Abstract
In this article, I develop a methodology for inferring the transmission rate and reproductive value of an epidemic on the basis of genotype data from a sample of infected hosts. The epidemic is modeled by a birth–death process describing the transmission dynamics in combination with an infinite-allele model describing the evolution of alleles. I provide a recursive formulation for the probability of the allele frequencies in a sample of hosts and a Bayesian framework for estimating transmission rates and reproductive values on the basis of observed allele frequencies. Using the Bayesian method, I reanalyze tuberculosis data from the United States. I estimate a net transmission rate of 0.19/year [0.13, 0.24] and a reproductive value of 1.02 [1.01, 1.04]. I demonstrate that the allele frequency probability under the birth–death model does not follow the well-known Ewens’ sampling formula that holds under Kingman's coalescent.
PATHOGENS evolve rapidly due to a short generation time and a high mutation rate. As a consequence, new alleles arise regularly, and in a population of infected individuals, a variety of alleles are present. Assuming a model for the spread of the pathogen to new hosts and a model for the mutation of a pathogen allele allows the estimation of key epidemiological parameters for a pathogen based on the sampled alleles in an epidemic (Tanaka et al. 2006; Luciani et al. 2008, 2009).
In this article, I consider the infinite-allele model (IAM) for the evolution of alleles; the constant rate birth–death model (BDM) is assumed for the epidemic spread of the pathogen. Under the infinite-allele model, new alleles arise in a host with a constant mutation rate θ. If a new allele arises, it has not appeared before. This means that there is no convergent evolution. Each infected host is characterized by an allele type. Under the BDM, the alleles spread with a constant transmission (birth) rate λ to new hosts, and infected hosts recover or die with a constant death rate μ. Note that through an estimated birth rate λ and death rate μ, the net transmission rate (λ−μ) and the reproductive value (λ/μ) are determined.
Assuming the IAM together with the BDM, the net transmission rate and reproductive value for tuberculosis have been estimated on the basis of the allele frequencies of the IS6110 marker (Tanaka et al. 2006) using an approximate Bayesian computation (ABC) approach (Pritchard et al. 1999; Beaumont et al. 2002; Marjoram et al. 2003). Bayesian methods infer the posterior distribution of parameters, whereas ABC methods infer the approximate posterior distribution of parameters. The quality of the approximation depends crucially on the choice of summary statistics (unless the full data are used, which is usually not feasible), and the speed of obtaining the approximation depends on the speed of the required simulation tools. Given efficient simulation tools, ABC methods might be faster than Bayesian methods; however, this comes with a cost in accuracy.
Bayesian methods require the knowledge of the likelihood of the data (here the allele frequencies). In this article, I derive the likelihood of the sampled allele frequency under the BDM with the IAM. The allele frequency likelihood is calculated recursively for the pure-birth process; i.e., μ = 0. Under the BDM with death, I calculate the allele frequency likelihood conditioned on the underlying birth–death tree structure. Integrating over all possible trees yields the allele frequency probability. However, when estimating parameters using a Bayesian approach, the integration is not necessary, as both parameters and trees can be sampled from the posterior distribution directly.
Using the Bayesian approach, I reanalyze the tuberculosis data of Small et al. (1994). I obtain significantly lower estimates than Tanaka et al. (2006) for the net transmission rate (0.19/year vs. 0.69/year) and the reproductive value (1.02 vs. 3.4). This demonstrates that summary statistics employed by ABC often do not yield a sufficiently good approximation of the posterior distribution. The approach presented here has further the advantage over the previous ABC approach in that it is much faster as no simulations of large trees are required and incomplete sampling is included into the likelihood directly (instead of considering complete trees as a temporary step).
Models and Methods
Framework for estimating birth and death rates on the basis of allele frequency data
I first introduce some definitions and notations that are used throughout this article. The model for the spread of an allele is based on the BDM, which starts with a single host infected at time tor in the past. Over time each host dies (or recovers) with a constant rate μ and infects a new host with a constant rate λ. One allele within a host is tracked. This allele can mutate to a new allele that has not appeared previously with a constant rate θ (IAM).
Let the number of hosts being sampled from the population be n. The allele types of the sampled hosts are summarized in a vector of allele frequencies a = (a1, a2, …, an) ε ℕn as follows. The number of hosts that share each allele is counted. The number ai is the number of alleles that are shared by exactly i hosts. Note that if for all j > k we have aj = 0, we simply write a = (a1,…, ak). Note that n = ∑jjaj. Let the number of sampled allele types (clusters) be c = ∑jaj.
An example for a vector of allele frequencies is a = (2, 3, 1), meaning that n = 2 × 1 + 3 × 2 + 1 × 3 = 11 hosts are sampled, which carry in total c = 2 + 3 + 1 = 6 different alleles, say alleles A1, … , A6. Alleles A1 and A2 appear only in one host; alleles A3, A4, and A5 appear each in two hosts; and allele A6 appears in three hosts.
In the following, the probability of observing a in a given sample of size n, ℙ[a], is determined. This probability can be used to estimate the model parameters on the basis of the data a. Throughout this section we define ei as a unit vector; i.e., it is a vector of only zeros but a 1 at position i.
Pure-birth model
First, a special case of the BDM, namely a pure-birth model, i.e., μ = 0, is considered. Under this model, a recursion for the probability of an allele frequency a is derived.
Complete sampling
First assume that the whole population of infected hosts is sampled. This is of course almost never the case. However, using the results under complete sampling, the probability of allele frequencies under incomplete sampling is derived in the next section.
Theorem 1. The probability of observing the allele frequency a under the pure-birth process and complete sampling is
| (1) |
A proof of the Theorem is found in the Appendix. The probability ℙ[a] can be calculated recursively in the following way. For a, a′ ε ℕn, define a > a′ if (i) or if (ii) and for all j < k, but . This defines a total order on the state space ℕn with the minimum amin = (1). Since a > a + ej−1 − ej and a > a − e1 − ej−1 + ej for j > 1, and > defines a total order, ℙ[a] can be calculated recursively using Equation 1, with the initial value ℙ[1] = 1. The probability ℙ[a] depends not on both parameters θ, λ, but only on their ratio θ/λ. I did not find a closed-form solution for ℙ[a]. In particular, Ewens’ sampling formula is not the solution. I calculated the probability ℙ[a] via the recursion for up to five individuals; see Table 1.
TABLE 1.
Probability of a for a small number of individuals with θ/λ = 1
| No. taxa | a | Probability (fraction) | Probability (float) |
| 2 | 2, 0 | 1/2 | 0.5 |
| 0, 1 | 1/2 | 0.5 | |
| 3 | 3, 0, 0 | 3/10 | 0.3 |
| 1, 1, 0 | 9/20 | 0.45 | |
| 0, 0, 1 | 1/4 | 0.25 | |
| 4 | 4, 0, 0, 0 | 13/70 | 0.1857 |
| 2, 1, 0, 0 | 13/35 | 0.3714 | |
| 0, 2, 0, 0 | 3/40 | 0.075 | |
| 1, 0, 1, 0 | 17/70 | 0.2429 | |
| 0, 0, 0, 1 | 1/8 | 0.125 | |
| 5 | 5, 0, 0, 0, 0 | 293/2520 | 0.1163 |
| 3, 1, 0, 0, 0 | 293/1008 | 0.2907 | |
| 1, 2, 0, 0, 0 | 317/2520 | 0.1258 | |
| 2, 0, 1, 0, 0 | 1013/5040 | 0.2010 | |
| 0, 1, 1, 0, 0 | 19/280 | 0.0679 | |
| 1, 0, 0, 1, 0 | 137/1008 | 0.1359 | |
| 0, 0, 0, 0, 1 | 1/6 | 0.0625 |
I did not find a pattern in these numbers.
The probability ℙ[a] is the likelihood of the data, and therefore maximum-likelihood or Bayesian methods can be employed to estimate birth and death rates on the basis of allele frequencies.
Incomplete sampling
Now I consider the scenario that a population of size N evolved under the pure-birth process, and then n of these N individuals are sampled uniformly at random. The probability of obtaining the allele frequency a when sampling n individuals of N individuals is calculated recursively,
| (2) |
with ℙN[a] = ℙ[a] for
Note that Ewens’ sampling formula is invariant toward sampling; i.e., ℙN[a] = ℙn[a] for n ≠ N (see also Discussion). However, under the pure-birth model, inspection of Equations 1 and 2 reveals ℙN[a] ≠ ℙn[a] for n ≠ N.
Birth–death model
Introducing a death rate μ for each individual yields, analogous to that above,
The recursion cannot be evaluated as above, since there are states a′ on the right-hand side with a < a′.
One solution would be to introduce a cutoff, i.e., assign probability 0 to all states with N >> n individuals. N has to be chosen so large that the probability of getting back to the final stage with n individuals is very small. However, even with this cutoff, the above recursion becomes computationally very time consuming, in particular with incomplete sampling, as the underlying number of individuals N can be very large.
I therefore introduce a Bayesian approach to estimate the birth and death rates. The idea is based on deriving a closed-form solution for the probability ℙ[a|], where is the tree that leads to the observed data, the allele frequencies.
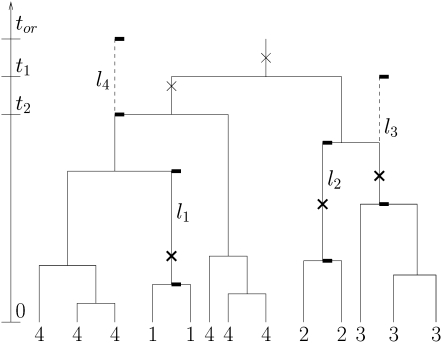
I first define formally. The BDM is producing a binary tree where the first infected host appeared at time tor ago. It is assumed that sampling of infected hosts is uniformly at random; i.e., each infected host at time tor (after the first infected host appeared) is sampled with probability ρ. All nonsampled and extinct lineages are suppressed from the tree. Let the tree induced in this way be T, and let the number of leaves be n. Mutations of the allele occur on the tree edges with constant rate θ. The tree T together with c − 1 edges where at least one mutation occurs such that the allele frequency a with c different alleles is induced is denoted by . The lengths of the c − 1 edges with mutations are l1, … , lc−1. The time from the origin of the process to the most recent common ancestor of the individuals that are not descendants of the c−1 edges with mutations is defined as lc. Note that mutations during this time lc do not change the allele frequency a. An example of a tree is shown in Figure 1.
Figure 1.—
A transmission tree induced under the constant rate birth-death model (BDM) as a model for transmission. Mutations of an allele are modeled using the infinite-allele model (IAM). The four allele types of the leaves are labeled by 1−4, and the allele frequency is a = (0, 2, 10, 0, 1). The root edge lengths l1, … , l4 of the four clusters are included. Note that at least one mutation must occur on the root edges of clusters 1−3 (marked as an x in boldface type). Mutations on the root edge of cluster 4 may or may not occur (x in regular type). The time of origin tor and the two oldest branching times t1, t2 are marked; the 3rd through 12th branching times are not marked for easier readability.
First note that ℙ[λ, μ, θ, , tor|a] = ℙ[λ, μ, θ, |a] as tor is specified by . With Bayes’ theorem, we can write
which is proportional to
since ℙ[a] is a normalizing constant. The probability ℙ[λ, μ, θ, tor] is a prior on λ, μ, θ, tor. We determine the quantity ℙ[|λ, μ, θ, tor] in the following.
Theorem 2. The probability density ℙ[|λ, μ, θ, tor] is
| (3) |
where t1, … , tn−1 are the branching times in , and
Proof. To derive ℙ[|λ, μ, θ, tor], we split the tree with c different alleles into c subtrees in the following way. We sequentially choose an edge with a mutation that has no mutated descending edge, and define this edge with all its descendants as a subtree, and delete this subtree from . After c − 1 subtrees are removed from in this way, the resulting cth subtree of has age tor and the most ancient edge may or may not have a mutation, while all other edges do not have a mutation.
In each of the subtrees, no edge descending from the first diversification event (the root) has a mutation. In the first c −1 subtrees, the edge above the root (root edge) has at least one mutation. In the cth subtree, a mutation might or might have not happened above the root (a mutation simply means that the ancestor allele is lost in the sample).
The probability density of a tree T with n leaves and bifurcation times t1, … , tn−1 given the age t0 is
(Stadler 2010). At least one mutation is required on the root edges of c − 1 subtrees, and the probability of observing a mutation on the first c − 1 root edges is
The sum of edge lengths where no mutation is allowed to occur is , and the probability that no mutation occurs during this time is
Therefore, the probability density of a tree is
Equation 3 allows us to infer the posterior distribution for λ, μ, which is done in the next section for tuberculosis data.
Application of the Bayesian approach to tuberculosis data
I implemented a Markov chain Monte Carlo (MCMC) approach using the Metropolis–Hastings algorithm (Metropolis et al. 1953; Hastings 1970) to sample from the posterior distribution
ℙ[|λ, μ, θ, tor] is provided in Equation 3. ℙ[λ, μ, θ, tor] is the prior distribution. I assume a uniform prior for the net diversification rate λ − μ on [0.01, 10] per year, a uniform prior for μ/λ on [0, 1], and a uniform prior for tor on [0, 100] years.
I fixed θ = 0.198. This is the major difference in prior assumptions compared to Tanaka et al. (2006). In the previous study, the prior for θ was a normal distribution with mean 0.198 and standard deviation 0.06735. However, under an IAM in combination with the BDM, the parameters λ, μ, tor, θ give rise to the same process as the time-scaled parameters λ/s, μ/s, tors, θ/s (recall that under the pure-birth process we already observed the invariance of the likelihood for θ/λ being constant). For example, if the original parameters were in units of years, the scaled parameters with s = 365 are in units of days. I provide all estimates in units of years, assuming θ = 0.198. For a different estimate of θ, the values can then be transformed to new variables using s = 0.198/θ. This scaling in parameters is also apparent in Tanaka et al. (2006): Figure 3 shows the net transmission rate for varying θ priors. The peak of the net transmission rate estimates correlates linearly with the mean θ.
There are two minor differences to Tanaka et al. (2006): First, in Tanaka et al. (2006), the priors were uniform for λ, μ on [0, ∞] with λ > μ. Note that, since there is a one-to-one mapping (bijection) between (λ, μ) and (λ − μ, μ/λ), the priors in Tanaka et al. (2006) are equivalent to uniform priors for λ − μ on [0, ∞] and μ/λ on [0, 1]. Since in my analysis and in Tanaka et al. (2006) the estimates for λ − μ are at least 10-fold smaller than the upper bound 10, the different upper bounds do not bias the posterior samples. Second, the tree in the previous study was stopped when a fixed number N of infected was reached, and from this tree the observed number n was sampled. I assume each individual is sampled with probability n/N from the big tree and condition on the number of sampled individuals being n. I cannot obtain a likelihood function accounting for the sampling procedure in Tanaka et al. (2006); however, my sampling procedure introduced only some random noise to the original tree size N, which should not bias the posterior distribution.
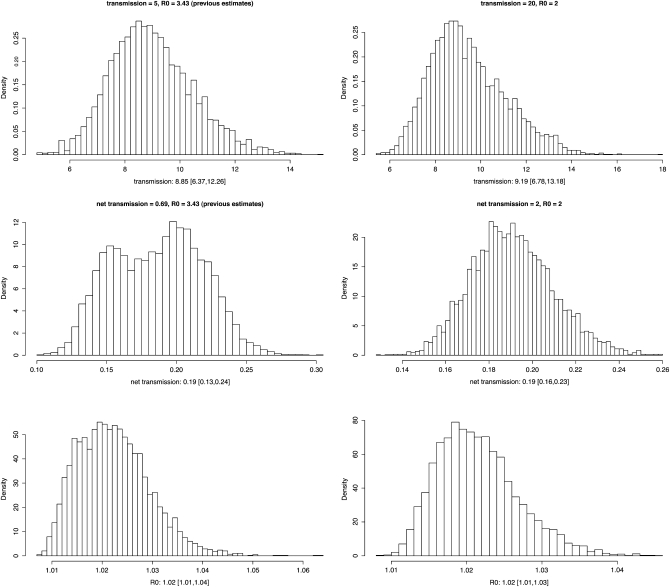
The MCMC chain after 8 million steps and neglecting the first 25% as burn-in returned a median net transmission rate (λ − μ) of 0.19/year with 95% credible interval [0.13, 0.24] and a reproductive value R (λ/μ) of 1.02 [1.01, 1.04]; for the posterior distribution see Figure 2 (left). The initial state was chosen to be the estimates from the previous study based on an ABC approach (Tanaka et al. 2006) (0.69 for net transmission rate, 3.4 for R). A further run of the MCMC with the initial state λ = 4 and μ = 2 yielded the same posterior distributions; see Figure 2 (right).
Figure 2.—
Posterior distribution of the transmission rate λ (unit: per year), the net transmission rate λ − μ (unit: per year) and reproductive value λ/μ for tuberculosis allele frequency data obtained through MCMC chains with different starting values. The numbers below each histogram are the median and those in brackets are the lower and upper 95% credible intervals.
The source code in R is available from the author on request.
Discussion
My estimate of the net transmission rate of tuberculosis is ∼3.5 times lower and the reproductive value is ∼3 times lower than the previous estimate based on the same data (Tanaka et al. 2006). The presented estimates challenge the statement of Tanaka et al. (2006, p. 1518), saying that “the genetic information (as interpreted with the methods in this study) supports a faster spread of tuberculosis, at least for the data of Small et al. (1994).” As Tanaka et al. (2006) used the same data, the same model assumption, and basically the same prior distributions as I used in the present study, the difference must come from the method. I obtained the same posterior distribution for different starting values; therefore I claim that the differences in my estimates from the previous estimates are due to the approximation of the ABC not being accurate enough. The quality of the approximation in an ABC analysis depends crucially on the summary statistics used, but unfortunately there is no straightforward way to determine whether a summary statistic is good. Clearly, for a given sample, the allele frequency a is a sufficient statistic (as the likelihood depends only on a); however, the Bayesian approach in this article revealed that the one-dimensional summary statistic H = 1 − ∑jaj(j/n)2 (Tanaka et al. 2006) is not sufficient.
My net transmission rate estimate is slightly low compared to the net transmission rate for tuberculosis estimated in Porco and Blower (1998) (0.231−0.693). That study developed a detailed model of tuberculosis transmission and obtained an estimate of the net transmission rate through previous estimates in the literature of other tuberculosis parameters like the number of infections per year and the progression rate to tuberculosis. The net transmission rate correlates linearly with the mutation rate. I assumed a mutation rate of 0.198/year following Tanaka et al. (2006). However, the estimates in the literature vary and assuming a mutation rate that is 50% larger (as estimated, e.g., in Rosenberg et al. 2003) yields a net transmission rate interval that largely overlaps with the interval of Porco and Blower (1998).
The estimated reproductive value is close to 1 (1.02); i.e., each infected individual infects only one further individual in expectation. This low number is likely due to the fact that the tuberculosis epidemic is in the equilibrium phase (after an initial exponential expansion). This means that we estimated the actual reproductive number (Amundsen et al. 2004), which should not be confused with the basic reproductive number (Anderson and May 1979, 1992). The basic reproductive number quantifies the expected number of individuals infected by a single infected individual in a fully susceptible population, while the actual reproductive number accounts for the fact that a fraction of the population is infected. Estimation of the basic reproductive number using a BDM requires knowledge of the early phase of the epidemic.
In classic population genetics, the spread of an allele is modeled with Kingman's coalescent under a constant population size (Kingman 1982a,b,c) instead of the BDM. The probability of a sampled allele frequency is Ewens’ sampling formula (Ewens 1972),
| (4) |
This formula can be used to estimate the mutation rate θ given an allele frequency a. A second scenario gives rise to Ewens’ sampling formula. Under the pure-birth process where new alleles are introduced via a constant migration rate η (instead of mutation), the allele frequency probability is also Ewens’ sampling formula (Joyce and Tavaré 1987) with parameter η/λ instead of θ (an extension that also includes death is discussed in Tavaré 1989 and Rannala 1996). Ewens’ sampling formula can be derived under both scenarios analogous to the recursive approach introduced in this article for the pure-birth model and mutation; the derivations are given in the Appendix. One convenient property of Ewens’ sampling formula is that if a subsample is chosen uniformly at random from a sample of alleles that are distributed according to Ewens’ sampling formula, the subsample again is distributed according to Ewens’ sampling formula.
Unfortunately, we cannot make use of the convenient properties of Ewens’ sampling formula in the framework of epidemiology: We cannot assume the coalescent since transmission and death rates are not parameterized and thus cannot be estimated (the coalescent captures only the population size). The BDM describes the spread of an allele in an epidemic by interpreting birth rates as transmission rates. In the context of epidemiology, new alleles evolve in hosts within a population; thus I chose the BDM with the IAM over the previously studied BDM with immigration.
An analog of Ewens’ sampling formula for the BDM in combination with the IAM is
with ℙ[] from Equation 3 and ℙ[tor] being a prior distribution for tor. An analytic integration for this expression seems not feasible (a hyperbolic function is involved), and therefore maximum-likelihood estimation of the birth and death rates or the mutation rate is not straightforward.
However, as I demonstrate, a Bayesian framework, which avoids the explicit integration, performs well. In particular, the Bayesian approach incorporates a sampling probability and therefore avoids considering trees that are larger than the actual sample size. This makes the approach attractive for sparsely sampled data, which is common in infectious diseases.
Using the BDM for the spread of tuberculosis, or any other epidemic, is the simplest epidemiological model and is appropriate when the epidemic is in the initial exponential phase (meaning λ >> μ) or in the equilibrium phase that is reached due to a reduced number of susceptibles (meaning λ ≈ μ). To model both phases simultaneously, SIR (susceptible-infected-recovered) models (Keeling and Rohani 2008) accounting for a declining number of susceptible individuals over time are required. While these models are well studied in a deterministic framework, they are not well understood in a probabilistic framework. However, a probabilistic formulation is required for a Bayesian analysis.
Acknowledgments
I thank the editor and two anonymous reviewers for very helpful comments.
APPENDIX
Proof of Theorem 1
Proof. Let B be the event that the ancestor state of a is followed by a bifurcation (i.e., transmission) event. Let M be the event that the ancestor state of a is followed by a mutation event. Let a′ be the allele frequency state before undergoing the most recent event that yields a. [For example, if a = (2, 3, 1) from above, and the most recent event is bifurcation, then a′ = (3, 2, 1) or a′ = (2, 4). If the most recent event is mutation, then a′ = (2, 3, 1) or a′ = (0, 4, 1) or a′ = (1, 2, 2) or a′ = (1, 3, 0, 1).] With these definitions, we obtain
We have
Assume a′ is followed by a bifurcation event. Let the bifurcation event be in a component of size j − 1, j = 2, … , n. Then,
Now assume a′ is followed by a mutation event. Let the mutation event be in a component of size j, j = 2, … , n. Then,
Let the mutation event be in a component of size 1. Then,
Together,
Therefore,
Derivation of Ewens’ Sampling Formula for the Coalescent With Mutation
We now determine the probability of the sampled allele frequency a under the coalescent. Let a′ be the state one event ancestral to a. Let B be the event that the present state evolved following a bifurcation event. Let M be the event that the present state evolved following a mutation event. Then,
Note that
Assume a evolved after a bifurcation event. Let the bifurcation event be in a component of size j − 1, j = 2, … , n. Then,
Now assume a evolved after a mutation event. Let the mutation event be in a component of size j, j = 2, … , n. Then,
Let the mutation event be in a component of size 1. Then,
Together,
Therefore,
Since a > a + ej−1 − ej and a > a − ej−1 + ej and > defines a total order with minimum (1) (as explained in the main text), we can calculate ℙ[a] recursively, with the initial value ℙ[(1)] = 1. The solution of this recursion is Ewens’ sampling formula (Equation 4 of main text), which can be easily proved by induction.
Derivation of Ewens' Sampling Formula for the Pure-Birth Process With Migration
We again have a pure-birth process for the population dynamics. Novel alleles migrate at a constant rate η. We assume that we sample the whole population. Since Ewens’ sampling formula is invariant to random sampling, the derivation of Ewens’ sampling formula for the whole population implies that also a subsample is distributed according to Ewens’ sampling formula.
Let M be the event that a′ is followed by a migration event. With the other notation as introduced above, we have again
We have
Assume a′ is followed by a bifurcation event. Let the bifurcation event be in a component of size j − 1, j = 2, … , n. Then,
Now assume a′ is followed by a migration event. Then,
Together,
Again, since a > a + ej−1 − ej and a > a − e1 − ej−1 + ej and > defines a total order with minimum (1), we can calculate ℙ[a] recursively, with the initial value ℙ[(1)] = 1. Here, the solution of the recursion is again Ewens’ sampling formula (Equation 4 of main text), which can be proved by a simple induction.
Literature Cited
- Amundsen E., Stigum H., Roettingen J., Aalen O., 2004. Definition and estimation of an actual reproduction number describing past infectious disease transmission: application to HIV epidemics among homosexual men in Denmark, Norway and Sweden. Epidemiol. Infect. 132: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R., May R., 1979. Population biology of infectious diseases: Part I. Nature 280: 361–367 [DOI] [PubMed] [Google Scholar]
- Anderson R., May R., 1992. Infectious Diseases of Humans: Dynamics and Control. Oxford University Press, New York [Google Scholar]
- Beaumont M., Zhang W., Balding D., 2002. Approximate Bayesian computation in population genetics. Genetics 162: 2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewens W., 1972. The sampling theory of selectively neutral alleles. Theor. Popul. Biol. 3: 87–112 [DOI] [PubMed] [Google Scholar]
- Hastings W., 1970. Monte Carlo sampling methods using Markov chains and their applications. Biometrika 57: 97 [Google Scholar]
- Joyce P., Tavaré S., 1987. Cycles, permutations and the structure of the Yule process with immigration. Stoch. Proc. Appl. 25: 309–314 [Google Scholar]
- Keeling M., Rohani P., 2008. Modeling Infectious Diseases in Humans and Animals. Princeton University Press, Princeton, NJ [Google Scholar]
- Kingman J. F. C., 1982a. The coalescent. Stoch. Proc. Appl. 13: 235–248 [Google Scholar]
- Kingman J. F. C., 1982b. Exchangeability and the evolution of large populations, pp. 97–112 in Exchangeability in Probability and Statistics, edited by G. Koch and F. Spizzichino North-Holland Publishing, Amsterdam [Google Scholar]
- Kingman J. F. C., 1982c. On the genealogy of large populations. J. Appl. Probab. 19A: 27–43 [Google Scholar]
- Luciani F., Francis A., Tanaka M., 2008. Interpreting genotype cluster sizes of Mycobacterium tuberculosis isolates typed with IS6110 and spoligotyping. Infect. Genet. Evol. 8: 182–190 [DOI] [PubMed] [Google Scholar]
- Luciani F., Sisson S., Jiang H., Francis A., Tanaka M., 2009. The epidemiological fitness cost of drug resistance in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 106: 14711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marjoram P., Molitor J., Plagnol V., Tavaré S., 2003. Markov chain Monte Carlo without likelihoods. Proc. Natl. Acad. Sci. USA 100: 15324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metropolis N., Rosenbluth A., Rosenbluth M., Teller A., Teller E., et al. , 1953. Equation of state calculations by fast computing machines. J. Chem. Phys. 21: 1087 [Google Scholar]
- Porco T., Blower S., 1998. Quantifying the intrinsic transmission dynamics of tuberculosis. Theor. Popul. Biol. 54: 117–132 [DOI] [PubMed] [Google Scholar]
- Pritchard J., Seielstad M., Perez-Lezaun A., Feldman M., 1999. Population growth of human Y chromosomes: a study of Y chromosome microsatellites. Mol. Biol. Evol. 16: 1791. [DOI] [PubMed] [Google Scholar]
- Rannala B., 1996. The sampling theory of neutral alleles in an island population of fluctuating size. Theor. Popul. Biol. 50: 91. [DOI] [PubMed] [Google Scholar]
- Rosenberg N., Tsolaki A., Tanaka M., 2003. Estimating change rates of genetic markers using serial samples: applications to the transposon IS6110 in Mycobacterium tuberculosis. Theor. Popul. Biol. 63: 347–363 [DOI] [PubMed] [Google Scholar]
- Small P., Hopewell P., Singh S., Paz A., Parsonnet J., et al. , 1994. The epidemiology of tuberculosis in San Francisco—a population-based study using conventional and molecular methods. N. Engl. J. Med. 330: 1703. [DOI] [PubMed] [Google Scholar]
- Stadler T., 2010. Sampling-through-time in birth-death trees. J. Theor. Biol. 267: 396–404 [DOI] [PubMed] [Google Scholar]
- Tanaka M., Francis A., Luciani F., Sisson S., 2006. Using approximate Bayesian computation to estimate tuberculosis transmission parameters from genotype data. Genetics 173: 1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavaré S., 1989. The genealogy of the birth, death, and immigration process. Math. Evol. Theory 41: 56 [Google Scholar]