ugt-23(RNAi) embryo, and 2/6 gad-2(RNAi) embryos, suggesting that these genes may affect endoderm specification as well. Genes given new names here are listed as gad- or gadr-, with the corresponding sequence name in parentheses. Expression data are from Rabbitts et al. 2008 for glo-3, and Robertson et al. 2004 for sdz genes.
Nonhomologous genetic redundancies have been found in C. elegans before (Culotti et al. 1981; Johnson et al. 1981; Ferguson and Horvitz 1989; Davies et al. 1999, for examples). One well characterized C. elegans nonhomologous redundancy is the synthetic multivulval (SynMuv) system (Ferguson et al. 1987; Ferguson and Horvitz 1989; for review, see Fay and Han 2000; Fay et al. 2002). We identified several genes that could only be implicated in gastrulation in specific genetic backgrounds, and not in wild-type worms. We refer to such a synthetic gastrulation phenotype as SynGad, or Gadr. The nonredundant Gad and redundant Gadr phenotype categories are not definitive: Gad genes have detectable gastrulation defects when knocked down alone, but could also be partially redundant. For example, loss of function of these genes could produce more severe, synergistic gastrulation defects in combinations with each other or with loss of function of other genes. Nonredundant roles in gastrulation have not been found for Gadr genes, but it is possible that for some, null alleles will show some gastrulation defects in nonsensitized backgrounds.
Predicted roles for some of the new genes involved in C. elegans gastrulation
Many of the genes we have identified encode proteins of unknown function in C. elegans but have specific, predicted protein domains (Table 1). For example, sdz-23 encodes a predicted transmembrane protein with an EGF domain, expressed in the early E lineage (Robertson et al. 2004), and kin-33 encodes a predicted kinase (Manning 2005). tbx-11 encodes a putative T-box transcription factor the resembles proteins of the Tbx2 subfamily, and a function for tbx-11 had not been reported previously. T-box transcription factors play roles in cell fate specification and morphogenetic movements in diverse organisms including C. elegans, Xenopus, zebrafish, mouse, and human (Lustig et al. 1996; Chisholm and Hardin 2005; Naiche et al. 2005; Amack et al. 2007). In Xenopus, one of the T-box proteins, Xombi/VegT, was first identified on the basis of its ability to induce invagination in an overexpression screen, suggesting that this protein has a direct or indirect role in morphogenesis (Lustig et al. 1996). How tbx-11 contributes to C. elegans gastrulation is not yet clear.
glo-3, which is expressed specifically in endoderm progenitors as early as the 2E cell stage, has been proposed to function later in vesicle trafficking to the embryonic gut granules, which are lysosome-related organelles (Rabbitts et al. 2008). GLO-3 protein is likely to play a direct role in regulating the formation, maturation, and/or stability of gut granules, since glo-3 is required for gut granule formation, and a rescuing GLO-3::GFP fusion is localized to the gut granule membrane. apy-1 encodes a predicted apyrase, a membrane-bound enzyme that catalyzes the hydrolysis of nucleoside triphosphates and diphosphates. apy-1 mutant worms abnormally accumulate intestinal autofluorescence, which has been interpreted as a lysosomal traffic defect also associated with aging (Uccelletti et al. 2008). Taken together, these results suggest the possibility that normal lysosomal trafficking in the early E lineage cells might play a specific role in successful gastrulation.
Several of the proteins implicated in our screens are predicted to regulate proteolysis, and two of these were shown here to be able to interact with ubiquitin ligase complex members when expressed in human cells. An expression screen in Xenopus for proteins degraded near gastrulation revealed that regulated proteolysis plays a role in gastrulation in this system. Xom is a homeobox transcriptional repressor of dorsally expressed genes, and it is degraded early in gastrulation, allowing the dorsal side of the embryo to develop properly (Zhu and Kirschner 2002). If the putative substrate recognition proteins we identified can be confirmed to function in regulated proteolysis in vivo, it will be of interest to identify potential targets whose degradation might contribute to normal gastrulation. Such targets might include the other gene products we identified here, as well as previously identified proteins that function in gastrulation, for example PAR proteins (Nance et al. 2005).
Acknowledgments
We thank members of the Goldstein lab for discussions, and R. Baugh for help with re-analyzing his microarray data. This work was supported by a National Institutes of Health (NIH) Developmental Biology training grant, by NIH grant R01GM083071 to B.G., and NIH grant R01GM074212 to E.T.K.
Literature Cited
- Abascal F., Zardoya R., Posada D., 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21: 2104–2105 [DOI] [PubMed] [Google Scholar]
- Amack J. D., Wang X., Yost H. J., 2007. Two T-box genes play independent and cooperative roles to regulate morphogenesis of ciliated Kupffer’s vesicle in zebrafish. Dev. Biol. 310: 196–210 [DOI] [PubMed] [Google Scholar]
- Anderson D. C., Gill J. S., Cinalli R. M., Nance J., 2008. Polarization of the C. elegans embryo by RhoGAP-mediated exclusion of PAR-6 from cell contacts. Science 320: 1771–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh L. R., Hill A. A., Claggett J. M., Hill-Harfe K., Wen J. C., et al. , 2005. The homeodomain protein PAL-1 specifies a lineage-specific regulatory network in the C. elegans embryo. Development 132: 1843–1854 [DOI] [PubMed] [Google Scholar]
- Beitel G. J., Krasnow M. A., 2000. Genetic control of epithelial tube size in the Drosophila tracheal system. Development 127: 3271–3282 [DOI] [PubMed] [Google Scholar]
- Boutros M., Ahringer J., 2008. The art and design of genetic screens: RNA interference. Nat. Rev. Genet. 9: 554–566 [DOI] [PubMed] [Google Scholar]
- Bowerman B., Eaton B. A., Priess J. R., 1992. skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryo. Cell 68: 1061–1075 [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., et al. , 2009. BLAST+: architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenna R., Sugawara H., Koike T., Lopez R., Gibson T. J., et al. , 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31: 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm A. D., Hardin J., 2005. Epidermal morphogenesis, in WormBook, edited by The C. elegans Research Community. WormBook, doi/10.1895/wormbook.1.35.1, http://www.wormbook.org
- Conant G. C., Wagner A., 2003. Asymmetric sequence divergence of duplicate genes. Genome Res. 13: 2052–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M., Raich W., Agbunag C., Leung B., Hardin J., et al. , 1998. A putative catenin-cadherin system mediates morphogenesis of the Caenorhabditis elegans embryo. J. Cell. Biol. 141: 297–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culotti J. G., Von Ehrenstein G., Culotti M. R., Russell R. L., 1981. A second class of acetylcholinesterase-deficient mutants of the nematode Caenorhabditis elegans. Genetics 97: 281–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies A. G., Spike C. A., Shaw J. E., Herman R. K., 1999. Functional overlap between the mec-8 gene and five sym genes in Caenorhabditis elegans. Genetics 153: 117–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRenzo C., Reese K. J., Seydoux G., 2003. Exclusion of germ plasm proteins from somatic lineages by cullin-dependent degradation. Nature 424: 685–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman M., Gomes J. E., O’Rourke S., Bowerman B., 2009. Using RNA interference to identify specific modifiers of a temperature-sensitive, embryonic-lethal mutation in the Caenorhabditis elegans ubiquitin-like Nedd8 protein modification pathway E1-activating gene rfl-1. Genetics 182: 1035–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C., 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar L. G., McGhee J. D., 1988. DNA synthesis and the control of embryonic gene expression in C. elegans. Cell 53: 589–599 [DOI] [PubMed] [Google Scholar]
- Ellertsdóttir E., Lenard A., Blum Y., Krudewig A., Herwig L., et al. , 2010. Vascular morphogenesis in the zebrafish embryo. Dev. Biol. 341: 56–65 [DOI] [PubMed] [Google Scholar]
- Fay D. S., Han M., 2000. The synthetic multivulval genes of C. elegans: functional redundancy, Ras-antagonism, and cell fate determination. Genesis 26: 279–284 [DOI] [PubMed] [Google Scholar]
- Fay D. S., Keenan S., Han M., 2002. fzr-1 and lin-35/Rb function redundantly to control cell proliferation in C. elegans as revealed by a nonbiased synthetic screen. Genes Dev. 16: 503–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay D. S., Yochem J., 2007. The SynMuv genes of Caenorhabditis elegans in vulval development and beyond. Dev. Biol. 306: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix M. A., Wagner A., 2008. Robustness and evolution: concepts, insights and challenges from a developmental model system. Heredity 100: 132–140 [DOI] [PubMed] [Google Scholar]
- Feng H., Zhong W., Punkosdy G., Gu S., Zhou L., et al. , 1999. CUL-2 is required for the G1-to-S-phase transition and mitotic chromosome condensation in Caenorhabditis elegans. Nat. Cell Biol. 1: 486–492 [DOI] [PubMed] [Google Scholar]
- Ferguson E. L., Sternberg P. W., Horvitz H. R., 1987. A genetic pathway for the specification of the vulval cell lineages of Caenorhabditis elegans. Nature 326: 259–267 [DOI] [PubMed] [Google Scholar]
- Ferguson E. L., Horvitz H. R., 1989. Identification and characterization of 22 genes that affect the vulval cell lineages of the nematode Caenorhabditis elegans. Genetics 110: 17–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushige T., Hawkins M. G., McGhee J. D., 1998. The GATA-factor elt-2 is essential for formation of the Caenorhabditis elegans intestine. Dev. Biol. 198: 286–302 [PubMed] [Google Scholar]
- Grana T. M., Cox E. A., Lynch A. M., Hardin J., 2010. SAX-7/L1CAM and HMR-1/cadherin function redundantly in blastomere compaction and non-muscle myosin accumulation during Caenorhabditis elegans gastrulation. Dev. Biol. 344: 731–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X., 2003. Evolution of duplicate genes vs. genetic robustness against null mutations. Trends Genet. 19: 354–356 [DOI] [PubMed] [Google Scholar]
- Gu Z., Steinmetz L. M., Gu X., Scharfe C., Davis R. W., et al. , 2003. Role of duplicate genes in genetic robustness against null mutations. Nature. 421: 63–66 [DOI] [PubMed] [Google Scholar]
- Guindon S., Gascuel O., 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Sys. Biol. 52: 696–704 [DOI] [PubMed] [Google Scholar]
- Harrell J. R., Goldstein B., 2011. Internalization of multiple cells during C. elegans gastrulation depends on common cytoskeletal mechanisms but different cell polarity and cell fate regulators. Dev. Biol. 350: 1?12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen R. C., Baillie D. L., 1997. Mutation, in C. elegans II, edited by Riddle D. L., Blumenthal T., Meyer B. J., R. Priess J., pp. 79–95 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- Johnson C. D., Duckett J. G., Culotti J. G., Herman R. K., Meneely P. M., et al. , 1981. An acetylcholinesterase-deficient mutant of the nematode Caenorhabditis elegans. Genetics 97: 261–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen E. M., Mango S. E., 2002. The art and design of genetic screens: Caenorhabditis elegans. Nat. Rev. Genet. 3: 356–369 [DOI] [PubMed] [Google Scholar]
- Kamath R. S., Martinez-Campos M., Zipperlen P., Fraser A. G., Ahringer J., 2001. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2: RESEARCH0002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R. S., Ahringer J., 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30: 313–321 [DOI] [PubMed] [Google Scholar]
- Karabinos A., Bussing I., Schulze E., Wang J., Weber K., et al. , 2003. Functional analysis of the single calmodulin gene in the nematode Caenorhabditis elegans by RNA interference and 4-D microscopy. Eur. J. Cell. Biol. 82: 557–563 [DOI] [PubMed] [Google Scholar]
- Kennedy S., Wang D., Ruvkun G., 2004. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature 427: 645–649 [DOI] [PubMed] [Google Scholar]
- Kiehart D. P., Ketchum A., Young P., Lutz D., Alfenito M. R., et al. , 1990. Contractile proteins in Drosophila development. Ann. N Y Acad. Sci. 582: 233–251 [DOI] [PubMed] [Google Scholar]
- Kipreos E. T., 2005. Ubiquitin-mediated pathways in C. elegans, in WormBook, edited by The C. elegans Research Community. WormBook, doi/10.1895/wormbook.1.36.1, http://www.wormbook.org
- Knight J. K., Wood W. B., 1998. Gastrulation initiation in Caenorhabditis elegans requires the function of gad-1, which encodes a protein with WD repeats. Dev. Biol. 198: 253–265 [PubMed] [Google Scholar]
- Kumar S., Dudley J., Nei M., Tamura K., 2008. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinformatics 9: 299–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbe J. C., Pacquelet A., Marty T., Gotta M., 2006. A genomewide screen for suppressors of par-2 uncovers potential regulators of PAR protein-dependent cell polarity in Caenorhabditis elegans. Genetics 174: 285–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. Y., Goldstein B., 2003. Mechanisms of cell positioning during C. elegans gastrulation. Development. 130: 307–320 [DOI] [PubMed] [Google Scholar]
- Lee J. Y., Marston D. J., Walston T., Hardin J., Halberstadt A., et al. , 2006. Wnt/Frizzled signaling controls C. elegans gastrulation by activating actomyosin contractility. Curr. Biol. 16: 1986–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig K. D., Kroll K. L., Sun E. E., Kirschner M. W., 1996. Expression cloning of a Xenopus T-related gene (Xombi) involved in mesodermal patterning and blastopore lip formation. Development 122: 4001–4012 [DOI] [PubMed] [Google Scholar]
- Maduro M. F., Hill R. J., Heid P. J., Newman-Smith E. D., Zhu J., et al. , 2005. Genetic redundancy in endoderm specification within the genus Caenorhabditis. Dev. Biol. 284: 509–522 [DOI] [PubMed] [Google Scholar]
- Manning G., 2005. Genomic overview of protein kinases, in WormBook, edited by The C. elegans Research Community. WormBook, doi/10.1895/wormbook.1.60.1, http://www.wormbook.org
- Marchler-Bauer A., Anderson J. B., Chitsaz F., Derbyshire M. K., DeWeese-Scott C., et al. , 2009. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37: D205–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maybeck V., Röper K., 2009. A targeted gain-of-function screen identifies genes affecting salivary gland morphogenesis/tubulogenesis in Drosophila. Genetics 181: 543–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta R., Steinkraus K. A., Sutphin G. L., Ramos F. J., Shamieh L. S., et al. , 2009. Proteasomal regulation of the hypoxic response modulates aging in C. elegans. Science. 324: 1196–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger R. J., Krasnow M. A., 1999. Genetic control of branching morphogenesis. Science 284: 1635–1639 [DOI] [PubMed] [Google Scholar]
- Naiche L. A., Harrelson Z., Kelly R. G., Papaioannou V. E., 2005. T-box genes in vertebrate development. Annu. Rev. Genet. 39: 219–239 [DOI] [PubMed] [Google Scholar]
- Nance J., Munro E. M., Priess J. R., 2003. C. elegans PAR-3 and PAR-6 are required for apicobasal asymmetries associated with cell adhesion and gastrulation. Development 130: 5339–5350 [DOI] [PubMed] [Google Scholar]
- Nance J., Priess J. R., 2002. Cell polarity and gastrulation in C. elegans. Development. 129: 387–397 [DOI] [PubMed] [Google Scholar]
- Nance J., Lee J. Y., Goldstein B., 2005. Gastrulation in C. elegans, in WormBook, edited by The C. elegans Research Community. WormBook, doi/10.1895/wormbook.1.23.1, http://www.wormbook.org
- Newman S. A., Comper W. D., 1990. ‘Generic’ physical mechanisms of morphogenesis and pattern formation. Development 110: 1–18 [DOI] [PubMed] [Google Scholar]
- O’Rourke S. M., Dorfman M. D., Carter J. C., Bowerman B., 2007. Dynein modifiers in C. elegans: light chains suppress conditional heavy chain mutants. PLoS Genet. 3: e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbitts B. M., Ciotti M. K., Miller N. E., Kramer M., Lawrenson A. L., et al. , 2008. glo-3, a novel Caenorhabditis elegans gene, is required for lysosome-related organelle biogenesis. Genetics 180: 857–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson S. M., Shetty P., Lin R., 2004. Identification of lineage-specific zygotic transcripts in early Caenorhabditis elegans embryos. Dev. Biol. 276: 493–507 [DOI] [PubMed] [Google Scholar]
- Rocheleau C. E., Downs W. D., Lin R., Wittmann C., Bei Y., et al. , 1997. Wnt signaling and an APC-related gene specify endoderm in early C. elegans embryos. Cell 90: 707–716 [DOI] [PubMed] [Google Scholar]
- Rochlin K., Yu S., Roy S., Baylies M. K., 2010. Myoblast fusion: when it takes more to make one. Dev. Biol. 341: 66–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh-Johnson M., Goldstein B., 2009. In vivo roles for Arp2/3 in cortical actin organization during C. elegans gastrulation. J. Cell. Sci. 122: 3983–3993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrschneider M. R., Nance J., 2009. Polarity and cell fate specification in the control of Caenorhabditis elegans gastrulation. Dev. Dyn. 238: 789–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford S. L., 2000. From genotype to phenotype: buffering mechanisms and the storage of genetic information. Bioessays 22: 1095–1105 [DOI] [PubMed] [Google Scholar]
- Sawyer J. M., Harrell J. R., Shemer G., Sullivan-Brown J., Roh-Johnson M., et al. , 2010. Apical constriction: a cell shape change that can drive morphogenesis. Dev. Biol. 341: 5–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel H. S., Rockman M. V., Kruglyak L., 2008. Widespread genetic incompatibility in C. elegans maintained by balancing selection. Science 319: 589–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson A. F., Baillie D. L., Bowerman B., 2002. A Formin homology protein and a profilin are required for cytokinesis and Arp2/3-independent assembly of cortical microfilaments in C. elegans. Curr. Biol. 12: 2066–2075 [DOI] [PubMed] [Google Scholar]
- Shaham S., Reddien P. W., Davies B., Horvitz H. R., 1999. Mutational analysis of the Caenorhabditis elegans cell-death gene ced-3. Genetics 153: 1655–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starostina N. G., Lim J. M., Schvarzstein M., Wells L., Spence A. M., et al. , 2007. A CUL-2 ubiquitin ligase containing three FEM proteins degrades TRA-1 to regulate C. elegans sex determination. Dev. Cell 13: 127–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starostina N. G., Simpliciano J. M., McGuirk M. A., Kipreos E. T., 2010. CRL2(LRR-1) targets a CDK inhibitor for cell cycle control in C. elegans and actin-based motility regulation in human cells. Dev. Cell 19: 753–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starz-Gaiano M., Montell D. J., 2004. Genes that drive invasion and migration in Drosophila. Curr. Opin. Genet. Dev. 14: 86–91 [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Schierenberg E., White J. G., Thomson J. N., 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100: 64–119 [DOI] [PubMed] [Google Scholar]
- Szabo-Rogers H. L., Smithers L. E., Yakob W., Liu K. J., 2010. New directions in craniofacial morphogenesis. Dev. Biol. 341: 84–94 [DOI] [PubMed] [Google Scholar]
- Tenlen J. R., Molk J. N., London N., Page B. D., Priess J. R., 2008. MEX-5 asymmetry in one-cell C. elegans embryos requires PAR-4- and PAR-1-dependent phosphorylation. Development 135: 3665–3675 [DOI] [PubMed] [Google Scholar]
- Thomas C., DeVries P., Hardin J., White J., 1996. Four-dimensional imaging: computer visualization of 3D movements in living specimens. Science 273: 603–607 [DOI] [PubMed] [Google Scholar]
- Thorpe C. J., Schlesinger A., Carter J. C., Bowerman B., 1997. Wnt signaling polarizes an early C. elegans blastomere to distinguish endoderm from mesoderm. Cell 90: 695–705 [DOI] [PubMed] [Google Scholar]
- Timmons L., Fire A., 1998. Specific interference by ingested dsRNA. Nature 395: 854. [DOI] [PubMed] [Google Scholar]
- Uccelletti D., Pascoli A., Farina F., Alberti A., Mancini P., et al. , 2008. APY-1, a novel Caenorhabditis elegans apyrase involved in unfolded protein response signalling and stress responses. Mol. Biol. Cell 19: 1337–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S., Starostina N. G., Kipreos E. T., 2007. The Caenorhabditis elegans cell-cycle regulator ZYG-11 defines a conserved family of CUL-2 complex components. EMBO Rep. 8: 279–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieschaus E. F., 1997. From molecular patterns to morphogenesis: the lessons from Drosophila. In: Nobel Lectures in Physiology or Medicine 1991–1995, edited by Ringertz N., Vol. 7 World Scientific Publishing, Singapore [Google Scholar]
- Woollard A., 2005. Gene duplications and genetic redundancy in C. elegans. WormBook, edited by The C. elegans Research Community. WormBook, doi/10.1895/wormbook.1.2.1, http://www.wormbook.org
- Wu Y. C., Horvitz H. R., 1998. C. elegans phagocytosis and cell-migration protein CED-5 is similar to human DOCK180. Nature. 392: 501–504 [DOI] [PubMed] [Google Scholar]
- Zhu J., Hill R. J., Heid P. J., Fukuyama M., Sugimoto A., et al. , 1997. end-1 encodes an apparent GATA factor that specifies the endoderm precursor in Caenorhabditis elegans embryos. Genes Dev. 11: 2883–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Kirschner M., 2002. Regulated proteolysis of Xom mediates dorsoventral pattern formation during early Xenopus development. Dev. Cell. 3: 557–568 [DOI] [PubMed] [Google Scholar]
- Zohn I. E., Anderson K. V., Niswander L., 2005. Using genomewide mutagenesis screens to identify the genes required for neural tube closure in the mouse. Birth Defects Res. A Clin. Mol. Teratol. 73: 583–590 [DOI] [PubMed] [Google Scholar]