Abstract
Functional diversification across the left/right axis is a common feature of many nervous systems. The genetic programs that control left/right asymmetric neuron function and gene expression in the nervous system are, however, poorly understood. We describe here the molecular characterization of two phenotypically similar mutant Caenorhabditis elegans strains in which left/right asymmetric gene expression programs of two gustatory neurons, called ASEL and ASER, are disrupted such that the differentiation program of the ASER neuron is derepressed in the ASEL neuron. We show that in one mutant strain the LIM homeobox gene lim-6 is defective whereas in another strain a novel member of a nematode-specific, fast-evolving family of C2H2 zinc-finger transcription factors, lsy-27, is mutated, as revealed by whole-genome sequencing. lsy-27 is broadly and exclusively expressed in the embryo and acts during the initiation, but not during the maintenance phase of ASE asymmetry control to assist in the initiation of lim-6 expression.
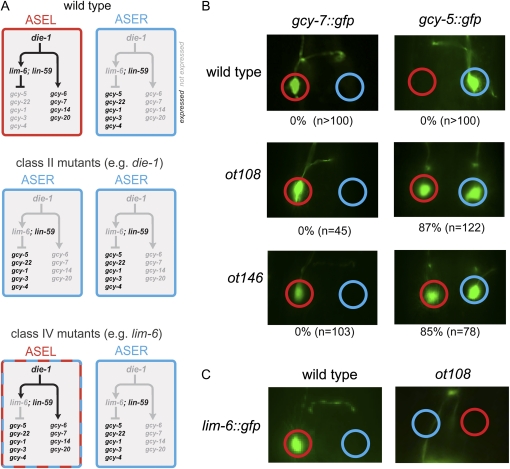
LEFT/RIGHT asymmetric gene expression patterns in the nervous system of invertebrate and vertebrates species have been described and are generally thought to be the foundation of the striking functional lateralization of many nervous systems (Hobert et al. 2002; Sun et al. 2005; Sun and Walsh 2006; Taylor et al. 2010). Yet it is not well understood how left/right gene expression patterns are regulated. In the nematode Caenorhabditis elegans, a class of putative chemoreceptors of the GCY family are expressed in a left/right asymmetric manner in a bilateral pair of functionally lateralized gustatory neurons, called ASEL and ASER (Yu et al. 1997; Ortiz et al. 2006). These gcy genes are required for the left/right asymmetric processing of chemosensory information by the two ASE neurons (Ortiz et al. 2009). Genetic mutant screens have revealed a number of genes (called “lsy genes” for laterally symmetric) that control the left/right asymmetric expression of gcy genes (Sarin et al. 2007). Phenotypic analysis of these mutants has revealed several distinct types of asymmetry mutants. In class I mutants, the gcy expression profile of the ASER neuron completely converts to that of the ASEL neuron (“2 ASEL” mutants). In class II mutants, the opposite occurs (“2 ASER” mutants; e.g., die-1 as shown in Figure 1A). In class III mutants, both ASEL and ASER gcy receptors are lost. In class IV mutants, the ASER-specific gcy genes are derepressed in ASEL, but the ASEL-specific gcy genes remain unaffected; or vice versa, ASEL-specific gcy genes are derepressed in ASER, but ASER-specific gcy genes remain unaffected (Sarin et al. 2007). Either the ASEL or ASER neurons therefore exist in a “mixed” state in class IV mutants (Figure 1A). Due to their more limited phenotypic effects, class IV genes would be expected to work downstream of class I and class II genes, and indeed, the analysis of the expression of class IV genes in class I or II mutant backgrounds confirmed this notion (Johnston et al. 2005, 2006) (Figure 1A).
Figure 1.—
lsy genes and mutant phenotypes. (A) A simplified version of the genetic pathway that controls left/right asymmetry in the ASE neurons. Loss of die-1, a Zn-finger transcription factor, results in a class II Lsy phenotype (in which ASEL fate markers are lost and ASER fate markers are gained in ASEL), and loss of lim-6, a LIM homeobox gene, results in a class IV Lsy phenotype (in which ASER fate markers are gained in ASEL, but ASEL fate markers unaffected) (Hobert et al. 1999; Chang et al. 2004). Loss of broadly expressed lin-59, a histone methyltransferase, also results in a class IV Lsy phenotype (Sarin et al. 2007, 2010). (B) Effect of lsy-27(ot108) and lim-6(ot146) mutant alleles on ASEL/ASER asymmetry markers. otIs3(gcy-7::gfp) labels ASEL and ntIs1(gcy-5::gfp) labels ASER. The phenotype is quantified in Table 1. (C) ot108 also affects lim-6::gfp (otIs114) expression. In 57.4% of animals, lim-6::gfp fails to be expressed, and in 27.7% of animals, expression is visible but weaker than in wild type (n = 47).
Class IV genes are essential for the appropriate function of the ASE neurons. This was first demonstrated through a detailed phenotypic analysis of animals that lack the ASEL-expressed lim-6 LIM homeobox gene and that therefore display a class IV phenotype in which ASEL-expressed gcy genes are unaffected, but ASER-expressed gcy genes are derepressed in ASEL (Figure 1A) (Hobert et al. 1999). Such mutant animals are unable to discriminate between ASEL- and ASER-sensed chemosensory cues (Pierce-Shimomura et al. 2001).
lim-6 is not the only gene with such a function. Three mutants retrieved from a previous large-scale mutagenesis screen for the asymmetry mutants ot104, ot108, and ot146 (Sarin et al. 2007) display a phenotype similar to lim-6 (Figure 1B and Table 1). ot104 was found to be an allele of the ubiquitously expressed ASH1-type histone methyltransferase lin-59 (Sarin et al. 2010), but the ot108 and ot146 alleles had not previously been molecularly characterized. We present their characterization in this Note.
Table 1.
Lsy phenotypes of lim-6 and lsy-27
| % animals with the following phenotypes (at 25°): |
||||||||
| ASEL only (%) | ASEL > ASER (%) | No expression (%) | ASEL = ASER (%) | ASEL < ASER (%) | ASER only (%) | n | % Lsy | |
| ASEL marker (gcy-7::gfp; otIs3) | ||||||||
| Wild type | 100 | 0 | 0 | 0 | 0 | 0 | >100 | 0 |
| lim-6(nr2073) | 100 | 0 | 0 | 0 | 0 | 0 | 35 | 0 |
| lim-6(ot146) | 100 | 0 | 0 | 0 | 0 | 0 | 103 | 0 |
| lsy-27(ot108) | 100 | 0 | 0 | 0 | 0 | 0 | 45 | 0 |
| lsy-27(tm593) | 100 | 0 | 0 | 0 | 0 | 0 | 66 | 0 |
| ASER marker (gcy-5::gfp; ntIs1) | ||||||||
| Wild type | 0 | 0 | 0 | 0 | 0 | 100 | >100 | 0 |
| lim-6(nr2073) | 0 | 0 | 0 | 89 | 5 | 6 | 82 | 94 |
| lim-6(ot146) | 0 | 0 | 0 | 8 | 77 | 15 | 78 | 85 |
| ot146/nr2073 | 0 | 0 | 0 | 0 | 56 | 44 | 50 | 56 |
| lsy-27(ot108) | 0 | 0 | 0 | 39 | 48 | 13 | 122 | 87 |
| lsy-27(tm593) | 0 | 0 | 0 | 0 | 62 | 38 | 117 | 62 |
| ot108/tm593 | 0 | 0 | 0 | 3 | 6 | 91 | 31 | 9 |
| ot108/+ | 0 | 0 | 0 | 0 | 0 | 100 | 56 | 0 |
ot146 is an allele of the LIM homeobox gene lim-6
ot146 mutant animals are viable and fertile and display no obvious morphological abnormalities. Their class IV Lsy phenotype is recessive. Due to its failure to complement what turned out to be a very unusual allele, called ot101, of the zinc (Zn)-finger transcription factor che-1, a terminal selector of ASEL and ASER neuron fate (Etchberger et al. 2009), we had assumed that ot146 was located on chromosome I, where che-1 is located (Sarin et al. 2007). However, subsequent mapping placed ot146 on chromosome X, where the lim-6 locus resides. We find that ot146 contains a C83Y change in the second LIM domain of lim-6 (supporting information, Figure S1). The mutated cysteine residue is 100% conserved in all LIM domains and is essential for the structural integrity of a LIM domain through the coordination of a Zn ion (Kadrmas and Beckerle 2004). The ot146 allele fails to complement the lim-6 null allele nr2073, and its Lsy phenotype is rescued by a genomic piece of DNA that contains the lim-6 locus (Table 2). We conclude that ot146 is an allele of lim-6. This is the first lim-6 allele retrieved from our mutant screen [the only previously characterized lim-6 allele, nr2073, is a reverse engineered allele (Hobert et al. 1999)].
Table 2.
Transformation rescue and RNAi analysis
| Genotype | Lsy phenotypea (%) | Wild-type phenotype (%) | n |
| Wild type | 0 | 100 | >100 |
| lim-6(ot146) | 85 | 15 | 78 |
| ot146; otEx3859 (Ex[lim-6 fosmid::yfp; rol-6(d)]) | 0 | 100 | 41 |
| lsy-27(ot108) | 86.9 | 13.1 | 122 |
| lsy-27(ot108); lsy-27(RNAi) | 2.5 | 97.5 | 200 |
| lsy-27(ot108); empty vector (RNAi) | 86.8 | 13.2 | 111 |
| lsy-27(RNAi) | 0 | 100 | 71 |
| lsy-27(ot108); Ex[lsy-27transl::gfp], line #1b | 18.2 | 81.8 | 44 |
| lsy-27(ot108); Ex[lsy-27transl::gfp], line #2 | 17.2 | 82.8 | 87 |
| lsy-27(ot108); Ex[fosmid], line #1 | 5.6 | 94.4 | 18 |
| lsy-27(ot108); Ex[fosmid], line #2 | 9.1 | 90.9 | 44 |
| lsy-27(ot108); Ex[lsy-27fosmid::yfp], line #1b | 0 | 100 | 70 |
| Genotype as above but array not transmitted from parental generationc | 0 | 100 | 11 |
| lsy-27(ot108); Ex[lsy-27fosmid::yfp], line #2 | 0 | 100 | 54 |
| Genotype as above but array not transmitted from parental generation | 21.1 | 78.9 | 19 |
The ot108 and ot146 control data are repeated from Table 1 for comparison purposes. RNAi experiments were done by feeding, using standard protocols with a double-stranded RNA clone obtained from Geneservice.
Scored as a gcy-5 reporter (ntIs1 or otIs220) derepressed in ASEL in first eleven rows or loss of lim-6::gfp (otIs114) in remaining four rows.
All expression constructs are shown in Figure 2A. See File S1 for details on the generation of the reporter constructs.
Arrays contain the elt-2::gfp injection marker. Animals derived from elt-2::gfp(+) parents that have lost this array as assessed by lack of intestinal gfp expression were scored.
ot108 affects a member of a C2H2 Zn-finger protein family
Like lim-6 mutant animals, ot108 mutant animals show derepression of the ASER marker gcy-5 in ASEL, while gcy-7 expression in ASEL is unaffected (Figure 1B and Table 1). Other than the Lsy phenotype, ot108 mutants animals are viable and fertile and display no obvious morphological abnormalities. Aside from the effect of ot108 on gcy-5 expression, ot108 animals also show a significant loss of lim-6 expression in ASEL, thereby providing an explanation of the lim-6-like phenotype of ot108 mutant animals (Figure 1C).
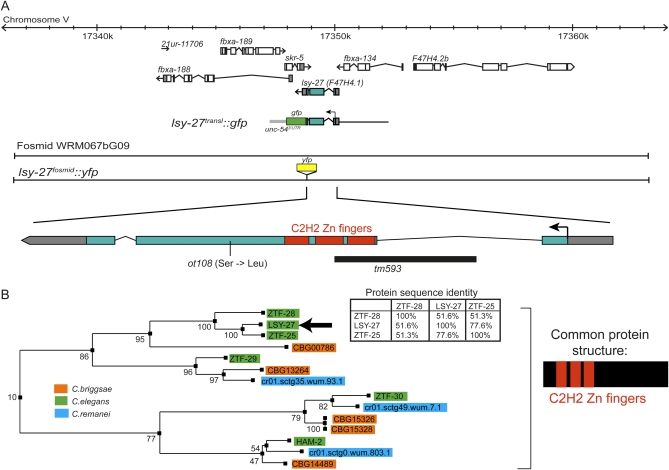
Upon isolation of ot108 mutant animals in our original Lsy screen (Sarin et al. 2007), we noted that ot108 fails to complement the derepression of ASER fate in the ASEL phenotype of a mutation in the die-1 Zn-finger transcription factor, an inducer of lim-6 expression in ASEL (a class II gene that also results in the loss of ASEL fate) (Figure 1A). Due to this lack of complementation, we had therefore initially considered ot108 to be an allele of die-1 (Sarin et al. 2007). However, our subsequent analysis revealed no mutation in the die-1 locus of ot108 mutant animals and, moreover, the ot108 mutant phenotype could not be rescued with a genomic piece of DNA that rescues a canonical die-1 allele (data not shown). Subsequent chromosomal linkage analysis showed that ot108 is linked to chromosome V, while die-1 maps to chromosome II. After mapping ot108 to the right arm of chromosome V using conventional SNP mapping (Wicks et al. 2001), we subjected the strain to whole-genome sequencing using an Illumina GAII genome analyzer (Sarin et al. 2008) and analyzed the data with MAQGene (Bigelow et al. 2009). Sequencing parameters and results are summarized in Table S1. In brief, within the genetically defined interval, we detected 22 sequence variants predicted to affect protein-coding genes (missense, non-sense or splice-site mutations). Nineteen of these variants were found in other whole-genome sequencing data sets that our lab has generated and were therefore considered background variants, leaving three protein-coding alterations. One of these alterations is a Ser-to-Leu change in the predicted C2H2 Zn-finger transcription factor F47H4.1 (Figure 2A and Figure S2). F47H4.1 is a member of C2H2 Zn-finger transcription factors with several paralogs in Caenorhabditis elegans and orthologs in other nematode species, but no apparent orthologs outside nematodes (Figure 2B and Figure S2). All members of this family contain three closely clustered C2H2 Zn fingers at the N terminus of the protein, but no other recognizable domains. The serine residue that is mutated in ot108 is phylogenetically conserved (Figure S2). The only gene in this family that had been previously characterized is the ham-2 transcription factor, which is involved in C. elegans HSN motor neuron specification (Baum et al. 1999).
Figure 2.—
lsy-27 is a new C2H2 Zn-finger protein. (A) Genomic position of lsy-27 and rescue and reporter gene constructs. See File S1 for details on the generation of the reporter constructs. (B) The lsy-27 gene family [modified from the TF317235 family tree generated by Treefam (http://www.treefam.org)] (Li et al. 2006).
Both a fosmid spanning the entire F47H4.1 locus plus neighboring genes and a genomic piece of DNA containing 2.6 kb upstream of F47H4.1 and the F47H4.1 locus (Figure 2A) rescue the ot108 mutant phenotype (Table 2). Animals carrying a deletion allele of F47H4.1, tm593 (kindly provided by the C. elegans knockout facility at Tokyo Women's Medical University School of Medicine) (Figure 2A), also display a class IV Lsy phenotype (Table 1). Also, like ot108 animals, tm593 animals are viable and fertile and display no obvious morphological abnormalities. Taken together, we conclude that it is the mutation in F47H4.1 that results in the class IV Lsy phenotype of ot108 mutant animals, and we therefore called this gene lsy-27 (Table S3 shows an updated numbering of lsy genes).
ot108 is an altered function allele
The tm593 deletion allele is a molecular null, as confirmed by RT-PCR analysis, which revealed that only very short (<37 amino acids), truncated forms of the protein are generated in tm593 animals, which do not contain any of the DNA-binding Zn-fingers (see File S1). We were therefore surprised to note that the Lsy phenotype of the tm593 deletion allele is notably milder than the ot108 missense allele in terms of both expressivity and penetrance (Table 1). We therefore considered the possibility that ot108 (which is recessive) is an altered function allele (Table 1). We tested this possibility by removing lsy-27 gene activity in ot108 mutant animals using RNA interference (RNAi) directed against lsy-27. We found that RNAi treatment completely reverted the ot108 phenotype (Table 2), suggesting that it is indeed altered lsy-27 function that explains the ot108 phenotype.
We noted that animals that carry one copy of the ot108 allele and one copy of the tm593 allele display a phenotype that is even milder than the phenotype of either allele alone (Table 1). One copy of the ot108 allele alone is therefore not enough to induce the altered function activity, but perhaps may be enough to provide some wild-type gene activity, thereby alleviating the tm593 phenotype. The need for sufficient ot108 dosage is also illustrated by the fact that the phenotype of ot108 mutant animals can be rescued through supplying wild-type copies of the locus (Table 1).
We considered the possibility that the complete removal of lsy-27 in tm593 animals may be mostly compensated for by lsy-27 paralogs, while the ot108 allele may interfere with the compensatory function of the paralogues. Through the use of deletion alleles of these loci (again kindly provided by the C. elegans knockout facility in Tokyo), we found that neither of the two most closely related lsy-27 paralogs, ztf-25 or ztf-28, either alone or in combination (i.e., ztf-25ztf-28 double nulls) displayed a Lsy phenotype (Table S2). ztf-28lsy-27 double-null mutant animals also display no Lsy phenotype. ztf-25lsy-27 double mutants could not be built due to close linkage of the two loci, and we therefore needed to resort to RNAi. lsy-27 RNAi in a ztf-28ztf-25 double-mutant background also did not result in a Lsy phenotype, but we note that even though lsy-27 RNAi does suppress the ot108 Lsy phenotype, it does not recapitulate the lsy-27(tm593) phenotype (Table 2), thereby allowing no firm conclusion about a triple loss of function of all three lsy-27 paralogs.
Expression pattern and timing of action of lsy-27
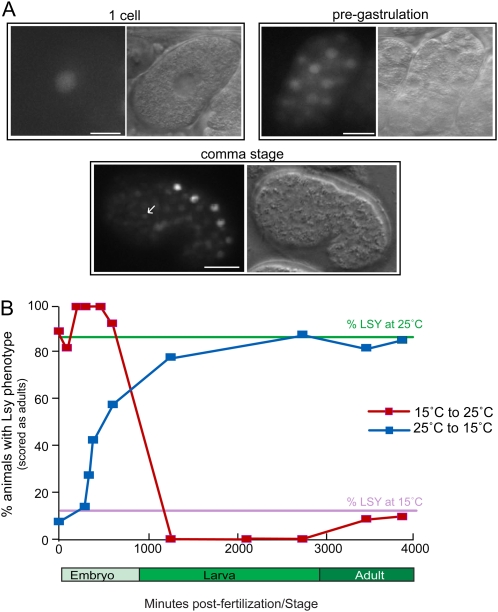
By recombineering yfp into the fosmid that contains the lsy-27 locus and that rescues the lsy-27 phenotype (Table 2), we generated a reporter with which we monitored lsy-27 expression (Figure 2A). We find that lsy-27 is expressed very broadly throughout the embryo (Figure 3A). Expression can already be observed in one-cell embryos and continues to about the comma stage, when expression starts to fade out (Figure 3A). By the comma stage, most neurons, including ASEL/ASER, have terminally divided and begun to terminally differentiate. No expression is observed after hatching in larvae or in adult animals. Through colocalizing expression of the lsy-27 reporter with an ASE-specific mCherry reporter, we confirmed that lsy-27 is expressed in both ASE neurons in the comma-stage embryo when ASE laterality is established. As assessed with translational gfp reporters that fuse the entire loci to gfp, the most closely related lsy-27 paralog, ztf-25, displays an essentially indistinguishable broad, embryo-restricted expression pattern (Figure S3), while the more distant paralog ztf-28 shows no expression in embryos and postembryonically is expressed only in the intestine (data not shown).
Figure 3.—
lsy-27 is expressed and acts during the initiation but not during the maintenance phase of left/right asymmetry control. (A) Expression pattern of lsy-27fosmid::yfp (shown in Figure 2A) at different embryonic stages. The embryos appear slightly deformed as they are squished together in the gonad of an adult animal. The white arrow indicates the ASEL neuron shortly after birth based on colocalization with a bilateral ASE-specific reporter otIs232(che-1::mCherry) (not shown). The lsy-27transl::gfp (shown in Figure 2A) shows a similar expression pattern except that, due to its failure to be expressed in the germline, we see only the onset of expression when zygotic gene expression starts in the early embryo. Bar, 10 μm. (B) Temperature-shift experiments with ot108; ntIs1 animals indicate that lsy-27 activity is required only during embryogenesis, but not during postembryonic stages. Animals were cultured for at least three generations at either 15° or 25°. Animals were analyzed by isolating two- to four-cell embryos and temperature shifts were performed at various developmental stages. All animals were scored as 3-day-old adults.
The expression pattern of lsy-27 suggests an embryonic role for the gene. We sought to corroborate this notion by exploiting the observation that the ot108 allele is strongly temperature sensitive (Figure 3B). At 25°, 87% of animals display a Lsy phenotype while 12% do at 15°. By altering lsy-27 gene activity at different stages through temperature shifts, we find that lsy-27 activity is required only during embryogenesis, but not during postembryonic stages (Figure 3B). This contrasts with the continuous requirement of other lsy genes during postembryonic stages (O’Meara et al. 2010) and demonstrates that laterality control can be divided into initiation and maintenance phases.
The maternal loading of LSY-27 protein into oocytes as well as the embryonic focus of action also prompted us to ask whether lsy-27 gene activity can be solely maternally supplied. Using transgenic lsy-27 mutant animals that carry the germline-expressed lsy-27 reporter fosmid, we assayed progeny that have lost the array and therefore contain only maternally supplied gene activity. In such animals, the Lsy phenotype is rescued (Table 2), corroborating maternal deposition of lsy-27 gene activity.
Concluding remarks
We have described here a member of a nematode-specific C2H2 Zn-finger transcription factor family, lsy-27, which functions in ASE laterality control. The lsy-27 mutant phenotype is similar to that of the ASEL-restricted LIM homeobox gene lim-6, as well as the ubiquitously expressed lin-59 histone methyltransferase. We found that lsy-27 not only affects the terminal gcy gene markers in a manner similar to lim-6, but also affects lim-6 expression. The embryo-restricted expression and function of lsy-27 contrasts with the expression of lim-6, which is expressed continuously throughout the life of the ASEL neuron. We propose that the function of lsy-27 is restricted to triggering the initial onset of lim-6 expression. lsy-27 may cooperate with ASEL-expressed die-1 to trigger lim-6 expression in the embryo. Once lim-6 is turned on, lsy-27 is no longer required to control laterality. This maintenance role is carried out by die-1 (O'Meara et al. 2010) in conjunction with lim-6, which positively autoregulates (Johnston et al. 2005). Interestingly, lsy-27 is not involved in conveying other die-1 functions, such as the induction of ASEL fate markers (e.g., gcy-7), since those are affected only in die-1, but not in lsy-27 mutants.
With the molecular identification of ot108 and ot146, we have identified all but one gene retrieved from our large-scale screening of left/right asymmetry mutants (summarized in Table S4). Due to some adjustments in allele assignments as described here and elsewhere (Etchberger et al. 2009; Sarin et al. 2009; Flowers et al. 2010), we have recalculated saturation using various models (Sarin et al. 2007) and retain our previous conclusion that the screen has not yet reached saturation. Future genetic screens are likely to provide further insights into the control of lateralized gene expression in the nervous system.
Acknowledgments
We thank the C. elegans knockout consortia directed by Shohei Mitani at Tokyo Women’s Medical University School of Medicine for the tm573, tm593, and tm630 alleles; Sumeet Sarin for help in recalculating genetic saturation; Qi Chen for expert assistance in generating transgenic strains; and members of the Hobert lab for comments on the manuscript. This work was funded by the National Institutes of Health (grants R01NS039996-05 and R01NS050266-03). O.H. is an Investigator of the Howard Hughes Medical Institute.
Literature Cited
- Baum P. D., Guenther C., Frank C. A., Pham B. V., Garriga G., 1999. The Caenorhabditis elegans gene ham-2 links Hox patterning to migration of the HSN motor neuron. Genes Dev. 13: 472–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigelow H., Doitsidou M., Sarin S., Hobert O., 2009. MAQGene: software to facilitate C. elegans mutant genome sequence analysis. Nat. Methods 6: 549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S., Johnston R. J., Frokjaer-Jensen C., Lockery S., Hobert O., 2004. MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode. Nature 430: 785–789 [DOI] [PubMed] [Google Scholar]
- Etchberger J. F., Flowers E. B., Poole R. J., Bashllari E., Hobert O., 2009. Cis-regulatory mechanisms of left/right asymmetric neuron-subtype specification in C. elegans. Development 136: 147–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers E. B., Poole R. J., Tursun B., Bashllari E., Pe’er I., et al. , 2010. The Groucho ortholog UNC-37 interacts with the short Groucho-like protein LSY-22 to control developmental decisions in C. elegans. Development 137: 1799–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O., Tessmar K., Ruvkun G., 1999. The Caenorhabditis elegans lim-6 LIM homeobox gene regulates neurite outgrowth and function of particular GABAergic neurons. Development 126: 1547–1562 [DOI] [PubMed] [Google Scholar]
- Hobert O., Johnston R. J., Jr., Chang S., 2002. Left-right asymmetry in the nervous system: the Caenorhabditis elegans model. Nat. Rev. Neurosci. 3: 629–640 [DOI] [PubMed] [Google Scholar]
- Johnston R. J., Jr., Chang S., Etchberger J. F., Ortiz C. O., Hobert O., 2005. MicroRNAs acting in a double-negative feedback loop to control a neuronal cell fate decision. Proc. Natl. Acad. Sci. USA 102: 12449–12454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston R. J., Jr., Copeland J. W., Fasnacht M., Etchberger J. F., Liu J., et al. , 2006. An unusual Zn-finger/FH2 domain protein controls a left/right asymmetric neuronal fate decision in C. elegans. Development 133: 3317–3328 [DOI] [PubMed] [Google Scholar]
- Kadrmas J. L., Beckerle M. C., 2004. The LIM domain: from the cytoskeleton to the nucleus. Nat. Rev. Mol. Cell Biol. 5: 920–931 [DOI] [PubMed] [Google Scholar]
- Li H., Coghlan A., Ruan J., Coin L. J., Heriche J. K., et al. , 2006. TreeFam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 34: D572–D580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Meara M. M., Zhang F., Hobert O., 2010. Maintenance of neuronal laterality in Caenorhabditis elegans through MYST histone acetyltransferase complex components LSY-12, LSY-13 and LIN-49. Genetics 186: 1497–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz C. O., Etchberger J. F., Posy S. L., Frokjaer-Jensen C., Lockery S., et al. , 2006. Searching for neuronal left/right asymmetry: genomewide analysis of nematode receptor-type guanylyl cyclases. Genetics 173: 131–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz C. O., Faumont S., Takayama J., Ahmed H. K., Goldsmith A. D., et al. , 2009. Lateralized gustatory behavior of C. elegans is controlled by specific receptor-type guanylyl cyclases. Curr. Biol. 19: 996–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce-Shimomura J. T., Faumont S., Gaston M. R., Pearson B. J., Lockery S. R., 2001. The homeobox gene lim-6 is required for distinct chemosensory representations in C. elegans. Nature 410: 694–698 [DOI] [PubMed] [Google Scholar]
- Sarin S., O’Meara M. M., Flowers E. B., Antonio C., Poole R. J., et al. , 2007. Genetic screens for Caenorhabditis elegans mutants defective in left/right asymmetric neuronal fate specification. Genetics 176: 2109–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarin S., Prabhu S., O’Meara M. M., Pe'er I., Hobert O., 2008. Caenorhabditis elegans mutant allele identification by whole-genome sequencing. Nat. Methods 5: 865–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarin S., Antonio C., Tursun B., Hobert O., 2009. The C. elegans Tailless/TLX transcription factor nhr-67 controls neuronal identity and left/right asymmetric fate diversification. Development 136: 2933–2944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarin S., Bertrand V., Bigelow H., Boyanov A., Doitsidou M., et al. , 2010. Analysis of multiple ethyl methanesulfonate-mutagenized Caenorhabditis elegans strains by whole-genome sequencing. Genetics 185: 417–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T., Walsh C. A., 2006. Molecular approaches to brain asymmetry and handedness. Nat. Rev. Neurosci. 7: 655–662 [DOI] [PubMed] [Google Scholar]
- Sun T., Patoine C., Abu-Khalil A., Visvader J., Sum E., et al. , 2005. Early asymmetry of gene transcription in embryonic human left and right cerebral cortex. Science 308: 1794–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R. W., Hsieh Y. W., Gamse J. T., Chuang C. F., 2010. Making a difference together: reciprocal interactions in C. elegans and zebrafish asymmetric neural development. Development 137: 681–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicks S. R., Yeh R. T., Gish W. R., Waterston R. H., Plasterk R. H., 2001. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28: 160–164 [DOI] [PubMed] [Google Scholar]
- Yu S., Avery L., Baude E., Garbers D. L., 1997. Guanylyl cyclase expression in specific sensory neurons: a new family of chemosensory receptors. Proc. Natl. Acad. Sci. USA 94: 3384–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]